Abstract
Background
Understanding nosocomial pathogen transmission is restricted by culture limitations. Novel platforms, such as PCR-based electron spray ionization-time-of-flight-mass spectrometry (ESI-TOF-MS), may be useful as investigational tools.
Methods
Traditional clinical microbiology (TCM) and PCR/ESI-TOF-MS were used to recover and detect microorganisms from the hands and personal protective equipment of 10 burn intensive care unit (ICU) healthcare workers providing clinical care at a tertiary care military referral hospital. High-use environmental surfaces were assessed in 9 burn ICU and 10 orthopedic patient rooms. Clinical cultures during the study period were reviewed for pathogen comparison with investigational molecular diagnostic methods.
Results
From 158 samples, 142 organisms were identified by TCM and 718 by PCR/ESI-TOF-MS. The molecular diagnostic method detected more organisms (4.5 ± 2.1 vs. 0.9 ± 0.8, p < 0.01) from 99% vs. 67% of samples (p < 0.01). TCM detected S. aureus in 13 samples vs. 21 by PCR/ESI-TOF-MS. Gram-negative organisms were less commonly identified than gram-positive by both methods; especially by TCM. Among all detected bacterial species, similar percentages were typical nosocomial pathogens (18-19%) for TCM vs. PCR/ESI-TOF-MS. PCR/ESI-TOF-MS also detected mecA in 112 samples, vanA in 13, and KPC-3 in 2. MecA was associated (p < 0.01) with codetection of coagulase negative staphylococci but not S. aureus. No vanA was codetected with enterococci; one KPC-3 was detected without Klebsiella spp.
Conclusions
In this pilot study, PCR/ESI-TOF-MS detected more organisms, especially gram-negatives, compared to TCM, but the current assay format is limited by the number of antibiotic resistance determinants it covers. Further large-scale assessments of PCR/ESI-TOF-MS for hospital surveillance are warranted.
Keywords: PCR/ESI-TOF-MS, Ibis, Microbiology, Contamination, Environment
Background
Healthcare-associated infections (HAI) account for substantial morbidity and mortality worldwide [1]. These occur both in epidemics, with a common pathogen, and in endemic settings, where no clusters or common pathogens are identified. Numerous reservoirs for epidemiologically significant organisms have been demonstrated in healthcare settings. These include high-use environmental surfaces, such as door handles and handrails; patient care items such as bedside tables, bedrails, and intravenous fluid (IV) pumps; healthcare provider protective clothing such as lead aprons; and plumbing structures including drains and faucet heads, and computer equipment, among many others [2-7]. Contamination of personal protective equipment (PPE) during patient care is a mechanism for transient colonization in healthcare workers (HCW) after doffing PPE [8,9]. However, in any healthcare environment, identification of a reservoir for endemic transmission of pathogens is the exception rather than the rule. Identifying reservoirs is limited by the sensitivity of traditional clinical microbiology (TCM), especially since many pathogens establish biofilms, which are recalcitrant to TCM, on environmental surfaces [10,11]. More accurate identification and speciation of environmental pathogens should assist infection prevention efforts and mitigate excess patient morbidity and mortality.
Molecular techniques are increasingly used for microbial detection; however, these methods often focus on a single pathogen, such as methicillin-resistant Staphylococcus aureus (MRSA), or are used only after initial growth of bacteria in culture [12,13]. Ideal molecular methods would include the ability to screen samples for numerous species rapidly and simultaneously. The Ibis T5000 (PCR electron spray ionization-time-of-flight-mass spectrometry; PCR/ESI-TOF-MS) technology is based on the determination of the ratios of the four nucleotide bases (A, T, G and C) in multiple (n = 16) PCR amplicons that target conserved bacterial genes (including the 16S rDNA gene). Using a triangulation algorithm based on multiple independent amplicon mass determinations, it can identify and speciate all eubacterial species present in a complex sample that are present at greater than 3% of the microbial burden [14]. The technology has been recently reviewed in detail [15-17]. It has been used in outbreak investigations of Streptococcus pyogenes and Acinetobacter spp., to characterize and genotype a diverse collection of S. aureus isolates, and to characterize orthopedic infections [18-23]. However, no previous study using this technology has evaluated recovery of endemic pathogens in a healthcare environment. This pilot study uses TCM and PCR/ESI-TOF-MS to compare contamination of HCW hands and PPE used in the care of patients on the burn intensive care unit (ICU), and contamination of high-use surfaces in the burn ICU and the orthopedic ward. Additionally, we explored whether results obtained from either TCM or PCR/ESI-TOF-MS reflected contemporaneous clinical cultures obtained from hospitalized patients on the study units.
Methods
Isolates tested
Sample acquisition was planned from 20 occupied single-bed patient care rooms, ten from the burn ICU (burn unit rooms were designed with anterooms and universal gowns and gloves are used) and ten from the orthopedic ward. Nine rooms in the burn ICU had sample acquisition completed due to patient census. In the burn ICU, one HCW for each selected patient room was also enrolled for screening. Two HCW completed patient care in the same room in one instance due to patient census. Two swabs (one for TCM and one for PCR/ESI-TOF-MS; Fisherfinest Transport Swabs with Liquid Stuarts) were obtained using a standard rolling technique from: the door handle exiting the room, sink faucet, bedrail, IV pump, in-room computer keyboard, and in-room computer mouse where available. In rooms where any of these items was unavailable, these data were omitted. Bandage shears from 10 orthopedic surgeons were also swabbed.
HCW screening
Two swabs (Fisherfinest Transport Swabs with Liquid Stuarts) were obtained (using the standard rolling technique) from subjects’ hands. HCW donned PPE (gowns and gloves) and managed their patients in single patient room. Upon return, the surfaces of gloves, the waistline of the gown, and the hands after glove removal and before hand hygiene were swabbed. One swab was tested using TCM techniques and the other by PCR/ESI-TOF-MS.
Clinical culture data
A summary of de-identified clinical culture and Clostridium difficile toxin assay results (included due to its significance as a HAI bacterial pathogen, inability to isolate by routine clinical culture, and in order to correlate against any PCR/ESI-MS-TOF C. difficile results obtained) obtained during routine patient care from the burn ICU and orthopedics ward during the study period was retrospectively collated via the patient’s electronic medical records. Clinical cultures (and C. difficile toxin assay results) were included if performed from t-14 through t + 14 days with respect to the dates of room sampling for that unit, which took place from May-July 2010. No concurrent chart review was performed for hospital length of stay, definitions of infections, or any other clinical criteria since the hospital microbial ecology was the outcome of interest, and no potentially duplicate isolates from the same patient were excluded. Organisms were considered potentially clinically relevant if isolated on at least five occasions from separate clinical cultures during the study period and they were not common skin contaminants. For the purposes of statistical comparisons, coagulase negative staphylococci (CNS) were excluded, and aerobic gram-negative rods other than Escherichia coli, Acinetobacter spp., Klebsiella spp., Pseudomonas spp., or Enterobacter spp. were coalesced into one category.
PCR/ESI-TOF-MS
Methods for genotypic characterization of bacterial and fungal isolates, and genetic resistance elements (mecA, vanA, and KPC-3) using the commercially available Ibis T5000 (Ibis Biosciences) have been described elsewhere [21,24]. Swabs were frozen at −80°C and shipped on dry ice for batched PCR/ESI-TOF-MS testing. Following thawing of the swabs they were placed into sterile microcentrifuge tubes containing 270 μl of ATL Lysis buffer (Qiagen, Germantown, MD, cat# 19076) and 30 μl proteinase K (Qiagen, cat# 19131). Samples were incubated at 56°C for one hour. One hundred μl of a mixture containing 50 μl each of 0.1 mm and 0.7 mm Zirconia beads (Biospec cat# 11079101z, 11079107zx respectively) were added to the samples which were then homogenized for 10 min at 25 Hz using a Qiagen Tissuelyser. Nucleic acid from the lysed sample was then extracted using the Qiagen DNeasy kit (Qiagen cat# 69506). 10 μl of each sample was loaded per well onto the BAC detection PCR plate (Abbott Molecular, cat# PN 05 N13-01). The BAC detection plate is a 96 well plate which contains 16 primers that survey all bacterial organisms by using multiple omnipresent loci (e.g. 16S rDNA sequences) and multiple pluripresent loci (e.g. the tufB gene). This has been validated against 613 organisms, meaning it correctly identified them when presented with unknowns. The system also detects the presence of several key antibiotic resistance markers: vanA and vanB (vancomycin resistance) in Enterococcus spp., KPC-3 (carbapenem resistance) in gram-negative bacteria, and mecA (methicillin resistance) in Staphylococcus spp. An internal calibrant of synthetic nucleic acid template is also included in each assay, controlling for false negatives (e.g. from PCR inhibitors) and enabling a semi-quantitative analysis of the amount of template DNA present. PCR amplification was carried out as per Ecker et al [25]. The PCR products were then desalted in a 96-well plate format and sequentially electrosprayed into a time-of-flight mass spectrometer. The spectral signals were processed to determine the masses of each of the PCR products present with sufficient accuracy that the base composition of each amplicon could be unambiguously deduced. Using combined base compositions from multiple PCRs, the identities of the pathogens and a semi-quantitative determination of their relative concentrations in the starting sample were established by using a proprietary algorithm to interface with the Ibis database of known organisms.
Semi-quantitative data was obtained from all PCR/ESI-TOF-MS analyses as each well of each assay is seeded with a DNA template that contains the appropriate primer binding sites for the primers in that well. These primer binding sites flank a synthetic DNA sequence of known composition. By comparing the amount of each species’ amplimer produced in a well to the amount of the amplimer resulting from the synthetic template the number of genomes/well of each bacterial species can be approximated. However, given the exploratory nature of the study, semi-quantitative data were not analyzed here.
TCM
Clinical microbiology swabs were transferred to brain heart infusion (BHI) broth medium and this was incubated 48 h at 35–37°C. If the BHI demonstrated turbidity, the inoculated broth was subcultured onto sheep’s blood agar plates (BBL, Cockeysville, MD, USA) and MacConkey agar plates (BBL, Cockeysville, MD, USA). All colony forming units were worked up with no minimum threshold for evaluation. Organisms and antimicrobial resistance testing were performed using standard clinical microbiology techniques including semi-automated mechanisms for gram-negative isolates (Siemens WalkAway 40 System; Siemens Healthcare Diagnostics, Deerfield, IN, USA).
Human subject protection
The protocol was reviewed and approved by the Brooke Army Medical Center Institutional Review Board and human subjects provided informed consent.
Statistical analysis
Descriptive statistics were used to summarize findings. Analysis was performed using existing software (SPSS, version 19.0; IBM SPSS). Categorical variables were compared by chi-squared test, and t-test for normal continuous variables. Paired tests were applied when comparing two methods of testing from the same sample; McNemar’s test was used for nonparametric paired testing. Means and standard deviations are expressed throughout as mean ± SD. All p-values are two-tailed and statistical significance represented by p < 0.05.
Results
Samples were taken from 158 sites; 40 from HCW (10 pre-patient care hands, 10 gloves, 10 gowns, 10 post-patient care hands), 19 from door handles, sink faucets, IV pumps and bedrails, 17 from keyboards, 15 from computer mice, and 10 from orthopedic shears. From these sites, 142 organisms were recovered by TCM and 718 by PCR/ESI-TOF-MS. At all sites, compared to TCM, PCR/ESI-TOF-MS recovered a larger number of organisms (4.5 ± 2.1 vs. 0.9 ±0.8, p <0.01) from a greater proportion of samples (99% vs. 67%, p <0.01; Table 1). HCW hands revealed more organisms by PCR/ESI-TOF-MS than TCM before care (3.9 ± 2.0 vs. 0.4 ± 0.5, p < 0.01) and after care (3.8 ±1.6 vs. 0.6 ± 0.5, p < 0.01). PCR/ESI-TOF-MS also recovered a greater number of organisms than TCM among used gowns (2.8 ± 1.1 vs. 0.6 ± 0.7, p < 0.01), but not gloves (3.1 ± 2.3 vs. 1.3 ± 1.6, p = 0.10).
Table 1.
PCR/Electron spray ionization-time-of-flight-mass spectrometry (PCR/ESI-TOF-MS) versus traditional clinical microbiology (TCM) for detection of organisms contaminating high-use surfaces, healthcare worker hands, and personal protective equipment in a burn intensive care unit (ICU) and an orthopedic ward
| |
Burn ICU # sites with at least one organism recovered (# organisms recovered) |
Orthopedic ward # sites with at least one organism recovered (# organisms recovered) |
||||
|---|---|---|---|---|---|---|
| Screened | PCR/ESI-TOF-MS | TCM | Screened | PCR/ESI-TOF-MS | TCM | |
| Bedrails |
9 |
9 (44) |
7 (11) |
10 |
10 (53) |
8 (11) |
| Door handles |
9 |
9 (34) |
6 (7) |
10 |
10 (48) |
3 (3) |
| Sink faucets |
9 |
9 (41) |
7 (8) |
10 |
10 (56) |
9 (11) |
| IV pumps |
9 |
8 (34) |
6 (7) |
10 |
10 (53) |
5 (6) |
| Keyboards |
9 |
9 (48) |
9 (16) |
8 |
8 (50) |
8 (11) |
| Mouse |
9 |
9 (38) |
6 (8) |
6 |
6 (35) |
4 (6) |
| Shears |
|
|
|
10 |
10 (48) |
7 (8) |
| Hands pre-care |
10 |
10 (39) |
4 (4) |
|
|
|
| Gloves |
10 |
10 (31) |
6 (13) |
|
|
|
| Gowns |
10 |
9 (28) |
5 (6) |
|
|
|
| Hands post-care |
10 |
10 (38) |
6 (6) |
|
|
|
| Total | 94 | 92 (375) | 62 (86) | 64 | 64 (343) | 44 (56) |
Total number of isolates recovered.
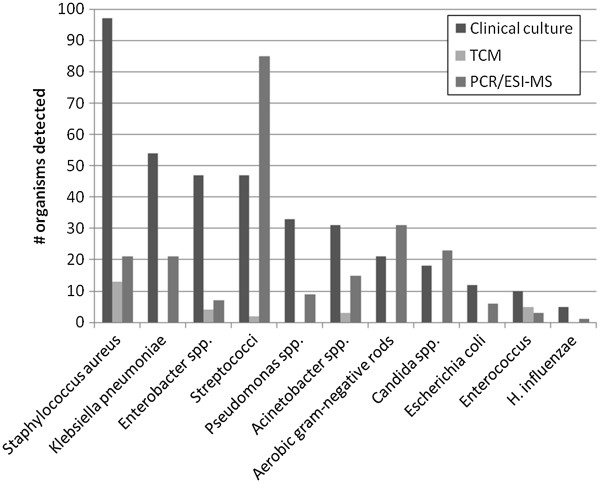
Organisms recovered from 393 clinical cultures included S. aureus, Klebsiella pneumoniae, Enterobacter spp., Streptococcus spp. (68% viridans group), Pseudomonas aeruginosa, and Acinetobacter baumannii-calcoaceticus complex; the proportions of these organisms detected by PCR/ESI-TOF-MS and TCM are depicted in Figure 1. The most common clinical culture sources included respiratory (29%), wound (22%), body fluid (19%), and blood (16%). There were no positive toxin assay results for C. difficile. Twelve isolates of CNS were recovered, 8 from blood cultures. By the study definition of potentially clinically relevant organisms, and combining less commonly recovered aerobic gram-negative rods (e.g. Serratia, Morganella, Stenotrophomonas spp.), 84% of clinical cultures were potentially clinically relevant. There was no difference in the proportion of potentially clinically relevant organisms detected by TCM vs. PCR/ESI-TOF-MS (18 vs. 19%, p = 0.77). Including streptococci, which were the third most commonly recovered organisms among clinical cultures, 19% of TCM organisms recovered were of potential clinical significance vs. 31% for PCR/ESI-TOF-MS (p < 0.01). Comparison of samples positive for a potentially clinically relevant organism revealed consistently higher proportions detected by PCR/ESI-TOF-MS (Table 2). This was statistically significant by McNemar’s test with or without inclusion of streptococci, and remained significant even when comparing only samples positive for the most commonly cultured bacteria (S. aureus, K. pneumoniae, Enterobacter spp., Pseudomonas spp., Acinetobacter spp., and E. coli).
Figure 1.
Number of pathogens of potential clinical relevance detected by clinical culture versus numbers detected on healthcare worker hands, personal protective equipment and environmental surfaces by traditional clinical microbiology (TCM) and PCR/Electron spray ionization-time-of-flight-mass spectrometry (PCR/ESI-TOF-MS) in the burn unit and orthopedic ward. “Aerobic gram-negative rods:” Clinical culture: 1 Achromobacter xylosoxidans, 1 Aeromonas sobria, 4 Citrobacter koseri, 4 Morganella morganii, 5 Providencia rettgeri, 2 Serratia marcescens, 8 Stenotrophomonas maltophilia. PCR/ESI-TOF-MS: 1 Actinobacillus sp., 1 Azoarcus sp., 4 Bordetella avium, 1 Bordetella bronchiseptica, 1 Bordetella parapertussis, 2 Bordetella petri, 2 Burkholderia cenocepacia, 4 Burkholderia thailandensis, 1 Campylobacter sp., 1 Caulobacter sp., 1 Leptothrix cholodnii, 1 Nitrosomonas europaea, 2 Novosphingobium aromaticivorans, 1 Raoultella ornithinolytica, 3 S. marcescens, 1. S. flexneri, 1 Sphingomonas sp., 1 Vibrio rumoiensis, 1 Vibrio vulnificus, 1 Xanthomonas oryza.
Table 2.
PCR/Electron spray ionization-time-of-flight-mass spectrometry (PCR/ESI-TOF-MS) versus traditional clinical microbiology (TCM) for detection of pathogens of potential clinical relevance on healthcare worker hands/personal protective equipment and high-use surfaces (n = 158)
| PCR/ESI-TOF-MS # sites with at least one organism recovered (%) | TCM # sites with at least one organism recovered (%) | p value | ||
|---|---|---|---|---|
| Any potentially clinically relevant organism* |
Including streptococci |
123 (77.8) |
20 (12.7) |
<0.01 |
| |
Not including streptococci |
94 (59.4) |
19 (12.8) |
<0.01 |
| Six most common bacteria recovered from clinical cultures** | 58 (36.7%) | 16 (10.1) | <0.01 |
*S. aureus, K. pneumoniae, Enterobacter spp., Pseudomonas spp., Acinetobacter spp., Aerobic gram-negative rods (see Figure 1), Candida spp., E. coli, Enterococcus spp., and Haemophilus influenzae.
**S. aureus, K. pneumoniae, Enterobacter spp., Pseudomonas spp., Acinetobacter spp., and E. coli.
Distribution of potentially clinically relevant organisms, plus CNS, recovered from HCW hands/PPE, and the hospital environment, are presented in Tables 3 and 4 respectively. Most organisms recovered by either mechanism were gram-positive. Eight-six total CNS isolates were recovered by TCM and 214 by PCR/ESI-TOF-MS; 13 S. aureus by TCM and 21 by PCR/ESI-TOF-MS. Gram-negative organisms were less commonly identified, especially by TCM. There were 3 Acinetobacter spp. recovered by TCM and 15 by PCR/ESI-TOF-MS; 4 Enterobacter spp. by TCM and 7 by PCR/ESI-TOF-MS; no E. coli by TCM and 6 by PCR/ESI-TOF-MS, no Klebsiella spp. by TCM and 21 by PCR/ESI-TOF-MS, no Pseudomonas spp. by TCM and 9 by PCR/ESI-TOF-MS. These five gram-negative rod species (GNR) contributed 177 of 389 (45%) clinical cultures during the study period, however only 7 (5%) of the environmental samples were positive for these organisms by TCM, and 58 (8%) by PCR/ESI-TOF-MS. TCM contributed to only 11% of total detections of these GNR, vs. 38% of all S. aureus detections, a difference that was statistically significant (p <0.01).
Table 3.
PCR/Electron spray ionization-time-of-flight-mass spectrometry (PCR/ESI-TOF-MS) versus traditional clinical microbiology (TCM) for detection of most frequently recovered potentially clinically relevant pathogens, plus coagulase negative staphylococci (CNS), contaminating healthcare workers and personal protective equipment
| |
PCR/ESI-TOF-MS (n = 77) |
TCM (n = 23) |
||||||
|---|---|---|---|---|---|---|---|---|
| Organism | Hands pre-care | Gloves | Gowns | Hands post-care | Hands pre-care | Gloves | Gowns | Hands post -care |
| CNS |
12 |
6 |
8 |
12 |
4 |
4 |
3 |
3 |
|
Staphylococcus aureus |
|
3 |
3 |
1 |
|
3 |
1 |
1 |
| Streptococci* |
9 |
4 |
3 |
1 |
|
1 |
|
|
|
Acinetobacter spp.** |
|
3 |
1 |
|
|
1 |
|
|
|
Enterobacter spp.*** |
|
1 |
|
|
|
2 |
|
|
|
Escherichia coli |
|
1 |
1 |
|
|
|
|
|
|
Klebsiella pneumoniae |
1 |
3 |
1 |
1 |
|
|
|
|
|
Pseudomonas spp.**** |
1 |
|
|
1 |
|
|
|
|
| Candida spp.***** | 2 | |||||||
*TCM: 1 Group D Streptococcus; PCR/ESI-TOF-MS: 7 S. agalactiae, 10 viridans group streptococci.
**TCM: 1 A. baumannii; PCR/EIS-MS: 2 A. baumannii, 2 A. calcoaceticus.
***TCM: 2 Enterobacter aerogenes; PCR/ESI-TOF-MS: 1 E. aerogenes.
****PCR/ESI-TOF-MS: 1 Pseudomonas fluorescens, 1 Pseudomonas mendocina.
*****PCR/ESI-TOF-MS: 1 C. albicans, 1 Candida glabrata.
Table 4.
PCR/Electron spray ionization-time-of-flight-mass spectrometry (PCR/ESI-TOF-MS) versus traditional clinical microbiology (TCM) for detection of most frequently recovered potentially clinically relevant organisms, plus coagulase negative staphylococci (CNS), contaminating high-use surfaces
|
Bacteria |
PCR/ESI-TOF-MS (# isolates) n = 305 |
TCM (# isolates) n = 89 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bedrail | Door handle | Faucet | IV pump | Keyboard | Mouse | Shears | Bedrail | Door handle | Faucet | IV pump | Keyboard | Mouse | Shears | |
| CNS |
24 |
31 |
28 |
23 |
30 |
22 |
19 |
11 |
6 |
14 |
9 |
16 |
9 |
7 |
| Enterococci* |
|
2 |
|
|
|
|
1 |
2 |
1 |
|
1 |
|
|
|
|
S. aureus |
4 |
|
1 |
4 |
2 |
3 |
|
3 |
|
1 |
1 |
1 |
2 |
|
| Streptococci** |
7 |
8 |
12 |
12 |
17 |
7 |
5 |
|
|
|
|
|
|
1 |
|
Acinetobacter spp.*** |
2 |
1 |
2 |
6 |
|
|
|
|
1 |
|
1 |
|
|
|
|
Enterobacter spp.**** |
3 |
1 |
1 |
1 |
|
|
|
1 |
1 |
|
|
|
|
|
|
E. coli |
2 |
1 |
|
1 |
|
|
|
|
|
|
|
|
|
|
|
K. pneumoniae |
7 |
|
1 |
3 |
1 |
3 |
|
|
|
|
|
|
|
|
|
Pseudomonas spp.***** |
4 |
|
2 |
|
1 |
|
|
|
|
|
|
|
|
|
| Candida spp.****** | 7 | 3 | 5 | 3 | 1 | 2 | ||||||||
*TCM: “Enterococcus spp.”; PCR/ESI-TOF-MS: 2 Enterococcus faecalis, 1 Enterococcus faecium.
**TCM: 1 viridans group streptococci; PCR/ESI-TOF-MS: 1 Group D Streptococcus, 24 S. agalactiae, 8 Streptococcus pneumoniae, 1 S. pyogenes, 1 “Streptococcus spp.”, 33 viridans group streptococci.
***TCM: 2 A. baumannii; PCR/ESI-TOF-MS: 10 A. baumannii, 1 “Acinetobacter spp.”
****TCM: 2 Enterobacter cloacae; PCR/ESI-TOF-MS: 4 E. cloacae; 2 Enterobacter sakazakii.
*****PCR/ESI-TOF-MS: 1 P. aeruginosa, 2 Pseudomonas entomophila/putida, 3 P. fluorescens, 1 “Pseudomonas spp.”
******PCR/ESI-TOF-MS: 10 C. albicans, 4 C. glabrata, 1 Candida kefyr, 1 Candida kruseii, 1 Candida parapsilosis, 1 Candida rugosa, 3 Candida tropicalis.
In addition to detection of bacteria, PCR/ESI-TOF-MS detected the mecA gene in 112 samples. The majority of these codetected CNS with no S. aureus present (93; 83%). The remainder were comprised of S. aureus alone (9); S. aureus and CNS together (8), or no staphylococci (2). MecA detection was statistically associated with CNS codetection (p <0.01) but not with either S. aureus detection or MRSA growth on culture (p = 0.22) from the same samples. Of 13 S. aureus isolates recovered by TCM, 9 were MRSA. Seven of 9 MRSA cultured also had mecA and S. aureus detected by PCR/ESI-TOF-MS from the same sample (one detected mecA with no S. aureus and the other detected neither). Of the two samples for which mecA but no staphylococci were recovered, one grew Acinetobacter spp. and enterococci by TCM. PCR/ESI-TOF-MS detected A. baumannii, Candida albicans, and Polynucleobacter spp. The other sample was TCM negative with both P. acnes and Streptococcus thermophilus detected by PCR/ESI-TOF-MS.
There were 13 samples in which vanA was detected. None of these had enterococci codetected by PCR/ESI-TOF-MS, and no cultures grew vancomycin-resistant enterococci (VRE); one grew susceptible enterococci. There was a significant association with lactobacilli codetection; 6 of 20 lactobacillus detections had vanA codetected (p < 0.01). However, 7 samples were positive for vanA with neither lactobacilli nor enterococci codetections. There were no clear trends among the other organisms codetected with vanA, but 12 of 13 vanA samples also had a mecA codetected.
KPC-3 was detected in 2 samples, in one of which K. pneumoniae was codetected (along with Clostridium perfringens, Saccharomyces cerevisiae, S. thermophilus, CNS, and mecA). The other KPC-3 positive sample codetected CNS, Streptococcus agalactiae, Propionibacterium acnes, Lactobacillus salivarius, Nocardia asteroides, Bordetella bronchiseptica, and vanA.
There were a number of rare or unexpected microorganisms detected in the hospital environment by PCR/ESI-TOF-MS, including Shigella, Vibrio and Bartonella spp. Selected unusual or less commonly detected organisms are presented in Table 5.
Table 5.
Selected rare organisms detected by PCR/Electron spray ionization-time-of-flight-mass spectrometry (ESI-TOF-MS) in the hospital environment
|
Microbiology |
# PCR/ESI-TOF-MS detections |
||
|---|---|---|---|
| HCW/PPE | Burn ICU rooms | Orthopedic rooms/Shears | |
| Gram-positive |
|
|
|
| Cocci |
|
|
|
| Leuconostoc spp. |
2 |
0 |
5 |
| S. pyogenes |
|
|
1 |
| S. pneumoniae |
|
1 |
7 |
| Bacilli |
|
|
|
| Clostridium spp. |
1 |
1 |
2 |
| Clostridium tetani |
|
1 |
1 |
| Listeria monocytogenes |
2 |
|
|
| Nocardia spp. |
3 |
1 |
6 |
| Gram-negative |
|
|
|
| Cocci |
|
|
|
| Neisseria spp. |
|
1 |
2 |
| Aerobic Bacilli |
|
|
|
| Bordetella spp. |
3 |
1 |
4 |
| Burkholderia spp. |
1 |
2 |
3 |
| Polynucleobacter spp. |
7 |
11 |
7 |
| Shigella flexneri |
|
|
1 |
| Vibrio spp. |
|
1 |
1 |
| Anaerobic Bacilli |
|
|
|
| Bacteroides spp. |
1 |
1 |
|
| Other bacteria |
|
|
|
| Bartonella spp. |
1 |
|
|
| Borrelia turicatae |
|
|
1 |
| Mycobacterium abscessus |
|
|
1 |
| Mycoplasma hominis |
1 |
|
|
| Fungi |
|
|
|
| Alternaria |
2 |
1 |
4 |
| Saccharomyces cerevisiae |
1 |
3 |
17 |
| Other fungi* | 1 | 3 | |
HCW healthcare worker, PPE personal protective equipment, ICU intensive care unit.
* One of each: Botryosphaeria rhodina, Macroventuria spp., Cochliobolus spp., Arthrographis cuboidea.
Few clinically relevant pathogens were detected by TCM and not by PCR/ESI-TOF-MS. Altogether, PCR/ESI-TOF-MS failed to detect 35 of the 142 isolates from TCM, most of which were identified as Micrococcus spp. and CNS. There were five Enterococcus spp. isolated by TCM which went undetected by PCR/ESI-TOF-MS; additional clinically relevant organisms included S. aureus (2), Enterobacter spp. (2), and Acinetobacter spp. (1).
Discussion
Endemic transmission of nosocomial pathogens, especially gram-negative organisms, in a hospital environment is often poorly defined and difficult to control. While some studies have demonstrated that previous occupancy of an ICU room by a patient with multidrug-resistant (MDR) gram-negative bacteria is a risk factor for acquisition by subsequent occupants, others have infrequently recovered gram-negatives from the hospital environment [26,27]. Additionally, while contact precautions have been demonstrated to have efficacy in control of transmission of MRSA, VRE and C. difficile, the evidence is less robust for gram-negative organisms [28,29]. Although gram-negatives are the predominant pathogens in cases of ventilator-associated pneumonia and in ICU HAIs, where death from HAI is most likely to occur, existing guidelines pertaining to control of MDR pathogens either exclude gram-negatives or acknowledge limitations in recommendations pertaining to these organisms [30-32]. Increased ability to detect environmental reservoirs of these organisms should lead to improvements in targeted control efforts. Prior studies have evaluated (by TCM) the frequency of microorganisms on high-use surfaces, HCW, PPE, and other items in the healthcare environment, with widely differing results depending on the site sampled and organism of interest. Many have focused on one organism in the immediate surroundings of a patient known to be colonized with that organism [3,9,26,33-35]. Apart from this context, studies in non-outbreak settings often evaluate epidemiologically significant pathogens for one site of interest per study. One evaluation of MDR A. baumannii contamination in a medical intensive care unit found none except in colonized patients’ immediate surroundings [5]. In our institution, an evaluation (by TCM) was made of protective lead garments at various sites, with only 5 of 182 samples positive for any bacteria, all normal skin flora; another assessment of computer keyboards/mice recovered S. aureus, Acinetobacter spp., or Pseudomonas spp. on 17% of tested surfaces [2,4]. One study demonstrated a majority of bedside charts in the ICU were contaminated with MDR bacteria [36].
Against this backdrop, we sought to determine the burden and spectrum of microorganisms detected from HCW, PPE, and a variety of high-use hospital environmental surfaces by PCR/ESI-TOF-MS compared to TCM, and to compare the two methodologies in reference to the most commonly identified microorganisms detected among clinical cultures. This study is the first to our knowledge to evaluate selected sections of the hospital microbiome, comparing TCM and the unbiased T-5000-based PCR/ESI-TOF-MS method, in an effort to understand potential reservoirs of endemic nosocomial bacteria. Compared to traditional culture, PCR/ESI-TOF-MS detected more microbes, including more pathogens of potential clinical relevance, from a greater number of surfaces, hands of HCW, and PPE. PCR/ESI-TOF-MS also disproportionately recovered more gram-negative organisms missed by culture than S. aureus. As would be expected, both TCM and PCR/ESI-TOF-MS also detected many clinically less important organisms. Potentially clinically relevant pathogens accounted for similar proportions of all results (an estimated “signal-to-noise” ratio), unless streptococci, which are not typically considered major HAI pathogens, were included. However, compared to TCM, PCR/ESI-TOF-MS was able to detect potentially clinically relevant pathogens from 3-6x the number of sites screened, depending upon the inclusivity of the definition of “clinically relevant”. There is no standard definition for potential clinical relevance, and lower-virulence organisms, such as coagulase-negative staphylococci, can present major problems for patients with orthopedic or other implanted devices, or severely compromised patients. The study definition was chosen in order to reflect the most common pathogens seen clinically at the time and to include organisms for which there is high concern for virulence and/or drug resistance [32,37]. Additionally, much greater microorganism diversity was detected by PCR/ESI-TOF-MS, including unexpected organisms with high virulence or outbreak potential (e.g. Clostridium tetani, S. pyogenes). All surfaces, hands, and PPE samples demonstrated large numbers of recoverable pathogens, but without clear trends related to number of organisms by site. Based on these data, no obvious target for increased infection control efforts was seen in the study units. As this study was designed as a pilot study with a small number of sites/samples tested, there are clear limitations to the generalizability of the results to an entire hospital microbiome. It is also difficult to draw conclusions about organisms such as Shigella flexneri and Borrelia turicatae recovered from the hospital environment in the absence of known clinical cases during the study period. It is possible that these represent misidentifications of related organisms, as with hundreds of identified organisms, even 99% specificity would lead to several misidentifications. However, previous characterizations of this technology with 405 unique bacterial species have demonstrated accurate characterization in 95% of instances, with the remaining 5% unresolved species all accurate to the genus level [15]. Other limitations of the use of PCR/ESI-TOF-MS in this context include the possibility of detection of nonviable organisms, cost, the semi-quantitative nature of the data, and inability to recover specific strains linked to a patient or outbreak isolate. In this study, PCR/ESI-TOF-MS was not used to detect specific strains of bacteria detected, though it can be and has been used specifically for rapid genotyping of A. baumannii[21], S. aureus[22] and Streptococcus pneumoniae[38]. Thus, this technology may yet prove useful in outbreak investigations using environmental sampling, especially since it detected 4–5 fold higher numbers of pathogens per site without adversely affecting the ratio of clinically irrelevant microbes.
Interestingly, PCR/ESI-TOF-MS detected widespread resistance elements throughout the hospital environment. In the case of mecA, most were associated with CNS codetections rather than S. aureus. As mecA elements are widely distributed in CNS, this is not surprising [39]. VanA was not detected with enterococcus by PCR/ESI-TOF-MS, but did have an association with the presence of Lactobacillus spp. While this organism is largely intrinsically resistant to vancomycin, previous studies have demonstrated that this is not related to the presence of vanA, which to our knowledge has not been demonstrated in lactobacilli [40,41]. It is interesting that one of the samples positive for vanA, but without enterococci by PCR/ESI-TOF-MS, grew a susceptible Enterococcus spp. on clinical culture. Furthermore, none of the five samples positive for enterococci by TCM had this organism codetected by PCR/ESI-TOF-MS, generating a question of whether there might have been specific Enterococcus spp. detection problems, which have not been previously described. Overall, there were no attempts to resolve discordant results from TCM and PCR/ESI-TOF-MS, given that essentially every sample showed discordance, at least in greater number of pathogens isolated by the latter method. The PCR/ESI-TOF-MS technology does not screen for all possible resistance genes, and as applied here only detects the resistance element, without reporting whether it is incorporated into the genome of an organism. If more than one organism is present in the test sample along with the resistance element, it was not clear with which organism the element might be associated, if any. It is possible that free-floating or promiscuous plasmids are responsible for some of these detections, which also has significance for infection transmission in recent literature [42]. Given the difficulty of performing plasmid genetic analysis and whole genome sequencing of unculturable organisms, it is likely that horizontal transmission and intergenus transfer of antimicrobial resistance elements plays a larger role in healthcare-associated transmission of gram-negative MDR pathogens than has yet been described [43]. The ability of PCR/ESI-TOF-MS to screen for a broad spectrum of genetic elements may be a starting point for hypothesis development related to horizontal transmission.
Conclusions
In summary, PCR/ESI-TOF-MS detected larger numbers and a greater diversity of organisms from a higher proportion of environmental surfaces in the hospital pilot study, particularly pathogenic gram-negative organisms, without adversely affecting the “signal-to-noise” ratio of common skin contaminants detected. This may prove to be a useful technology for investigations of hospital outbreaks. However, though PCR/ESI-TOF-MS has the capacity to genotype organisms, its use in this screening context did not provide for further information about strain or antimicrobial resistance. Additionally, further investigation is warranted in reference to the frequent detection of resistance elements, particularly vanA, in the absence of known host species for these resistance elements. PCR/ESI-TOF-MS may be a useful adjunct among infection control investigational tools for understanding transmission of endemic pathogens.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Study concept and design: HY, JW, CM. Acquisition of samples: HY, MC. Laboratory analyses: RK, GE, CG, TS, JC. Analysis and interpretation of data: HY, RK, GE, CM. Drafting of the manuscript: HY, CM. Critical revision of the manuscript for important intellectual content: RK, GE, HC, KC, JW, JH, JC, KM. Statistical analysis: HY. All authors read and approved the final manuscript.
Authors’ information
We give thanks for the life of collaborator, coauthor and friend J. William Costerton, who passed away during the final preparation of this manuscript.
Disclaimer
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or reflecting the views of the Department of the Army, Department of the Air Force, Department of Defense or the U.S. Government. This work was prepared as part of their official duties and, as such, there is no copyright to be transferred. All authors report no conflicts of interest relevant to this article.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Heather C Yun, Email: heather.yun@amedd.army.mil.
Rachael E Kreft, Email: rkreft1@wpahs.org.
Mayra A Castillo, Email: mayra.castillo@amedd.army.mil.
Garth D Ehrlich, Email: gehrlich@wpahs.org.
Charles H Guymon, Email: charles.guymon@us.army.mil.
Helen K Crouch, Email: helen.crouch@us.army.mil.
Kevin K Chung, Email: kevin.chung@us.army.mil.
Joseph C Wenke, Email: joseph.wenke@us.army.mil.
Joseph R Hsu, Email: joe.hsu@us.army.mil.
Tracy L Spirk, Email: tspirk@wpahs.org.
J William Costerton, Email: wcostert@wpahs.org.
Katrin Mende, Email: katrin.mende@us.army.mil.
Clinton K Murray, Email: clinton.murray@amedd.army.mil.
Acknowledgements
This work was supported by the US Army Institute of Surgical Research and the Armed Forces Health Surveillance Center including the Global Emerging Infectious System.
References
- Pittet D, Donaldson L. Clean care is safer care: a worldwide priority. Lancet. 2005;366:1246–1247. doi: 10.1016/S0140-6736(05)67506-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PL, Siu LK, Chen TC, Ma L, Chiang WG, Chen YH, Lin SF, Chen TP. Methicillin-resistant Staphylococcus aureus and Acinetobacter baumannii on computer interface surfaces of hospital wards and association with clinical isolates. BMC Infect Dis. 2009;9:164. doi: 10.1186/1471-2334-9-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees M, Snydman DR, Schmid CH, Barefoot L, Hansjosten K, Vue PM, Cronin M, Nasraway SA, Golan Y. Antibiotic exposure and room contamination among patients colonized with vancomycin-resistant enterococci. Infect Control Hosp Epidemiol. 2008;29:709–715. doi: 10.1086/589582. [DOI] [PubMed] [Google Scholar]
- Grogan BF, Cranston WC, Lopez DM, Furbee C, Murray CK, Hsu JR. Do protective lead garments harbor harmful bacteria? Orthopedics. 2011;34:e765–e767. doi: 10.3928/01477447-20110922-09. [DOI] [PubMed] [Google Scholar]
- Levin AS, Gobara S, Mendes CM, Cursino MR, Sinto S. Environmental contamination by multidrug-resistant Acinetobacter baumannii in an intensive care unit. Infect Control Hosp Epidemiol. 2001;22:717–720. doi: 10.1086/501852. [DOI] [PubMed] [Google Scholar]
- Weber DJ, Rutala WA, Miller MB, Huslage K, Sickbert-Bennett E. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J Infect Control. 2010;38:S25–S33. doi: 10.1016/j.ajic.2010.04.196. [DOI] [PubMed] [Google Scholar]
- Bures S, Fishbain JT, Uyehara CF, Parker JM, Berg BW. Computer keyboards and faucet handles as reservoirs of nosocomial pathogens in the intensive care unit. Am J Infect Control. 2000;28:465–471. doi: 10.1067/mic.2000.107267. [DOI] [PubMed] [Google Scholar]
- Morgan DJ, Rogawski E, Thom KA, Johnson JK, Perencevich EN, Shardell M, Leekha S, Harris AD. Transfer of multidrug-resistant bacteria to healthcare workers' gloves and gowns after patient contact increases with environmental contamination. Crit Care Med. 2012;40:1045–1051. doi: 10.1097/CCM.0b013e31823bc7c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder GM, Thom KA, Furuno JP, Perencevich EN, Roghmann MC, Strauss SM, Netzer G, Harris AD. Detection of methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci on the gowns and gloves of healthcare workers. Infect Control Hosp Epidemiol. 2008;29:583–589. doi: 10.1086/588701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti S, Rodriguez-Bano J, Catel-Ferreira M, Jouenne T, Vila J, Seifert H, De E. Biofilm formation at the solid–liquid and air-liquid interfaces by Acinetobacter species. BMC Res Notes. 2011;4:5. doi: 10.1186/1756-0500-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Hunter IS. Efficacy of common hospital biocides with biofilms of multi-drug resistant clinical isolates. J Med Microbiol. 2008;57:966–973. doi: 10.1099/jmm.0.47668-0. [DOI] [PubMed] [Google Scholar]
- Jonas D, Speck M, Daschner FD, Grundmann H. Rapid PCR-based identification of methicillin-resistant Staphylococcus aureus from screening swabs. J Clin Microbiol. 2002;40:1821–1823. doi: 10.1128/JCM.40.5.1821-1823.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamper PD, Cai M, Howard T, Speser S, Carroll KC. Clinical validation of the molecular BD GeneOhm StaphSR assay for direct detection of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in positive blood cultures. J Clin Microbiol. 2007;45:2191–2196. doi: 10.1128/JCM.00552-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle MS, Mostow E, Mukherjee P, Hu FZ, Melton-Kreft R, Ehrlich GD, Dowd SE, Ghannoum MA. Characterization of bacterial communities in venous insufficiency wounds by use of conventional culture and molecular diagnostic methods. J Clin Microbiol. 2011;49:3812–3819. doi: 10.1128/JCM.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker DJ, Sampath R, Massire C, Blyn LB, Hall TA, Eshoo MW, Hofstadler SA. Ibis T5000: a universal biosensor approach for microbiology. Nat Rev Microbiol. 2008;6:553–558. doi: 10.1038/nrmicro1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A. Mass spectrometry for species or strain identification after culture or without culture: past, present, and future. J Clin Microbiol. 2006;44:2677–2680. doi: 10.1128/JCM.00971-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wall Street Journal 2009 Technology Innovation Awards. 2009. http://online.wsj.com/article/SB10001424052970203440104574399714096167656.html.
- Costerton JW, Post JC, Ehrlich GD, Hu FZ, Kreft R, Nistico L, Kathju S, Stoodley P, Hall-Stoodley L, Maale G, James G, Sotereanos N, DeMeo P. New methods for the detection of orthopedic and other biofilm infections. FEMS Immunol Med Microbiol. 2011;61:133–140. doi: 10.1111/j.1574-695X.2010.00766.x. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Conti SF, DeMeo PJ, Nistico L, Melton-Kreft R, Johnson S, Darabi A, Ehrlich GD, Costerton JW, Kathju S. Characterization of a mixed MRSA/MRSE biofilm in an explanted total ankle arthroplasty. FEMS Immunol Med Microbiol. 2011;62:66–74. doi: 10.1111/j.1574-695X.2011.00793.x. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Ehrlich GD, Sedghizadeh PP, Hall-Stoodley L, Baratz ME, Altman DT, Sotereanos NG, Costerton JW, Demeo P. Orthopaedic biofilm infections. Curr Orthop Pract. 2011;22:558–563. doi: 10.1097/BCO.0b013e318230efcf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JA, Massire C, Hall TA, Ranken R, Pennella TT, Agasino Ivy C, Blyn LB, Hofstadler SA, Endy TP, Scott PT, Lindler L, Hamilton T, Gaddy C, Snow K, Pe M, Fishbain J, Craft D, Deye G, Riddell S, Milstrey E, Petruccelli B, Brisse S, Harpin V, Schink A, Ecker DJ, Sampath R, Eshoo MW. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J Clin Microbiol. 2006;44:2921–2932. doi: 10.1128/JCM.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA, Sampath R, Blyn LB, Ranken R, Ivy C, Melton R, Matthews H, White N, Li F, Harpin V, Ecker DJ, McDougal LK, Limbago B, Ross T, Wolk DM, Wysocki V, Carroll KC. Rapid molecular genotyping and clonal complex assignment of Staphylococcus aureus isolates by PCR coupled to electrospray ionization-mass spectrometry. J Clin Microbiol. 2009;47:1733–1741. doi: 10.1128/JCM.02175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother. 2006;50:4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannis JC, Manalili SM, Hall TA, Ranken R, White N, Sampath R, Blyn LB, Ecker DJ, Mandrell RE, Fagerquist CK, Bates AH, Miller WG, Hofstadler SA. High-resolution genotyping of Campylobacter species by use of PCR and high-throughput mass spectrometry. J Clin Microbiol. 2008;46:1220–1225. doi: 10.1128/JCM.02158-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker DJ, Massire C, Blyn LB, Hofstadler SA, Hannis JC, Eshoo MW, Hall TA, Sampath R. Molecular genotyping of microbes by multilocus PCR and mass spectrometry: a new tool for hospital infection control and public health surveillance. Methods Mol Biol. 2009;551:71–87. doi: 10.1007/978-1-60327-999-4_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmen SW, Hafner H, Zolldann D, Stanzel S, Lutticken R. Distribution of multi-resistant Gram-negative versus Gram-positive bacteria in the hospital inanimate environment. J Hosp Infect. 2004;56:191–197. doi: 10.1016/j.jhin.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Nseir S, Blazejewski C, Lubret R, Wallet F, Courcol R, Durocher A. Risk of acquiring multidrug-resistant Gram-negative bacilli from prior room occupants in the intensive care unit. Clin Microbiol Infect. 2011;17:1201–1208. doi: 10.1111/j.1469-0691.2010.03420.x. [DOI] [PubMed] [Google Scholar]
- Harris AD, McGregor JC, Furuno JP. What infection control interventions should be undertaken to control multidrug-resistant gram-negative bacteria? Clin Infect Dis. 2006;43(Suppl 2):S57–S61. doi: 10.1086/504479. [DOI] [PubMed] [Google Scholar]
- Strausbaugh LJ, Siegel JD, Weinstein RA. Preventing transmission of multidrug-resistant bacteria in health care settings: a tale of 2 guidelines. Clin Infect Dis. 2006;42:828–835. doi: 10.1086/500408. [DOI] [PubMed] [Google Scholar]
- Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto CA, Jernigan JA, Ostrowsky BE, Richet HM, Jarvis WR, Boyce JM, Farr BM. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. Infect Control Hosp Epidemiol. 2003;24:362–386. doi: 10.1086/502213. [DOI] [PubMed] [Google Scholar]
- Siegel JD, Rhinehart E, Jackson M, Chiarello L. Management of multidrug-resistant organisms in health care settings, 2006. Am J Infect Control. 2007;35:S165–S193. doi: 10.1016/j.ajic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Morgan DJ, Liang SY, Smith CL, Johnson JK, Harris AD, Furuno JP, Thom KA, Snyder GM, Day HR, Perencevich EN. Frequent multidrug-resistant Acinetobacter baumannii contamination of gloves, gowns, and hands of healthcare workers. Infect Control Hosp Epidemiol. 2010;31:716–721. doi: 10.1086/653201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiefel U, Cadnum JL, Eckstein BC, Guerrero DM, Tima MA, Donskey CJ. Contamination of hands with methicillin-resistant Staphylococcus aureus after contact with environmental surfaces and after contact with the skin of colonized patients. Infect Control Hosp Epidemiol. 2011;32:185–187. doi: 10.1086/657944. [DOI] [PubMed] [Google Scholar]
- Zachary KC, Bayne PS, Morrison VJ, Ford DS, Silver LC, Hooper DC. Contamination of gowns, gloves, and stethoscopes with vancomycin-resistant enterococci. Infect Control Hosp Epidemiol. 2001;22:560–564. doi: 10.1086/501952. [DOI] [PubMed] [Google Scholar]
- Panhotra BR, Saxena AK, Al-Mulhim AS. Contamination of patients' files in intensive care units: an indication of strict handwashing after entering case notes. Am J Infect Control. 2005;33:398–401. doi: 10.1016/j.ajic.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- Massire C, Gertz RE Jr, Svoboda P, Levert K, Reed MS, Pohl J, Kreft R, Li F, White N, Ranken R, Blyn LB, Ecker DJ, Sampath R, Beall B. Concurrent serotyping and genotyping of pneumococci by use of PCR and electrospray ionization mass spectrometry. J Clin Microbiol. 2012;50:2018–2025. doi: 10.1128/JCM.06735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryffel C, Tesch W, Birch-Machin I, Reynolds PE, Barberis-Maino L, Kayser FH, Berger-Bachi B. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene. 1990;94:137–138. doi: 10.1016/0378-1119(90)90481-6. [DOI] [PubMed] [Google Scholar]
- Klein G, Hallmann C, Casas IA, Abad J, Louwers J, Reuter G. Exclusion of vanA, vanB and vanC type glycopeptide resistance in strains of Lactobacillus reuteri and Lactobacillus rhamnosus used as probiotics by polymerase chain reaction and hybridization methods. J Appl Microbiol. 2000;89:815–824. doi: 10.1046/j.1365-2672.2000.01187.x. [DOI] [PubMed] [Google Scholar]
- Patel R. Enterococcal-type glycopeptide resistance genes in non-enterococcal organisms. FEMS Microbiol Lett. 2000;185:1–7. doi: 10.1111/j.1574-6968.2000.tb09032.x. [DOI] [PubMed] [Google Scholar]
- Mathers AJ, Cox HL, Kitchel B, Bonatti H, Brassinga AK, Carroll J, Scheld WM, Hazen KC, Sifri CD. Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals Intergenus KPC carbapenemase transmission through a promiscuous plasmid. MBio. 2011;2:e00204–e00211. doi: 10.1128/mBio.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussmann M, Hu FZ, Richter M, de Beer D, Preisler A, Jorgensen BB, Huntemann M, Glockner FO, Amann R, Koopman WJ, Lasken RS, Janto B, Hogg J, Stoodley P, Boissy R, Ehrlich GD. Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 2007;5:e230. doi: 10.1371/journal.pbio.0050230. [DOI] [PMC free article] [PubMed] [Google Scholar]