Abstract
The Type VII protein translocation/secretion system, unique to Gram-positive bacteria, is a key virulence determinant in Staphylococcus aureus. We aim to characterize the architecture of this secretion machinery and now describe the present study of S. aureus EssB, a 52 kDa bitopic membrane protein essential for secretion of the ESAT-6 (early secretory antigenic target of 6 kDa) family of proteins, the prototypic substrate of Type VII secretion. Full-length EssB was heterologously expressed in Escherichia coli, solubilized from the bacterial membrane, purified to homogeneity and shown to be dimeric. A C-terminal truncation, EssB∆C, and two soluble fragments termed EssB-N and EssB-C, predicted to occur on either side of the cytoplasmic membrane, have been successfully purified in a recombinant form, characterized and, together with the full-length protein, used in crystallization trials. EssB-N, the 25 kDa N-terminal cytoplasmic fragment, gave well-ordered crystals and we report the structure, determined by SAD (single-wavelength anomalous diffraction) targeting an SeMet (selenomethionine) derivative, refined to atomic (1.05 Å; 1 Å=0.1 nm) resolution. EssB-N is dimeric in solution, but crystallizes as a monomer and displays a fold comprised of two globular domains separated by a cleft. The structure is related to that of serine/threonine protein kinases and the present study identifies that the Type VII secretion system exploits and re-uses a stable modular entity and fold that has evolved to participate in protein–protein interactions in a similar fashion to the catalytically inert pseudokinases.
Keywords: early secretory antigenic target of 6 kDa system 1 (ESX-1), Gram-positive bacterium, protein kinase, protein secretion, pseudokinase, X-ray crystallography
Abbreviations: BAP, biotin-acceptor peptide; BCG, Bacille Calmette–Guérin; BN-PAGE, Blue native PAGE; CV, column volume; DDM, dodecyl maltoside; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; DTT, dithiothreitol; ESAT-6, early secreted antigenic target of 6 kDa; ESI–Q–TOF-MS, electrospray ionization–quadrupole–time-of-flight MS; ESX-1, ESAT-6 system 1; ess, ESX-1 secretion system; IPTG, isopropyl-β-D-thiogalactopyranoside; LB, Luria–Bertani; MALDI–TOF-MS, matrix-assisted laser-desorption ionization–time-of-flight MS; MWCO, molecular-mass cut-off; PEG3350, poly(ethylene glycol) 3350; rmsd, root-mean-square deviation; SAD, single-wavelength anomalous diffraction; SeMet, selenomethionine; SPR, surface plasmon resonance; TEV, tobacco etch virus; T7SS, Type VII secretion system
INTRODUCTION
The T7SS (Type VII secretion system) is a specialized and complex protein-secretion pathway first identified in Mycobacterium tuberculosis, the infectious agent that causes tuberculosis [1]. T7SS gene clusters are widespread among Gram-positive bacteria of the phyla Actinobacteria and Firmicutes [2]. A characteristic of these clusters is the presence of genes encoding ESAT-6 (early secreted antigenic target of 6 kDa) proteins. These small highly immunogenic polypeptides represent the prototype substrates for the T7SS [3]. The clusters also encode an integral membrane protein containing domains related to both the FtsK cell division protein from Escherichia coli [4] and the stage III sporulation protein SpoIIIE from Bacillus subtilis [5]. Other genes in the T7SS genomic loci are less well conserved, although in general they are essential for a functional secretion system to be formed in organisms of each phylum [1,6].
The T7SS is a major virulence determinant in Gram-positive bacteria, indeed it is the loss of T7SS genes that attenuates pathogenicity of the vaccine strain Mycobacterium bovis BCG (Bacille Calmette–Guérin) [7]. Deletion of the relevant genomic region in M. tuberculosis mimics the BCG attenuation [8] and in the reverse control experiment BCG partially regains virulence upon complementation [9]. In Staphylococcus aureus, a versatile opportunistic pathogen and major cause of nosocomial and community-acquired infections [10], mutants compromised in their ability to secrete the ESAT-6-family proteins EsxA and EsxB display a 2–4-log reduction in virulence in a murine pathogenicity model [6]. This is consistent with the observations made in Mycobacteria. However, the presence of T7SS gene clusters in non-pathogenic bacteria, for example Streptomyces coelicolor and B. subtilis, suggests this secretion system is not a specific adaption to virulence itself, but may perhaps contribute to survival fitness.
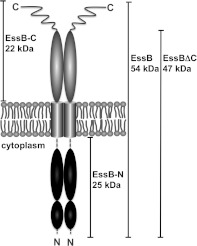
Little is known about the structure of the T7SS, or individual components or substrates of this secretion system. To address this deficiency we set out to investigate the T7SS system of S. aureus. The disruption of individual genes in the ess [ESX-1 (ESAT-6 system 1) secretion system] locus has identified three secreted proteins. These are EsaC and the two ESAT-6 family members EsxA and EsxB [6,11]. A total of four integral membrane proteins, EssA, EssB, EsaD and the Ftsk-SpoIII type ATPase EssC, have been shown to be necessary for translocation of ESAT-6 proteins across the bacterial envelope [6,12]. In the present study we targeted EssB, a 52 kDa bitopic membrane-bound protein presenting two segments of similar size on either side of the membrane (Figure 1). The protein is conserved among T7SS gene clusters in Firmicutes, but is of unknown function. A total of four expression constructs were prepared (Figure 1), heterologously expressed, the products purified, quaternary structure investigated and crystallization trials carried out. Of these constructs, three generated crystals, which although of good appearance were poorly ordered. Fortunately, EssB-N, the 25 kDa N-terminal fragment predicted to reside in the cytoplasm, gave well-ordered crystals and the structure was solved using SAD (single-wavelength anomalous diffraction) phasing then refined to 1.05 Å (1 Å=0.1 nm) resolution. The structural similarity of this EssB-N to serine/threonine protein kinases suggested a functional role in mediating protein–protein interactions. Biophysical and bacterial two-hybrid methods were used to initiate a search for potential binding partners from known components of the T7SS.
Figure 1. Schematic diagram of EssB predicted topology and domain borders.
Summary of constructs used in the present study. EssB is a 52-kDa integral membrane protein with a single predicted transmembrane segment (grey cylinders) spanning Trp229–Ser251. EssBΔC denotes the construct C-terminally truncated by 43 residues, which are predicted to be disordered. EssB-N, the 25-kDa N-terminal cytoplasmic fragment, is predicted to reside in the cytoplasm owing to the absence of a N-terminal signal sequence. Broken lines indicate EssB-N residues not defined by electron density. The 22-kDa C-terminal fragment is predicted to be on the trans-side of the cytoplasmic membrane.
EXPERIMENTAL
Expression and purification
The S. aureus subsp. aureus EssB (UniProt code Q2G185) coding sequence [codon optimized for E. coli K12 (Genscript)] was cloned into the SalI/XhoI site of a modified pET27b vector (Novagen) using the forward primer 5′-GCGCGCGTCGACAAAAAACCATAACCCGAAAAACG-3′ and the reverse primer 5′-GCGCGCCTCGAGCTATTTTTTGCGTTCCGCTTCCTGGCG-3′ (restriction sites are in bold). A gene fragment encoding an EssB construct truncated by 43 C-terminal residues (EssBΔC) was cloned likewise, but using 5′-GCGCGCCTCGAGCTATTTATCCAGAATATCCTGCAGTTTATCGG-3′ (restriction site is in bold) as the reverse primer. The plasmids produce an N-terminal His6-tagged protein followed by a TEV (tobacco etch virus) protease cleavage site.
The coding sequence spanning amino acid residues 12–226, the predicted cytoplasmic N-terminal fragment EssB-N, was cloned using the forward primer 5′-GCGCGCGTCGACACAGGATATGCTGACCCCGCTGGATG-3′ and the reverse primer 5′-GCGCGCCTCGAGCTACACGGTATGGCCCACTTTGCGCAC-3′ (restriction sites are in bold). The DNA encoding residues 257–444, the C-terminal fragment EssB-C, was cloned using 5′-GCGCGCGTCGACAGAACGCATTGAAAAAGGCTATCAGGC-3′ and 5′-GCGCGCCTCGAGCTATTTTTTGCGTTCCGCTTCCTGGCG-3′ as the forward and reverse primers respectively (restriction sites are in bold).
The gene fragments encoding EssB-N and EssB-C were expressed and the recombinant proteins purified using the same protocols. Freshly transformed E. coli BL21(DE3) cells were cultivated at 37°C in 20 ml of LB (Luria–Bertani) medium containing 50 μg/ml kanamycin and 0.2% glucose, which provided inoculum of the 1 litre main culture (grown at 37°C in LB medium supplemented with 1 mM MgCl2, 0.5 mM CaCl2 using baffled 5 litre Erlenmeyer flasks). The temperature was lowered to 25°C when an attenuance of 0.6 at λ=600 nm was attained and gene expression was induced with 1 mM IPTG (isopropyl β-D-thiogalactopyranoside). Cells were harvested after 14 h, resuspended in buffer A [50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 1 mM DTT (dithiothreitol) and 10% (w/v) glycerol, supplemented with one protease inhibitor cocktail tablet (complete, EDTA-free, Roche)] and broken by two passages through a French press (Thermo). The homogenate was centrifuged (150000 g for 60 min at 4°C) and the supernatant was passed through a 0.45-μm filter, supplemented with 25 mM imidazole and then loaded on to a 5 ml CV (column volume) His-Trap HP column (GE Healthcare) previously charged with Ni2+. After a washing step (4 CV) in the same buffer, the samples were eluted applying a linear imidazole gradient (25–250 mM over 18 CV). The buffer was exchanged using a spin concentrator [10 kDa MWCO (molecular-mass cut-off); Sartorius] to buffer B [50 mM Tris/HCl (pH 7.8), 2 mM DTT and 10% (w/v) glycerol]. After a 14 h incubation step with His-tagged TEV protease (at a molar ratio of 1:20 TEV/recombinant protein) the sample was passed through a HisTrap HP column equilibrated with buffer A to remove the protease, cleaved peptide and non-cleaved material. The cleaved product retains a glycine-alanine-serine sequence at the N-terminus.
The flow through was collected, concentrated and passed through a size-exclusion chromatography column (HR 30/100 GL, Superdex 75, GE Healthcare) equilibrated with buffer C {10 mM Tris/HCl (pH 7.5), 10 mM NaCl and 0.5 mM TCEP [tris-(2-carboxyethyl)phosphine)]}. All size-exclusion chromatography columns were calibrated with molecular mass standards (thyroglobulin, 670 kDa; γ-globulin, 158 kDa; serum albumin, 67 kDa; ovalbumin; 44 kDa, myoglobin, 17 kDa; and vitamin B12, 1 kDa).
The preparation of full-length EssB and EssBΔC followed the same protocol except that E. coli LEMO21(DE3) cells [13] were used in medium supplemented with 100 μM L-rhamnose and gene expression was induced with 400 μM IPTG. Membranes were isolated by centrifugation (150000 g for 60 min at 4°C), resuspended in buffer A and solubilized at room temperature (20°C) with DDM (dodecyl maltoside) at a protein/detergent mass ratio of 1:3 for 2 h. Solid material was removed by centrifugation and the supernatant purified as described above except that all buffers were supplemented with 0.02% DDM and 1 μg/ml DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine; a gift from Lipoid AG) and for size-exclusion chromatography a Superdex 200 column (GE Healthcare) was used.
An SeMet (selenomethionine) derivative of EssB-N was obtained using the protocol described above with full incorporation achieved by metabolic inhibition [14] and confirmed by MALDI–TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight MS) analysis.
Blue native gel electrophoresis
Linear 4–16% gradient Native Bis-Tris gels (Novex®, Life Technologies) were run and destained according to the manufacturer's protocol. Samples comprising 3 μg of protein were loaded. Membrane protein samples were supplemented with 0.1% DDM. Lactate dehydrogenase (146 kDa), BSA (66 kDa), and soya bean trypsin inhibitor (20 kDa) were used as the standard proteins.
MS
ESI–Q–TOF-MS (electrospray ionization–quadrupole–time-of-flight MS) and MALDI–TOF-MS analysis was performed at the University of Dundee ‘Fingerprints’ Proteomics Facility (Dundee, U.K.) using an Applied Biosystems DE-STR and a Agilent 6520 QTOF spectrometer respectively. Crystals of EssB-N were harvested by centrifugation (18000 g for 2 min at 20°C and dissolved in 5 mM Tris/HCl (pH 7.8). Residual PEG3350 [poly(ethylene glycol) 3350] from the mother liquor was removed using a spin concentrator (10 kDa MWCO; Sartorius).
Crystallization and structure determination
Isomorphous crystals of native and SeMet-labelled EssB-N were obtained by hanging-drop vapour diffusion at 20°C in 3 μl drops of a 1:1 ratio of protein solution [20 mg/ml EssB-N in 10 mM Tris/HCl (pH 7.5), 10 mM NaCl and 0.5 mM TCEP] with the reservoir [0.18 M ammonium citrate and 20% (w/v) PEG3350]. Orthorhombic blocks (100–300 μm) with space group P212121 appeared after 2 days. The crystals were transferred to LV-oil (MiTeGen) before flash cooling in a stream of gaseous nitrogen at −173°C and then characterized in-house using a Rigaku 007 rotating anode X-ray generator coupled to an RAXIS IV++ image plate detector. Suitable samples were stored and subsequently used to measure full datasets on station ID29 at the ESRF (European Synchrotron Radiation Facility; Grenoble, France). Intensity measurements were recorded on an ADSC Q315R Charge Couple Device detector, integrated and processed using XDS [15]. Initial phases were calculated to 1.4 Å by SAD methods in CRANK [16] as implemented in the CCP4 suite of programs [17].
Two selenium sites (out of the three possible) were located and gave an initial figure-of-merit of 0.35, which increased to 0.75 after density modification. An initial model, consisting of residues 31–204, was built using ARP/wARP [18] and gave Rwork/Rfree values of 0.178 and 0.211 respectively.
High- and low-resolution data sets, with resolution limits of 1.05 Å and 2.5 Å respectively (Table 1), were collected using different translations of a single native crystal. The datasets were merged using SCALA [19] and the phases determined by molecular replacement with PHASER MR [20] using the SeMet structure as the search model. The refinement was initiated in REFMAC5 [21] and then ported to SHELXL [22]. Several rounds of model-map inspection and re-building in COOT [23] interspersed with least-squares optimization and isotropic thermal parameter refinement gave Rwork and Rfree values of 0.217 and 0.246 respectively. The addition of water molecules, hydrogen atoms and inclusion of anisotropic B-factor refinement lowered Rwork/Rfree to 0.138 and 0.158 respectively.
Table 1. Crystallographic statistics.
Values in parentheses refer to the highest resolution bin. Rmerge=ΣhΣi‖(h,i)−<I(h)> ΣhΣi I(h,i). Rwork=Σhkl‖Fo|−|Fc‖/Σ|Fo|, where Fo is the observed structure factor amplitude and Fc is the structure–factor amplitude calculated from the model. Rfree is the same as Rwork except only calculated using a subset, 5%, of the data that are not included in any refinement calculations.
| Parameter | SeMet | Native |
|---|---|---|
| Diffraction data | ||
| Wavelength (Å) | 0.9788 | 0.9814 |
| Resolution (Å) | 47.6–1.4 (1.44–1.37) | 58.74–1.05 (1.15–1.05) |
| Unit cell lengths (Å) | 38.7, 58.6, 95.3 | 38.7, 58.7, 95.1 |
| Unique reflections | 46296 (6595) | 100245 (13554) |
| Completeness (%) | 99.8 (98.8) | 99.0 (92.8) |
| <I/σ(I)> | 46.3 (10.4) | 18.0 (3.6) |
| Redundancy | 26.3 (15.0) | 7.1 (4.6) |
| Rmerge | 0.050 (0.236) | 0.061 (0.409) |
| Refinement statistics | ||
| Rwork/Rfree | 13.8/15.8 | |
| Protein residues | 176 | |
| Waters | 376 | |
| rmsd from ideal geometry (Å) | ||
| Bond lengths | 0.015 | |
| Angular distances | 0.033 | |
| Thermal parameter (B) values (Å2) | ||
| Wilson B | 6.1 | |
| Mean B over all atoms/protein/water | 18.4/13.8 /25.6 | |
| Ramachandran favoured/allowed/outliers (%) | 98.4 /1.6/0 |
Macroscopically different rounded EssB-N crystals were grown using a reservoir condition containing a high phosphate concentration [0.1 M Hepes (pH 7.5), 0.8 M NaH2PO4 and 0.8 M KH2PO4]. Diffraction data were collected on beam line I03 at the Diamond Light Source (Harwell, U.K.). Processing the data, using XDS and SCALA, indicated that the two crystal forms, although distinct in appearance, are in fact isomorphous. The structure was determined to 1.6 Å (results not shown). A total of three iterative cycles of inspection and manual fitting in COOT and refinement in REFMAC5 gave Rwork/Rfree values of 0.167 and 0.199 respectively. Comparisons indicate that this model is essentially the same as that determined at atomic resolution and so no further details are given.
Model geometry and the fit to electron-density maps were monitored with MOLPROBITY [24] and the validation tools in COOT. Figures were prepared using PyMOL (http://www.pymol.org). The DALI server was used to search the PDB for structural homologues and structural superimpositions were calculated using DALILITE [26]. Multiple sequence alignments were obtained using CLUSTALW2 [27] and edited with ALINE [28]. The conservation of residues at the protein surface was investigated using CONSURF [29]. Potential EssB orthologues were excluded from the analysis if the sequence identities were less than 19%. Altogether 54 EssB-homologues were used.
Computational analysis of the sequence
A transmembrane segment of EssB was predicted using TMHMM v2.0 [30]. The design of constructs was guided by secondary structure prediction using JPRED [31] and the PHYRE fold-recognition server [32].
Bacterial two-hybrid system
The bacterial two-hybrid screening for EssB-N interactions was performed on MacConkey agar plates containing 1% maltose, essentially following an established protocol [33]. The S. aureus subsp. aureus coding sequences for EsxA, EsxB, EsaB and EssB-N [codon optimized for expression in E. coli K12 (Genscript)] were cloned into the XhoI/HindIII or BamHI/KpnI sites of a modified pT18 or pT25 plasmid [34] respectively (for primers see Supplementary Table S1 at http://www.biochemj.org/bj/449/bj4490469add.htm). The genes on pT25narJ and pT18narG express proteins that form a heterodimer [34] provided a positive control. E. coli strain BTH101 cells were co-transformed with the pT18 and pT25 constructs, plated and cultured for 2 days at 25°C. Colonies were re-streaked on to fresh MacConkey medium and incubated for 36 h at 25°C.
SPR (surface plasmon resonance) spectroscopy
For the SPR experiments EssB-N was immobilized via a C-terminally fused BAP (biotin-acceptor peptide) [35]. Biotin-protein ligase BirA was used for site-specific monobiotinylation of this 15-amino-acid peptide (GLNDIFEAQKIEWHE). The covalently attached biotin binds streptavidin with high affinity [36].
The E. coli K12 BirA (P06709) coding sequence stretch was PCR-amplified from genomic copy and cloned into the Nco1/Xho1 site of a modified pET27b vector (Novagen) fusing a C-terminal His6-tag-coding sequence with the reverse primer. Expression followed the same protocol as described for EssB-N. Harvested cells were resuspended in 50 mM Tris/HCl (pH 7.5), 300 mM NaCl, 1 mM DTT and 10% (w/v) glycerol and broken by two passages through a French press (Thermo). The homogenate was centrifuged at 150000 g for 60 min at 4°C. The resulting supernatant was passed through a 0.45-μm filter, supplemented with 25 mM imidazole and loaded on to a 5 ml HisTrap HP column (GE Healthcare). After a 4 CV washing step in the same buffer, the recombinant protein was eluted applying a linear imidazole gradient (25–250 mM over 18 CV). The buffer of the late elution fractions was exchanged using a spin concentrator (10 kDa MWCO; Sartorius) to 10 mM Tris/HCl (pH 7.5), 200 mM KCl and 0.5 mM DTT and the protein concentration was adjusted to 10 mg/ml.
EsaB, EsxA and BAP-tagged EssB-N were cloned, expressed and purified essentially as described for EssB-N (for the primers see Supplementary Table S2 at http://www.biochemj.org/bj/449/bj4490469add.htm). BAP-tagged EssB-N (20 μM) was incubated at 37°C for 2 h in buffer C [10 mM Tris/HCl (pH 7.5), 200 mM KCl, 5 mM MgCl2 and 100 μM d-biotin) containing 500 μM ATP and 1 μM His6-tagged BirA. The sample was passed through a HisTrap HP column equilibrated with buffer A to remove BirA. The flow through was collected, concentrated and passed through a size-exclusion chromatography column (HR 30/100 GL, Superdex 75 prep grade, GE Healthcare) equilibrated with buffer C to remove free biotin. The incorporation of biotin was monitored by MALDI–TOF-MS.
SPR analysis was performed using a Biacore 3000 instrument (GE Healthcare) using 20 mM Tris/HCl (pH 7.8), 150 mM NaCl, 0.1 mg/ml BSA and 0.005% Tween 20 as a running buffer. BAP-tagged EssB-N was captured on an immobilized streptavidin surface at a density of 3500 resonance units. Binding of EsxA and EsaB was measured at 25°C and 4°C in the reported concentrations (Supplementary Figure S1 at http://www.biochemj.org/bj/449/bj4490469add.htm). Curves for binding to a blank reference chip were subtracted using the software package Biaevaluation (GE Healthcare).
Data deposition
The atomic co-ordinates and structure factors have been deposited in the PDB under code 4ANN.
RESULTS AND DISCUSSION
Characterization of membrane-bound EssB
Two expression constructs of membrane-bound EssB (Figure 1) were prepared, the encoded proteins characterized and crystallization trials carried out. Full-length EssB was solubilized from the cytoplasmic membrane and purified to homogeneity with a yield of approximately 1–1.5 mg/l of culture. A protein truncated by 43 C-terminal residues, EssBΔC, which retains membrane-binding capacity, was obtained at a similar yield. The deleted residues, from Lys401 to the terminal Lys444, form a highly basic segment with a theoretical pI of 9.4. This stretch is highly divergent, indeed absent in some EssB orthologues, as evidenced by a multiple sequence alignment (Supplementary Figure S2 at http://www.biochemj.org/bj/449/bj4490469add.htm). Secondary structure prediction and fold analysis suggested a heptad-repeat on the basis of α-helical propensity from Glu421 to Gln438 together with disorder in other regions. It appeared sensible therefore to omit the C-terminal tail to improve the chances of crystallization.
The EssBΔC construct has a theoretical mass of 47 kDa, but in gel filtration eluted as a single species with an apparent molecular mass of 198 kDa (Supplementary Figure S3 at http://www.biochemj.org/bj/449/bj4490469add.htm). This suggests a dimeric state taking into consideration the detergent micelle around the transmembrane helices. The average micellar size of the detergent used, DDM, is 50–70 kDa. The 52 kDa full-length EssB elutes with a lower retention volume and an apparent molecular mass of 254 kDa. The shift in retention volume is larger than expected from the mass difference of 5 kDa per monomer. A possible explanation is a change in the overall shape of the protein and thus its hydrodynamic radius, when the potentially flexible C-terminal residues are present. A small peak at 0.38 CV in the chromatogram (Supplementary Figure S3) indicates the presence of aggregated material in the full-length EssB preparation, material that is absent from the EssBΔC chromatogram. The dimeric state of EssB was furthermore corroborated by BN-PAGE (Blue native PAGE) analysis (Supplementary Figure S4 at http://www.biochemj.org/bj/449/bj4490469add.htm).
Extensive crystallization trials were undertaken. EssBΔC formed needles, of length 100 μm, in the presence of DDM, DOPC and DHPC (1,2-diheptanoyl-sn-glycero-3-phosphocholine), but no useful diffraction was observed even when tested on a microfocus-beam line (ID23) at the ESRF, merely smears between 20 and 35 Å resolution.
Characterization of soluble EssB fragments
Recombinant proteins corresponding to the N-terminal fragment, EssB-N, predicted to be localized in the cytoplasm and the C-terminal fragment, EssB-C, predicted to reside on the trans-side of the membrane, were purified with yields of 3–4 mg/l and 6–7 mg/l of bacterial culture respectively. EssB-N and EssB-C have predicted masses of 25 kDa and 22 kDa respectively. In BN-PAGE analysis the fragments migrate with apparent masses of 58 kDa for EssB-N and 42 kDa for EssB-C (Supplementary Figure S4). This indicates that each forms dimeric species in solution. EssB-N runs as a double band on the BN-PAGE most probably reflecting the 1 kDa C-terminal degradation observed by MS (discussed below).
Although EssB-N eluted at an apparent mass of 54 kDa in analytical gel filtration the observed retention volume for EssB-C was lower than expected for a protein of mass 44 kDa (Supplementary Figure S5 at http://www.biochemj.org/bj/449/bj4490469add.htm). This is in agreement with the mass discrepancy observed when comparing full-length EssB and EssBΔC (see above) and it appears that the highly basic potentially flexible C-terminal segment does indeed influence the protein shape. Expression of a construct encoding a polypeptide truncated at the C-terminus of EssB-C, in a similar fashion to EssBΔC, led only to the formation of inclusion bodies. Rectangular plate crystals of EssB-C, measuring up to 80 μm after optimization, appeared after 3 weeks, but only diffracted to about 7 Å (Supplementary Figure S6 at http://www.biochemj.org/bj/449/bj4490469add.htm) and were not studied further. EssB-N crystallized readily and high-resolution diffraction data were obtained.
Structure of EssB-N
The crystal structure of S. aureus EssB-N was solved by SAD methods exploiting a SeMet derivative and produced an atomic-resolution model. The asymmetric unit consists of a monomer and the model comprises 176 residues from Leu30 to Tyr205. All residues display φ/ψ combinations within allowed regions of a Ramachandran plot (Table 1). The model is missing 39 residues due to a combination of proteolysis (accounting for about 30 residues) and diffuse electron density at N- and C-termini, which we were unable to interpret. ESI–Q–TOF-MS analysis of the EssB-N sample prior to crystallization revealed a mass difference of 1038 Da (Supplementary Figure S7 at http://www.biochemj.org/bj/449/bj4490469add.htm), which would correspond to nine missing C-terminal residues from Tyr218 onwards (YVRKVGHTV). EssB-N crystals were subjected to SDS/PAGE and MALDI–TOF-MS analysis after tryptic digest of the excised 24 kDa band. An intact N-terminal fragment was identified and the last C-terminal tryptic fragment covered the sequence from Lys207 to Ala217 (KQEQDYSQNYA) (Supplementary Table S3 at http://www.biochemj.org/bj/449/bj4490469add.htm), which is proximal to the peptide identified as missing by the ESI–Q–TOF-MS data. The loss of only nine C-terminal residues, not 39, was thus confirmed, ruling out the possibility that proteolysis occurred during crystallization and indicting therefore that degradation of the C-terminus stretch occurred in the expression host.
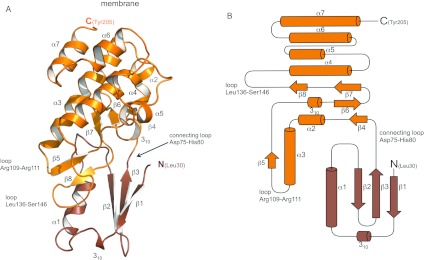
The EssB-N structure is cylindrical with a length of approximately 53 Å and diameter of 33 Å, and consists of two domains (Figure 2 and Supplementary Figure S8 at http://www.biochemj.org/bj/449/bj4490469add.htm). The smaller N-terminal domain consists of a three-stranded antiparallel β-sheet with a single turn of 310-helix and α1, interspersed between β1 and β2. Adjacent to β1 there is diffuse electron density, which may represent an additional N-terminal β-strand, but we were unable to model this satisfactorily. β3 leads into a loop (Asp75–His80) connecting the domains. The C-terminal domain structure is dominated by a helical bundle formed from two antiparallel helix pairs α3/α4 and α7/α8. The connecting loop leads to β4–α2 and a sequence stretch (Thr110–Ser146) that forms two short β-strands (β6–β7) that with β4 creates a β-sheet tucked in below the helical bundle. A buried turn of 310-helix forms an acute bend that changes the direction of the polypeptide from β5 to β6 and supports the alignment of β6 with β4. The model terminates with α7. The amino acid sequence is predicted to form a longer α-helix extending into the outer membrane.
Figure 2. Overall structure of EssB-N.
(A) Cartoon representation. (B) Topology diagram. The N- and C-terminal domains of EssB-N are coloured maroon and orange respectively. Labelled loop regions are discussed in the text.
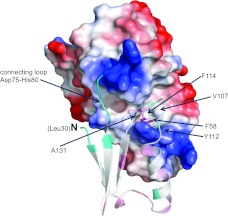
A deep cleft, approximately 30 Å, is formed between the two domains of EssB-N. The cleft is closed off at one side by the interdomain connecting loop (Asp75–His80). Situated next to this loop, that joins the neighbouring strand β3 of the N-terminal sheet to β4, a further interdomain contact is made by a loop interspersed between α1 and β2. Phe58 contributes to this structural feature by forming a hydrophobic pocket together with Val107, Tyr112, Phe114 and Ala131 (Figure 3). A number of conserved residues (Thr113, Pro118, Glu120, Arg134 and Gly134) cluster around the C-terminal side of the cleft (Supplementary Figure S9 at http://www.biochemj.org/bj/449/bj4490469add.htm). This structure is formed by two back-to-back β-sheet motifs between α3 and α4. A two-stranded (β5 and β8) sheet is located at the mouth of the cleft. The 310-helix, preceding β6, is a key structural element and features the highly conserved residues Pro118 and Glu120. Residues lining the N-terminal domain side of the cleft are less well conserved but this serves to highlight the structural importance of the 310-helix carrying conserved residues (Ser38 and Ile40) that forms a lining to the cleft.
Figure 3. van der Waals surface representation of the EssB-N C-terminal domain.
The PyMOL calculated electrostatic potential is mapped on to the EssB-N C-terminal domain. The N-terminal domain is in cartoon representation. Phe58 is drawn in stick representation. Residues forming a hydrophobic pocket accommodating Phe58 are labelled.
The core of EssB-N is extremely well-ordered. However, the three loops (Asp75–His80, Arg109–Arg111 and Leu137–Ser145), which create part of the inter domain cleft, display higher B-factor values (26.4 Å2, 25.1 Å2 and 21.5 Å2 respectively) than the overall average (13.8 Å2). This suggests a greater flexibility of the loops compared with the core of the structure.
EssB-N crystallizes with a single molecule in the asymmetric unit yet forms a stable dimer in solution. Highly ordered crystals with a different morphology were grown in high phosphate concentrations at pH 7.5. We thought it possible that this might represent a different oligomeric state, but the crystals were isomorphous to those obtained in the presence of citrate and gave a virtually identical structural model comprising residues Leu30–Gln204. In these crystals there are four interfaces formed by EssB-N with symmetry related molecules and these have solvent-accessible surface areas in the range 100–560 Å2. The surface area of EssB-N is approximately 9890 Å2 so the maximum interface represents less than 6% of the monomer surface area. Such a low value is unlikely to be thermodynamically stable in solution and the discrepancy may simply reflect the different conditions under which quaternary structure has been analysed.
The EssB-N structure is reminiscent of a pseudokinase
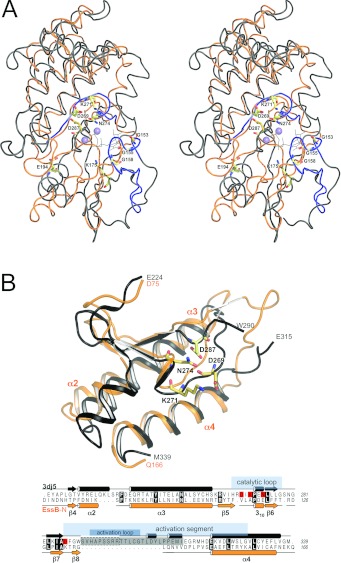
The structure of EssB-N resembles the bilobal structure typical of protein kinases, i.e. an N-terminal domain or N-lobe with a β-sheet packed against an intersecting α-helix, and a predominantly α-helical C-terminal domain (C-lobe, Figure 2). In serine/threonine protein kinases the cleft formed between the two lobes is occupied by the ATP cofactor and protein substrates. Many protein kinases are regulated by phosphorylation of an activation segment, which produces an extended active state permissive for substrate binding [37]. EssB-N displays closest structural similarity to inactive protein kinases that result from a specific orientation of the kinase activation segment. The most similar orthologue is the catalytic domain, in an inhibited form, of the mammalian serine/threonine kinase Aurora-A (PDB code 3DJ5) with a Z-score 9.4 and rmsd (root-mean-square deviation) of 4.5 Å for the alignment of 170 Cα atoms (Figure 4). Similarity is measured by DALI Z-scores and significant similarities give values greater than 2; such values usually correspond to related folds [38]. The sequence identity is only 12%, nevertheless these statistics suggest an evolutionary relationship in which a significant degree of structural homology is retained. Comparison with the active conformation of Aurora-A kinase (PDB code 2C6D) gives a lower Z-score of 7.9 and slightly higher rmsd of 4.8 Å compared with the inactive state. The major difference is the orientation of the EssB-N sequence stretch from Lys132–Ser146 (Figure 2A and Supplementary Figure S8) that corresponds to the activation segment of serine/threonine protein kinases (Figure 4). In EssB-N the N-terminal domain is more similar to the conformation observed in inactive protein kinases rather than the extended active conformation.
Figure 4. Comparison with kinases.
(A) Stereo view of rigid body superimposition of EssB-N (orange) and Aurora-A kinase catalytic domain (grey) is depicted in ribbon representation. Prosite serine/threonine protein kinase signature motifs PS00107 ([LIV]-G-{P}-G-{P}-[FYWMGSTNH]-[SGA]-{PW}-[LIVCAT]-{PD}-x-[GSTACLIVMFY]-x(5,18)-[LIVMFYWCSTAR]-[AIVP]-[LIVMFAGCKR]-K) and PS00108 ([LIVMFYC]-x-[HY]-x-D-[LIVMFY]-K-x(2)-N-[LIVMFYCT](3)) are highlighted in black. Highly conserved serine/threonine kinase residues are drawn as sticks. ATP (shown as black lines) and two Mn2+ ions (lilac spheres) were modelled by superimposition of the ATP-bound structure of protein kinase A (PDB code 1ATP). (B) View of the C-terminal side of the cleft, which features the highest degree of structural similarity. A partial structural alignment over 86 residues (EssB Asp75–Gln166 and Aurora-A kinase Glu224–M339) is shown. Aurora-A kinase invariant active-site residues Asp269, Lys271 and Asn274, and the activation segment residue Asp287 are in sticks and highlighted in red in the underlying sequence alignment. Aurora-A kinase residues Ser291–Glu315 (grey box in the sequence alignment), including seven residues of the activation loop, which is not resolved in the structure (boxed in a broken line), have been omitted for clarity. Secondary structure elements and known functional serine/threonine protein-kinase segments deduced from [47] are indicated.
EssB lacks the protein kinase ATP-binding signature motif situated in the N-lobe (Figure 4A); an observation that is consistent with failure of EssB to bind the ATP-analogue 2′-3′-O-(N′-methylanthraniloyl) adenosine-5′-O-triphosphate (results not shown). EssB also lacks the serine/threonine protein kinase active-site signature, which is located in the C-lobe and includes a catalytic aspartate (Asp269 in Aurora-A kinase; Figure 4). However, on the inner surface of the EssB cleft, residues Pro118–Ile130, a remarkable degree of structural similarity is observed (Figure 4). This involves the turn of 310-helix and the β-sheet motif at the rear of the cleft, a sequence stretch that in the structural alignment corresponds to the localization of the serine/threonine protein kinase active-site signature in Aurora-A. Helices α2, α3 and the central α4 are similar in size and orientation in the two structures and these help to align the two domains with respect to each other. These similarities are reflected in a significantly lower rmsd of 2.9 Å (Z-score 7.5) for a partial structural alignment (Figure 4B) over 86 residues (from Asp75–Gln166) of the C-terminal domain.
The topology of secondary structure elements that facilitate the interdomain contact area is similar and this involves the regions important for polypeptide substrate recognition by protein kinases [39]. Moreover, this part of the EssB-N structure displays the highest degree of sequence conservation in orthologues (Supplementary Figures S2 and S9). The implication is that EssB-N uses the interdomain cleft to bind a polypeptide partner in a similar fashion to serine/threonine kinases binding to substrates. The absence of obvious catalytic residues renders it unlikely that EssB-N exhibits kinase activity itself and it is therefore reminiscent of pseudokinases; a sub-group of the protein kinase family that facilitate protein–protein interactions [40,41]. A single bacterial pseudokinase has been reported, MviN. This multi-domain integral membrane protein forms a complex that controls cell wall synthesis in M. tuberculosis upon phosphorylation by the serine/threonine kinase PknB [42]. A structural alignment of 160 Cα positions of the MviN pseudokinase domain (PDB code 3UQC [42]) with EssB-N indicates a structural relationship at a similar level as with active kinases (Z-score 8.2 and rmsd 4.7 Å). This appears sensible since pseudokinases have retained the overall fold, whereas the active-site signature motifs are modified or absent. Eukaryotic pseudokinases participate in various biological processes exerting scaffolding and/or regulatory functions [40] and the structural similarity displayed by EssB-N may reflect a capacity to act as a protein-binding module to provide a scaffold as part of the functional multi-subunit T7SS.
There are also intriguing similarities between EssB and eukaryote-like serine/threonine kinases that occur in bacteria, such as PknB and PknG. M. tuberculosis PknB and PknG are localized to the cytoplasmic membrane positioned with the C-terminal lobe of the kinase domain directed into the membrane leading to an extracellular domain [43]. Ligand-induced dimerization is important in these systems and hints that in EssB the fluxional behaviour we observe may be an intrinsic property of the protein and one that could be influenced by the presence of a molecular partner. Such a partner might be substrate for the T7SS or a component of the secretion apparatus itself.
EssB interaction studies
The structure suggested a role in binding partner proteins, and to address the hypothesis that EssB-N is a scaffold or hub of the secretion machine, a bacterial two-hybrid assay was established to test for potential interactions with other proteins encoded within the T7SS gene cluster. The assay depends on reconstitution of adenylate cyclase activity from two non-interacting Bordetella pertussis CyaA (bifunctional haemolysin-adenylate cyclase) fragments called T18 and T25 [44,45]. cAMP synthesis triggers the transcriptional activation of maltose and lactose catabolic operons and E. coli cya BTH101 colonies fermenting these carbohydrates acidify the medium and produce a red phenotype on MacConkey/maltose indicator plates.
EssB-N was fused to the T18 fragment, and T25 fusions of EsxA, EsxB, EsaB and EsaC were co-expressed (Supplementary Figure S10 at http://www.biochemj.org/bj/449/bj4490469add.htm). Colonies remained white except for EsxA and EsaB, which adopted a faint red colour with a later onset compared with the positive control (Supplementary Figure S10). SPR methods were used to test if purified EsxA, or EsaB, a small soluble protein that appears to regulate the production of the T7SS substrate protein EsaC [11], bind to immobilized EssB-N (Supplementary Figure S1). However, no binding was observed. In fact significant negative responses were obtained after subtracting responses from the reference flow cell due to binding to the reference. These results hint to a non-specific interaction, which is conceivable given the highly polar surface of the EsxA dimer [46] and this would classify the weak signals observed in the bacterial two-hybrid system as false positives.
Numerous factors can complicate detection of protein–protein interactions in a multi-component secretion apparatus. For example, more than two binding partners might be necessary to stabilize protein–protein interactions. A further issue is that the expression levels of membrane-bound proteins may be insufficient for the two-hybrid assay. Also, that isolated fragments might in some cases poorly reflect their protein–protein interaction characteristics, especially if transmembrane domains participate in binding. Furthermore in vitro protein–protein interaction studies might be hampered by the presence of a detergent that poorly mimics a lipidic environment. In future studies homologous expression in Staphylococcus mutants should allow for the isolation of T7SS protein complexes to shed light on the architecture of the entire secretion system.
Concluding remarks
The monotopic membrane protein EssB from S. aureus and constructs encoding the isolated soluble domains have been expressed in recombinant form, the products purified to homogeneity and their quaternary structure determined to be dimeric. The cytoplasmic fragment EssB-N yielded highly ordered crystals and the structure was determined to atomic resolution.
EssB-N exhibits an intriguing structural relationship to serine/threonine protein kinases. The lack of conserved catalytic residues rules out EssB-N as an enzyme, but rather suggests a function as a protein–protein interaction module using the stable modular entity of the protein kinase fold. In effect EssB resembles a membrane inset pseudokinase. The biological role may be as an adaptor protein; to recruit or position other components of the secretion apparatus at the cytoplasmic membrane and in so doing form the functional machine. Future studies in this area will now be required to elucidate the precise contribution of this protein to the T7SS and the virulence of an important human pathogen.
Online data
AUTHOR CONTRIBUTION
Tracy Palmer and William Hunter conceived the project. All aspects of sample preparation, characterization and crystallization were carried out by Martin Zoltner. Crystallographic studies involved Martin Zoltner, Paul Fyfe and William Hunter. Martin Zoltner and William Hunter were primarily responsible for interpretation of the results. Martin Zoltner contributed most to preparation of the paper with help from all other authors.
ACKNOWLEDGEMENTS
We thank the DLS (Diamond Light Source) and ESRF (European Synchrotron Radiation Facility) for synchrotron beam time and excellent support, and Mark Agacan, Grant Buchanan and Sarah Coulthurst for assistance and materials.
FUNDING
This work was supported by the Biotechnology and Biological Sciences Research Council [grant number H007571], the Medical Research Council [grant number G117/519] and the Wellcome Trust [grant numbers 082596, 083481 and 094090].
References
- 1.Abdallah A. M., Gey van Pittius N. C., Champion P. A. D., Cox J., Luirink J., Vandenbroucke-Grauls C. M. J. E., Appelmelk B. J., Bitter W. Type VII secretion: mycobacteria show the way. Nat. Rev. Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 2.Pallen M. J. The ESAT-6/WXG100 superfamily: and a new Gram-positive secretion system? Trends Microbiol. 2002;10:209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- 3.Champion P. A. D., Stanley S. A., Champion M. M., Brown E. J., Cox J. S. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science. 2006;313:1632–1636. doi: 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
- 4.Begg K. J., Dewar S. J., Donachie W. D. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu L. J., Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 6.Burts M. L., Williams W. A., DeBord K., Missiakas D. M. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahairas G. G., Sabo P. J., Hickey M. J., Singh D. C., Stover C. K. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis K. N., Liao R., Guinn K. M., Hickey M. J., Smith S., Behr M. A., Sherman D. R. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette–Guérin attenuation. J. Infect. Dis. 2003;187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majlessi L., Brodin P., Brosch R., Rojas M.-J., Khun H., Huerre M., Cole S. T., Leclerc C. Influence of ESAT-6 secretion system 1 (RD1) of Mycobacterium tuberculosis on the interaction between mycobacteria and the host immune system. J. Immunol. 2005;174:3570–3579. doi: 10.4049/jimmunol.174.6.3570. [DOI] [PubMed] [Google Scholar]
- 10.Lowy F. D. Staphylococcus aureus infections. New Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 11.Burts M. L., DeDent A. C., Missiakas D. M. EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol. Microbiol. 2008;69:736–746. doi: 10.1111/j.1365-2958.2008.06324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson M., Chen Y. H., Butler E. K., Missiakas D. M. EsaD, a secretion factor for the Ess pathway in Staphylococcus aureus. J. Bacteriol. 2011;193:1583–1589. doi: 10.1128/JB.01096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner S., Klepsch M. M., Schlegel S., Appel A., Draheim R., Tarry M., Högbom M., van Wijk K. J., Slotboom D. J., Persson J. O., de Gier J.-W. Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14371–14376. doi: 10.1073/pnas.0804090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doublie S. Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol. Biol. 2007;363:91–108. doi: 10.1007/978-1-59745-209-0_5. [DOI] [PubMed] [Google Scholar]
- 15.Kabsch W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ness S., de Graaff R., Abrahams J. Crank: new methods for automated macromolecular crystal structure solution. Structure. 2004;12:1753–1761. doi: 10.1016/j.str.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 18.Perrakis A., Morris R. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 19.Evans P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 20.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheldrick G. M., Schneider T. R. SHELXL: high-resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Lovell S. C., Davis I. W., Arendall W. B., de Bakker P. I. W., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. Structure validation by Cα geometry: phi, psi and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 25.Reference deleted
- 26.Holm L., Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 27.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 28.Bond C. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 29.Landau M., Mayrose I., Rosenberg Y. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krogh A., Larsson B., Heijne, von G., Sonnhammer E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 31.Cole C., Barber J. D., Barton G. J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelley L. A., Sternberg M. J. E. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 33.Handford J. I., Ize B., Buchanan G., Butland G. P., Greenblatt J., Emili A., Palmer T. Conserved network of proteins essential for bacterial viability. J. Bacteriol. 2009;191:4732–4749. doi: 10.1128/JB.00136-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karimova G., Pidoux J., Ullmann A., Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckett D., Kovaleva E., Schatz P. J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber P. C., Ohlendorf D. D., Wendolowski J. J., Salemme F. R. Structural origins of high affinity biotin binding to streptavidin. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 37.Rajakulendran T., Sicheri F. Allosteric protein kinase regulation by pseudokinases: insights from STRAD. Sci. Signaling. 2010;3:pe8. doi: 10.1126/scisignal.3111pe8. [DOI] [PubMed] [Google Scholar]
- 38.Holm L., Kääriäinen S., Rosenström P., Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knighton D., Zheng J., Eyck, Ten L., Ashford V., Xuong N., Taylor S., Sowadski J. Crystal structure of the catalytic subunit of cyclic adenosine monophosphatedependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 40.Boudeau J., Miranda-Saavedra D., Barton G. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 41.Scheeff E. D., Eswaran J., Bunkoczi G., Knapp S., Manning G. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure. 2009;17:128–138. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gee C. L., Papavinasasundaram K. G., Blair S. R., Baer C. E., Falick A. M., King D. S., Griffin J. E., Venghatakrishnan H., Zukauskas A., Wei J. R., et al. A phosphorylated pseudokinase complex controls cell wall synthesis in Mycobacteria. Sci. Signaling. 2012;5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pereira S. F. F., Goss L., Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 2011;75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karimova G., Ullmann A. Protein–protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 2001;3:73–82. [PubMed] [Google Scholar]
- 45.Ize B., Coulthurst S. J., Hatzixanthis K., Caldelari I., Buchanan G., Barclay E. C., Richardson D. J., Palmer T., Sargent F. Remnant signal peptides on non-exported enzymes: implications for the evolution of prokaryotic respiratory chains. Microbiology. 2009;155:3992–4004. doi: 10.1099/mic.0.033647-0. [DOI] [PubMed] [Google Scholar]
- 46.Sundaramoorthy R., Fyfe P. K., Hunter W. N. Structure of Staphylococcus aureus EsxA suggests a contribution to virulence by action as a transport chaperone and/or adaptor protein. J. Mol. Biol. 2008;383:603–614. doi: 10.1016/j.jmb.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pike A. C. W., Rellos P., Niesen F. H., Turnbull A., Oliver A. W., Parker S. A., Turk B. E., Pearl L. H., Knapp S. Activation segment dimerization: a mechanism for kinase autophosphorylation of non-consensus sites. EMBO J. 2008;27:704–714. doi: 10.1038/emboj.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.