Abstract
Background
The clinical significance of variants in genes associated with inherited cardiomyopathies can be difficult to determine due to uncertainty regarding population genetic variation and a surprising amount of tolerance of the genome even to loss of function variants. We hypothesized that genes associated with cardiomyopathy might be particularly resistant to the accumulation of genetic variation.
Methods and Results
We analyzed the rates of single nucleotide genetic variation in all known genes from the exomes of >5,000 individuals from the National Heart, Lung, and Blood Institute’s Exome Sequencing Project (ESP), as well as the rates of structural variation from the Database of Genomic Variants. Most variants were rare, with over half unique to one individual. Cardiomyopathy associated genes exhibited a rate of nonsense variants 96.1% lower than other Mendelian disease genes. We tested the ability of in-silico algorithms to distinguish between a set of variants in MYBPC3, MYH7, and TNNT2 with strong evidence for pathogenicity and variants from the ESP data. Algorithms based on conservation at the nucleotide level (GERP, PhastCons) did not perform as well as amino acid level prediction algorithms (Polyphen-2, SIFT). Variants with strong evidence for disease causality were found in the ESP data at prevalence higher than expected.
Conclusions
Genes associated with cardiomyopathy carry very low rates of population variation. The existence in population data of variants with strong evidence for pathogenicity suggests that even for Mendelian disease genetics, a probabilistic weighting of multiple variants may be preferred over the ‘single gene’ causality model.
Keywords: cardiomyopathy, genetic heart disease, genetic variation, genomics, genetic testing
Background
New DNA sequencing technologies are poised to transform the genetic evaluation of patients. Soon the availability of genetic information will no longer be a barrier to our understanding of the genetic basis of disease. Rather it will be our ability to understand and interpret the data that will be paramount. The interpretation of clinical genetic testing is a complex process that requires an appreciation of factors establishing causality as well as a detailed understanding of the ‘tolerated’ genetic variation present in human genomes of different ethnicities. Until recently, much of the genetic variation in human populations was unknown. With large scale population sequencing projects such as the 1000 Genomes Project1, the true extent of this variation is now becoming clear. Indeed, recent analyses indicate a surprising prevalence of tolerated genetic variation.2-4
Clinical genetic testing is increasingly available for conditions such as hypertrophic cardiomyopathy, where it is used for predictive family testing, and long QT syndrome, where it may alter management as well as impact family screening.5-7 The yield from genetic testing, however, can be variable. Evidence for or against a variant’s role is assembled from previous reports in the literature, co-segregation, the likelihood that the variant disrupts the reading frame (weighted more towards nonsense variants, small insertion-deletion variants, or splice site variants) and algorithmic predictions based on conservation, constraint, or protein motif disruption. Despite such resources, a large number of variants found through clinical genetic testing remain of unclear significance. Greatly lacking is knowledge of the population genetic variation in these and other genes, which is needed for the interpretation of variants not just in Mendelian diseases, but also for common disease risk assessment8,9 and pharmacogenomics.10-12
One recent project to catalog population scale single nucleotide variant (SNV) data has been the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP)13,14. This large scale effort is aimed at sequencing the exome, consisting of the protein coding regions (exons) of the human genome, from members of several different cohorts followed throughout the country for the purpose of defining the genetic components of complex diseases. In contrast with the 1000 Genomes study, which has low coverage of hundreds of genomes, the NHLBI exomes study has high coverage (average >100x), high quality sequencing data for >5000 individuals of Caucasian and African American ethnicity. Thus, it represents a valuable comparison dataset for variants thought to cause monogenic Mendelian disease. One limitation of both of these datasets, however, is the absence of structural variants. These may be particularly important because of their tendency to disrupt the reading frame. The Database of Genomic Variants (DGV)15 is a curated repository of structural variation (consisting of insertions, deletions, and copy-number variants) which serves a similar purpose as the above for structural variation.
Using these sources of population genetic variation, we sought to characterize the tolerance of the human genome to variation in genes associated with Mendelian diseases with a specific focus on those that have been associated with inherited cardiomyopathy.
Methods
NHLBI Exome Sequencing Project Data
Data from the NHLBI ESP5400 dataset was accessed on December 12th, 2011 and downloaded for analysis. This data is the accumulation of called variants from the exomes of 5,379 individuals from multiple cohorts of the ESP, including the Women’s Health Initiative, Framingham Heart Study, Jackson Heart Study, Multi-Ethnic Study of Atherosclerosis, Atherosclerosis Risk in Communities, Coronary Artery Risk Development in Young Adults, Cardiovascular Health Study, Genomic Research on Asthma in the African Diaspora, Lung Health Study, Pulmonary Arterial Hypertension population, Acute Lung Injury cohort, and the Cystic Fibrosis cohort. The primary purpose of the ESP is to sequence the exomes of a large number of individuals selected for the extremes of primarily complex traits from these cohorts. While these exomes may not represent a true sample of the general population, they do represent a phenotyped cohort that is unlikely to be enriched for Mendelian disease, with the possible exception of cystic fibrosis. Resulting SNV calls were filtered for depth and base call thresholds and were annotated for quality using a support vector machine algorithm by the NHLBI ESP data analysis group. Only calls that passed all quality filters were used for downstream analysis. Further information regarding alignment, variant calls, and filtering, as well as the entirety of this data is available at http://evs.gs.washington.edu/EVS/.
1000 Genomes Data
Data from the 1000 Genomes Phase 1 March 2012 release1 was retrieved and the subset of small insertion/deletion (1-50 bp) calls were used for analysis (http://www.1000genomes.org/).
Database of Genomic Variants
For evaluation of structural variation, the November 2010 data release from the Database of Genomic Variants (http://projects.tcag.ca/variation), aligned to the hg19 version of the human genome, was accessed. This includes data from 42 separate studies evaluating for structural variation involving segments of DNA greater than 1kb, as well as smaller insertions/deletions (indels). The data is collected from small individual genome and population level studies without known enrichment for disease.
Gene Annotation
Gene annotation data was accessed from the Online Mendelian Inheritance in Man database16(http://www.ncbi.nlm.nih.gov/omim). All genes as annotated in the NCBI Reference Sequence Database (RefSeq)17 via the University of California, Santa Cruz (UCSC) Genome Browser18 (including alternate isoforms) were divided into subgroups by OMIM annotation and/or literature review according to their known association with: i) cardiomyopathy, specifically HCM or DCM ii) any other Mendelian disease, or iii) neither of the above. After accounting for alternate isoforms, there were 120 isoforms of 46 separate genes associated with inherited cardiomyopathy, 5,764 isoforms of 2,831 separate genes with other Mendelian disease association, and 25,437 isoforms of 16,102 separate genes without known Mendelian disease association for which variant data from the ESP5400 dataset was available.
Analysis of Population Variation Data
Variants from the ESP5400 dataset were grouped by gene into the three categories previously described and minor allele frequencies for each variant were extracted. Variant subtypes were analyzed by predicted functional effect (synonymous, missense, nonsense, splice) and the sum of minor allele frequencies across known isoforms was used to come up with a raw count of expected number of variants per type per transcript. For synonymous, missense, and nonsense variants, this number was then normalized for transcript length based on data from RefSeq. For splice site variants, this number was normalized by number of known exons per transcript.
In order to evaluate the distribution of small indels (1-50 bp) which were notably absent from the public release of the ESP5400 dataset, the subset of called indels from the 1000 Genomes Phase 1 March 2012 release was retrieved and annotated using ANNOVAR19 software against the NCBI RefSeq database17 to determine the subset in coding regions of genes with any disease association as above.
Curation of Known Variants
We manually curated a set of variants in MYH7, MYBPC3, and TNNT2 with strong evidence for causing cardiomyopathy. This set comprised of missense variants seen in patients at the Stanford Center for Inherited Cardiovascular Disease from September 2010 to December 2011 and considered likely or very likely disease causing. To supplement this list, we selected variants from a publicly available repository of sarcomeric variants20 with the highest number of independent citations. These variants were then manually curated and any variants we considered likely or very likely disease causing were included in our high confidence set. Curation relied on published data, cases from our clinical cohort, and case or control data from commercial genetic testing laboratories. Classification was based on segregation data, presence in multiple unrelated cases, absence in controls, and availability of compelling animal model or in vitro data. Variants were considered very likely disease causing only if strong segregation data and/or animal model data was available.
Algorithmic Prediction of Variant Pathogenicity
All missense variants from the NHLBI ESP5400 dataset as well as variants from our curated list of known pathogenic variants in HCM were scored using GERP21, a measure of evolutionary constraint at a nucleotide base level utilizing a rejected substitution score, and PhastCons22, another measure of evolutionary conservation at the nucleotide base level utilizing multiple sequence alignment, using the SeattleSeq SNP Annotation server (http://snp.gs.washington.edu/SeattleSeqAnnotation). Polyphen-223(http://genetics.bwh.harvard.edu/pph2/) and SIFT24 (http://sift.jcvi.org/) scores, both predictions of pathogenicity of missense variants based on the effects of the predicted resulting amino acid substitution, were obtained from their respective servers.
Structural Variation Analysis
Structural variants from DGV were grouped on the basis of Mendelian disease association. The average number of structural variants per gene was computed. Due to the varying size of both structural variants and the transcripts they affect, we normalized by evaluating only structural variants affecting protein coding regions of genes and calculating the percent of each gene’s coding region based on transcript length affected by a deletion in DGV.
Statistical Analysis
All data analysis was carried out using the R Statistical Programming Language. Tests for statistical significance between groups were non-parametric tests without assumption of the underlying distribution. These included the Wilcoxon rank-sum test for direct comparison between two groups, the Kruskal-Wallis test for analysis of variance, and Spearman’s rank order for correlation. Given that most genes are not in linkage with each other, linkage between genes does not affect the results of the Kruskal-Wallis test significantly.
For the analysis of the exonic distribution of pathogenic and ESP variants, Fisher’s exact test was used. While Fisher’s test does assume independence of events which may not necessarily be true for the distribution of variants in a gene due to linkage disequilibrium, given the overall rarity of most variants analyzed (almost all less than 1% minor allele frequency and the majority being unique) it is unlikely that a rare variant in one exon significantly affects the probability of a variant in another exon.
Results
Most Genetic Variation is Rare
Most variants in the population data were not shared between many individuals. Private variants, those that were found only in one person, were abundant. Out of the 9,974 total variants called in the NHLBI exomes distributed amongst 46 separate cardiomyopathy associated genes, 9,103 (91%) had minor allele frequencies less than 1%. Of these rare variants, 5,448 (60%) were private. This predominance of rare variants was almost identical in other genes, whether or not they were associated with Mendelian disease. Common variants (minor allele frequency > 5%) comprised only 5% of all genetic variation in the coding regions of human genes.
We found many genes for which a large amount of genetic variation was not only expected, but likely serves a critical purpose. Among Mendelian disease associated genes, the five with the highest rates of missense variation were all HLA loci (Supplementary Table 2), where high rates of polymorphism are thought to be selectively maintained.25 Another well-recognized gene locus with very high missense variation was the ABO blood group locus. Among non-Mendelian disease associated genes, those with the most variation included many of the olfactory receptor genes, consistent with the survival advantage of a sophisticated sensing system for environmental odorant molecules.26
Missense variant and nonsense variant rates did not appear correlated when looking across all genes (Spearman’s rho=0.36). This remained true when looking at the subset of genes with Mendelian disease association or the subset without Mendelian disease association.
Mendelian Disease Genes Exhibit Lower Rates of Genetic Variation
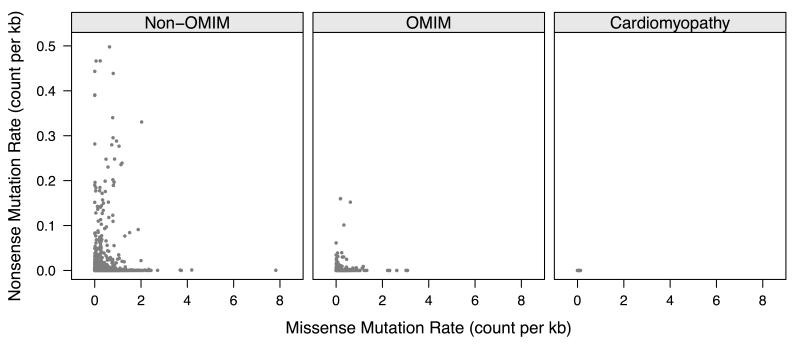
We found significantly lower levels of variation in genes associated with Mendelian disease as compared to genes without a known association (Table 1). In general, this reduction was much stronger for types of genetic variation that would be predicted to have more impact on the resulting protein product, such as splice site or nonsense variants. Mendelian disease genes were noted to have a 67.3% lower rate of nonsense variants as compared to genes without known disease association (p=9.6×10−6). These were even more rare in cardiomyopathy associated genes (Figure 1), which exhibited a 98.7% lower nonsense variant rate as compared to non-disease associated genes and a 96.1% lower rate as compared to the remaining Mendelian disease associated genes (p=5.7 × 10−7). Similarly lower variant rates were seen for both missense and splice site variants as well. Interestingly, this was reversed with respect to synonymous variation, with cardiomyopathy specific genes having slightly higher rates of variation (116.4 variants per megabase of coding region per chromosome in cardiomyopathy genes vs. 90.8 and 95.1 variants per megabase of coding region per chromosome for non-OMIM and OMIM genes, respectively, p=2.7×10−3).
Table 1.
Average rates of variation by subtype across genes without Mendelian disease association(non-OMIM), genes with annotated Mendelian disease association (OMIM), and genes associated with inherited cardiomyopathies. For synonymous, missense, and nonsense variant rates, units are counts per 1×106 base pairs of coding region per chromosome. For rates of splice site variants, units are counts per exon per chromosome. 1st Qu. = 1st quartile, 3rd Qu. = 3rd quartile. P-values were computed using non-parametric Kruskal-Wallis test for analysis of variance.
| Synonymous | p-value | |||
|---|---|---|---|---|
| Non-OMIM Genes | OMIM Genes | Cardiomyopathy Genes | 2.7×10−3 | |
| 1st Qu. | 4.2 | 5.2 | 4.8 | |
| Median | 30.1 | 35.2 | 44.8 | |
| Mean | 90.8 | 95.1 | 116.4 | |
| 3rd Qu. | 122.5 | 121.1 | 151.6 | |
| Missense | ||||
|---|---|---|---|---|
| Non-OMIM Genes | OMIM Genes | Cardiomyopathy Genes | ||
| 1st Qu. | 2.2 | 2.5 | 1.5 | 1.8×10−2 |
| Median | 13.5 | 16.2 | 11.4 | |
| Mean | 85.5 | 76.6 | 27.6 | |
| 3rd Qu. | 89.9 | 86.2 | 46.8 | |
| Nonsense | ||||
|---|---|---|---|---|
| Non-OMIM Genes | OMIM Genes | Cardiomyopathy Genes | ||
| 1st Qu. | 0.000 | 0.000 | 0.000 | 5.7×10−7 |
| Median | 0.000 | 0.000 | 0.000 | |
| Mean | 0.794 | 0.266 | 0.011 | |
| 3rd Qu. | 0.048 | 0.037 | 0.012 | |
| Splice | ||||
|---|---|---|---|---|
| Non-OMIM Genes | OMIM Genes | Cardiomyopathy Genes | ||
| 1st Qu. | 0.000 | 0.000 | 0.000 | 8.8×10−8 |
| Median | 0.000 | 0.000 | 0.000 | |
| Mean | 0.094 | 0.072 | 0.004 | |
| 3rd Qu. | 0.000 | 0.003 | 0.000 | |
Figure 1.
Plot of missense and nonsense variant rates for all known human gene transcripts calculated from the exomes of 5,379 persons in the NHLBI Exome Sequencing Project. Non-OMIM = genes without known association with a Mendelian disease. OMIM = genes with a known association with a Mendelian disease in the Online Mendelian Inheritance in Man (OMIM) database. Cardiomyopathy = genes with known association with a familial cardiomyopathy. Variant rates are in units of counts per 1,000 base pairs of coding region per transcript per chromosome.
Nonsense Variants are Extremely Rare in Cardiac Structural and Sarcomere Genes
Single nucleotide variants thought to have the most effect on protein function are ones that result in a premature stop codon, i.e. nonsense variants. We looked at cardiomyopathy-associated genes in the NHLBI exome data to evaluate for the overall prevalence of this type of variation in a population without known inherited cardiomyopathy. Overall, we found that nonsense variants were extremely rare in these genes. In fact, in the subset of genes that are routinely sequenced for clinical purposes in HCM, we found only one nonsense variant each in MYH7 and MYBPC3. Nonsense variants were completely absent in the sarcomeric genes ACTC1, TNNT2, TNNI3, MYL2, MYL3, and TPM1. While the nonsense variant in MYH7 has not been reported previously, the nonsense variant found in MYBPC3 (p.Trp1214Ter) has been associated with hypertrophic cardiomyopathy in one published report in an Asian Indian population27.
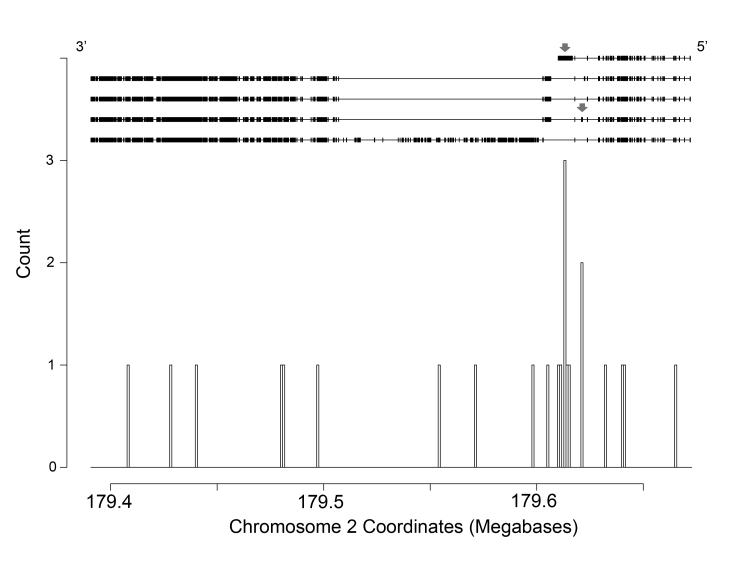
Among cardiomyopathy-associated genes, the gene with the greatest number of nonsense variants in the ESP5400 exomes data was the very large gene titin (TTN), which has been implicated in familial DCM. This may be largely due to its immense size, as the coding region of titin consists of upwards of 100 kilobases. In total we noted 23 predicted nonsense variants in titin in the NHLBI exome data. The majority of these nonsense variants seemed to be distributed evenly throughout the length of the gene, although there were two notable clusters of nonsense variants near the 5′ end of the gene (Figure 2). This is in direct contrast to a recent report of a high burden of variants in the A band of the titin protein (corresponding to a group of exons near the 3′ end of the transcript) associated with dilated cardiomyopathy (DCM)28. Both clusters of nonsense variants in our analysis were in exons that are specific to the novex alternate splice isoforms of titin, the first in the terminal exon (exon 46) of the novex-3 isoform (NM_133379) and the other in exon 44 of the novex-2 isoform (NM_133437). Neither of these are the major cardiac isoforms of titin, which may explain why nonsense variants in these regions may be more tolerated.
Figure 2.
Location of nonsense variants found in the large sarcomeric gene titin (TTN). The structure of 5 known isoforms is displayed at the top of the figure oriented by location on chromosome 2, with the 5′ end of the transcript on the right and the 3′ end on the left. Red arrows depict exons in which clusters of nonsense variants were noted.
On the other hand, DMD, which has been implicated in Duchenne and Becker muscular dystrophy as well as X-linked familial cardiomyopathy,29,30 was noted to manifest an extremely low rate of nonsense variants despite its enormous size. Of all human genes, DMD spans the largest region of the genome: encompassing 2.4 million bases, with a coding region consisting of about 14 kb spread over greater than 70 exons. The NHLBI dataset, however, contained only one predicted nonsense variant within this gene.
Prediction of Pathogenicity of Missense Variants Remains Challenging
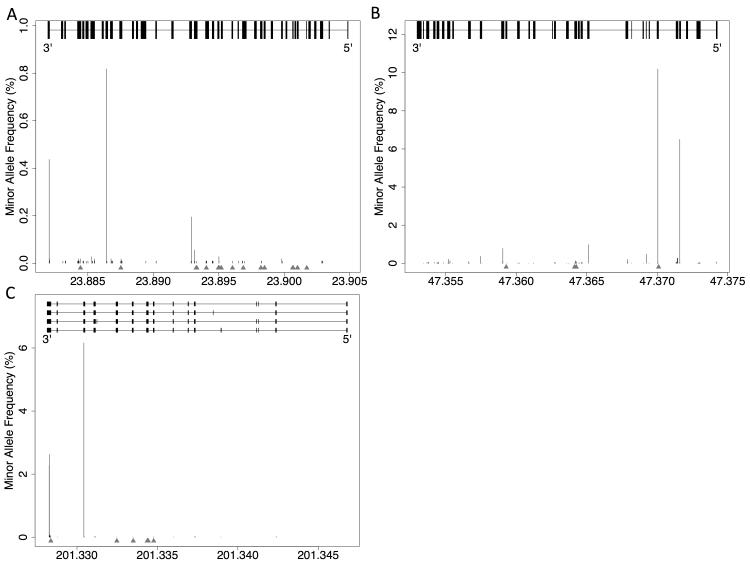
We collected 46 variants, 40 of which were missense, with particularly strong evidence of causality from three genes most often found to be causal in HCM (MYBPC3, MYH7, and TNNT2) (Supplemental Table 3). Given a large amount of ambiguity over the effects of missense variants in the genome, we compared the missense variants from this pathogenic list to missense variants from the NHLBI exome data within the same genes. These 40 pathogenic missense variants were generally located in regions within these three genes that were notable for very low variant frequencies in the population data, suggesting that these are regions with vital functions that do not tolerate high rates of variation (Figure 3).
Figure 3.
Plot of minor allele frequency of non-synonymous coding variants from the NHLBI ESP data set over the distribution of the known exons of A) MYH7 (chromosome 14), B) MYBPC3 (chromosome 11), and C) TNNT2 (chromosome 1). Red arrows denote locations of pathogenic variants from a curated list from clinical experience at our institution and literature reports. X-axis is genomic coordinates in megabases. 5′ and 3′ refer to start and end of transcript, respectively.
Furthermore, 10/26 pathogenic missense variants in MYH7 and 6/10 of the pathogenic missense variants in TNNT2 were found in exons that were notable for a complete absence of non-synonymous likely benign variation (Supplemental Table 4). These exons in MYH7 (exons 6,7,9, 13, and 19 of NM_000257) and in TNNT2 (exon 10 of NM_000364) thus likely encode critical functional domains in the resultant peptide. In support of this, the above noted exons in MYH7 all encode for portions of the functional head and neck domains31. In addition, the above mentioned exon in TNNT2 encodes a portion of a tropomyosin binding site, with induced variants in this exon previously shown to strongly reduce binding efficacy32,33. In general, exonic distribution was strikingly different between the pathogenic variants and ESP5400 variants in MYH7 (p=.0059) and TNNT2 (p=.013). This difference was not statistically significant in MYBPC3, which may be due to the low number of pathogenic missense variants in this gene in our collection, consistent with reports that the majority of disease-causing variants in this gene tend to be frameshift, splice, or nonsense variants rather than missense34,35.
Of note, 4 of the 46 variants with good evidence of pathogenicity were present in the NHLBI exome data. The individual incidences of these variants were very low, with almost all found in only 1 individual each, except for one variant in TNNT2, p.Arg278Cys that was found in 6 individuals in the NHLBI exome cohort. No phenotype information was available to us for these individuals. These variants were removed from the NHLBI ESP variant list for any further analysis.
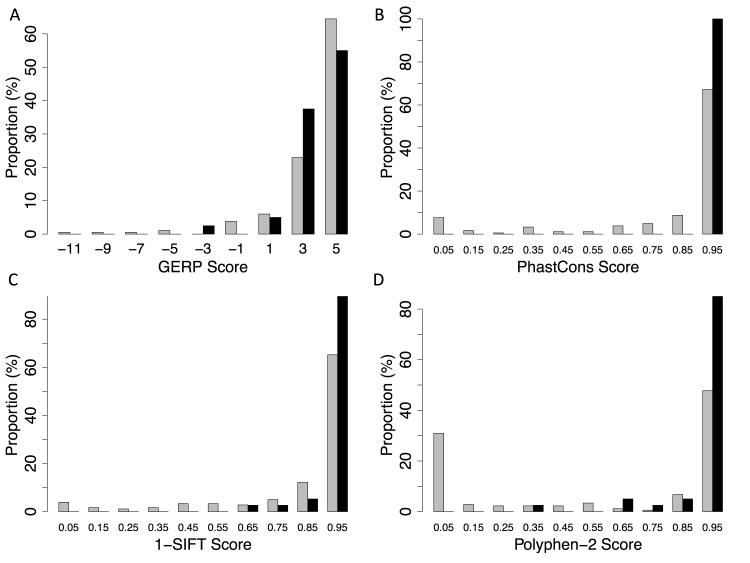
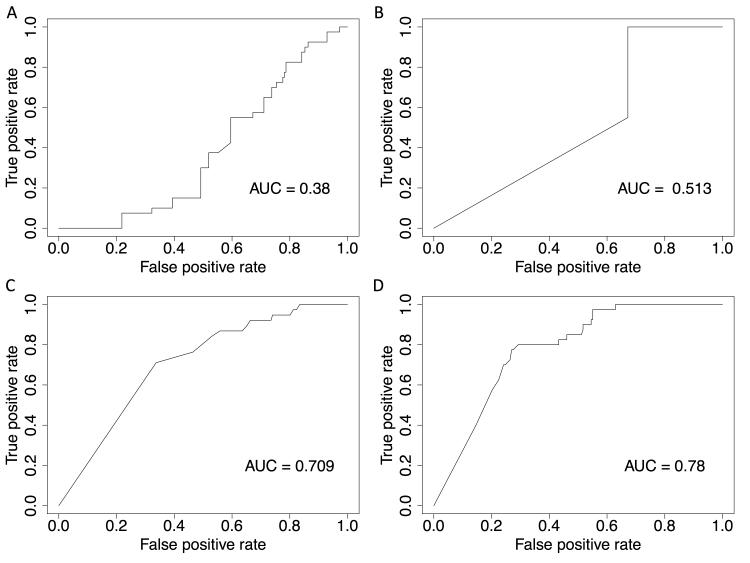
We used widely accepted variant classification algorithms to predict pathogenicity of missense variants. We found the evolutionary constraint based algorithms GERP and PhastCons to be poorly predictive of variant pathogenicity in this data. Notably, GERP scores appeared on the whole to be higher in the NHLBI ESP variant set (Figure 4), the opposite of what would be expected. While PhastCons predicted scores of > 0.95 (max score of 1) for all the variants in our curated causative variant list, the majority of presumably tolerated missense variants (67%) from the NHLBI exome data set were also noted to have a similarly high PhastCons score, resulting in a c-statistic for classification of 0.52, akin to no discriminatory power (Figure 5).
Figure 4.
Relative distribution of missense variants from the NHLBI ESP and a curated pathogenic variant list in the genes MYH7, MYBPC3, and TNNT2, as scored by A) GERP, B) PhastCons, C) SIFT, and D) Polyphen-2. Grey bars denote variants from NHLBI ESP data, black bars denote variants from the pathogenic list. For panel C, 1 – SIFT score was used to preserve consistency between panels, with far right predicted to be more pathogenic and far left predicted to be less pathogenic.
Figure 5.
Receiver operator curves for A) GERP, B) PhastCons, C) SIFT, and D) Polyphen-2 for the classification of collected missense variants from the NHLBI ESP data set and a curated pathogenic missense variant list in the genes MYH7, MYBPC3, and TNNT2. AUC = area under the curve.
The use of algorithms based on amino acid substitution gave much better results. SIFT had modest discriminatory power with a c-statistic of 0.70. Polyphen-2, which also utilizes information about peptide structure and interaction, performed the best with a c-statistic of 0.77. It should be noted however that Polyphen-2 is based on a machine-learning algorithm that was trained on variants that may have included some of those from our curated list.
Cardiomyopathy Genes Exhibit Less Structural Variation
We attempted to recapitulate these findings in other types of genetic variation by evaluating the distribution of small indels in data from the 1000 Genomes Project. There were notably only 5,969 indels from this dataset in coding regions, of which 868 were in Mendelian disease associated genes and 26 were in cardiomyopathy associated genes. This gave total rates of 17 indels per 1,000 exons in non-Mendelian disease genes, 10 indels per 1,000 exons in Mendelian disease genes, and 9 indels per 1,000 exons in cardiomyopathy genes. However the overall low number of these types of variants in this data limited any further statistical analysis.
We then used data from DGV to query on a per gene basis the number of all structural variants that have been reported as well as the overall extent of the coding region of genes that are covered by known structural variants. We found that the total number, per gene, of all structural variants and only structural variants affecting coding regions did not differ between genes associated with Mendelian disease and those that are not (Table 2). However we did note a 53% reduction of coding region covered by reported deletion type structural variants in genes that are specifically associated with cardiomyopathy as compared to genes without Mendelian disease association (p-value=0.02).
Table 2.
Average counts of structural variants (SVs) and percent of transcript affected by known SVs in the Database of Genomic Variants. Non-OMIM – genes without Mendelian disease association. OMIM – genes with known Mendelian disease association. Numbers are averages of per gene counts or percents for all genes within that classification.
| Non-OMIM | OMIM | Cardiomyopathy | |
|---|---|---|---|
| Average Number of SVs Affecting Gene | 2.8 | 3.1 | 3.2 |
| Average Number of SVs Affecting Gene in Coding Regions | 1.5 | 1.6 | 1.2 |
| Average % of Coding Regions Affected by Known SVs | 32% | 31% | 15%* |
Denotes statistically significant difference between cardiomyopathy associated genes and genes without Mendelian disease association (p=.02 by Wilcoxon rank-sum test).
Discussion
Recent studies have suggested a surprising rate of tolerance to genetic variation within the human genome. Here, we show that this tolerance does not extend to genes associated with cardiomyopathy, especially structural and sarcomere genes. This observation fits with a systems model of organism function where some genes are disproportionately intolerant of variation because their function has less redundancy. In addition, in describing population variation data for these genes, we note the presence of a surprising number of disease-associated variants in a population without enrichment for cardiomyopathy.
In contrast to the high rate of genetic variation found in genes dependent on diversity for effective function such as the olfactory receptor loci, we found that population genetic variation, especially variation expected to affect protein function, was rare in Mendelian disease associated genes. We hypothesized that genes essential for cardiac function might be among the genes most intolerant of variation. Not only was this the case, but the strength of these associations was also found to be dependent on the severity of the predicted alteration of protein function, exemplified by the extreme rarity of nonsense variants in cardiomyopathy-specific genes. These findings extended to structural variants as well, specifically in regards to the percent of the coding transcript that is involved in deletion type structural variants in individuals without disease.
One strength of our study is in the practical application to clinical genetic testing, which relies on data from unaffected individuals to judge the likely pathogenicity of novel variants. As our understanding of human genetic variation has improved, it has become clear that even rare genetic variation can be normal and well tolerated, representing a challenge in linking genotypes to phenotypes. One recent study has estimated, using 1000 Genomes data, that the average person has as many as 100 loss of function variants per genome2. This population level of variation has implications for the interpretation of results of clinical genetic testing. However our results indicate that this variation is not evenly distributed and genes for which associations with Mendelian disease have been established have much lower levels of such variation, likely representing the effects of purifying selection.
Why genes associated with cardiomyopathy show even lower rates of genetic variation than other Mendelian disease associated genes is not self-evident but there are many possibilities as to why this may be the case. One study has suggested that Mendelian disease genes may not necessarily be the hubs of gene networks36 (because to manifest disease a variant cannot be fatal). However, genes associated with cardiomyopathy may be an exception given their essential functions within the sarcomere and the heart’s unique position in serving all other organs. Variants in these highly structured peptides with molecular motor functions that operate constantly throughout life would be expected to be heavily selected against in the general population. The finding of a slight increase in synonymous variants in cardiomyopathy-associated genes is unexpected. It is possible that this represents a decrease in codon use bias in these genes relative to others, which may in turn reflect a decreased need of efficiency of translation of these structural proteins, but why this may be the case is not evident.
One intriguing finding in cardiomyopathy genetics is the contrast between disease causing variants found in MYBPC3 and those in MYH7, the two genes with the highest number of HCM-causing variants. Indeed, the high rate of nonsense pathogenic variants found in MYBPC337 is in contrast with the almost universal missense nature of those found in MYH7. The extreme rarity of nonsense variants in cardiomyopathy genes in the data presented here suggests that a high probability for pathogenicity for such variants found in MYBPC3 in patients would be appropriate. The absence of disease-causing nonsense variants in MYH7 is curious. It may be that MYH7 haploinsufficiency may not be tolerated at all. We do note that predisposition of genes towards one type of variation versus another is not uncommon given the poor correlation between rates of different types of variation noted in our data, which may be driven by the resulting effects of such variants (dominant-negative effects in missense variants versus haploinsufficiency states in nonsense variants).
Missense variants remain among the most difficult to interpret in a clinical context. Without a large number of affected and unaffected family members to show co-segregation of variant with disease, it is often difficult to determine if a missense variant truly is pathogenic. Much has been made of the use of measures of evolutionary conservation to prioritize missense variants. Our analysis shows that while these measures can help exclude variants at positions in the genome that do not show conservation, they are unable to efficiently discriminate between likely causative and non-causative variants. While evolutionary conservation at the nucleotide base level appears to be a necessary characteristic of a pathogenic variant, it is not sufficient in and of itself to classify a variant as causative. Algorithms using the predicted effects of the resulting amino acid substitution showed much better classification potential although this may in part reflect the use of cardiomyopathy causative variants as training data for these classifiers.
Our analysis also confirms recent evidence that the overwhelming majority of variation in the human genome is rare (i.e. affecting < 1% of the population). Interestingly, more than half of variants analyzed were private (found in only one person). In fact, taking all 8 commonly sequenced genes for HCM together (ACTC1, TNNT2, TNNI3, MYL2, MYL3, TPM1, MYH7, MYBPC3), we found 159 private missense variants, 3 private splice site variants, and 2 private nonsense variants for a total of 164 private variants that would have the potential to affect the resulting protein. Assuming that none of these variants was found in the same person, this would imply that 3% of a general population sample who were to be sequenced today would have candidate variants not seen previously on a small HCM disease genetic testing panel. This highlights the continued importance of co-segregation and other supporting data in deciding whether or not a novel variant is causative of disease.
It was also surprising to find 4 of 46 “gold standard” pathogenic variants present in this population sample, with a total pathogenic allele count of 9 among 5,379 individuals. These data would imply a background prevalence of variants believed causative of HCM of approximately 0.2% (based on 46 variants in 3 genes, and thus likely a substantial underestimate). However, this is much higher than expected in a general population where the prevalence of HCM is estimated to be 0.2% in multiple populations,38-40 when considering that the yield of genetic testing is far from 100%. This is consistent with other recently published studies finding higher than expected prevalence of genetic variants associated with other Mendelian cardiovascular diseases such as familial DCM14 and long QT syndrome41, though the burden of evidence of pathogenicity for variants in these studies was variable.
While it remains possible that some individuals within these cohorts may harbor undiagnosed HCM given that phenotype data for these individuals is not publicly available, the genetic prevalence rate would still be expected to be much lower than that observed in this data. Based on this genetic variant prevalence data, estimates of the incidence of HCM would have to be underestimated by a factor of at least 2 for our current models of HCM disease inheritance to be true. Given that these estimates of HCM disease prevalence were based on multi-modality screening in diverse populations, it seems likely that some proportion of the variants thought to be causal of HCM under a single gene model cannot be. Alternatively, we posit that the idea of a single gene disorder with variable penetrance is likely an artifact of a limited genomic window, and that what has commonly been perceived as a single gene disorder may in fact be the result of a combination of multiple genetic variants each contributing a portion of the variance, with variants contributing differently in different individuals. Just as some have suggested that a number of rare variants with strong effect size may be the driver of the inherited component in many common diseases8,42,43, so too might this be the case for what have historically been perceived as monogenic disorders.
Limitations
Our study has limitations. No individual phenotype data for the cohorts in NHLBI-ESP, 1000 Genomes, or DGV is publicly available, so it is not possible for us at this time to determine if those individuals with variants from our curated set may have features of an undiagnosed cardiomyopathy. While the accumulated set of variants from these 5,379 individuals is available, individual exomes cannot be reconstructed so it is not possible to determine which variants may be shared on the same chromosome. Also the family structure of the individuals within the NHLBI ESP data was also unknown. It is thus possible that a rare variant could be overrepresented if many members of the same family were sequenced.
Conclusion
In conclusion, using publicly available exome-wide sequencing data from thousands of individuals, we found that genes associated with Mendelian diseases show much lower rates of protein-altering genetic variation, including missense, nonsense, and splice-site variation, with an extreme intolerance of variation noted specifically in cardiomyopathy-associated genes. Cardiomyopathy-associated genes specifically showed intolerance to structural variation as well. Nonsense variants in genes that have been recurrently linked to hypertrophic cardiomyopathy were extremely rare, and our results suggest that such variants in these genes found on clinical testing have a very high likelihood of being pathogenic. In contrast, novel missense variants were present in at least 3% of individuals, and thus the careful interpretation of missense variants found on clinical genetic testing is critical. Current in silico classification schemes for predicting the pathogenicity of missense variants unfortunately have low power in classifying cardiomyopathy variants. Finally, we note a much higher than expected prevalence of variants with strong evidence for pathogenicity. This suggests that, using the power of genome sequencing, a new framework for heterogeneous Mendelian disorders such as inherited cardiomyopathies needs to be developed where variants found in patients and family members are viewed probabilistically on a spectrum from unlikely to likely contributors of variable individual magnitude. While this model challenges the classic ‘single variant in a single gene disorder’ view, it may also begin to explain some of the significant variability in disease expression found in family members with the same ‘causal’ variant.
Supplementary Material
Supplemental Table I: List of genes determined to have association with cardiomyopathy. Associations were noted from the Online Mendelian Inheritance in Man (OMIM) database or through literature review.
Supplemental Table 2: Top 20 genes in each category with highest variant rate by subtype.
Supplemental Table 3. Manually curated high confidence pathogenic variants. a: total number of unrelated individuals with the variant from published data, our clinical cohort, and clinical laboratory data provided in genetic test report. b: strength of segregation data based on largest number of affected individuals with the variant within a single kindred. >5 – strong, 4-5 – moderate, 2-3 – weak. c: total number of controls the variant was not observed in from published data and clinical laboratory data.
Supplemental Table 4: Distribution of missense variants amongst the exons of MYBPC3, MYH7, and TNNT2. The canonical isoform was used in the case of multiple isoforms. P-values represent results of a Fisher’s Exact Test for independence of distributions between pathogenic variants and the variants from the Exome Sequencing Project (ESP) dataset.
Recent studies have revealed a high degree of genetic variation in human populations, some of which would be predicted to cause loss of function of the genes encoded. These studies provide a challenge to the careful interpretation of the results from genetic testing and raise concerns about our ability to distinguish between benign and pathogenic variants. We analyzed data from more than 5,000 participants in the NHLBI Exome Sequencing Project. To study tolerance of genetic variation in different genes, we derived rates of genetic variation within: 1) genes without Mendelian disease association, 2) genes associated with Mendelian disease, and 3) genes associated with inherited cardiomyopathies. We found that genes associated with Mendelian diseases exhibit markedly lower rates of genetic variation. This was even more marked for genes associated with cardiomyopathy. Nonsense variants were extremely rare in most cardiomyopathy genes, suggesting that when such variants are found, they are likely to be pathogenic. We also compared known pathogenic variants in MYH7, MYBPC3, and TNNT2 with those in the population data. We found neither rarity nor nucleotide evolutionary conservation helpful in distinguishing benign from pathogenic variants in these genes. However, the exon distribution of pathogenic and benign variants in MYH7 and TNNT2 was significantly different. Rates of pathogenic variants in population data were higher than would be anticipated, suggesting that a single gene/variant model may not be sufficient to explain many cases of inherited cardiomyopathy. These findings highlight the continued importance of co-segregation and other supporting data in determining variant pathogenicity.
Acknowledgments
The authors would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
Funding Sources: Stephen Pan is supported by NIH grant 5T15LM007033. This work was also supported in part by NIH grants DP2OD004613, R01HL105993, UL1RR029890 (Euan Ashley).
Footnotes
Conflict of Interest Disclosures: Euan Ashley reports equity and consulting in relation to Personalis Inc.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Vinckenbosch N, Tian G, Huerta-Sanchez E, Jiang T, Jiang H, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 4.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011;124:2761–2796. doi: 10.1161/CIR.0b013e318223e230. [DOI] [PubMed] [Google Scholar]
- 6.Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8:1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Wheeler M, Pavlovic A, DeGoma E, Salisbury H, Brown C, Ashley EA. A New Era in Clinical Genetic Testing for Hypertrophic Cardiomyopathy. J Cardiovasc Transl Res. 2009;2:381–391. doi: 10.1007/s12265-009-9139-0. [DOI] [PubMed] [Google Scholar]
- 8.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 9.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewey FE, Chen R, Cordero SP, Ormond KE, Caleshu C, Karczewski KJ, et al. Phased Whole-Genome Genetic Risk in a Family Quartet Using a Major Allele Reference Sequence. PLoS Genet. 2011;7:e1002280. doi: 10.1371/journal.pgen.1002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan S, Dewey FE, Perez MV, Knowles JW, Chen R, Butte AJ, et al. Personalized Medicine and Cardiovascular Disease: From Genome to Bedside. Curr Cardiovasc Risk Rep. 2011;5:542–551. [Google Scholar]
- 13.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, et al. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norton N, Robertson PD, Rieder MJ, Züchner S, Rampersaud E, Martin E, et al. Evaluating Pathogenicity of Rare Variants From Dilated Cardiomyopathy in the Exome Era. Circ Cardiovasc Genet. 2012;5:167–174. doi: 10.1161/CIRCGENETICS.111.961805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res. 2006;115:205–214. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- 16. [Accessed December 11, 2011];Online Mendelian Inheritance in Man, OMIM®. Online Mendelian Inheritance in Man. Available at: http://omim.org.
- 17.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, et al. The UCSC genome browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genomics of Cardiovascular Development, Adaptation, and Remodeling. NHLBI Program for Genomic Applications, Harvard Medical School; [Accessed January 20, 2012]. Available at: http://www.cardiogenomics.org. [Google Scholar]
- 21.Cooper GM. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 25.Hughes AL, Yeager M. Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet. 1998;32:415–435. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- 26.Menashe I, Man O, Lancet D, Gilad Y. Different noses for different people. Nat Genet. 2003;34:143–144. doi: 10.1038/ng1160. [DOI] [PubMed] [Google Scholar]
- 27.Bashyam MD, Purushotham G, Chaudhary AK, Rao KM, Acharya V, Mohammad TA, et al. A low prevalence of MYH7/MYBPC3 mutations among familial hypertrophic cardiomyopathy patients in India. Mol Cell Biochem. 2012;360:373–382. doi: 10.1007/s11010-011-1077-x. [DOI] [PubMed] [Google Scholar]
- 28.Herman DS, Lam L, Taylor MRG, Wang L, Teekakirikul P, Christodoulou D, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Politano L, Nigro V, Nigro G, Petretta VR, Passamano L, Papparella S, et al. Development of cardiomyopathy in female carriers of Duchenne and Becker muscular dystrophies. JAMA. 1996;275:1335–1338. [PubMed] [Google Scholar]
- 30.Sylvius N, Tesson F, Gayet C, Charron P, Bénaïche A, Peuchmaurd M, et al. A new locus for autosomal dominant dilated cardiomyopathy identified on chromosome 6q12-q16. Am J Hum Genet. 2001;68:241–246. doi: 10.1086/316929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Driest SL, Jaeger MA, Ommen SR, Will ML, Gersh BJ, Tajik AJ, et al. Comprehensive Analysis of the Beta-Myosin Heavy Chain Gene in 389 Unrelated Patients With Hypertrophic Cardiomyopathy. J Am Coll Cardiol. 2004;44:602–610. doi: 10.1016/j.jacc.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 32.Jin J-P, Chong SM. Localization of the two tropomyosin-binding sites of troponin T. Arch. Biochem. Biophys. 2010;500:144–150. doi: 10.1016/j.abb.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palm T, Graboski S, Hitchcock-DeGregori SE, Greenfield NJ. Disease-causing mutations in cardiac troponin T: identification of a critical tropomyosin-binding region. Biophys J. 2001;81:2827–2837. doi: 10.1016/S0006-3495(01)75924-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen PS, Havndrup O, Hougs L, Srensen KM, Jensen M, Larsen LA, et al. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Hum. Mutat. 2009;30:363–370. doi: 10.1002/humu.20862. [DOI] [PubMed] [Google Scholar]
- 35.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 36.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erdmann J, Daehmlow S, Wischke S, Senyuva M, Werner U, Raible J, et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clin Genet. 2003;64:339–349. doi: 10.1034/j.1399-0004.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 38.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults: echocardiographic analysis of 4111 subjects in the CARDIA study. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 39.Zou Y, Song L, Wang Z, Ma A, Liu T, Gu H, et al. Prevalence of idiopathic hypertrophic cardiomyopathy in China: a population-based echocardiographic analysis of 8080 adults. Am J Med. 2004;116:14–18. doi: 10.1016/j.amjmed.2003.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 41.Refsgaard L, Holst AG, Sadjadieh G, Haunsø S, Nielsen JB, Olesen MS. High prevalence of genetic variants previously associated with LQT syndrome in new exome data. Eur J Hum Genet. 2012;20:905–908. doi: 10.1038/ejhg.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table I: List of genes determined to have association with cardiomyopathy. Associations were noted from the Online Mendelian Inheritance in Man (OMIM) database or through literature review.
Supplemental Table 2: Top 20 genes in each category with highest variant rate by subtype.
Supplemental Table 3. Manually curated high confidence pathogenic variants. a: total number of unrelated individuals with the variant from published data, our clinical cohort, and clinical laboratory data provided in genetic test report. b: strength of segregation data based on largest number of affected individuals with the variant within a single kindred. >5 – strong, 4-5 – moderate, 2-3 – weak. c: total number of controls the variant was not observed in from published data and clinical laboratory data.
Supplemental Table 4: Distribution of missense variants amongst the exons of MYBPC3, MYH7, and TNNT2. The canonical isoform was used in the case of multiple isoforms. P-values represent results of a Fisher’s Exact Test for independence of distributions between pathogenic variants and the variants from the Exome Sequencing Project (ESP) dataset.