Abstract
Aims
The transcriptional coactivator peroxisome proliferator-activated receptor-γ coactivator-1 α (PPARGC1A or PGC-1α) is a powerful controller of cell metabolism and assures the balance between the production and the scavenging of pro-oxidant molecules by coordinating mitochondrial biogenesis and the expression of antioxidants. However, even though a huge amount of data referring to the role of PGC-1α is available, the molecular mechanisms of its regulation at the transcriptional level are not completely understood. In the present report, we aim at characterizing whether the decrease of antioxidant glutathione (GSH) modulates PGC-1α expression and its downstream metabolic pathways. Results: We found that upon GSH shortage, induced either by its chemical depletion or by metabolic stress (i.e., fasting), p53 binds to the PPARGC1A promoter of both human and mouse genes, and this event is positively related to increased PGC-1α expression. This effect was abrogated by inhibiting nitric oxide (NO) synthase or guanylate cyclase, implicating NO/cGMP signaling in such a process. We show that p53-mediated PGC-1α upregulation is directed to potentiate the antioxidant defense through nuclear factor (erythroid-derived 2)-like2 (NFE2L2)-mediated expression of manganese superoxide dismutase (SOD2) and γ-glutamylcysteine ligase without modulating mitochondrial biogenesis. Innovation and Conclusions: We outlined a new NO-dependent signaling axis responsible for survival antioxidant response upon mild metabolic stress (fasting) and/or oxidative imbalance (GSH depletion). Such signaling axis could become the cornerstone for new pharmacological or dietary approaches for improving antioxidant response during ageing and human pathologies associated with oxidative stress. Antioxid. Redox Signal. 18, 386–399.
Introduction
The transcriptional coactivator peroxisome proliferator-activated receptor-γ coactivator-1 α (PPARGC1A or PGC-1α) is a powerful and very versatile transcriptional coactivator promoting the expression of a wide set of metabolic genes in response to nutritional and physiological stimuli and metabolic stress (15). PGC-1α is a master controller of mitochondrial biogenesis, as it induces mitochondrial genes both at the level of nuclear and mitochondrial genome (2, 35). Besides stimulating mitochondrial activity, which per se represents the principal cellular source of reactive oxygen species (ROS), it coactivates the expression of many antioxidant/detoxifying enzymes (40, 41). The downstream effect of PGC-1α upregulation is generally linked to protection against metabolic (39) and oxidative stress (40, 41, 43) and is likely implicated in the extension of lifespan (34). However, even though a huge amount of data referring to the role of PGC-1α is available, the molecular mechanisms of its regulation at the transcriptional level are not completely understood.
Innovation.
Bodies of evidence are now cumulating that cell fate depends on its redox status. Here, we show that oxidative and mild metabolic stresses are able to activate the same nitric oxide-dependent redox signaling pathway. Our study pinpoints a crucial intermediate in the process of redox control, that is, the p53-dictated PGC-1α antioxidant response. We show that the transcriptional complex in charge of antioxidants expression, the nuclear factor (erythroid-derived 2)-like2, is a key component of this system.
The tumour suppressor p53 was firstly recognized as having a key role in the control of cell cycle and DNA repair pathways, thus maintaining genomic stability (9, 31). More recent discoveries reveal new and opposite functions for p53 in the regulation of various aspects of cellular metabolism, senescence, and apoptosis (7, 13, 36, 46). Among the genes that are directly modulated by p53, antioxidants, mitochondrial, and glycolytic genes are included (16, 17, 36, 42). Another important key modulator of cellular metabolism is nuclear factor (erythroid-derived 2)-like2 (NFE2L2 or NRF2), a bZIP transcription factor controlling the expression of genes coding for detoxifying and antioxidant enzymes, thus maintaining ROS homeostasis (10). For these reasons, p53, NFE2L2, and PGC-1α may have overlapping functions as regulators of the antioxidant defense system. Intriguingly, NFE2L2, PGC-1α, and p53 can be activated by oxidative stress through the same redox-dependent pathways, including those managed by the nitric oxide (NO)/cGMP (1, 8, 23). Then, it could be hypothesized that a cross-talk exists between them upon redox imbalance.
We recently demonstrated that the disruption of the cellular redox buffer controlled by glutathione (GSH) increases endogenous physiological flux of NO in neuronal cells, leading to an upregulation of p53 by the NO/cGMP signaling pathway (1). GSH is the major cellular nonenzymatic antioxidant (21, 48) playing important roles in nutrient metabolism, and regulation of many cellular events (48). Decrement of GSH levels contributes to oxidative stress associated with ageing and many pathological states, including neurodegeneration, inflammation, and infections (6, 22, 25). PGC-1α expression is enhanced by energetic stress (i.e., fasting/calorie restriction and physical exercise), a condition that may also affect GSH content (30, 37, 45). On the basis of this evidence, in the present report, we investigated whether redox-mediated p53 activation could intersect with PGC-1α-regulated pathways and modulate cellular adaptive response to redox and metabolic imbalance.
Results
NO increase induced by GSH deficiency is associated with p53 and PGC-1α upregulation in SH-SY5Y cells
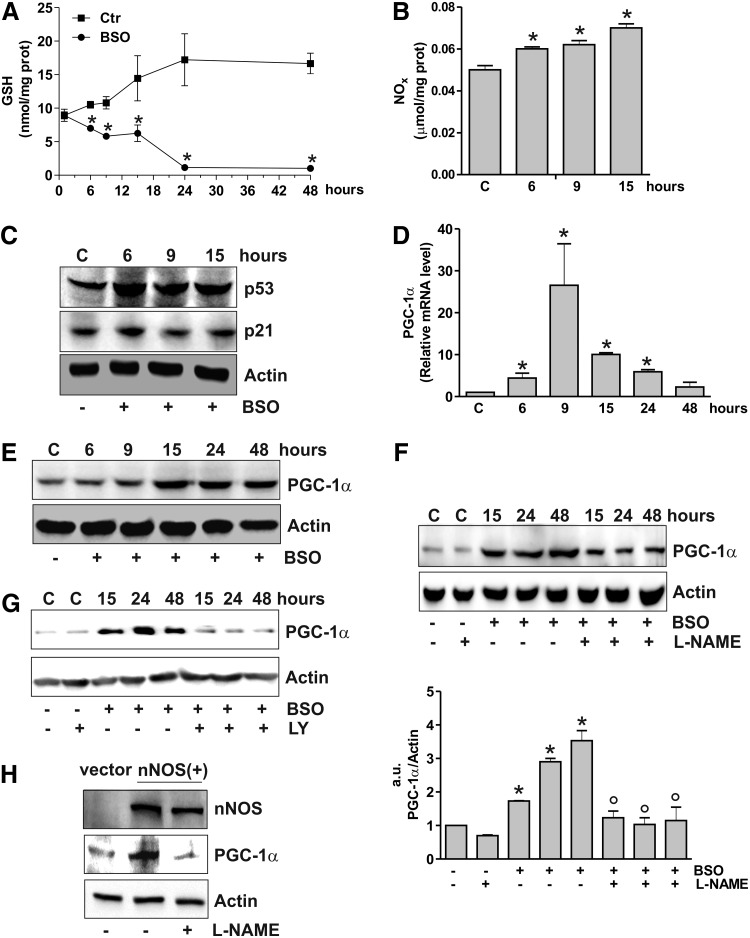
We have previously showed that in SH-SY5Y neuroblastoma cells GSH depletion obtained by l-buthionine sulfoximine (BSO) treatment results in the NO- extracellular-regulated kinase 1/2 (ERK1/2)-mediated increase of p53 at early stages of treatment (starting at 3 h up to 15 h) (1). Here, we investigated whether this event was associated with changes in PGC-1α expression. According to what reported before, BSO treatment was effective in decreasing GSH content (Fig. 1A), increasing NOx concentration (Fig. 1B), and p53 protein levels (Fig. 1C). The content of p21, which represents a canonical downstream target of p53, was not changed (Fig. 1C), in line with the absence of cell cycle arrest and apoptosis up to 15 h (1). Conversely, reverse transcription–quantitative polymerase chain reaction (RT-qPCR) (Fig. 1D) and Western blot analysis (Fig. 1E) of PGC-1α show that GSH deficiency increased PGC-1α both in terms of its mRNA (starting at 6 h) and protein levels (starting at 15 h). Either the inhibition of NO synthase (NOS) by l-NG-nitroarginine methyl ester (L-NAME) (Fig. 1F) or guanylate cyclase by LY-83583 (Fig. 1G) was able to inhibit PGC-1α upregulation elicited by BSO, suggesting that NO signaling is implicated in the modulation of PGC-1α expression. Indeed, the sole overexpression of neuronal NOS (nNOS) in SY-SY5Y cells was effective in increasing PGC-1α, and L-NAME was able to hamper this event (Fig. 1H).
FIG. 1.
GSH depletion leads to NO/p53-dependent PGC-1α upregulation. (A) SH-SY5Y cells were treated with BSO (1 mM) for the indicated times. GSH content was assayed by HPLC, and data are expressed as means±SD (n=4, *p<0.001 vs. control). (B) Nitrite plus nitrate (NOx) in the culture medium were determined by the Griess reaction. Data are reported as micromoles per milligram of protein and expressed as means±SD (n=4, *p<0.001 vs. control). (C) Cells were lysed, and 20 μg of proteins was loaded for Western blot analysis of p53 and p21. (D) Total RNA was isolated, and relative mRNA level of PGC-1α was analyzed by RT-qPCR. Data are expressed as means±SD (n=6, *p<0.001 vs. control). (E) Cells were lysed, and 20 μg of proteins was loaded for Western blot analysis of PGC-1α. (F) L-NAME (0.1 mM) was added 1 h before BSO treatment and maintained throughout the experiment. Cells were lysed, and 20 μg of proteins was loaded for Western blot analysis of PGC-1α. Density of immunoreactive bands was calculated using the software Quantity one (Bio-Rad), and data are shown as the ratio of PGC-1α/actin. Data are expressed as means±SD (n=4, *p<0.001 vs. control, °p<0.001 vs. BSO-treated cells). (G) LY (0.002 mM) was added 1 h before BSO treatment and maintained throughout the experiment. Cells were lysed, and 20 μg of proteins was loaded for Western blot analysis of PGC-1α. (H) SH-SY5Y cells were transfected with an empty vector (vector) or with nNOS cDNA [nNOS(+)]. L-NAME (0.1 mM) was added 1 h before BSO treatment (15 h) and maintained throughout the experiment. Cells were lysed, and 20 μg of proteins was loaded for Western blot analysis of nNOS and PGC-1α. All the immunoblots reported are from one experiment representative of four that gave similar results. Actin was used as loading control. BSO, l-buthionine sulfoximine; L-NAME, l-NG-nitroarginine methyl ester; NOS, nitric oxide synthase; RT-qPCR, reverse transcription–quantitative polymerase chain reaction; nNOS, neuronal NOS; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator-1 α.
PGC-1α expression is associated with increased binding of p53 to human PPARGC1A promoter
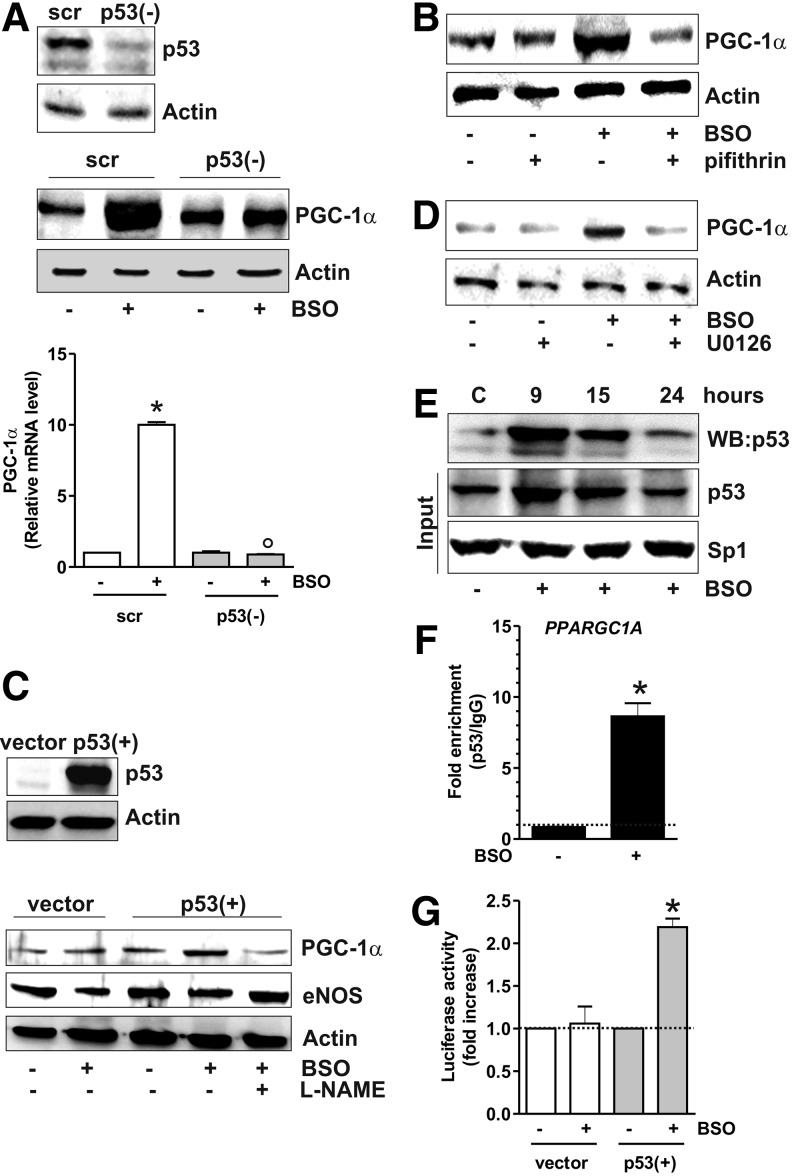
The data obtained show that the increase of p53 precedes PGC-1α induction. Thus, we further investigated the role of p53 as upstream modulator of PGC-1α expression by downregulating p53 through RNA interference (RNAi). p53 downregulation was efficiently achieved in SH-SY5Y cells and proved to be successful in inhibiting the BSO-mediated increase of PGC-1α mRNA and protein levels (Fig. 2A). The same results were obtained by treating cells with pifithrin α, a chemical compound having the ability to suppress p53-mediated transactivation (20) (Fig. 2B). We then used human lung cancer NCI-H1299 cells that are null for p53 and are able to produce NO via endothelial NOS (29). GSH depletion in these cells failed to induce PGC-1α unless wild-type p53 was transfected (Fig. 2C and Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/ars). L-NAME prevented PGC-1α induction in p53-transfected cells, confirming the central role of p53 in the signaling pathway that leads to upregulation of PGC-1α upon increased NO. We previously demonstrated that upon BSO treatment, ERK1/2 is upstream of p53 activation (1). Figure 2D shows that suppression of ERK1/2 activity by U0126 led to a significant reduction of BSO-mediated PGC-1α accumulation, demonstrating that PGC-1α is a downstream target of the ERK1/2-p53 signaling axis.
FIG. 2.
p53 positively influences PGC-1α transcription. (A) SH-SY5Y cells were transfected with scramble (scr) or p53 siRNA [p53(−)] and treated with BSO for 15 h. Cells were lysed, and 20 μg of proteins was loaded for Western blot analysis of p53 and PGC-1α. Total RNA was isolated, and the relative mRNA level of PGC-1α was analyzed by RT-qPCR. Data are expressed as means±SD (n=6, *p<0.001 vs. BSO-untreated scr, °p<0.001 vs. BSO-treated scr). (B) Pifithrin (0.02 mM) was added 1 h before BSO treatment (15 h) and maintained throughout the experiment. Cells were lysed, and 20 μg of proteins was loaded for Western blot analysis of PGC-1α. (C) NCI-H1299 cells was transfected with pcDNA 3.1 empty vector (vector) or pcDNA 3.1 wild-type p53 cDNA [p53(+)]. L-NAME (0.01 mM) was added 1 h before BSO treatment (15 h) and maintained throughout the experiment. Cells were lysed, and 20 μg of proteins was loaded for Western blot analysis of eNOS and PGC-1α. (D) U0126 (260 nM) was added 1 h before BSO treatment (15 h) in SH-SY5Y cells and maintained throughout the experiment. Cells were lysed, and 20 μg of proteins was loaded for Western blot analysis of PGC-1α. (E) SH-SY5Y cells were treated with BSO treatment for the indicated times. Five hundred μg of nuclear protein extracts was subjected to oligo-pulldown assay by using the biotinylated oligonucleotide representing the putative p53RE on the PPARGC1A promoter and bound p53 was detected by Western blot. Twenty micrograms of nuclear proteins (input) was used for Western blot analysis of p53 and Sp1. (F) After 9 h from BSO treatment, ChIP assay was carried out on cross-linked nuclei from SH-SY5Y cells using p53 antibody followed by qPCR analysis of p53RE. Dashed line indicates the value of IgG control. Data are expressed as means±SD (n=3, *p<0.001 vs. control). (G) NCI-H1299 cells were transfected with pGL3 Vector containing the PPARGC1A promoter (+1/-1600 bp) together with pcDNA 3.1 empty vector (vector) or pcDNA 3.1 wild-type p53 cDNA [p53(+)]. Normalized luciferase activities were expressed as fol increase with respect to the values from control, which were set arbitrarily at 1. Data are expressed as means±SD (n=3, *p<0.001 vs. control). All the immunoblots reported are from one experiment representative of four that gave similar results. Actin was used as loading control. ChIP, chromatin immunoprecipitation assay.
By analyzing the human PPARGC1A promoter, Irrcher et al. identified a unique putative p53-response element (p53RE) at −1237 (19). Our effort was therefore directed to study whether p53 could bind this promoter region. SH-SY5Y nuclear extracts were incubated with a biotinylated oligonucleotide representing p53RE, and Western blot analysis of p53 was carried out after the oligo-pulldown. As reported in Figure 2E, p53 protein significantly accumulated within the nucleus, and this event was associated with its increased DNA-binding activity, especially after 9 h from BSO addition. We performed a chromatin immunoprecipitation assay (ChIP) assay to confirm the increased binding of p53 to such p53RE. qPCR analysis revealed that the PPARGC1A promoter is not bound by p53 in control cells. However, a 10-fold increase in the occupancy of p53RE in BSO-treated cells was observed (Fig. 2F). The capacity of p53 to bind the PPARGC1A promoter was also investigated by performing a gene reporter assay. In particular, the PPARGC1A promoter region spanning from +1 to −1600 bp, and comprising the p53RE located at −1237, was cloned into a pGL3 vector upstream the luciferase gene and transfected either in NCI-H1299 p53-null or NCI-H1299-p53(+) cells. Figure 2G shows that BSO is not able to increase luciferase activity in NCI-H1299 cells that are null for p53. Conversely, in NCI-H1299 p53(+) cells, about a twofold increase in luciferase activity was observed after BSO administration, confirming that PPARGC1A promoter expression is under the control of p53.
p53-mediated PGC-1α expression results in NFE2L2-mediated antioxidant response
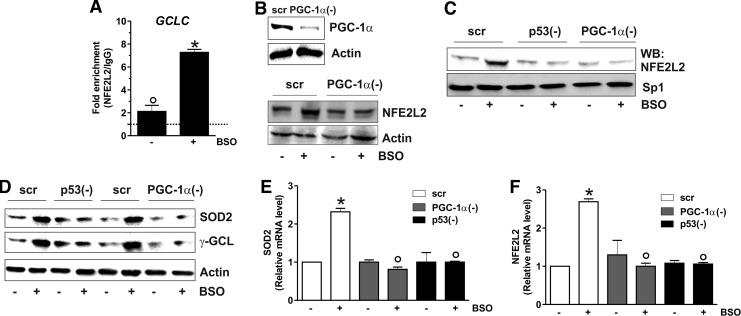
Our previous data indicated that depletion of GSH was not associated with increased mitochondrial biogenesis; indeed, the content of mitochondrial proteins such as Hsp60, cytochrome c oxidase, and cytochrome c remained unchanged after BSO treatment (1). To deeply investigate on this matter, we looked at canonical markers of PGC-1α-induced mitochondrial biogenesis, that is, nuclear respiratory factor-1 (NRF-1) and mitochondrial transcription factor A (TFAM) (38). Figure 3A and B show that NRF-1 and TFAM levels were not altered both in terms of protein and mRNA, supporting the evidence that mitochondrial biogenesis is not operative upon GSH depletion. PGC-1α has also a pivotal role in coordinating a large part of the antioxidant defense system and has complementary and often-overlapping functions with NFE2L2 (10). The analysis of the mRNA and protein levels of NFE2L2 (Fig. 3C) demonstrated that it was significantly induced by BSO treatment starting at 15 h. We then evaluated whether the PGC-1α- and/or NFE2L2-governed antioxidant responses were enhanced. Figure 3 shows that the protein (Fig. 3D, E) and mRNA (Fig. 3F, G) levels of manganese superoxide dismutase (SOD2) and γ-glutamylcysteine ligase (γ-GCL) were significantly increased starting from 15 up to 48 h after BSO administration. Conversely, the level of Trx1, catalase, and SOD1 was not affected (Fig. 3D). To verify whether the binding activity of NFE2L2 was modulated by BSO, NFE2L2 association with the promoter of the human GCLC gene, coding for the catalytic subunit of γ-GCL, was assayed. Both oligo-pulldown (Supplementary Fig. S2A) and ChIP analyses (Fig. 4A) indicate that NFE2L2 binding activity was enhanced, with an eightfold increase in the occupancy of NFE2L2RE in the GCLC promoter at 15 h.
FIG. 3.
PGC-1α upregulation is associated with NFE2L2-mediated SOD2 and γ-GCL increase. (A) SH-SY5Y cells were treated with BSO for the indicated times. Total RNA was isolated, and the relative mRNA level of NRF-1 was analyzed by RT-qPCR. Data are expressed as means±SD (n=4). Cells were lysed, and 20 μg of proteins was used for Western blot analysis of NRF-1 (B) Total RNA was isolated, and the relative mRNA level of TFAM was analyzed by RT-qPCR. Data are expressed as means±SD (n=5). Cells were lysed, and 20 μg of proteins was used for Western blot analysis of TFAM. (C) Total RNA was isolated, and the relative mRNA level of NFE2L2 was analyzed by RT-qPCR. Data are expressed as means±SD (n=4, *p<0.001 vs. control). Cells were lysed, and 20 μg of proteins was used for Western blot analysis of NFE2L2. Cells were lysed, and 20 μg of proteins was used for Western blot analysis of SOD2, Trx1, catalase, SOD1 (D), and γ-GCL (E). Total RNA was isolated, and relative mRNA level of SOD2 (F) and γ-GCL (G) was analyzed by RT-qPCR. Data are expressed as means±SD (n=4, *p<0.001 vs. control). All the immunoblots reported are from one experiment representative of four that gave similar results. Actin was used as loading control. γ-GCL, γ-glutamylcysteine ligase; NRF-1, nuclear respiratory factor-1; SOD2, manganese superoxide dismutase; NFE2L2, nuclear factor (erythroid-derived 2)-like2; TFAM, mitochondrial transcription factor A.
FIG. 4.
p53/PGC-1α/NFE2L2 axis induces SOD2 and γ-GCL. (A) After 15 h from BSO treatment, ChIP assay was carried out on cross-linked nuclei from SH-SY5Y cells by using NFE2L2 antibody followed by qPCR analysis of ARE sequence on the GCLC promoter. Dashed line indicates the value of IgG control. Data are expressed as means±SD (n=3, *p<0.001 vs. control; °p<0.01 vs. IgG). (B) SH-SY5Y cells were transfected with scramble (scr) or PGC-1α siRNA [PGC-1α(−)] and treated with BSO for 15 h. PGC-1α and NFE2L2 were detected by Western blot. (C) SH-SY5Y cells were transfected with scramble (scr) or PGC-1α siRNA [PGC-1α(−)] or p53 siRNA [p53(−)] and treated with BSO for 15 h. Nuclear protein extracts (500 μg) were subjected to oligo-pulldown assay using the biotinylated oligonucleotide representing the NFE2L2 consensus sequence on the GCLC promoter. (D) SH-SY5Y cells were transfected with scramble (scr) or PGC-1α siRNA [PGC-1α(−)] or p53 siRNA [p53(−)] and treated with BSO for 15 h. Cells were lysed, and 20 μg of proteins was used for Western blot analysis of SOD2 and γ-GCL. (E, F) SH-SY5Y cells were transfected with scramble (scr) or PGC-1α siRNA [PGC-1α(−)] or p53 siRNA [p53(−)] and treated with BSO for 15 h. Total RNA was isolated, and relative mRNA level of SOD2 (E) and NFE2L2 (F) was analyzed by RT-qPCR. Data are expressed as means±SD (n=4, *p<0.001 vs. BSO-untreated scr, °p<0.001 vs. BSO-treated scr). All the immunoblots reported are from one experiment representative of five that gave similar results. Actin or Sp1 were used as loading control.
To characterize the sequence of the signaling events leading to the antioxidant response, we monitored the level of NFE2L2 after downregulation of PGC-1α through RNAi. Figure 4B shows that PGC-1α is efficiently downregulated (upper panel), and this results in the abrogation of BSO-induced NFE2L2 expression (bottom panel). Moreover, the silencing of either p53 or PGC-1α led to a significant abrogation of NFE2L2 binding to the GCLC promoter upon GSH depletion as demonstrated by the oligo-pulldown assay reported in Figure 4C. A parallel decrease in the SOD2 and γ-GCL protein content was also observed upon downregulation of p53 or PGC-1α by RNAi (Fig. 4D). Figure 4E and F show that either p53 or PGC-1α downregulation was effective in inhibiting NFE2L2 as well as SOD2 mRNA expression, demonstrating that they are upstream modulators of the NFE2L2-mediated antioxidant response.
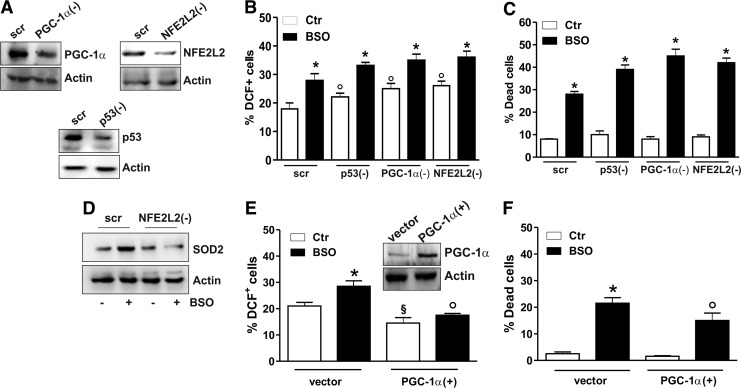
We previously reported that BSO was able to increase the level of ROS/RNS in SH-SY5Y cells (3), and prolonged treatment with BSO (48 h) resulted in the induction of a caspase-independent apoptotic program (1); therefore, we wonder whether the antioxidant response downstream of the p53/PGC-1α pathway could modulate cell fate. To this end, we downregulated p53, PGC-1α, or NFE2L2 protein by RNAi (Fig. 5A), and we monitored the production of ROS/ONOO˙− and the rate of dead cells after BSO treatment. Figure 5B shows an augmented production of ROS/ONOO˙− in BSO-treated cells, which is further enhanced after downregulation of p53, PGC-1α, or NFE2L2. As expected, p53, PGC-1α, or NFE2L2 downregulation strongly enhanced cell death induced by BSO at 48 h (Fig. 5C). The role of NFE2L2 in promoting the antioxidant response is evident in NFE2L2-downregulating cells. In particular, upon NFE2L2 downregulation through RNAi, BSO treatment was not more able to elicit SOD2 upregulation (Fig. 5D). In line with the protective role of PGC-1α against oxidative stress, overexpression of PGC-1α completely abrogated the increase of ROS/ONOO˙− levels after BSO treatment (Fig. 5E), resulting in a significant reduction of cell death commitment (Fig. 5F).
FIG. 5.
PGC-1α prevents oxidative stress and cell death mediated by the loss of GSH. (A–C) SH-SY5Y cells were transfected with scramble (scr) or PGC-1α siRNA [PGC-1α(−)], p53 siRNA [p53(−)], or NFE2L2 siRNA [NFE2L2(−)] and treated with BSO for 15 h. (A) PGC-1α, p53, and NFE2L2 downregulation was detected by Western blot analysis. (B) Cells were treated with BSO for 15 h and assayed for ROS/ONOO− production by cytofluorimetric analysis after DCF-DA staining. ROS/ONOO− level was reported as percentage of DCF-positive cells and expressed as means±SD (n=4, *p<0.001 vs. BSO-untreated scr; °p<0.01 vs. BSO-treated and untreated scr). (C) Cells were treated with BSO for 48 h, and dead cells were counted by Trypan blue exclusion. Data are expressed as means±SD (n=5, *p<0.001 vs. BSO-untreated scr). (D) SH-SY5Y cells were transfected with scramble (scr) or NFE2L2 siRNA [NFE2L2(−)] and treated with BSO for 15 h. Cells were lysed, and 20 μg of proteins was used for Western blot analysis of SOD2. (E) SH-SY5Y cells were transfected with empty vector (vector) or with PGC-1α cDNA [PGC-1α(+)] and treated with BSO for 15 h. PGC-1α upregulation (inset) was detected by Western blot. Cells were assayed for ROS/ONOO− production by cytofluorimetric analysis after DCF-DA staining. ROS/ONOO− level was reported as percentage of DCF-positive cells and expressed as means±SD (n=4, *p<0.01 vs. vector-untreated cells; °p<0.001 vs. BSO-treated vector, §p<0.01 vs. BSO-untreated vector). (F) Cells were treated with BSO for 48 h, and dead cells were counted by Trypan blue exclusion. Data are expressed as means±SD (n=5, *p<0.001 vs. vector-untreated cells; °p<0.01 vs. BSO-treated vector). All the immunoblots reported are from one experiment representative of four that gave similar results. Actin was used as loading control. ROS, reactive oxygen species.
p53 binding to the mouse ppargc1a promoter is positively related to PGC-1α increase in GSH-depleted murine models
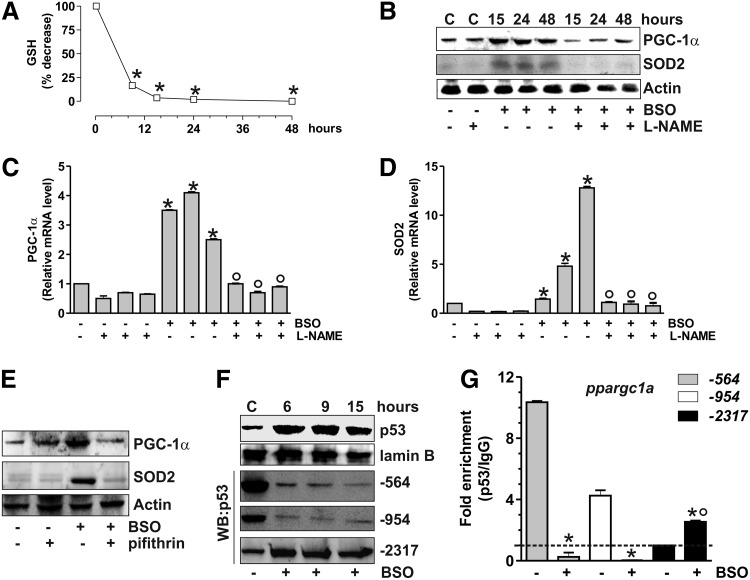
Recently, three p53 putative binding sites on the mouse ppargc1a promoter were identified. The sites located at −954 and −564 were demonstrated to function as p53 repressive regions that inhibit PGC-1α expression (36). Therefore, we investigated whether p53 could modulate PGC-1α expression also in murine cells. To this end, C2C12 myoblasts were treated with BSO that was efficient in decreasing GSH content (Fig. 6A). Also, in this case, GSH decrement results in the increase of PGC-1α and SOD2 content both in terms of protein (Fig. 6B) and mRNA (Fig. 6C, D). L-NAME completely abolished such event (Fig. 6B), confirming that NO increase was the triggering factor. Moreover, p53 inhibition by pifithrin-α was able to block PGC-1α and SOD2 upregulation (Fig. 6E and Supplementary Fig. S1B).
FIG. 6.
p53 modulates PGC-1α transcription in murine C2C12 cells upon GSH depletion. (A) C2C12 cells were treated with BSO (1 mM) for the indicated times. GSH content was assayed by HPLC, and data are expressed as percentage of decrease with respect to controls and reported as means±SD (n=4, *p<0.001 vs. control). (B) C2C12 cells were treated with BSO and/or L-NAME (0.1 mM) for the indicated times. Cells were lysed, and 20 μg of proteins was used for Western blot analysis of PGC-1α and SOD2. Total RNA was isolated, and relative mRNA level of PGC-1α (C) and SOD2 (D) was analyzed by RT-qPCR. Data are expressed as means±SD (n=4, *p<0.001 vs. control, °p<0.001 vs. BSO and NAME-treated cells). (E) Pifithrin (0.02 mM) was added 1 h before BSO treatment (15 h) and maintained throughout the experiment. Cells were lysed, and 20 μg of proteins was loaded for Western blot analysis of PGC-1α and SOD2. (F) After BSO treatment, C2C12 nuclear protein extracts (500 μg) were subjected to Western blot analysis of p53 and oligo-pulldown assay by using the biotinylated oligonucleotides representing the three p53REs on the ppargc1a promoter (−564, −954, and −2317). (G) After 15 h from BSO treatment, ChIP assay was carried out on cross-linked nuclei from C2C12 cells by using p53 antibody, followed by qPCR analysis of p53REs on the ppargc1a promoter (−564, −954, and −2317). Dashed line indicates the value of IgG control (n=3, *p<0.001 vs. control; °p<0.01 vs. IgG).
We performed oligo-pulldown assays by using the two identified p53 repressive regions (−954 and −564) and a third region located at −2317, which was hypothesized to be an additional p53-binding site (36). Consistent with the PGC-1α upregulation that we observed after BSO treatment, the two repressive regions were rapidly released by p53 (Fig. 6F). The oligo-pulldown assay reported in Figure 6F reveals that the p53 binding to the −2317 region was concomitantly enhanced. ChIP analyses of all the three p53-binding sites were carried out to confirm the regulatory role of p53 on the murine ppargc1a promoter. The qPCR analysis shows that the −2317 region is not bound by p53 at the basal level, in line with the results reported by Sahin et al. (36). However, a significant increase (threefold) in p53 occupancy of the −2317 region was observed after BSO treatment, while the −564 and −954 regions show a complete loss of p53 binding (Fig. 6G).
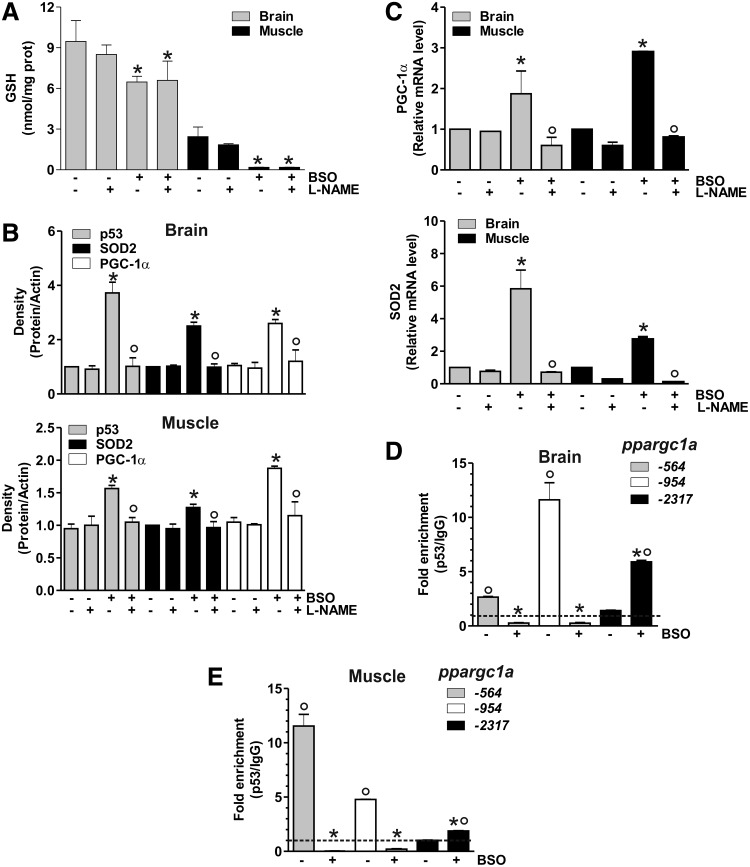
The proof that the signaling pathway discovered in human cells is also operative in murine cells prompted us to move to study the in vivo system. In particular, we induced GSH deficiency in mice by adding BSO in drinking water. Mice were sacrificed after 5 weeks of treatment, and GSH content was analyzed by HPLC in the brain and skeletal muscle. Among the metabolically active organs, the brain was selected on the basis of its reported resistance to GSH depletion by BSO (12) and protection from the energetic imbalance caused by fasting (14). Skeletal muscle was chosen because of the well-known dependence of its metabolism on PGC-1α activity (33). As shown in Figure 7A, a significant GSH depletion in tissues from BSO-treated mice was obtained that was more marked in skeletal muscle with respect to the brain. Densitometric analyses of p53, PGC-1α, and SOD2 levels (Fig. 7B) detected by Western blot (Supplementary Fig. S3) demonstrated a significant increase of these proteins after BSO treatment. Concomitant administration of L-NAME completely abolished the response to GSH deficiency (Fig. 7B and Supplementary Fig. S3). In line with these results, an increase of PGC-1α and SOD2 mRNA was also observed upon BSO supplementation, and L-NAME was able to abolish such increase (Fig. 7C).
FIG. 7.
p53 positively regulates PGC-1α transcription in mice upon GSH depletion. (A) C57/BL6 mice were treated with BSO (20 mM) and/or L-NAME (4 mM) for 5 weeks in drinking water. Brains and skeletal muscles were homogenized, and GSH content was assayed by HPLC. Data are expressed as nmoles of GSH/mg of proteins and reported as means±SD (n=5, *p<0.001 vs. control). (B) Brains and skeletal muscles were homogenized, and 50 μg of proteins was used for Western blot analysis of p53, SOD2 and PGC-1α. Density of immunoreactive bands was calculated using the software Quantity one (Bio-Rad), and data are shown as a ratio of protein/actin. Data are expressed as means±SD (n=5, *p<0.01 vs. control, °p<0.01 vs. BSO-treated cells). (C) Total RNA was isolated, and relative mRNA level of PGC-1α and SOD2 were analyzed by RT-qPCR. Data are expressed as means±SD (n=4, *p<0.01 vs. control, °p<0.01 vs. BSO-treated cells). ChIP assay was carried out on cross-linked nuclei from the brain (D) and skeletal muscle (E) by using p53 antibody followed by qPCR analysis of p53REs on the ppargc1a promoter (−564, −954, and −2317). Dashed line indicates the value of IgG control (n=3, *p<0.001 vs. control; °p<0.01 vs. IgG). All the immunoblots reported are from one experiment representative of four that gave similar results. Actin was used as loading control.
ChIP assay was carried out to analyze the binding of p53 to the ppargc1a promoter. As reported in Figure 7D and E, BSO induced a release of p53 from −564 and −954 regions, and a concomitant increase of p53 binding to −2317 region both in brain and skeletal muscle, respectively. Different degrees of p53 binding to these promoter regions are operative under basal conditions, with the binding of p53 more efficient at −954 in the brain, and −564 in skeletal muscle and C2C12 cells.
GSH decrease promoted by fasting is accompanied by the upregulation of the p53/PGC-1α pathway
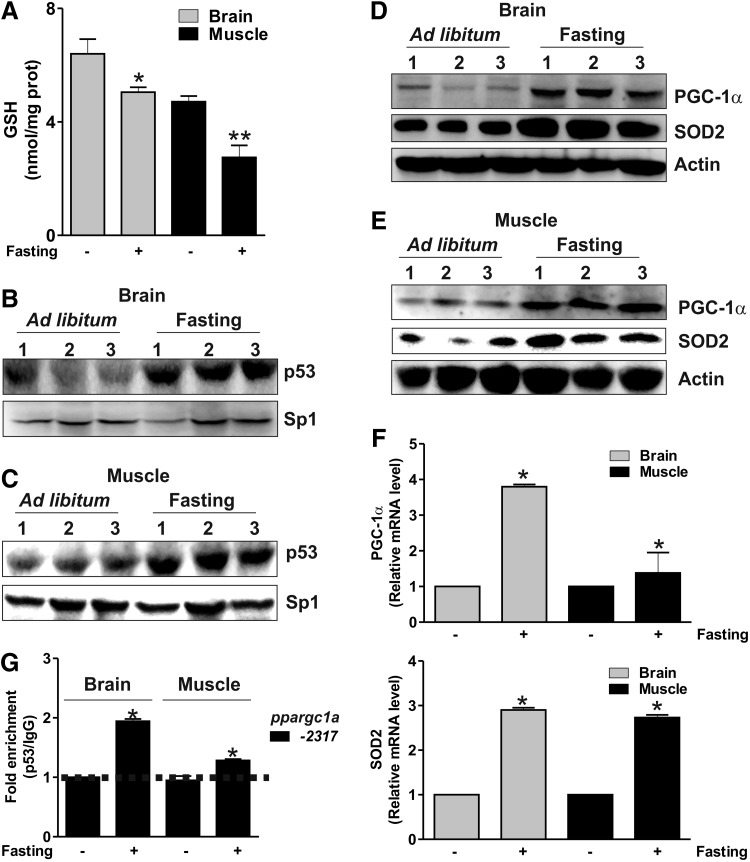
PGC-1α is regulated transcriptionally by different pathways upon starvation (11, 24). Therefore, we asked whether the binding of p53 to the ppargc1a promoter could be influenced upon fasting. We first investigated whether fasting was able to affect GSH content in the mouse brain and skeletal muscle. Mice were starved for 24 h, and GSH level was assayed in tissue homogenates. Figure 8A shows that GSH was significantly decreased in the brain and skeletal muscle, and this phenomenon was associated with an increase of p53 protein (Fig. 8B, C). Fasted mice also displayed augmented protein (Fig. 8D, E) and mRNA (Fig. 8F) levels of PGC-1α and SOD2 in both the brain and skeletal muscle. Nuclear extracts from the brain and skeletal muscle of ad libitum fed or fasted mice were subjected to analysis of p53 binding to the ppargc1a promoter. Oligo-pulldown (Supplementary Fig. S2B, C) and ChIP analyses (Fig. 8G) demonstrated that fasting was associated with the release of p53 from the repressive regions (Supplementary Fig. S2B, C) and the increase of p53 binding activity on the −2317 site. ChIP analysis also confirmed that the −2317 region is not bound by p53 under resting condition (as the same value was observed in sample immunoprecipitated with anti-p53 with respect to IgG isotype control). On the contrary, the p53 binding to such region is significantly enhanced during fasting (2-fold and 1.3-fold with respect to IgG isotype control in the brain and muscle, respectively) (Fig. 8G).
FIG. 8.
p53 binding to the ppargc1a promoter is modulated by fasting. (A) CD1 mice were either fed ad libitum or fasted for 24 h. Brains and skeletal muscles were homogenized, and GSH content was assayed by HPLC. Data are expressed as nmoles of GSH/mg of proteins and reported as means±SD (n=4, *p<0.001 vs. control). Nuclear proteins (50 μg) from the brain (B) and skeletal muscle (C) of ad libitum fed or fasted mice were subjected to Western blot analysis of p53. Twenty micrograms of total brain (D) or skeletal muscle (E) proteins from three ad libitum fed or fasted mice was loaded for the detection of PGC-1α and SOD2 by Western blot. (F) Total RNA was isolated, and relative mRNA level of PGC-1α and SOD2 was analyzed by RT-qPCR. Data are expressed as means±SD (n=6, *p<0.001 vs. control). (G) ChIP assay was carried out on cross-linked nuclei from the brain and skeletal muscle from ad libitum fed or fasted mice by using p53 antibody, followed by qPCR analysis of p53RE on the ppargc1a promoter (−2317). Dashed line indicates the value of IgG control (n=3, *p<0.001 vs. ad libitum fed). All the immunoblots reported are from one experiment representative of four that gave similar results. Actin or Sp1 were used as loading control.
Discussion
Here we show that p53, PGC-1α, and NFE2L2 are part of the same redox signaling pathway that positively coordinates the expression of SOD2 and γ-GCL in response to GSH decrement. GSH depletion—obtained by BSO-mediated inhibition of the rate-limiting enzyme of its synthesis (γ-GCL)—represents the primum movens of such a cellular response. The most important finding is that p53 is able to bind the −1237 p53RE on the human PPARGC1A promoter, and is fundamental to assure the expression of PGC-1α. Indeed, the downregulation of p53 was associated with the failure of PGC-1α mRNA increase. We also show for the first time that NFE2L2 expression is controlled by the p53/PGC-1α axis, thus giving a mechanistic effort to their overlapping functions and/or synergistic actions in regulating the antioxidant system (10, 47). During the progressive decline of GSH, the activated PGC-1α/NFE2L2 pathway stimulates a sort of adaptive response that buffers the potential harmful effect of ROS/RNS, resulting in mild oxidative stress and protection against cell death. The statement that PGC-1α plays a central role in modulating the NFE2L2-mediated antioxidant response comes from the data demonstrating an enhanced ROS/RNS concentration and cell death and a failure in the upregulation of SOD2 upon p53, PGC-1α, or NFE2L2 downregulation. In this context, we have strong evidence that PGC-1α is in some way directly involved in the regulation of the NFE2L2 expression/activity. Even though a molecular interaction between NFE2L2 and PGC-1α has not been investigated in the present work, it could be interesting to establish whether besides modulating transcription factors such as NRF-1, -2 and CREB, PGC-1α could also coactivate NFE2L2, thus assisting the induction of antioxidant genes. This matter is currently under investigation in our laboratory. The NO-mediated p53 upregulation culminates in the expression of PGC-1α and SOD2 also in murine cells and mice, revealing that the transduction of the redox signaling triggered by GSH deficiency is of general application. Even if PGC-1α is significantly upregulated, mitochondrial biogenesis is not induced, according to a recent report showing that GSH is a pivotal factor in the activation of mitochondrial biogenesis. In particular, GSH positively regulates the activity of the most important modulator of mitochondrial biogenesis in yeast, that is, the transcriptional coactivator Hap4p (49). It is likely that a reducing environment is fundamental for the function of PGC-1α as a coactivator of mitochondrial genes. Conversely, it is possible that a decreased redox state triggered by GSH deficiency may switch PGC-1α activity toward the antioxidant response.
We found that, in GSH-deficient murine cells and mice, PGC-1α and SOD2 upregulation is associated with p53 release from the two repressive regions in line with the suggested negative role exerted by p53 on ppargc1a transcription (36). Interestingly, we have also demonstrated that a third p53RE on the mouse ppargc1a promoter, which is not bound by p53 under resting conditions, is engaged by p53 upon GSH deficiency. On the basis of all these observations, we can suggest that in mice, PGC-1α transcription is controlled by p53 at two different levels. The first level consists in the negative regulation by p53 through its binding to the two repressive regions as previously suggested (36); the second level is represented by the binding of p53 at a unique activating region. In our mouse experimental model, the enhanced expression of PGC-1α is likely the result of the release of p53 from the repressive regions and the parallel increased binding of p53 at the activating site. The diverse behavior of p53 in recognizing the p53REs on the mouse ppargc1a promoter may be dependent on redox modification (e.g., S-nitrosylation) of critical p53 amino acids that differently affect (enhance/inhibit) its DNA-binding activity to specific consensus sequences. The regulation of PGC-1α expression appears to be more intricate in mouse than in human, wherein the two p53 repressive regions have not been conserved during evolution. Indeed, PPARGC1A is one of the 49 identified human accelerated regions, which means that such gene is conserved throughout vertebrate evolution, but is strikingly different in humans (28). Indeed, the pairwise alignment of the PPARGC1A promoter region with that from mouse results in a poor identity (49%). Even if no apparent structural similarities can be observed between the mouse and the human p53RE, our evidence indicates that the −2317 region on the mouse ppargc1a promoter is the functional homolog of the −1237 on the human PPARGC1A promoter, and its binding by p53 positively correlates with enhanced PGC-1α transcription.
We show that 1-day fasting in mice is able to reduce GSH levels in the brain and skeletal muscle, and this event is associated with the detachment of p53 from the repressive regions and the increased binding activity of p53 to the activating region on the ppargc1a promoter. We also documented the effective activation of the antioxidant response via the upregulation of SOD2 during fasting. It was proposed that the increase of NO levels was essential for many beneficial effects of dietary limitation (26, 27), and the decrease of GSH content could further enhance NO imbalance. Although the effective increase of NO concentration and the strict connection between GSH decrement, oxidative stress, and the activation of p53/PGC-1α remain to be investigated in our experimental model of fasting, the observation that the ppargc1a promoter undergoes the same regulatory mechanisms with respect to chemical GSH depletion strongly indicates the intervention of NO/cGMP signaling.
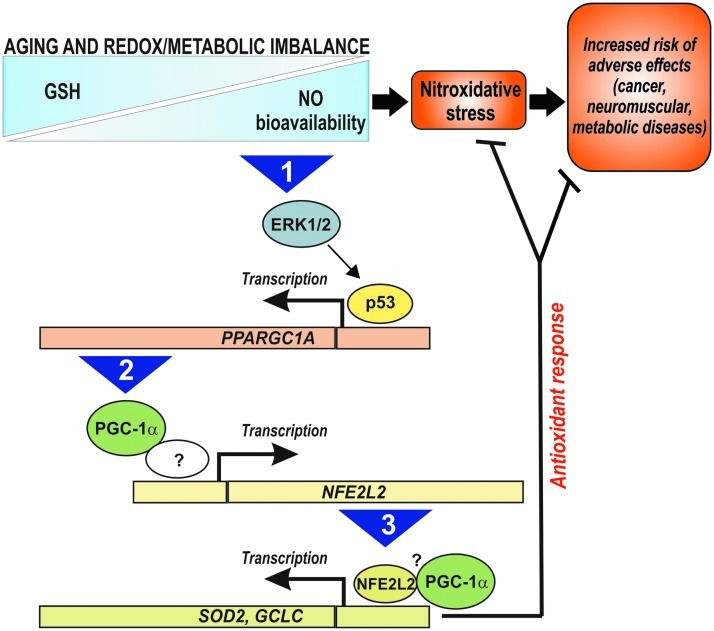
We previously showed that increased NO bioavailability is the primary mediator of neurotoxicity upon GSH depletion in vitro, resulting in a substantial nitration and S-nitrosylation of cellular proteins (1). However, the results obtained in the present work show that a parallel and compensative NO-dependent induction of antioxidants via a p53/PGC-1α/NFE2L2 pathway is induced, which is directed to dampen the detrimental effect of GSH decrement. Then, it can be hypothesized that the decline of GSH level occurring with normal ageing in many organisms (32) is a beneficial rather than detrimental event and may serve as a molecular signal inducing an endogenous antioxidant adaptive response that culminates in increased resistance to environmental/pathological stress and possibly in the extension of lifespan. Dramatic pathological variations of GSH content and/or the inability to activate such stress response could thus increase the risk of adverse effects leading to age-related pathologies (e.g., cancer, neuromuscular, and metabolic diseases). Therefore, the pharmacological targeting (e.g., through BSO treatment) or metabolic modulation (e.g., through fasting or exercise) of the NO/p53/PGC-1α/NFE2L2 pathway promises to be an effective therapeutic approach that can safeguard health, therefore favoring longevity (Fig. 9).
FIG. 9.
Schematic model of the signaling pathway activated by redox/metabolic imbalance and its possible positive impact on human lifespan. Question marks represent possible still-undiscovered molecular mechanisms/factors regulating the transcription and activity of NFE2L2. The numbers in the arrows indicate the sequence of events. GSH, glutathione; ERK1/2, extracellular-regulated kinase 1/2; GCLC, γ-glutamylcysteine ligase, catalytic subunit.
Materials and Methods
Cell cultures, transfections, and ROS/RNS assay
Human SH-SY5Y neuroblastoma cells and murine C2C12 myoblasts were purchased from the European Collection of Cell Cultures (Salisbury, United Kingdom) and cultured as suggested by the supplier. NCI-H1299 cells lacking of p53 were a kind gift of Prof. Cesareni (Department Biology, University of Rome Tor Vergata).
BSO, a highly selective and potent inhibitor of the enzyme γ-GCL, was added in the culture medium at a concentration of 1 mM. The NOS inhibitor L-NAME and the guanylate cyclase inhibitor LY-83583 were used at a concentration of 0.1 mM and 0.002 mM, respectively. Treatments with cell-permeable MEK1/2 inhibitor U0126 and p53 inhibitor pifithrin α were done at a concentration of 260 nM and 0.02 mM, respectively.
Plasmids were transfected by electroporation as previously described (2) using pSV-PGC-1α (Addgene, Cambridge, MA), pcDNA3.1-nNOS, or pcDNA3.1-p53 (kindly donated by Dr. Yvonne Sun, The Cancer Institute of New Jersey, NJ). PGC-1α and p53 siRNA oligonucleotides are reported in Table 1. ROS/RNS levels were assayed by means of DCF-DA staining as previously reported (44).
Table 1.
List of siRNAs Used for RNA Interference
| PGC-1α | 5′-AAGACCAGCCUCUUUGCCCAG-3′ |
| p53 | 5′-GACUCCAGUGGUAAUCUACTT-3′ |
PGC-1α, peroxisome proliferator-activated receptor-γ coactivator-1α.
Animals
C57/BL/6J and CD1 (5 weeks of age) were obtained from Harlan Laboratories S.r.l. (Urbino, Italy). Twenty C57/BL/6J mice were randomly divided into four experimental groups: mice treated with BSO (20 mM); mice treated with L-NAME (4 mM), mice treated with BSO together with L-NAME, or with unsupplemented water. BSO and L-NAME were administered for 5 weeks in drinking water. Mice had free access to food and water.
For fasting experiments, eight mice were divided randomly into two groups: ad libitum fed and fasting (24 h without food). We conducted all mouse experiments in accordance with the accepted standard of humane animal care and with the approval by relevant national (Ministry of Welfare) and local (Institutional Animal Care and Use Committee, Tor Vergata University) committees.
Determination of GSH
Intracellular GSH level was measured by HPLC as previously described (4).
Preparation of cell lysates and Western blot analysis
Cell lysates were obtained and Western blot analyses were performed as previously described (1).
RT-qPCR analysis
Total RNA was extracted using TRI reagent (Sigma-Aldrich, St. Louis, MO). Three micrograms of RNA was used for retrotranscription with M-MLV (Promega, Madison, WI). qPCR was performed in triplicates by using validated qPCR primers (BLAST), Ex TAq qPCR Premix (Lonza Sales, Basel, Switzerland), and the Real-Time PCR LightCycler II (Roche Diagnostics, Indianapolis, IN). mRNA levels were normalized to actin mRNA, and the relative mRNA levels were determined by using the 2−ΔΔCt method. The primer sequences are listed in Table 2.
Table 2.
List of Primers Used for Reverse Transcription–Quantitative Polymerase Chain Reaction and Chromatin Immunoprecipitation Assay Analysis
| PGC-1α | RV 5′-ACTCGGATTGCTCCGGCCCT-3′ |
| FW 5′-ACTGACGGCCTAACTCCACCCA-3′ | |
| NRF-1 | RV 5′-AGCCACATGGACCTGCTGCACT-3′ |
| FW 5′-GCGCAGCCGCTCTGAGAACTTCAT-3′ | |
| NFE2L2 | RV 5′-GGAACAAGTGACTGAAACGTAGCCG-3′ |
| FW 5′-GCCAACTACTCCCAGGTTGCCC-3′ | |
| γ-GCL | RV 5′-GCAACATGCTGGGCCAGGAGA-3′ |
| FW 5′-AGGAGCGAGGACTGGAGCCAT-3′ | |
| SOD2 | RV 5′-ACTGAAGGTAGTAAGCGTGC-3′ |
| FW 5′-TCTTCAGCCTGCACTGAAGT-3′ | |
| TFAM | RV 5′-TCCGCCCTATAAGCATCTTG-3′ |
| FW 5′-CCGAGGTGGTTTTCATCTGT-3′ | |
| Actin | RV 5′-GGGACTTCCTGTTAACAACGCA-3′ |
| FW 5′-GGCCGAGGACTTTGATTGCA-3′ | |
| PPARGC1A (−1237) | RV 5′-CACCTACCTGCATTAGCCCT-3′ |
| FW 5′-TGGGGGATTGTTTTCAGGTA-3′ | |
| GCLC | RV 5′-GTGAATGAAAGGTGATTGTGTG-3′ |
| FW 5′-GTTATGCTAGAAGCAAAGAGGG-3′ | |
| ppargc1a (−954) | RV 5′-AGAACGGAGTAGTTAACTTTCAA-3′ |
| FW 5′-AGGTGAAAAATCCAAGACATTAT-3′ | |
| ppargc1a (−564) | RV 5′-CCATGAGGTATTGACCATCTCTC-3′ |
| FW 5′-CCGGTATCTTGAGTTGTGTTAAA-3′ | |
| ppargc1a (−2317) | RV 5′-GAAATTCACCCTATTCTGATCCC-3′ |
| FW 5′-GATTCCCTGCACCCACATGATG-3′ |
NRF-1, nuclear respiratory factor-1; SOD2, manganese superoxide dismutase; NFE2L2, nuclear factor (erythroid-derived 2)-like2; TFAM, mitochondrial transcription factor A.
Oligo-pulldown assay
Oligo-pulldown assay was carried out as previously described (5) by using the p53RE at −1237 on the human PPARGC1A, or at −2317, −954, or −564 on mouse ppargc1a promoter, and ARE sequence on the human GCLC promoter (Table 3). Oligo-pulldown specificity was demonstrated with mutant oligonucleotides used as negative controls (data not shown).
Table 3.
List of Oligonucleotides Used for Oligo-Pull-Down Assay
| PPARGC1A (−1237) | 5′-(btn)AACATGTTTATTCACACAGA-3′ |
| 5′-(btn)TCTGTGTGAATAAACATGTT-3′ | |
| GCLC | 5′-(btn)TCACAGTGACGCAGCAGAATC-3′ |
| 5′-(btn)GATTCTGCTGAGTCACTGTGA-3′ | |
| ppargc1a (−2317) | 5′-(btn)CTCTAAATAAAAAATGTTATAC-3′ |
| 5′-(btn)GTATAACATTTTTTATTTAGAG-3′ | |
| ppargc1a (−954) | 5′-(btn)ACCTTTGCTTCTAAGTCATTG-3′ |
| 5′-(btn)CAATGACTTAGAAGCAAAGGT-3′ | |
| ppargc1a (−564) | 5′-(btn)CTGGATTTTGATAGTTTACTG-3′ |
| 5′-(btn)CAGTAAACTATCAAAATCC AG-3′ |
Chromatin immunoprecipitation assay
ChIP was carried out according to the protocol of Im et al. (18) with some modifications. Briefly, after cross-linking of nuclei extracted from SH-SY5Y cells were fragmented by ultrasonication using 4×15 pulse (output 10%, duty 30%). Samples were precleared with preadsorbed salmon sperm Protein G agarose beads (1 h, 4°C), and overnight immunoprecipitation using anti-p53 or control IgG antibody was carried out. After de-cross-linking (1% sodium dodecyl sulfate at 65°C for 3 h), qPCR was used to quantify the promoter binding with 30 cycles total (95°C, 1 s; 60°C, 30 s; 72°C, 60 s). Results are expressed as fold enrichment with respect to IgG control. The primers used are reported in Table 1.
Luciferase-based transcription assay
The PPARGC1A promoter region spanning from +1 to −1600 bp was cloned into a pGL3 Basic Vector (Promega). Reporter gene construct was transfected in NCI-H1299 cells together with pcDNA3.1-p53 [p53(+)] or pcDNA3.1 empty vector (vector) as above reported. Luciferase reporter gene activities were determined by using Luciferase Assay System (Promega). Light intensity was measured using a microplate luminometer (Perkin Elmer, Milan, Italy). Light intensity values from cell cultures transfected with pGL3 Basic Vector were used to correct for background. Normalized luciferase activities were expressed as fold increase with respect to the values from control (BSO-untreated), which were set arbitrarily at 1.
Statistical analysis
The results are presented as means±SD. Statistical evaluation was conducted by ANOVA, followed by the post-Student-Newman-Keuls. Differences were considered to be significant at p<0.05.
Supplementary Material
Abbreviations Used
- BSO
l-buthionine sulfoximine
- ChIP
chromatin immunoprecipitation assay
- ERK1/2
extracellular-regulated kinase ½
- GCLC
γ-glutamylcysteine ligase, catalytic subunit
- GSH
glutathione
- L-NAME
l-NG-nitroarginine methyl ester
- NFE2L2
nuclear factor (erythroid-derived 2)-like2
- NO
nitric oxide
- NOS
nitric oxide synthase
- nNOS
neuronal NOS
- NRF-1, −2
nuclear respiratory factor-1, -2
- PGC-1α or PPARGC1A
peroxisome proliferator-activated receptor-γ coactivator-1 α
- RNAi
RNA interference
- ROS
reactive oxygen species
- RT-qPCR
reverse transcription–quantitative polymerase chain reaction
- SOD2
manganese superoxide dismutase
- TFAM
mitochondrial transcription factor A
- γ-GCL
γ-glutamylcysteine ligase
Acknowledgments
We thank Dr. Marcello Giorgi (Dept. of Biology, University of Rome Tor Vergata) for his technical assistance in performing gene reporter assay experiments. We are also grateful to Prof. Fabrizio Loreni (Dept. of Biology, University of Rome Tor Vergata) for providing intellectual support. This work was partially funded by grants from MIUR and Fondazione Roma (Research Grant 2008).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Aquilano K. Baldelli S. Cardaci S. Rotilio G. Ciriolo MR. Nitric oxide is the primary mediator of cytotoxicity induced by GSH depletion in neuronal cells. J Cell Sci. 2011;124:1043–1054. doi: 10.1242/jcs.077149. [DOI] [PubMed] [Google Scholar]
- 2.Aquilano K. Vigilanza P. Baldelli S. Pagliei B. Rotilio G. Ciriolo MR. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285:21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aquilano K. Vigilanza P. Rotilio G. Ciriolo MR. Mitochondrial damage due to SOD1 deficiency in SH-SY5Y neuroblastoma cells: a rationale for the redundancy of SOD1. FASEB J. 2006;20:1683–1685. doi: 10.1096/fj.05-5225fje. [DOI] [PubMed] [Google Scholar]
- 4.Baldelli S. Aquilano K. Rotilio G. Ciriolo MR. Glutathione and copper, zinc superoxide dismutase are modulated by overexpression of neuronal nitric oxide synthase. Int J Biochem Cell Biol. 2008;40:2660–2670. doi: 10.1016/j.biocel.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Baldelli S. Aquilano K. Rotilio G. Ciriolo MR. Neuronal nitric oxide synthase interacts with Sp1 through the PDZ domain inhibiting Sp1-mediated copper-zinc superoxide dismutase expression. Int J Biochem Cell Biol. 2011;43:163–169. doi: 10.1016/j.biocel.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Ballatori N. Krance SM. Notenboom S. Shi S. Tieu K. Hammond CL. Glutathione dysregulation and the etiology and progression of human diseases. Biol Chem. 2009;390:191–214. doi: 10.1515/BC.2009.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensaad K. Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Borniquel S. Valle I. Cadenas S. Lamas S. Monsalve M. Nitric oxide regulates mitochondrial oxidative stress protection via the transcriptional coactivator PGC-1alpha. FASEB J. 2006;20:1889–1891. doi: 10.1096/fj.05-5189fje. [DOI] [PubMed] [Google Scholar]
- 9.Buettner VL. Nishino H. Haavik J. Knoll A. Hill K. Sommer SS. Spontaneous mutation frequencies and spectra in p53 (+/+) and p53 (−/−) mice: a test of the ‘guardian of the genome’ hypothesis in the Big Blue transgenic mouse mutation detection system. Mutat Res. 1997;379:13–20. doi: 10.1016/s0027-5107(97)00078-x. [DOI] [PubMed] [Google Scholar]
- 10.Clark J. Simon DK. Transcribe to survive: transcriptional control of antioxidant defense programs for neuroprotection in Parkinson's disease. Antioxid Redox Signal. 2009;11:509–528. doi: 10.1089/ars.2008.2241. [DOI] [PubMed] [Google Scholar]
- 11.Daitoku H. Yamagata K. Matsuzaki H. Hatta M. Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 12.Fekete I. Griffith OW. Schlageter KE. Bigner DD. Friedman HS. Groothuis DR. Rate of buthionine sulfoximine entry into brain and xenotransplanted human gliomas. Cancer Res. 1990;50:1251–1256. [PubMed] [Google Scholar]
- 13.Hafsi H. Hainaut P. Redox control and interplay between p53 isoforms: roles in the regulation of basal p53 levels, cell fate and senescence. Antioxid Redox Signal. 2011;15:1655–1667. doi: 10.1089/ars.2010.3771. [DOI] [PubMed] [Google Scholar]
- 14.Hakvoort TB. Moerland PD. Frijters R. Sokolovic A. Labruyere WT. Vermeulen JL. Ver Loren van Themaat E. Breit TM. Wittink FR. van Kampen AH. Verhoeven AJ. Lamers WH. Sokolovic M. Interorgan coordination of the murine adaptive response to fasting. J Biol Chem. 2011;286:16332–16343. doi: 10.1074/jbc.M110.216986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handschin C. Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 16.Holley AK. Dhar SK. St. Clair DK. Manganese superoxide dismutase vs. p53: regulation of mitochondrial ROS. Mitochondrion. 2010;10:649–661. doi: 10.1016/j.mito.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Hussain SP. Amstad P. He P. Robles A. Lupold S. Kaneko I. Ichimiya M. Sengupta S. Mechanic L. Okamura S. Hofseth LJ. Moake M. Nagashima M. Forrester KS. Harris CC. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer Res. 2004;64:2350–2356. doi: 10.1158/0008-5472.can-2287-2. [DOI] [PubMed] [Google Scholar]
- 18.Im H. Grass JA. Johnson KD. Boyer ME. Wu J. Bresnick EH. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol Biol. 2004;284:129–146. doi: 10.1385/1-59259-816-1:129. [DOI] [PubMed] [Google Scholar]
- 19.Irrcher I. Ljubicic V. Kirwan AF. Hood DA. AMP-activated protein kinase-regulated activation of the PGC-1alpha promoter in skeletal muscle cells. PLoS One. 2008;3:e3614. doi: 10.1371/journal.pone.0003614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komarova EA. Neznanov N. Komarov PG. Chernov MV. Wang K. Gudkov AV. p53 inhibitor pifithrin alpha can suppress heat shock and glucocorticoid signaling pathways. J Biol Chem. 2003;278:15465–15468. doi: 10.1074/jbc.C300011200. [DOI] [PubMed] [Google Scholar]
- 21.Liddell JR. Dringen R. Crack PJ. Robinson SR. Glutathione peroxidase 1 and a high cellular glutathione concentration are essential for effective organic hydroperoxide detoxification in astrocytes. Glia. 2006;54:873–879. doi: 10.1002/glia.20433. [DOI] [PubMed] [Google Scholar]
- 22.Liu H. Wang H. Shenvi S. Hagen TM. Liu RM. Glutathione metabolism during aging and in Alzheimer disease. Ann N Y Acad Sci. 2004;1019:346–349. doi: 10.1196/annals.1297.059. [DOI] [PubMed] [Google Scholar]
- 23.Liu XM. Peyton KJ. Ensenat D. Wang H. Hannink M. Alam J. Durante W. Nitric oxide stimulates heme oxygenase-1 gene transcription via the Nrf2/ARE complex to promote vascular smooth muscle cell survival. Cardiovasc Res. 2007;75:381–389. doi: 10.1016/j.cardiores.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y. Dentin R. Chen D. Hedrick S. Ravnskjaer K. Schenk S. Milne J. Meyers DJ. Cole P. Yates J., 3rd Olefsky J. Guarente L. Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nencioni L. De Chiara G. Sgarbanti R. Amatore D. Aquilano K. Marcocci ME. Serafino A. Torcia M. Cozzolino F. Ciriolo MR. Garaci E. Palamara AT. Bcl-2 expression and p38MAPK activity in cells infected with influenza A virus: impact on virally induced apoptosis and viral replication. J Biol Chem. 2009;284:16004–16015. doi: 10.1074/jbc.M900146200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nisoli E. Clementi E. Paolucci C. Cozzi V. Tonello C. Sciorati C. Bracale R. Valerio A. Francolini M. Moncada S. Carruba MO. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 27.Nisoli E. Tonello C. Cardile A. Cozzi V. Bracale R. Tedesco L. Falcone S. Valerio A. Cantoni O. Clementi E. Moncada S. Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 28.Pollard KS. Salama SR. Lambert N. Lambot MA. Coppens S. Pedersen JS. Katzman S. King B. Onodera C. Siepel A. Kern AD. Dehay C. Igel H. Ares M., Jr. Vanderhaeghen P. Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 29.Punathil T. Katiyar SK. Inhibition of non-small cell lung cancer cell migration by grape seed proanthocyanidins is mediated through the inhibition of nitric oxide, guanylate cyclase, and ERK1/2. Mol Carcinog. 2009;48:232–242. doi: 10.1002/mc.20473. [DOI] [PubMed] [Google Scholar]
- 30.Pyke S. Lew H. Quintanilha A. Severe depletion in liver glutathione during physical exercise. Biochem Biophys Res Commun. 1986;139:926–931. doi: 10.1016/s0006-291x(86)80266-2. [DOI] [PubMed] [Google Scholar]
- 31.Rashi-Elkeles S. Elkon R. Shavit S. Lerenthal Y. Linhart C. Kupershtein A. Amariglio N. Rechavi G. Shamir R. Shiloh Y. Transcriptional modulation induced by ionizing radiation: p53 remains a central player. Mol Oncol. 2011;5:336–348. doi: 10.1016/j.molonc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rebrin I. Sohal RS. Pro-oxidant shift in glutathione redox state during aging. Adv Drug Deliv Rev. 2008;60:1545–1552. doi: 10.1016/j.addr.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodgers JT. Lerin C. Gerhart-Hines Z. Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodgers JT. Lerin C. Haas W. Gygi SP. Spiegelman BM. Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 35.Safdar A. Little JP. Stokl AJ. Hettinga BP. Akhtar M. Tarnopolsky MA. Exercise increases mitochondrial PGC-1{alpha} content and promotes nuclear-mitochondrial cross-talk to coordinate mitochondrial biogenesis. J Biol Chem. 2011;286:10605–17. doi: 10.1074/jbc.M110.211466. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Sahin E. Colla S. Liesa M. Moslehi J. Muller FL. Guo M. Cooper M. Kotton D. Fabian AJ. Walkey C. Maser RS. Tonon G. Foerster F. Xiong R. Wang YA. Shukla SA. Jaskelioff M. Martin ES. Heffernan TP. Protopopov A. Ivanova E. Mahoney JE. Kost-Alimova M. Perry SR. Bronson R. Liao R. Mulligan R. Shirihai OS. Chin L. DePinho RA. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sastre J. Asensi M. Gasco E. Pallardo FV. Ferrero JA. Furukawa T. Vina J. Exhaustive physical exercise causes oxidation of glutathione status in blood: prevention by antioxidant administration. Am J Physiol. 1992;263:R992–R995. doi: 10.1152/ajpregu.1992.263.5.R992. [DOI] [PubMed] [Google Scholar]
- 38.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 39.Sen N. Satija YK. Das S. PGC-1alpha, a key modulator of p53, promotes cell survival upon metabolic stress. Mol Cell. 2011;44:621–634. doi: 10.1016/j.molcel.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 40.Spiegelman BM. Transcriptional control of energy homeostasis through the PGC1 coactivators. Novartis Found Symp. 2007;286:3–6. ; discussion 6–12, 162–163, 196–203. [PubMed] [Google Scholar]
- 41.St-Pierre J. Drori S. Uldry M. Silvaggi JM. Rhee J. Jager S. Handschin C. Zheng K. Lin J. Yang W. Simon DK. Bachoo R. Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 42.Tomko RJ., Jr. Bansal P. Lazo JS. Airing out an antioxidant role for the tumor suppressor p53. Mol Interv. 2006;6:23–25. doi: 10.1124/mi.6.1.5. [DOI] [PubMed] [Google Scholar]
- 43.Valle I. Alvarez-Barrientos A. Arza E. Lamas S. Monsalve M. PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc Res. 2005;66:562–573. doi: 10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 44.Vigilanza P. Aquilano K. Baldelli S. Rotilio G. Ciriolo MR. Modulation of intracellular glutathione affects adipogenesis in 3T3-L1 cells. J Cell Physiol. 2011;226:2016–2024. doi: 10.1002/jcp.22542. [DOI] [PubMed] [Google Scholar]
- 45.Vogt BL. Richie JP., Jr Fasting-induced depletion of glutathione in the aging mouse. Biochem Pharmacol. 1993;46:257–263. doi: 10.1016/0006-2952(93)90412-p. [DOI] [PubMed] [Google Scholar]
- 46.Vousden KH. Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 47.Wakabayashi N. Slocum SL. Skoko JJ. Shin S. Kensler TW. When NRF2 talks, who's listening? Antioxid Redox Signal. 2010;13:1649–1663. doi: 10.1089/ars.2010.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu G. Fang YZ. Yang S. Lupton JR. Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 49.Yoboue ED. Augier E. Galinier A. Blancard C. Pinson B. Casteilla L. Rigoulet M. Devin A. cAMP-induced mitochondrial compartment biogenesis: role of glutathione redox state. J Biol Chem. 2012;287:14569–14578. doi: 10.1074/jbc.M111.302786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.