Abstract
The microbes residing in and on the human body influence human physiology in many ways, particularly through their impact on the metabolism of xenobiotic compounds, including therapeutic drugs, antibiotics, and diet-derived bioactive compounds. Despite the importance of these interactions and the many possibilities for intervention, microbial xenobiotic metabolism remains a largely underexplored component of pharmacology. Here, we discuss the emerging evidence for both direct and indirect effects of the human gut microbiota on xenobiotic metabolism, and the initial links that have been made between specific compounds, diverse members of this complex community, and the microbial genes responsible. Furthermore, we highlight the many parallels to the now well-established field of environmental bioremediation, and the vast potential to leverage emerging metagenomic tools to shed new light on these important microbial biotransformations.
Keywords: Metagenomics, microbiome, microbiota, reduction, hydrolysis, xenobiotics
1. Introduction
Humans have co-evolved with trillions of microorganisms, whose aggregate genomes (the microbiome; see Table 1 for terms and definitions) extend our metabolic capabilities beyond those encoded by our own human genome. The largest collection of these microorganisms resides within our gastrointestinal tract, referred to as the gut microbiota, and plays an important role in human health and disease [1]. Some of the major influences of the gut microbiota on the host are changes to the activity, bioavailability, and toxicity of xenobiotics [2]. Fortunately, unlike the human genome, the structure and/or function of the gut microbiome has the potential to be shaped by a variety of environmental factors [3], such as antibiotics [4,5], diet [6–8], and probiotics (live microorganisms administered in food or capsules) [9]. This relative plasticity, coupled to its physiological relevance, makes the gut microbiome an attractive target for personalized medicine. A better understanding of microbial xenobiotic metabolism might also help explain some of the patient-to-patient variability in the response to therapeutic drugs.
Table 1.
Key terms.
| Gnotobiotics | Derived from the Greek roots gnostos ‘known’ and bios ‘life’; refers to animals that are maintained without exposure to microbes (germ-free) prior to microbial colonization. Frequently used study groups include mono-or bi-association (1–2 microorganisms), conventionally-raised (maintained outside an isolator in a standard mouse facility), conventionalized (i.e., ex-germ-free; colonized with a complex microbiota), or humanized (colonized with a human sample). |
| Metagenomics | A rapidly growing field that focuses on using culture-independent techniques to characterize the structure and function of microbial communities and their interactions with the environment. Metagenomic studies include (i) shotgun sequencing of microbial DNA isolated directly from a given environment, (ii) high-throughput screening of expression libraries, constructed from cloned community DNA (functional metagenomics), (iii) profiling of community-wide gene expression and protein abundance (meta-transcriptomics and meta-proteomics), and (iv) identification of a community’s metabolic network (metabolomics). |
| Microbiota | A microbial community, often including Bacteria, Archaea, small Eukaryotes, and viruses occupying a given habitat. |
| Microbiome | A term used to refer to the aggregate genomes present in members of a given microbiota, and the activities that they encode. |
| Xenobiotics | Compounds foreign to a living organism, used here to refer to therapeutic drugs, antibiotics, and diet-derived bioactive compounds. |
In this review we highlight some of the recent progress towards a more comprehensive view of xenobiotic metabolism encompassing both our human and microbial genomes. These important proof-of-principle studies include the cancer drug irinotecan [10], the widely used analgesic acetaminophen [11], and the dietary supplement phosphatidylcholine [12]. In total, over 40 therapeutic drugs have been linked to metabolism by the gut microbiota (Table 2) [2], and in nearly all cases the specific microbes, or microbial consortia, responsible are largely uncharacterized (see Figure 1 and Table 3 for some key gut microbial species and enzymes of interest). The elucidation of these mechanistic details and the identification of the enzymes responsible for crucial steps in xenobiotic metabolism could be a critical step towards developing combination treatments targeting key bacterial enzymes, as demonstrated for irinotecan [10].
Table 2.
Xenobiotics metabolized by the human gut microbiota.
| Reaction type | Substrate | Reference |
|---|---|---|
| Reduction | Digoxin* | [48] |
| Clonazepam | [101] | |
| Sulindac | [102] | |
| Sulfasalazine* | [50] | |
| Prontosil | [103] | |
| Neoprontosil | [103] | |
| Balsalazine | [104] | |
| Olsalazine | [51] | |
| Nitrazepam | [105] | |
| Clonazepam | [101] | |
| Sulfinpyrazone | [102] | |
| Sulindac | [102] | |
| Omeprazole | [106] | |
| Metronidazole | [107] | |
| Misonidazole | [108] | |
| Zonisamide | [109] | |
|
| ||
| Hydrolysis | Nitroglycerin | [110] |
| Sodium picosulphate | [2] | |
| Carbenoxolone | [111] | |
| Methotrexate | [112] | |
| Morphine glucuronide | [113] | |
| Sennosides | [114] | |
| Sorivudine | [115] | |
| Lactulose | [116] | |
|
| ||
| Functional group removal | Flucytosine | [117] |
| Methamphetamine | [118] | |
| Levodopa* | [54] | |
| Phenacetin | [119] | |
| Succinylsulfathiazole | [2] | |
|
| ||
| N-oxide cleavage | Ranitidine | [120] |
| Nizatidine | [121] | |
|
| ||
| Proteolysis | Insulin | [122] |
| Calcitonin | [122] | |
|
| ||
| Denitration | Isosorbide dinitrate | [123] |
| Glyceryl trinitrate | [110] | |
|
| ||
| Deconjugation | Indomethacin | [124] |
| Irinotecan** | [125] | |
|
| ||
| Amine formation/amide hydrolysis | Chloramphenicol | [126] |
|
| ||
| Thiazole ring-opening | Levamisole | [127] |
|
| ||
| Acetylation | 5-Aminosalicyclic acid | [128] |
|
| ||
| Isoxazole scission | Risperidone | [129] |
|
| ||
| Other | Azetirelin | [130] |
|
| ||
| Potassium oxonate | [131] | |
|
| ||
| Hesperidin | [132] | |
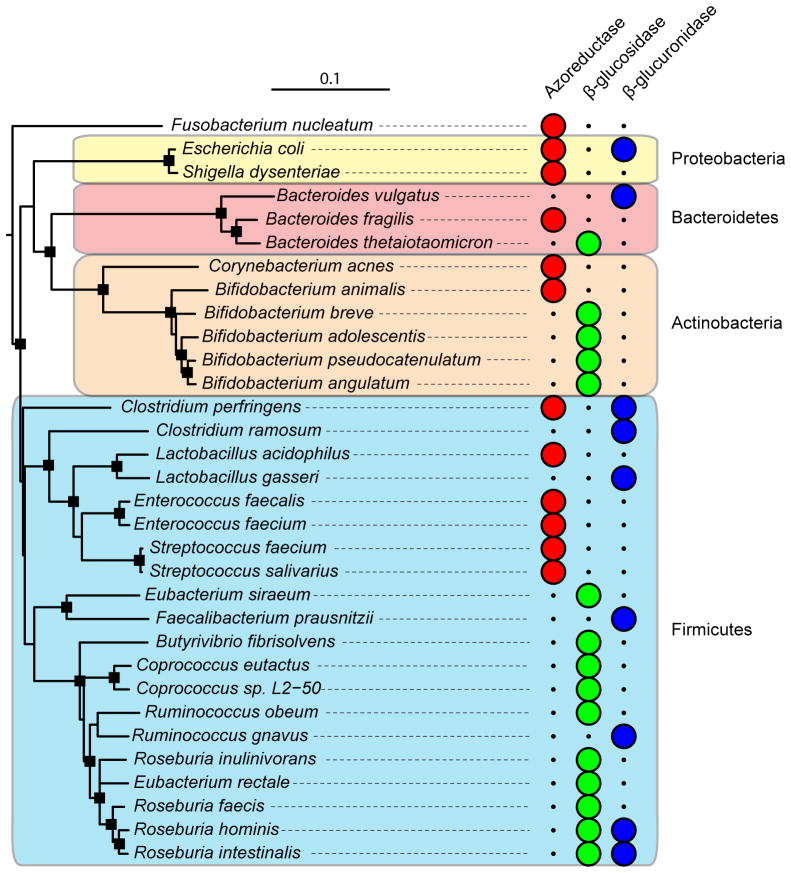
Figure 1. Phylogenetic distribution of cultured gut isolates with enzymatic activities relevant to xenobiotic metabolism.
Full-length aligned 16S rRNA gene sequences for bacteria of interest were retrieved from the Ribosomal Database Project website (Release 10, update 29) [99]. The “Tree Builder” tool was used with Fusobacterium nucleatum as the outgroup. The resulting tree was exported in Newick format and annotated using the Interactive Tree of Life website [100]. Major bacterial phyla are shown in colored boxes: Actinobacteria (orange), Bacteroidetes (red), Firmicutes (blue), and Proteobacteria (yellow). Large circles indicate the presence of a confirmed enzymatic activity within each bacterial species. Nodes with a bootstrap value >70 are indicated by black squares.
Table 3.
Known links between xenobiotics, members of the gut microbiota, and key reactions. See Figures 1 and 2 for more information.
| Xenobiotic compound | Metabolizing isolate(s) | Reaction | Reference |
|---|---|---|---|
| Digoxin | Eggerthella lenta | Reduction reaction | [48] |
| Irinotecan | Bacteroides, Clostridium, Escherichia | Deconjugation | [10] |
| Levamisole | Bacteroides, Clostridium | Thiazole ring-opening | [127] |
| Sorivudine | Various Bacteroides: B. vulgatus, B. thetaiotaomicron, B. fragilis, B. uniformis and B. eggerthii | Phosphorylase activity | [133] |
| Sulfasalazine | Many genera including: Bacteroides, Bifidobacterium, Lactobacillus, and Enterococcus | Azoreductase activity | [50] |
We will also discuss the various metagenomic methods that can now be harnessed to analyze these complex microbial communities, many of which were first applied to analogous environmental studies. For example, there are clear overlaps with the field of environmental bioremediation, wherein a combination of surveys of highly contaminated sites; detailed mechanistic studies of cultured isolates; the engineering of in vitro bioreactors; stable-isotope probing; and functional metagenomics have been applied successfully. In addition, a unique advantage of studying the human microbiome is the potential to use germ-free and intentionally colonized animal models (including “humanized” mice, formerly germ-free animals colonized with human microbial communities) [7,13,14], allowing extensive control over host genotype, environmental parameters, and microbial colonization. Together, these techniques have the potential to shed new light on the role of gut microbial communities in xenobiotic metabolism.
2. Lessons from the bioremediation literature
While the diversity of the microbes residing within the gastrointestinal tract is indeed impressive, it is thought that soil environments contain the most diverse microbial communities on Earth [15]. The study of soil microorganisms has revolutionized the pharmaceutical industry as many of the antibiotics and natural product therapeutics are originally derived from soil microbes [16]. Moreover, the microorganisms living in the soil have been integral in the biodegradation of anthropogenic compounds (i.e. pollutants) [17]. Indeed, the field of bioremediation has led the way in studying how microbial communities interact with xenobiotic compounds; importantly, findings from this work have been applied to improving the state of our environment in many cases [18]. A catalogue of microbial biocatalytic reactions on environmental pollutants currently lists almost 1,500 reactions carried out by 529 microorganisms affecting some 1,369 compounds [19]. Much of this knowledge is derived from studies of cultured organisms, and therefore does not even include the wealth of knowledge to be gained from the metagenomic study of whole communities.
Within the bioremediation literature there are a multitude of well-characterized enzymatic activities, and organisms that serve as tractable models for microbial xenobiotic metabolism. One well-studied enzyme class is a group called the dehalogenases that catalyze the cleavage of the halogen-carbon bond through a number of different mechanisms [20]. Halogenated compounds are one of the most abundant environmental pollutants due to their use in pesticides, herbicides, solvents, and many other sources. Interestingly, bacterial dehalogenase enzymes are thought to have evolved directly to degrade these xenobiotic halogenated compounds, as opposed to being a result of “catalytic promiscuity” that fortuitously results in dehalogenation [21]. Several dehalogenases have been studied at the level of protein crystal structure and this work has paved the way for the eventual rational design and control over the catalytic properties of enzymes that can degrade xenobiotics [22].
2.1. Established model organisms for bioremediation studies
A number of tractable model organisms have also emerged from bioremediation studies. Members of the genus Rhodococcus have been well-studied due to their robustness and ability to grow on a wide range of substrates that includes many xenobiotics, the best-known being polychlorinated biphenyls (PCBs) [23]. Researchers have obtained full genome sequences of several closely related Rhodococcus species, developed unmarked gene deletion techniques, and assembled an extensive set of plasmids and gene expression tools [24].
Organisms belonging to the genus Geobacter represent an example of a group playing an important role in biogeochemical cycling and the degradation of recalcitrant compounds, but are also quite amenable to culturing and manipulation in isolation [25]. Discovered just over 30 years ago, today both natural and engineered Geobacter strains are extensively used in bioremediation strategies. One example of this is the use of Geobacter and its unique ability to reductively precipitate uranium for the in situ decontamination of mining sites [26].
2.2. Microbial adaptation to xenobiotic exposure
Microorganisms living in the soil can adapt to the presence of xenobiotics in several different ways: (i) random mutations can increase the resistance to an otherwise toxic xenobiotic; (ii) mutations can also enhance the microbial ability to degrade a xenobiotic; and (iii) organisms can acquire genes encoding catabolic enzymes from other members in the environment [27]. Following these adaptations it is common to see the better-suited members of the community thrive and increase in relative abundance. A number of mechanisms have been studied that explain how the acquisition of foreign genes takes place; this typically involves plasmids, transposons, integrons, or phages as a vehicle for foreign genetic material. One of the first observations of bacteria adapting to anthropogenic compounds was the discovery of the spread of antibiotic resistance genes [28]; however, there is now indirect evidence of adaptation for many compounds.
Degradation of the widely used herbicide atrazine is accomplished by three major biotransformation reactions, each catalyzed by a different enzyme. Microbial consortia that were isolated from soil samples from around the world have demonstrated atrazine degradation capability; however, single organisms have also been shown to carry out the metabolism in pure culture [29]. Genetic characterization of the atrazine degrading genes in pure isolates showed that these catabolic genes are plasmid-linked, and that DNA similar to known insertion sequences flank the catabolic cluster [30]. Studies investigating the molecular mechanisms underlying the acquisition of catabolic genes have suggested that the horizontal gene transfer events can influence the de novo assembly of catabolic pathways.
2.3. Key techniques used in bioremediation studies
It is useful to consider the techniques utilized by researchers studying bioremediation, as they may also be applicable to studies involving the microbial metabolism of xenobiotics in the human gut. One example is the culture and sequence-independent approach of stable isotope probing (SIP). SIP involves the 13C or 14C-labeling of the compound of interest; organisms able to degrade the compound will assimilate the labeled carbon into their biomass (DNA, RNA, or fatty acids); this provides a way to get at sequence and functional information from candidate degraders [31]. Recent work has shown that SIP can be successful at monitoring the activity of specific members of the gut microbiota in response to changes in nutrient status by monitoring de novo RNA synthesis [32]. Another recent study demonstrated that 16S rRNA-based SIP can be used to assign metabolic activities such as glucose and starch fermentation to specific members of the gut microbiota [33,34].
Another widely used approach for isolating bacterial strains capable of degrading xenobiotics is to establish growth conditions where the compound of interest is provided as the sole source of carbon and/or nitrogen for complex microbial communities—a technique called selective enrichment. This approach was recently used to identify bacteria capable of subsisting on 18 antibiotics, belonging to 8 classes of both natural and synthetic origin [35]. These antibiotic consuming bacteria represented multiple bacteria phyla and were on average able to resist 17 of the 18 tested antibiotics at clinically relevant concentrations.
As a complementary approach to culture-based screens, researchers have constructed and screened small- and large-insert libraries in host organisms, such as Escherichia coli, for activities of interest (referred to as functional metagenomics). This has been frequently used to identify novel genes for antibiotic resistance, due to the widespread nature of these genes and the relatively straightforward nature of the assay: bacterial growth at inhibitory concentrations. This approach has been used to identify aminoglycoside, tetracycline, and β-lactamase resistance genes from soil samples [36,37]. Recently, functional metagenomics was used to identify resistance genes in human saliva and fecal samples [38].
A related strategy for screening microbial communities for genes involved in xenobiotic metabolism is a technique called substrate induced gene expression (SIGEX). SIGEX relies on the fact that catabolic genes are often transcriptionally activated by their substrate [39]. Briefly, community DNA is extracted from environmental samples, sheared to 5–10 kb in length, and then cloned into a green fluorescent protein (GFP) expression vector. FACS (fluorescence activated cell sorting) can then be used to screen the resulting metagenomic library for clones that induce GFP upon growth in media supplemented with xenobiotics. This approach was successfully used to isolate a previously characterized phenol degradation operon from Ralstonia eutropha, an organism isolated from sludge, and aromatic-hydrocarbon responsive operons from petroleum-contaminated groundwater [40].
Together, these studies demonstrate the insights that can be gleaned from a combination of culture-based and culture-independent approaches, with both functional and sequencing-based metagenomic tools. It will be interesting to determine if the human microbiota harbors similar xenobiotic metabolism powerhouses, as shown for Geobacter and Rhodococcus, that could be developed into genetically tractable models to elucidate the fundamental mechanisms at play in the gastrointestinal tract, the importance of horizontal gene transfer, and the selection pressures for or against these metabolic pathways. Furthermore, as demonstrated for contaminated environmental samples, it will be important to study the microbial communities associated with human subjects, or animal models, that have undergone long-term treatment with xenobiotic compounds.
3. Direct metabolism of orally administered compounds
The oral administration of therapeutic drugs is often preferred over other routes because of its convenience and minimal invasiveness. Compounds administered orally face a number of obstacles as they proceed to the body site for which they are intended. Some of the factors that contribute to determining the optimal route of administration include chemical stability, solubility, taste/odor, local irritation, absorption, blood/brain permeability (distribution), toxicity, and the potential for metabolism before reaching the desired body site [41].
While we have only recently begun to use metagenomic methods to characterize the diversity and function of the gut microbiota [42], their involvement in drug metabolism has been know about for decades [43]. Many of the early descriptions derived from experiments where the biotransformation was observed after incubating the drug with intestinal contents ex vivo (often from the cecum—a region of the proximal colon that is a large out-pouching in rodents) [2]. Other studies followed in vivo biotransformations in humans or animal models and noticed that the biotransformation was abolished after pre-treatment with broad-spectrum antimicrobials, or when comparing gnotobiotic versus conventionally-raised mice; in both cases the assumption is that the gut microbiota are primarily responsible for the biotransformation. However, it should be noted that studies where antimicrobials are applied often fail to account for the direct impact of antimicrobials on the host, as is the case for the inhibition of P-glycoprotein-mediated efflux of digoxin and its metabolites [44].
For drugs that are able to reach the distal small intestine or colon, the rich biochemical diversity of the gut microbiome has the potential to directly influence the trajectory of the compound. While many of the reactions contributing to drug metabolism from the host perspective are oxidation and conjugation reactions that take place in the liver, the reactions in the gut tend to be dominated by reduction or hydrolysis reactions [43]. In the following paragraphs, we will highlight three examples of the direct metabolism of xenobiotics: the inactivation of digoxin by Eggerthella lenta, the activation of sulfasalazine by microbial azoreductases, and the microbial activation of levodopa (L-DOPA) in the gut.
3.1. Inactivation of digoxin by Eggerthella lenta
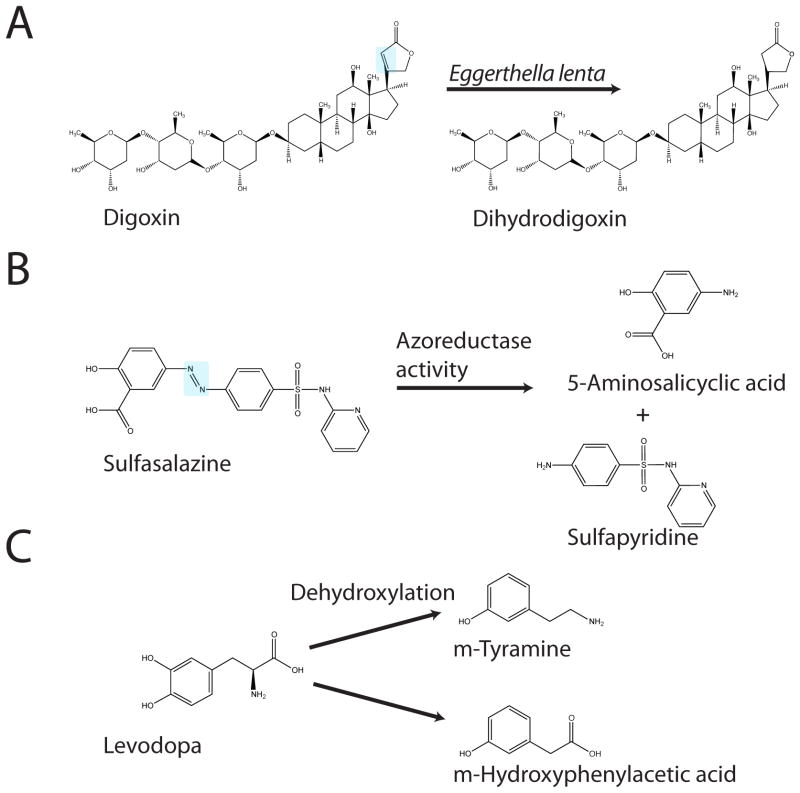
The cardiac glycoside digoxin is widely used as a therapeutic agent to treat atrial fibrillation and congestive heat failure. The positive effects of digoxin treatment are mediated through binding of the human Na+/K+ ATPase transporter in cardiac myocytes. Importantly, the well-characterized interaction between the glycoside and the Na+/K+ ATPase is almost completely abolished when the lactone ring portion of the molecule is in the reduced state [45]. The presence of the reduced form of digoxin (dihydrodigoxin) had puzzled investigators for years because this metabolite was not seen in all individuals receiving digoxin therapy [46]. A key finding was then uncovered; co-administration of a 5-day course of antibiotics prevented dihydrodigoxin excretion in patients that normally produce the metabolite [47], suggesting that the gut microbiota may be involved in this clinically relevant drug transformation. A large scale screening experiment of fecal isolates allowed Lindenbaum and colleagues to identify Eggerthella lenta as the sole microbe able to carry out the conversion of digoxin to dihydrodigoxin in isolation (Figure 2a) [48]. The case of digoxin metabolism by E. lenta is especially interesting not only for the clinical relevance of the biotransformation, but because it is one of the few examples where a relevant biotransformation has been demonstrated by a single bacterium in isolation (Table 3). The isolation of E. lenta sets the stage for more detailed mechanistic studies from the microbial perspective, and provides a valuable opportunity to study the factors that govern this transformation both in vitro and in animal models. The cardiac glycosides could also be an interesting test case to employ culture-independent methods, including functional metagenomics and microfluidics, to determine if there are any additional microorganisms capable of reducing digoxin, and if E. lenta has the ability to transfer this function to other bacteria.
Figure 2. Key examples of microbial biotransformations: (A) digoxin, (B) sulfasalazine, and (C) levodopa (L-DOPA).
Structures were obtained from the ChemBioDraw Ultra database (version 12.0.3.1216). The known organism or enzyme responsible is indicated for each transformation.
3.2. The activation of azo bond-containing prodrugs
One of the most common types of reduction reactions that take place in the gut is the reduction of the azo bond by azoreductases encoded by members of the gut microbiota (Figure 1). The azo bond is often used for the delivery of prodrugs whose pharmacological activity depends on gut localization. Prodrugs are compounds that are ingested in an inactive state, depending on an activation event to produce the pharmacologically active form of the compound [49]. A fascinating example of bacterial azoreductases playing an important role in therapeutics comes from the drug sulfasalazine, which is used to treat rheumatoid arthritis as well as inflammatory bowel diseases such as ulcerative colitis and Crohn’s disease [2]. Sulfasalazine consists of an anti-inflammatory 5-aminosalicylic acid (5-ASA) molecule linked by an azo bond to a sulfapyridine molecule (Figure 2b). The complete conversion of sulfasalazine into its constituent molecules was observed in the feces of conventional rats; however, the drug remained unchanged in antibiotic-treated and germ-free rats [50]. In this example, while the gut microbiota contribute to the activation of sulfasalazine, they also produce undesirable side effects as the sulfapyridine portion of the molecule is thought to cause toxicity. This problem can be circumvented by using the azo bond to fuse two 5-ASA molecules together in a drug called olsalazine [51].
3.3. Alteration of L-DOPA pharmacokinetics by the gut microbiota
To date the most effective drug-based treatment for Parkinson’s disease is the administration of the dopamine precursor L-DOPA [52]. Individuals suffering from Parkinson’s disease are temporarily relieved of their clinical symptoms after orally administered L-DOPA crosses the blood/brain barrier and is subjected to a decarboxylation reaction that results in the replenishment of dopamine in the central nervous system (CNS) [53]. Importantly, L-DOPA metabolism can also be demonstrated when the drug is incubated with rat cecal contents [54]. In this case the metabolism of L-DOPA by the gut microbiota (Figure 2c) could conceivably reduce the amount of the drug that arrives in the CNS and adversely affect therapy. Additionally, studies into the pharmacokinetics of orally administered L-DOPA revealed that the presence of Helicobacter pylori was associated with a decrease in the plasma levels of L-DOPA [55,56]. It was later shown using in vitro studies that L-DOPA is able to interact with the surface adhesins of H. pylori cells, potentially explaining this decrease in absorption and bioavailability [57], again titrating the drug away from its intended and therapeutically useful body site.
4. Deconjugation of drugs excreted in the bile
The process of enterohepatic circulation is an integral part of human digestion and drug metabolism, which can be broadly defined as the cycle consisting of the biliary excretion of a solute, followed by intestinal reabsorption [41]. Depending on the solute, the cycles may include hepatic conjugation and intestinal deconjugation, and while this cycling allows for the bile acids that help to metabolize dietary lipids to be reused, it also profoundly influences the metabolism of therapeutic drugs [58,59]. Xenobiotic compounds that enter into enterohepatic circulation effectively extend their exposure time to multiple body sites, which can lead to toxicity and other unintended consequences.
The extent to which enterohepatic circulation affects drug metabolism depends on several factors: the chemical properties and solubility of the drug; P-glycoprotein efflux and the expression of other transporters; metabolism in the gut wall; and metabolism by the intestinal microbiota [58]. Studies looking at the bulk enzymatic activity of the gut microbiota have shown that bacterial β-glucuronidases are involved in deconjugating xenobiotics in the gut, allowing them to be reabsorbed through the portal vein thus prolonging their time in the body. In some cases the local accumulation of deconjugated xenobiotics can have toxic side effects and potentially lead to carcinogenesis [60]. Another enzyme class, the β-glucosidases, can metabolize a wide range of plant-derived glucosides resulting in toxic or sometimes beneficial effects [61]. Of note, there is marked inter-individual variation in these various enzyme activities [62], and this disparity may be affected by dietary choices [63]. Indeed, an experiment monitoring the effects of dietary changes on β-glucuronidase, azoreductase, and nitroreductase activity found that an increase in meat consumption of rats prompted a subsequent increase in the activity of all three of these microbial enzyme activities; this same study also noted that enzyme activity also increased with the age of the animal [64]. It has also been shown that humans consuming a diet high in meat display a striking increase in fecal β-glucuronidase activity [65]. These early observations did not include microbial community analyses, but recent work suggests that the capacity for β-glucuronidase activity is widespread amongst members of the gut microbiota [66] (Figure 1).
4.1. The side effects of a chemotherapeutic drug are reduced by inhibiting a microbial enzyme
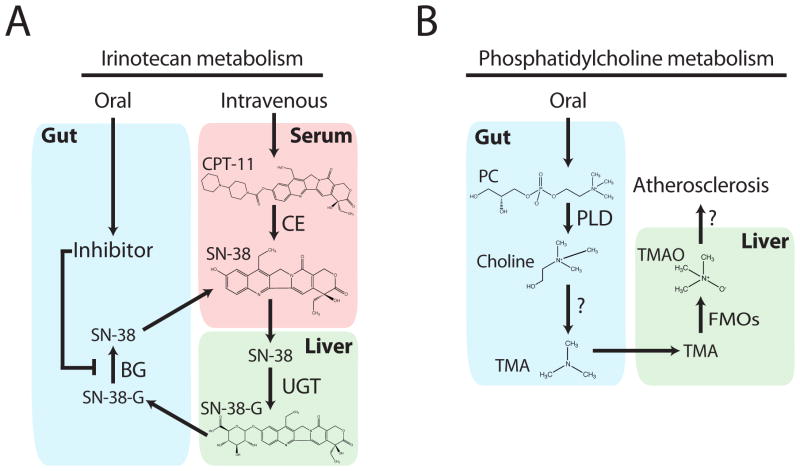
For a striking example of how a mechanistic understanding of the interactions between therapeutic drugs and the indigenous gut microbiota can affect clinical outcomes, we consider the complex metabolism of the chemotherapeutic agent irinotecan (CPT-11) (Figure 3a). Irinotecan is semi-synthetic derivative of the natural alkaloid camptothecin, and is used as a first-line therapy for colorectal cancers [67]. The drug is delivered intravenously and then metabolized by carboxylesterases in tissue and serum to form the active SN-38; the cytotoxic effects of SN-38 are mediated though the inhibition of topoisomerase I in tumor cells [67]. The active compound is then glucuronidated by hepatic UDP-glucuronosyltransferases to SN-38-G, prior to biliary excretion into the intestinal lumen. One of the major and often dose-limiting side effects of irinotecan chemotherapy is intestinal toxicity manifested by severe diarrhea, weight loss, and anorexia. These undesirable side effects were shown to result from the conversion of SN-38-G back into SN-38 by members of the gastrointestinal microbiota. Irinotecan-induced diarrhea has been linked to an increase in the expression of colonic β-glucuronidase, and also increases in β-glucuronidase-encoding microorganisms [68]. Antibiotic treatment to suppress the activity and growth of intestinal microbes drastically reduces the undesirable side effects of irinotecan [69]. While this type of co-therapy has improved irinotecan efficacy in the clinic [70], depleting ones gut microbiota with broad-spectrum antimicrobials is not ideal; and leaves patients open to other complications including infection by opportunistic pathogens such as Clostridium difficile [71].
Figure 3. The complex host-microbial metabolism of irinotecan and phosphatidylcholine.
(A) Irinotecan is administered intravenously in the inactive form (CPT-11), followed by activation and subsequent inactivation by host enzymes, and release into the gut via bile. Microbial enzymes can then reactivate these compounds; a process that can be blocked by orally administered inhibitors. (B) Phosphatidylcholine, consumed in many common foods or as a dietary supplement, is converted to choline followed by microbial production of TMA, the down-stream products of which can contribute to atheroschlerosis. Abbreviations: BG (microbial β-glucuronidase), CE (host carboxylesterases), CPT-11 (irinotecan), FMOs (hepatic flavin monooxygenases), PC (phosphatidylcholine), PLD (phospholipase D), TMA (trimethylamine), TMAO (trimethylamine oxide), and UGT (hepatic UDP-glucuronosyltransferases).
In a recent study investigators hypothesized that administering irinotecan with a specific inhibitor of microbial β-glucuronidase activity might yield a better clinical outcome. To this end, the researchers screened over 10,000 compounds for their ability to inhibit the E. coli β-glucuronidase enzyme [10]. Four positive hits were able to inhibit the enzyme both in vitro and in a whole cell assay and were thus chosen for further characterization. Comparing crystal structures of the E. coli and mammalian β-glucuronidases revealed a 17-residue loop unique to the bacterial enzyme that was key to the inhibitory properties of the lead compound [10]. Wallace and colleagues showed that the inhibitor did not reduce bacterial viability or affect the growth of prominent members of the gut microbiota. Importantly, there was also no effect on the viability of human cell lines. Finally, the authors demonstrated that co-administration of irinotecan with the inhibitor in a mouse model dramatically reduced the incidence of diarrhea, and protected intestinal tissues by showing a marked reduction in gut epithelial cell inflammation [10]. Recent findings have expanded the utility of this β-glucuronidase inhibitor as it was shown to prevent the formation of ulcers due to the administration of nonsteroidal anti-inflammatory drugs [72].
5. Microbial metabolism of dietary supplements
It is worth considering the microbial metabolism of micronutrients and dietary supplements as many of the enzymatic activities involved in these biotransformations may also be relevant for xenobiotic metabolism. These metabolic capabilities allow the conversion of many classes of phenolic compounds including flavonoids, isoflavonoids, lignans, phenolic acids, and tannins [73]. For example, the conjugated hydroxycinnamate chlorogenic acid is one of the most abundant polyphenols in the human diet, found in coffee, fruits, and vegetables [74]. Following ingestion, these compounds typically reach the gut in an unaltered state where they are subject to hydrolysis by esterases encoded by the gut microbiota. Hydrolysis of conjugated hydroxycinnimates gives rise to free acids including ferulic, p-coumaric, and caffeic acid, all of which possess antioxidant properties [75]. Individual bacterial strains able to carry out these hydrolysis reactions have been isolated from human fecal samples; these isolates belong to the genera Bifidobacterium, Escherichia, and Lactobacillus [76]. Studies involving the ex vivo incubation of fecal samples have demonstrated a striking variation across individuals in both the metabolism [77] and production [78] of bioactive phenolic compounds.
5.1. Equol production from soy-derived isoflavonoids
One biotransformation of considerable interest is the production of equol from isoflavonoids found in soy-containing foods (i.e. daidzein) [79]. Equol has received considerable attention because of several proposed health benefits including potent antioxidant activity [80] and a potential role in the prevention of prostate cancer [81]. Interestingly, there is significant inter-individual variation in equol production, with one study classifying ~36% of individuals as high equol producers [79], and another demonstrating both inter-individual variation and temporal changes within individuals [82]. A number of bacterial isolates have been identified that possess the ability to produce equol from daidzein in pure culture, these include members of the genus Lactococcus [83], and Slackia [84,85]; recent work has also isolated the bacterial genes responsible for the conversion [86,87]. More work is needed to investigate the factors that affect the colonization of equol producers and the expression of the genes relevant for the conversion.
5.2. A link between the microbial metabolism of choline metabolites and cardiovascular disease
While the microbial metabolism of bioactive components from our diet have beneficial effects in the case of equol production, recent work investigating a dietary link to cardiovascular disease (CVD) demonstrates the potential for gut microorganisms to contribute to disease. A study by Wang et al. (2011) began with a metabolomics screen looking for biomarkers predictive of heart disease. This screen revealed that metabolites of the lipid phosphatidylcholine, choline, trimethylamine N-oxide (TMAO), and betaine, are associated with increased risk for the development of CVD. In a mouse model prone to developing atherosclerosis, dietary choline supplementation triggered an increase in plasma TMAO and a subsequent increase in plaque development. Since gastrointestinal microorganisms play a role in the conversion of choline to trimethylamine (a compound that is subsequently converted to TMAO in the liver; see Figure 3b) [88]; it was therefore proposed that choline-induced atherosclerotic development might be prevented if the microbes were inhibited. Indeed, the administration of broad-spectrum antimicrobials not only prevented the formation of TMAO, but abolished the choline-induced atherosclerosis [12].
6. Impact of the gut microbiota on host xenobiotic metabolism
In addition to the direct metabolism of xenobiotics, the human gut microbiota is capable of indirectly affecting xenobiotic metabolism through host-microbial interactions. Recent studies, discussed in detail below, have demonstrated that these interactions result in altered host gene expression and the inhibition of host enzymatic activity by microbial metabolites [11,89].
6.1. Gut microbes can alter the expression of host genes involved in xenobiotic metabolism
One study used microarrays to examine the expression in the liver of genes that are known to play a role in xenobiotic metabolism and compared expression patterns between germ-free and conventionally-raised mice [89]. This work uncovered that in germ-free mice there is an increase in some of the key genes involved in xenobiotic metabolism, including the constitutively active androstane receptor, which serves as a master regulator of hepatic xenobiotic metabolism. This same study also showed that these differences were physiologically relevant as the metabolism of the anesthetic pentobarbital was expedited in the germ-free group when compared to the colonized group of mice [89].
6.2. Metabolites produced by gut microbes can affect the host’s capacity to process xenobiotics
Clayton and colleagues took a different approach by monitoring the pre- and postdose metabolite profiles of individuals receiving the common drug acetaminophen. Conjugated acetaminophen metabolites were quantified to assess the extent to which the drug was metabolized by the two major phase II pathways the drug is subject to: O-sulfonation and glucuronidation. A correlation between high predose levels of the microbial metabolite p-Cresol correlated with a low post dose ratio of acetaminophen sulfate to acetaminophen glucuronide [11]. p-Cresol is an amino acid derivative that, like acetaminophen, is a substrate for hepatic sulfotransferase enzymes [90]. Thus, it is proposed that microbial p-Cresol diminishes the host’s capacity to metabolize acetaminophen by titrating away sulfotransferase activity. These findings might explain some acetaminophen hepatotoxicity reported in the literature, but importantly may have far-reaching effects on a more broad range of xenobiotics, hormones, and neurotransmitters that depend on O-sulfonation [11].
7. Future directions
Given the tremendous progress in our understanding of the organismal and genetic diversity found within the human gut microbiome in the past few years [91–94], we now have the essential tools and experimental framework to conduct a more in-depth exploration of the contributions of our microbial communities to xenobiotic metabolism. Many important questions remain unanswered, including a fundamental understanding of the microbial and genetic underpinnings of key biotransformations, and their interactions with host and environmental risk factors.
One fascinating, if challenging to answer, question is why gut microbes have evolved multiple pathways for xenobiotic metabolism. As previously shown for environmental microorganisms in the context of bioremediation and antibiotic resistance, some of these metabolic pathways have likely evolved to avoid environmental or microbial toxins. Alternatively, they may represent evolutionary “accidents”, whereby enzymes used to activate naturally occurring compounds of dietary, microbial, or human origins exhibit cross-reactivity with xenobiotics. To address these evolutionary questions it will be important to shed new light on the potential selection pressures that may have favored the propagation of these genes; it will be critical to determine if and how microorganisms are able to use various xenobiotics as a source of carbon, nitrogen, or energy, and how important these processes are relative to minimizing toxicity.
One of the potential applications of a mechanistic understanding of microbial drug metabolism is the attempt to co-opt these metabolic pathways to rationally design compounds for release in specific locations within the gastrointestinal tract. For example, previous studies have demonstrated that azo bond cleavage by the colonic microbiota can be harnessed to generate novel prodrugs, preventing efficient absorption in the proximal gut [95]. This principal has been further extended to colon-targeted urethane-based coating linked together by azo bonds [96].
Recent studies have demonstrated the vast potential for manipulating the structure and function of our gastrointestinal microbiota, suggesting that it could be possible to affect pharmacokinetics through non-invasive strategies. These options include (i) dietary changes [6,7], (ii) probiotics [9], and (iii) drugs targeting the gut microbiota [10]. It may also be possible to deliver novel genes by harnessing the frequent exchange of genetic information between distantly-related microorganisms [97], as recently described for the transfer of genes for the metabolism of algal polysaccharides from diet-derived microorganisms into common members of the Japanese microbiota [98].
Finally, it will be important to move towards high-throughput assays for testing the impact of gut microorganisms on a wide variety of xenobiotics (and vice versa), including detailed structure-function analyses. These types of screens would provide critical information revealing which drugs are most interesting for detailed mechanistic analysis ranging from in vitro gut models, to animal models, to large-scale surveys of patients before, during, and after treatment. Substantial progress will require efforts across a wide range of disciplines, spanning microbiology, genomics, bioinformatics, pharmacology, and chemistry, among others. Together, these studies will help us move towards a more comprehensive, metagenomic view of xenobiotic metabolism, with potentially profound implications for the treatment of disease, and for our understanding of human and microbial metabolism.
Acknowledgments
HJH is supported by a postdoctoral fellowship from the Canadian Institutes of Health Research (MFE-112991). PJT is supported by a grant from the National Institutes of Health (P50 GM068763).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Turnbaugh PJ, Stintzi A. Human health and disease in a microbial world. Front Microbiol. 2011;2:190. doi: 10.3389/fmicb.2011.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008;363:1–25. doi: 10.1016/j.ijpharm.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: An ecological perspective. Sci Transl Med. 2012;4:137rv5. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108 (Suppl 1):4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–23. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–30. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, Redinbo MR. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–5. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106:14728–33. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faith JJ, Rey FE, O’Donnell D, Karlsson M, McNulty NP, Kallstrom G, Goodman AL, Gordon JI. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010;4:1094–8. doi: 10.1038/ismej.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gootenberg DB, Turnbaugh PJ. Companion animals symposium: Humanized animal models of the microbiome. J Anim Sci. 2011;89:1531–7. doi: 10.2527/jas.2010-3371. [DOI] [PubMed] [Google Scholar]
- 15.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–90. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 16.Davies J. How to discover new antibiotics: Harvesting the parvome. Curr Opin Chem Biol. 2011;15:5–10. doi: 10.1016/j.cbpa.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Galvão TC, Mohn WW, de Lorenzo V. Exploring the microbial biodegradation and biotransformation gene pool. Trends in Biotechnology. 2005;23:497–506. doi: 10.1016/j.tibtech.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Paliwal V, Puranik S, Purohit HJ. Integrated perspective for effective bioremediation. Appl Biochem Biotechnol. 2012;166:903–24. doi: 10.1007/s12010-011-9479-5. [DOI] [PubMed] [Google Scholar]
- 19.Gao J, Ellis LB, Wackett LP. The University of Minnesota biocatalysis/biodegradation database: Improving public access. Nucleic Acids Res. 2010;38:D488–91. doi: 10.1093/nar/gkp771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fetzner S, Lingens F. Bacterial dehalogenases: Biochemistry, genetics, and biotechnological applications. Microbiol Rev. 1994;58:641–85. doi: 10.1128/mr.58.4.641-685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janssen DB, Dinkla IJ, Poelarends GJ, Terpstra P. Bacterial degradation of xenobiotic compounds: Evolution and distribution of novel enzyme activities. Environ Microbiol. 2005;7:1868–82. doi: 10.1111/j.1462-2920.2005.00966.x. [DOI] [PubMed] [Google Scholar]
- 22.Pavlova M, Klvana M, Prokop Z, Chaloupkova R, Banas P, Otyepka M, Wade RC, Tsuda M, Nagata Y, Damborsky J. Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate. Nat Chem Biol. 2009;5:727–33. doi: 10.1038/nchembio.205. [DOI] [PubMed] [Google Scholar]
- 23.McLeod MP, Warren RL, Hsiao WW, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, Dosanjh M, Hara H, Petrescu A, Morin RD, Yang G, Stott JM, Schein JE, Shin H, Smailus D, Siddiqui AS, Marra MA, Jones SJ, Holt R, Brinkman FS, Miyauchi K, Fukuda M, Davies JE, Mohn WW, Eltis LD. The complete genome of Rhodococcus sp. Rha1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci U S A. 2006;103:15582–7. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Geize R, Dijkhuizen L. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr Opin Microbiol. 2004;7:255–61. doi: 10.1016/j.mib.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, Aklujkar M, Butler JE, Giloteaux L, Rotaru AE, Holmes DE, Franks AE, Orellana R, Risso C, Nevin KP. Geobacter: The microbe electric’s physiology, ecology, and practical applications. Adv Microb Physiol. 2011;59:1–100. doi: 10.1016/B978-0-12-387661-4.00004-5. [DOI] [PubMed] [Google Scholar]
- 26.Yun J, Ueki T, Miletto M, Lovley DR. Monitoring the metabolic status of Geobacter species in contaminated groundwater by quantifying key metabolic proteins with Geobacter-specific antibodies. Appl Environ Microbiol. 2011;77:4597–602. doi: 10.1128/AEM.00114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Top EM, Springael D. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol. 2003;14:262–9. doi: 10.1016/s0958-1669(03)00066-1. [DOI] [PubMed] [Google Scholar]
- 28.Cohen ML. Epidemiology of drug resistance: Implications for a post-antimicrobial era. Science. 1992;257:1050–5. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 29.Mandelbaum RT, Allan DL, Wackett LP. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl Environ Microbiol. 1995;61:1451–7. doi: 10.1128/aem.61.4.1451-1457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Souza ML, Newcombe D, Alvey S, Crowley DE, Hay A, Sadowsky MJ, Wackett LP. Molecular basis of a bacterial consortium: Interspecies catabolism of atrazine. Appl Environ Microbiol. 1998;64:178–84. doi: 10.1128/aem.64.1.178-184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radajewski S, Ineson P, Parekh NR, Murrell JC. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403:646–9. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- 32.Reichardt N, Barclay AR, Weaver LT, Morrison DJ. Use of stable isotopes to measure the metabolic activity of the human intestinal microbiota. Appl Environ Microbiol. 2011;77:8009–14. doi: 10.1128/AEM.05573-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egert M, de Graaf AA, Maathuis A, de Waard P, Plugge CM, Smidt H, Deutz NE, Dijkema C, de Vos WM, Venema K. Identification of glucose-fermenting bacteria present in an in vitro model of the human intestine by RNA-stable isotope probing. FEMS Microbiol Ecol. 2007;60:126–35. doi: 10.1111/j.1574-6941.2007.00281.x. [DOI] [PubMed] [Google Scholar]
- 34.Kovatcheva-Datchary P, Egert M, Maathuis A, Rajilic-Stojanovic M, de Graaf AA, Smidt H, de Vos WM, Venema K. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ Microbiol. 2009;11:914–26. doi: 10.1111/j.1462-2920.2008.01815.x. [DOI] [PubMed] [Google Scholar]
- 35.Dantas G, Sommer MO, Oluwasegun RD, Church GM. Bacteria subsisting on antibiotics. Science. 2008;320:100–3. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- 36.Riesenfeld CS, Goodman RM, Handelsman J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol. 2004;6:981–9. doi: 10.1111/j.1462-2920.2004.00664.x. [DOI] [PubMed] [Google Scholar]
- 37.Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 2009;3:243–51. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 38.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–31. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchiyama T, Watanabe K. Substrate-induced gene expression (SIGEX) screening of metagenome libraries. Nat Protoc. 2008;3:1202–12. doi: 10.1038/nprot.2008.96. [DOI] [PubMed] [Google Scholar]
- 40.Uchiyama T, Abe T, Ikemura T, Watanabe K. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat Biotechnol. 2005;23:88–93. doi: 10.1038/nbt1048. [DOI] [PubMed] [Google Scholar]
- 41.Goodman LS, Brunton LL, Chabner B, Knollmann BC. Goodman & Gilman’s pharmacological basis of therapeutics. 12. New York: McGraw-Hill; 2011. p. 2084. [Google Scholar]
- 42.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldman P. Biochemical pharmacology of the intestinal flora. Annu Rev Pharmacol Toxicol. 1978;18:523–39. doi: 10.1146/annurev.pa.18.040178.002515. [DOI] [PubMed] [Google Scholar]
- 44.Hughes J, Crowe A. Inhibition of p-glycoprotein-mediated efflux of digoxin and its metabolites by macrolide antibiotics. J Pharmacol Sci. 2010;113:315–24. doi: 10.1254/jphs.10109fp. [DOI] [PubMed] [Google Scholar]
- 45.Bach EJ, Reiter M. The difference in velocity between the lethal and inotropic action of dihydrodigoxin. Naunyn Schmiedebergs Arch Exp Pathol Pharmakol. 1964;248:437–49. doi: 10.1007/BF00246890. [DOI] [PubMed] [Google Scholar]
- 46.Peters U, Falk LC, Kalman SM. Digoxin metabolism in patients. Arch Intern Med. 1978;138:1074–6. [PubMed] [Google Scholar]
- 47.Lindenbaum J, Rund DG, Butler VP, Jr, Tse-Eng D, Saha JR. Inactivation of digoxin by the gut flora: Reversal by antibiotic therapy. N Engl J Med. 1981;305:789–94. doi: 10.1056/NEJM198110013051403. [DOI] [PubMed] [Google Scholar]
- 48.Dobkin JF, Saha JR, Butler VP, Jr, Neu HC, Lindenbaum J. Digoxin-inactivating bacteria: Identification in human gut flora. Science. 1983;220:325–7. doi: 10.1126/science.6836275. [DOI] [PubMed] [Google Scholar]
- 49.Oz HS, Ebersole JL. Application of prodrugs to inflammatory diseases of the gut. Molecules. 2008;13:452–74. doi: 10.3390/molecules13020452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peppercorn MA, Goldman P. The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J Pharmacol Exp Ther. 1972;181:555–62. [PubMed] [Google Scholar]
- 51.Wadworth AN, Fitton A. Olsalazine. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in inflammatory bowel disease. Drugs. 1991;41:647–64. doi: 10.2165/00003495-199141040-00009. [DOI] [PubMed] [Google Scholar]
- 52.Vlaar A, Hovestadt A, van Laar T, Bloem BR. The treatment of early Parkinson’s disease: Levodopa rehabilitated. Pract Neurol. 2011;11:145–52. doi: 10.1136/practneurol-2011-000011. [DOI] [PubMed] [Google Scholar]
- 53.Goole J, Amighi K. Levodopa delivery systems for the treatment of Parkinson’s disease: An overview. Int J Pharm. 2009;380:1–15. doi: 10.1016/j.ijpharm.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 54.Goldin BR, Peppercorn MA, Goldman P. Contributions of host and intestinal microflora in the metabolism of L-DOPA by the rat. J Pharmacol Exp Ther. 1973;186:160–6. [PubMed] [Google Scholar]
- 55.Lyte M. Microbial endocrinology as a basis for improved L-DOPA bioavailability in parkinson’s patients treated for Helicobacter pylori. Med Hypotheses. 2010;74:895–7. doi: 10.1016/j.mehy.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 56.Pierantozzi M, Pietroiusti A, Brusa L, Galati S, Stefani A, Lunardi G, Fedele E, Sancesario G, Bernardi G, Bergamaschi A, Magrini A, Stanzione P, Galante A. Helicobacter pylori eradication and L-DOPA absorption in patients with pd and motor fluctuations. Neurology. 2006;66:1824–9. doi: 10.1212/01.wnl.0000221672.01272.ba. [DOI] [PubMed] [Google Scholar]
- 57.Niehues M, Hensel A. In vitro interaction of L-DOPA with bacterial adhesins of Helicobacter pylori: An explanation for clinicial differences in bioavailability? J Pharm Pharmacol. 2009;61:1303–7. doi: 10.1211/jpp/61.10.0005. [DOI] [PubMed] [Google Scholar]
- 58.Roberts MS, Magnusson BM, Burczynski FJ, Weiss M. Enterohepatic circulation: Physiological, pharmacokinetic and clinical implications. Clin Pharmacokinet. 2002;41:751–90. doi: 10.2165/00003088-200241100-00005. [DOI] [PubMed] [Google Scholar]
- 59.Groh H, Schade K, Horhold-Schubert C. Steroid metabolism with intestinal microorganisms. J Basic Microbiol. 1993;33:59–72. doi: 10.1002/jobm.3620330115. [DOI] [PubMed] [Google Scholar]
- 60.Gill CI, Rowland IR. Diet and cancer: Assessing the risk. Br J Nutr. 2002;88 (Suppl 1):S73–87. doi: 10.1079/BJN2002632. [DOI] [PubMed] [Google Scholar]
- 61.Manach C, Scalbert A, Morand C, Remesy C, Jimenez L. Polyphenols: Food sources and bioavailability. Am J Clin Nutr. 2004;79:727–47. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- 62.McBain AJ, Macfarlane GT. Ecological and physiological studies on large intestinal bacteria in relation to production of hydrolytic and reductive enzymes involved in formation of genotoxic metabolites. J Med Microbiol. 1998;47:407–16. doi: 10.1099/00222615-47-5-407. [DOI] [PubMed] [Google Scholar]
- 63.Wiseman H, Casey K, Bowey EA, Duffy R, Davies M, Rowland IR, Lloyd AS, Murray A, Thompson R, Clarke DB. Influence of 10 wk of soy consumption on plasma concentrations and excretion of isoflavonoids and on gut microflora metabolism in healthy adults. Am J Clin Nutr. 2004;80:692–9. doi: 10.1093/ajcn/80.3.692. [DOI] [PubMed] [Google Scholar]
- 64.Goldin B, Dwyer J, Gorbach SL, Gordon W, Swenson L. Influence of diet and age on fecal bacterial enzymes. Am J Clin Nutr. 1978;31:S136–S40. doi: 10.1093/ajcn/31.10.S136. [DOI] [PubMed] [Google Scholar]
- 65.Reddy BS, Weisburger JH, Wynder EL. Fecal bacterial beta-glucuronidase: Control by diet. Science. 1974;183:416–7. doi: 10.1126/science.183.4123.416. [DOI] [PubMed] [Google Scholar]
- 66.Dabek M, McCrae SI, Stevens VJ, Duncan SH, Louis P. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol. 2008;66:487–95. doi: 10.1111/j.1574-6941.2008.00520.x. [DOI] [PubMed] [Google Scholar]
- 67.Vanhoefer U, Harstrick A, Achterrath W, Cao S, Seeber S, Rustum YM. Irinotecan in the treatment of colorectal cancer: Clinical overview. J Clin Oncol. 2001;19:1501–18. doi: 10.1200/JCO.2001.19.5.1501. [DOI] [PubMed] [Google Scholar]
- 68.Stringer AM, Gibson RJ, Logan RM, Bowen JM, Yeoh AS, Keefe DM. Faecal microflora and beta-glucuronidase expression are altered in an irinotecan-induced diarrhea model in rats. Cancer Biol Ther. 2008;7:1919–25. doi: 10.4161/cbt.7.12.6940. [DOI] [PubMed] [Google Scholar]
- 69.Takasuna K, Hagiwara T, Hirohashi M, Kato M, Nomura M, Nagai E, Yokoi T, Kamataki T. Involvement of beta-glucuronidase in intestinal microflora in the intestinal toxicity of the antitumor camptothecin derivative irinotecan hydrochloride (CPT-11) in rats. Cancer Res. 1996;56:3752–7. [PubMed] [Google Scholar]
- 70.Flieger D, Klassert C, Hainke S, Keller R, Kleinschmidt R, Fischbach W. Phase II clinical trial for prevention of delayed diarrhea with cholestyramine/levofloxacin in the second-line treatment with irinotecan biweekly in patients with metastatic colorectal carcinoma. Oncology. 2007;72:10–6. doi: 10.1159/000111083. [DOI] [PubMed] [Google Scholar]
- 71.Stein A, Voigt W, Jordan K. Chemotherapy-induced diarrhea: Pathophysiology, frequency and guideline-based management. Ther Adv Med Oncol. 2010;2:51–63. doi: 10.1177/1758834009355164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.LoGuidice A, Wallace BD, Bendel L, Redinbo MR, Boelsterli UA. Pharmacologic targeting of bacterial beta-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J Pharmacol Exp Ther. 2012;341:447–54. doi: 10.1124/jpet.111.191122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Selma MV, Espin JC, Tomas-Barberan FA. Interaction between phenolics and gut microbiota: Role in human health. J Agric Food Chem. 2009;57:6485–501. doi: 10.1021/jf902107d. [DOI] [PubMed] [Google Scholar]
- 74.Olthof MR, Hollman PC, Zock PL, Katan MB. Consumption of high doses of chlorogenic acid, present in coffee, or of black tea increases plasma total homocysteine concentrations in humans. Am J Clin Nutr. 2001;73:532–8. doi: 10.1093/ajcn/73.3.532. [DOI] [PubMed] [Google Scholar]
- 75.Laparra JM, Sanz Y. Interactions of gut microbiota with functional food components and nutraceuticals. Pharmacol Res. 2010;61:219–25. doi: 10.1016/j.phrs.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 76.Couteau D, McCartney AL, Gibson GR, Williamson G, Faulds CB. Isolation and characterization of human colonic bacteria able to hydrolyse chlorogenic acid. J Appl Microbiol. 2001;90:873–81. doi: 10.1046/j.1365-2672.2001.01316.x. [DOI] [PubMed] [Google Scholar]
- 77.Russell WR, Scobbie L, Chesson A, Richardson AJ, Stewart CS, Duncan SH, Drew JE, Duthie GG. Anti-inflammatory implications of the microbial transformation of dietary phenolic compounds. Nutr Cancer. 2008;60:636–42. doi: 10.1080/01635580801987498. [DOI] [PubMed] [Google Scholar]
- 78.van Nuenen MH, Venema K, van der Woude JC, Kuipers EJ. The metabolic activity of fecal microbiota from healthy individuals and patients with inflammatory bowel disease. Dig Dis Sci. 2004;49:485–91. doi: 10.1023/b:ddas.0000020508.64440.73. [DOI] [PubMed] [Google Scholar]
- 79.Rowland IR, Wiseman H, Sanders TA, Adlercreutz H, Bowey EA. Interindividual variation in metabolism of soy isoflavones and lignans: Influence of habitual diet on equol production by the gut microflora. Nutr Cancer. 2000;36:27–32. doi: 10.1207/S15327914NC3601_5. [DOI] [PubMed] [Google Scholar]
- 80.Rufer CE, Kulling SE. Antioxidant activity of isoflavones and their major metabolites using different in vitro assays. J Agric Food Chem. 2006;54:2926–31. doi: 10.1021/jf053112o. [DOI] [PubMed] [Google Scholar]
- 81.Akaza H, Miyanaga N, Takashima N, Naito S, Hirao Y, Tsukamoto T, Fujioka T, Mori M, Kim WJ, Song JM, Pantuck AJ. Comparisons of percent equol producers between prostate cancer patients and controls: Case-controlled studies of isoflavones in Japanese, Korean and American residents. Jpn J Clin Oncol. 2004;34:86–9. doi: 10.1093/jjco/hyh015. [DOI] [PubMed] [Google Scholar]
- 82.Rafii F, Davis C, Park M, Heinze TM, Beger RD. Variations in metabolism of the soy isoflavonoid daidzein by human intestinal microfloras from different individuals. Arch Microbiol. 2003;180:11–6. doi: 10.1007/s00203-003-0551-6. [DOI] [PubMed] [Google Scholar]
- 83.Shimada Y, Takahashi M, Miyazawa N, Ohtani T, Abiru Y, Uchiyama S, Hishigaki H. Identification of two novel reductases involved in equol biosynthesis in Lactococcus strain 20–92. J Mol Microbiol Biotechnol. 2011;21:160–72. doi: 10.1159/000335049. [DOI] [PubMed] [Google Scholar]
- 84.Jin JS, Kitahara M, Sakamoto M, Hattori M, Benno Y. Slackia equolifaciens sp. Nov. a human intestinal bacterium capable of producing equol. Int J Syst Evol Microbiol. 2010;60:1721–4. doi: 10.1099/ijs.0.016774-0. [DOI] [PubMed] [Google Scholar]
- 85.Matthies A, Loh G, Blaut M, Braune A. Daidzein and genistein are converted to equol and 5-hydroxy-equol by human intestinal Slackia isoflavoniconvertens in gnotobiotic rats. J Nutr. 2012;142:40–6. doi: 10.3945/jn.111.148247. [DOI] [PubMed] [Google Scholar]
- 86.Tsuji H, Moriyama K, Nomoto K, Akaza H. Identification of an enzyme system for daidzein-to-equol conversion in Slackia sp. Strain natts Appl Environ Microbiol. 2012;78:1228–36. doi: 10.1128/AEM.06779-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shimada Y, Yasuda S, Takahashi M, Hayashi T, Miyazawa N, Sato I, Abiru Y, Uchiyama S, Hishigaki H. Cloning and expression of a novel NADP(H)-dependent daidzein reductase, an enzyme involved in the metabolism of daidzein, from equol-producing Lactococcus strain 20–92. Appl Environ Microbiol. 2010;76:5892–901. doi: 10.1128/AEM.01101-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.al-Waiz M, Mikov M, Mitchell SC, Smith RL. The exogenous origin of trimethylamine in the mouse. Metabolism. 1992;41:135–6. doi: 10.1016/0026-0495(92)90140-6. [DOI] [PubMed] [Google Scholar]
- 89.Bjorkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS ONE. 2009;4:e6958. doi: 10.1371/journal.pone.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 91.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT, Feldgarden M, Gevers D, Haas BJ, Madupu R, Ward DV, Birren BW, Gibbs RA, Methe B, Petrosino JF, Strausberg RL, Sutton GG, White OR, Wilson RK, Durkin S, Giglio MG, Gujja S, Howarth C, Kodira CD, Kyrpides N, Mehta T, Muzny DM, Pearson M, Pepin K, Pati A, Qin X, Yandava C, Zeng Q, Zhang L, Berlin AM, Chen L, Hepburn TA, Johnson J, McCorrison J, Miller J, Minx P, Nusbaum C, Russ C, Sykes SM, Tomlinson CM, Young S, Warren WC, Badger J, Crabtree J, Markowitz VM, Orvis J, Cree A, Ferriera S, Fulton LL, Fulton RS, Gillis M, Hemphill LD, Joshi V, Kovar C, Torralba M, Wetterstrand KA, Abouellleil A, Wollam AM, Buhay CJ, Ding Y, Dugan S, FitzGerald MG, Holder M, Hostetler J, Clifton SW, Allen-Vercoe E, Earl AM, Farmer CN, Liolios K, Surette MG, Xu Q, Pohl C, Wilczek-Boney K, Zhu D. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–9. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Consortium THMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van den Mooter G, Maris B, Samyn C, Augustijns P, Kinget R. Use of azo polymers for colon-specific drug delivery. J Pharm Sci. 1997;86:1321–7. doi: 10.1021/js9702630. [DOI] [PubMed] [Google Scholar]
- 96.Chavan MS, Sant VP, Nagarsenker MS. Azo-containing urethane analogues for colonic drug delivery: Synthesis, characterization and in vitro evaluation. J Pharm Pharmacol. 2001;53:895–900. doi: 10.1211/0022357011776063. [DOI] [PubMed] [Google Scholar]
- 97.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–4. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 98.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–12. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 99.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The ribosomal database project: Improved alignments and new tools for rrna analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Letunic I, Bork P. Interactive tree of life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–8. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Elmer GW, Remmel RP. Role of the intestinal microflora in clonazepam metabolism in the rat. Xenobiotica. 1984;14:829–40. doi: 10.3109/00498258409151481. [DOI] [PubMed] [Google Scholar]
- 102.Strong HA, Renwick AG, George CF, Liu YF, Hill MJ. The reduction of sulphinpyrazone and sulindac by intestinal bacteria. Xenobiotica. 1987;17:685–96. doi: 10.3109/00498258709043976. [DOI] [PubMed] [Google Scholar]
- 103.Gingell R, Bridges JW, Williams RT. The role of the gut flora in the metabolism of prontosil and neoprontosil in the rat. Xenobiotica. 1971;1:143–56. doi: 10.3109/00498257109044386. [DOI] [PubMed] [Google Scholar]
- 104.Chan RP, Pope DJ, Gilbert AP, Sacra PJ, Baron JH, Lennard-Jones JE. Studies of two novel sulfasalazine analogs, ipsalazide and balsalazide. Dig Dis Sci. 1983;28:609–15. doi: 10.1007/BF01299921. [DOI] [PubMed] [Google Scholar]
- 105.Takeno S, Sakai T. Involvement of the intestinal microflora in nitrazepam-induced teratogenicity in rats and its relationship to nitroreduction. Teratology. 1991;44:209–14. doi: 10.1002/tera.1420440209. [DOI] [PubMed] [Google Scholar]
- 106.Watanabe K, Yamashita S, Furuno K, Kawasaki H, Gomita Y. Metabolism of omeprazole by gut flora in rats. J Pharm Sci. 1995;84:516–7. doi: 10.1002/jps.2600840425. [DOI] [PubMed] [Google Scholar]
- 107.Koch RL, Goldman P. The anaerobic metabolism of metronidazole forms N-(2-hydroxyethyl)-oxamic acid. J Pharmacol Exp Ther. 1979;208:406–10. [PubMed] [Google Scholar]
- 108.Koch RL, Beaulieu BB, Jr, Goldman P. Role of the intestinal flora in the metabolism of misonidazole. Biochem Pharmacol. 1980;29:3281–4. doi: 10.1016/0006-2952(80)90304-4. [DOI] [PubMed] [Google Scholar]
- 109.Kitamura S, Sugihara K, Kuwasako M, Tatsumi K. The role of mammalian intestinal bacteria in the reductive metabolism of zonisamide. J Pharm Pharmacol. 1997;49:253–6. doi: 10.1111/j.2042-7158.1997.tb06790.x. [DOI] [PubMed] [Google Scholar]
- 110.Abu Shamat MS, Beckett AH. Glyceryl trinitrite: Metabolism by the intestinal flora. J Pharm Pharmacol. 1983:35. [Google Scholar]
- 111.Iveson P, Lindup WE, Parke DV, Williams RT. The metabolism of carbenoxolone in the rat. Xenobiotica. 1971;1:79–95. doi: 10.3109/00498257109044381. [DOI] [PubMed] [Google Scholar]
- 112.Valerino DM. Studies of the metabolism of methotrexate. Ii. Isolation and identification of several unconjugated aminopteridines as metabolites in the rat. Res Commun Chem Pathol Pharmacol. 1972;4:529–42. [PubMed] [Google Scholar]
- 113.Walsh CT, Levine RR. Studies of the enterohepatic circulation of morphine in the rat. J Pharmacol Exp Ther. 1975;195:303–10. [PubMed] [Google Scholar]
- 114.Kobashi K, Nishimura T, Kusaka M, Hattori M, Namba T. Metabolism of sennosides by human intestinal bacteria. Planta Med. 1980;40:225–36. doi: 10.1055/s-2008-1074963. [DOI] [PubMed] [Google Scholar]
- 115.Okuda H, Ogura K, Kato A, Takubo H, Watabe T. A possible mechanism of eighteen patient deaths caused by interactions of sorivudine, a new antiviral drug, with oral 5-fluorouracil prodrugs. J Pharmacol Exp Ther. 1998;287:791–9. [PubMed] [Google Scholar]
- 116.Elkington SG, Floch MH, Conn HO. Lactulose in the treatment of chronic portal-systemic encephalopathy. A double-blind clinical trial. N Engl J Med. 1969;281:408–12. doi: 10.1056/NEJM196908212810803. [DOI] [PubMed] [Google Scholar]
- 117.Harris BE, Manning BW, Federle TW, Diasio RB. Conversion of 5-fluorocytosine to 5-fluorouracil by human intestinal microflora. Antimicrob Agents Chemother. 1986;29:44–8. doi: 10.1128/aac.29.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Caldwell J, Hawksworth GM. The demethylation of methamphetamine by intestinal microflora. J Pharm Pharmacol. 1973;25:422–4. doi: 10.1111/j.2042-7158.1973.tb10043.x. [DOI] [PubMed] [Google Scholar]
- 119.Smith GE, Griffiths LA. Metabolism of N-acylated and O-alkylated drugs by the intestinal microflora during anaerobic incubation in vitro. Xenobiotica. 1974;4:477–87. doi: 10.3109/00498257409052100. [DOI] [PubMed] [Google Scholar]
- 120.Basit AW, Lacey LF. Colonic metabolism of ranitidine: Implications for its delivery and absorption. Int J Pharm. 2001;227:157–65. doi: 10.1016/s0378-5173(01)00794-3. [DOI] [PubMed] [Google Scholar]
- 121.Basit AW, Newton JM, Lacey LF. Susceptibility of the H2-receptor antagonists cimetidine, famotidine and nizatidine, to metabolism by the gastrointestinal microflora. Int J Pharm. 2002;237:23–33. doi: 10.1016/s0378-5173(02)00018-2. [DOI] [PubMed] [Google Scholar]
- 122.Tozaki H, Emi Y, Horisaka E, Fujita T, Yamamoto A, Muranishi S. Degradation of insulin and calcitonin and their protection by various protease inhibitors in rat caecal contents: Implications in peptide delivery to the colon. J Pharm Pharmacol. 1997;49:164–8. doi: 10.1111/j.2042-7158.1997.tb06773.x. [DOI] [PubMed] [Google Scholar]
- 123.Abu Shamat M. The role of gastrointestinal microflora in the metabolism of drugs. Int J Pharm. 1993;97:13. [Google Scholar]
- 124.Smith RV. Metabolism of drugs and other foreign compounds by intestinal microorganisms. World Rev Nutr Diet. 1978;29:60–76. doi: 10.1159/000400751. [DOI] [PubMed] [Google Scholar]
- 125.Tobin PJ, Dodds HM, Clarke S, Schnitzler M, Rivory LP. The relative contributions of carboxylesterase and beta-glucuronidase in the formation of SN-38 in human colorectal tumours. Oncol Rep. 2003;10:1977–9. [PubMed] [Google Scholar]
- 126.Holt R. The bacterial degradation of chloramphenicol. Lancet. 1967;1:1259–60. doi: 10.1016/s0140-6736(67)92720-1. [DOI] [PubMed] [Google Scholar]
- 127.Shu YZ, Kingston DG, Van Tassell RL, Wilkins TD. Metabolism of levamisole, an anti-colon cancer drug, by human intestinal bacteria. Xenobiotica. 1991;21:737–50. doi: 10.3109/00498259109039513. [DOI] [PubMed] [Google Scholar]
- 128.Dull BJ, Salata K, Goldman P. Role of the intestinal flora in the acetylation of sulfasalazine metabolites. Biochem Pharmacol. 1987;36:3772–4. doi: 10.1016/0006-2952(87)90034-7. [DOI] [PubMed] [Google Scholar]
- 129.Meuldermans W, Hendrickx J, Mannens G, Lavrijsen K, Janssen C, Bracke J, Le Jeune L, Lauwers W, Heykants J. The metabolism and excretion of risperidone after oral administration in rats and dogs. Drug Metab Dispos. 1994;22:129–38. [PubMed] [Google Scholar]
- 130.Sasaki I, Tamura T, Shibakawa T, Fujita T, Murakami M, Yamamoto A, Muranishi S. Metabolism of azetirelin, a new thyrotropin-releasing hormone (TRH) analogue, by intestinal microorganisms. Pharm Res. 1997;14:1004–7. doi: 10.1023/a:1012141025938. [DOI] [PubMed] [Google Scholar]
- 131.Yoshisue K, Masuda H, Matsushima E, Ikeda K, Nagayama S, Kawaguchi Y. Tissue distribution and biotransformation of potassium oxonate after oral administration of a novel antitumor agent (drug combination of tegafur, 5-chloro-2,4-dihydroxypyridine, and potassium oxonate) to rats. Drug Metab Dispos. 2000;28:1162–7. [PubMed] [Google Scholar]
- 132.Lee NK, Choi SH, Park SH, Park EK, Kim DH. Antiallergic activity of hesperidin is activated by intestinal microflora. Pharmacology. 2004;71:174–80. doi: 10.1159/000078083. [DOI] [PubMed] [Google Scholar]
- 133.Nakayama H, Kinouchi T, Kataoka K, Akimoto S, Matsuda Y, Ohnishi Y. Intestinal anaerobic bacteria hydrolyse sorivudine, producing the high blood concentration of 5-(E)-(2-bromovinyl)uracil that increases the level and toxicity of 5-fluorouracil. Pharmacogenetics. 1997;7:35–43. doi: 10.1097/00008571-199702000-00005. [DOI] [PubMed] [Google Scholar]