Abstract
Aims
Yeast, like other eukaryotes, contains a complete mitochondrial thioredoxin system comprising a thioredoxin (Trx3) and a thioredoxin reductase (Trr2). Mitochondria are a main source of reactive oxygen species (ROS) in eukaryotic organisms, and this study investigates the role of Trx3 in regulating cell death during oxidative stress conditions. Results: We have previously shown that the redox state of mitochondrial Trx3 is buffered by the glutathione redox couple such that oxidized mitochondrial Trx3 only accumulates in mutants simultaneously lacking Trr2 and a glutathione reductase (Glr1). We show here that the redox state of mitochondrial Trx3 is important for yeast growth and its oxidation in a glr1 trr2 mutant induces programmed cell death. Apoptosis is dependent on the Yca1 metacaspase, since loss of YCA1 abrogates cell death induced by oxidized Trx3. Our data also indicate a role for a mitochondrial 1-cysteine (Cys) peroxiredoxin (Prx1) in the oxidation of Trx3, since Trx3 does not become oxidized in glr1 trr2 mutants or in a wild-type strain exposed to hydrogen peroxide in the absence of PRX1. Innovation: This study provides evidence that the redox state of a mitochondrial thioredoxin regulates yeast apoptosis in response to oxidative stress conditions. Moreover, the results identify a signaling pathway, where the thioredoxin system functions in both antioxidant defense and in controlling cell death. Conclusions: Mitochondrial Prx1 functions as a redox signaling molecule that oxidizes Trx3 and promotes apoptosis. This would mean that under conditions where Prx1 cannot detoxify mitochondrial ROS, it induces cell death to remove the affected cells. Antioxid. Redox Signal. 18, 376–385.
Introduction
An oxidative stress occurs when reactive oxygen species (ROS) overwhelm the cellular antioxidant defense systems. Compartmentalization can protect against oxidative stress by allowing cells to independently maintain their regulatory systems within individual organelles. Mitochondrial redox regulation is particularly important in this context, especially, since during respiration, mitochondria are a primary source of ROS and mitochondrial thiols are major ROS targets (5). Glutathione (GSH) is a low molecular weight thiol compound, that is found at high concentrations in most organisms. It is synthesized in the cytosol and must be transported into mitochondria via an active energy requiring process (14). Once inside mitochondria, GSH provides reducing power, but the resulting oxidized glutathione (GSSG) is unable to exit this compartment and must be reduced by the glutathione reductase (28). The yeast glutathione reductase (Glr1) colocalizes to the cytosol and mitochondria (29, 35). Nuclear genes encode both forms of the enzymes in a mechanism that appears to be conserved for the glutathione reductase in mammalian cells. Mitochondrial GSH is particularly important, since GSH deficiency in mammalian cells results in widespread mitochondrial damage (26) and yeast strains lacking GSH are unable to grow by respiration due to an accumulation of oxidative damage to mitochondrial DNA (22). However, GSH is not thought to play a major function as an antioxidant in cells (21), although it is required to maintain the redox status of the mitochondrial thioredoxin system (13, 40)
Innovation.
Glutathione (GSH) was previously shown to maintain the redox state of mitochondrial Trx3 (40). Our current data suggest that GSH regulates Prx1 activity rather than acting directly on Trx3. This provides a mechanism to couple the redox state ofmitochondrial GSH with a redox sensor that regulates programmed cell death. Yeast Prx1 is highly conserved with the mammalian 1-cysteine (Cys) peroxiredoxin (Prx6) and both are active as peroxidases, which are reduced by GSH rather than thioredoxin (13, 18). The finding that Prx1 can regulate apoptosis through modulating the oxidation state of thioredoxin may therefore aid in understanding 1-Cys Prx reaction mechanisms in eukaryotic organisms.
Thioredoxins are key oxidoreductases, which have been implicated in regulating thiol homeostasis in diverse organisms (16). Yeast, like other eukaryotes, contains a complete mitochondrial thioredoxin system, comprising a thioredoxin (TRX3) and a thioredoxin reductase (TRR2) (30). The redox state of the cytoplasmic and mitochondrial thioredoxin systems appears to be independently maintained and cells can survive in the absence of both systems (40). Mitochondrial Trx3 is maintained in a reduced form in wild-type cells, but surprisingly, is unaffected by the loss of Trr2 (40). In comparison, loss of the cytoplasmic thioredoxin reductase (Trr1) causes cytoplasmic thioredoxins (Trx1, Trx2) to accumulate in an oxidized form (38). Unlike cytoplasmic thioredoxins, the redox state of mitochondrial Trx3 is buffered by the GSSG/2GSH redox couple, such that oxidized mitochondrial Trx3 only accumulates in mutants simultaneously lacking TRR2 and the glutathione reductase (GLR1) (40). This is important, since it suggests that the mitochondrial thioredoxin function can be regulated by the GSH system. However, the requirement for the yeast mitochondrial Trx3 is as yet unknown, since it is dispensable for growth under normal and oxidative stress conditions (30, 40). This is surprising, since mammalian mitochondrial Trx2 is required for normal development of the mouse embryo and lack of Trx2 results in embryonic lethality (27).
Yeast contains a single 1-cysteine (Cys) peroxiredoxin (Prx1), which localizes to mitochondria (31). Prx's are ubiquitous, thiol-specific proteins that have multiple functions in stress protection (41). They are divided into 1-Cys and 2-Cys Prx's, based on the number of Cys residues directly involved in catalysis (41). Typical 2-Cys Prx's are active as a dimer and contain two redox active Cys residues that are directly involved in enzyme activity. 1-Cys Prx's contain a peroxidatic Cys, but do not contain a resolving Cys residue. Since 1-Cys Prx's cannot form a disulfide, the reduction reaction is thought to require a thiol-containing electron donor (36, 41). Cys91 is the peroxidatic Cys residue in Prx1, which is highly conserved in 1-Cys Prx's from bacterial, plant, and mammalian species (13). The peroxidatic Cys91 residue is detected in a reduced form unless cells are subjected to oxidative stress. Hydroperoxides oxidize Cys91 to the sulfenic acid form, which can then be glutathionylated through a reaction with GSH (13). This mixed disulfide is a substrate for reduction by Trr2, in a reaction that proceeds through the formation of a Prx1-Trr2 disulfide-bonded intermediate, regenerating reduced active Prx1. An additional role has been suggested for mitochondrial glutaredoxin (Grx2) based on the observation that Grx2 can reduce glutathionylated Prx1 in vitro (32). Trx3 does not appear to be able to support Prx1 antioxidant activity in cells, although it can reduce oxidized Prx1 in vitro (13, 31). We have suggested that Trx3 can directly reduce the sulfenic acid form of the Prx1 peroxidatic Cys residue. This is similar to the reduction by thioredoxin of the sulfenic acid intermediate formed as part of the reaction mechanism of mammalian methionine sulfoxide reductase (20). This raises the question as to why the mitochondrial thioredoxin does not appear to support Prx1 activity in vivo. One possibility is that the activity of Trx3 with Prx1 may only become important during particular growth or stress conditions.
An indication of a possible requirement for yeast Trx3 has come from the finding that simultaneous loss of Trr2 and Glr1 causes slow growth under respiratory conditions; this growth defect is mediated by oxidized Trx3, since deletion of TRX3 improves the growth of the glr1 trr2 mutant (40). In this current study, we have characterized the slow growth caused by oxidized Trx3 and we show that oxidized Trx3 promotes programmed cell death (PCD) in a glr1 trr2 mutant. Furthermore, analysis of the Trx3 redox state in prx1 mutants shows that Prx1 is required to oxidize Trx3 and regulates cell death by oxidizing Trx3.
Results
Oxidized Trx3 promotes apoptosis
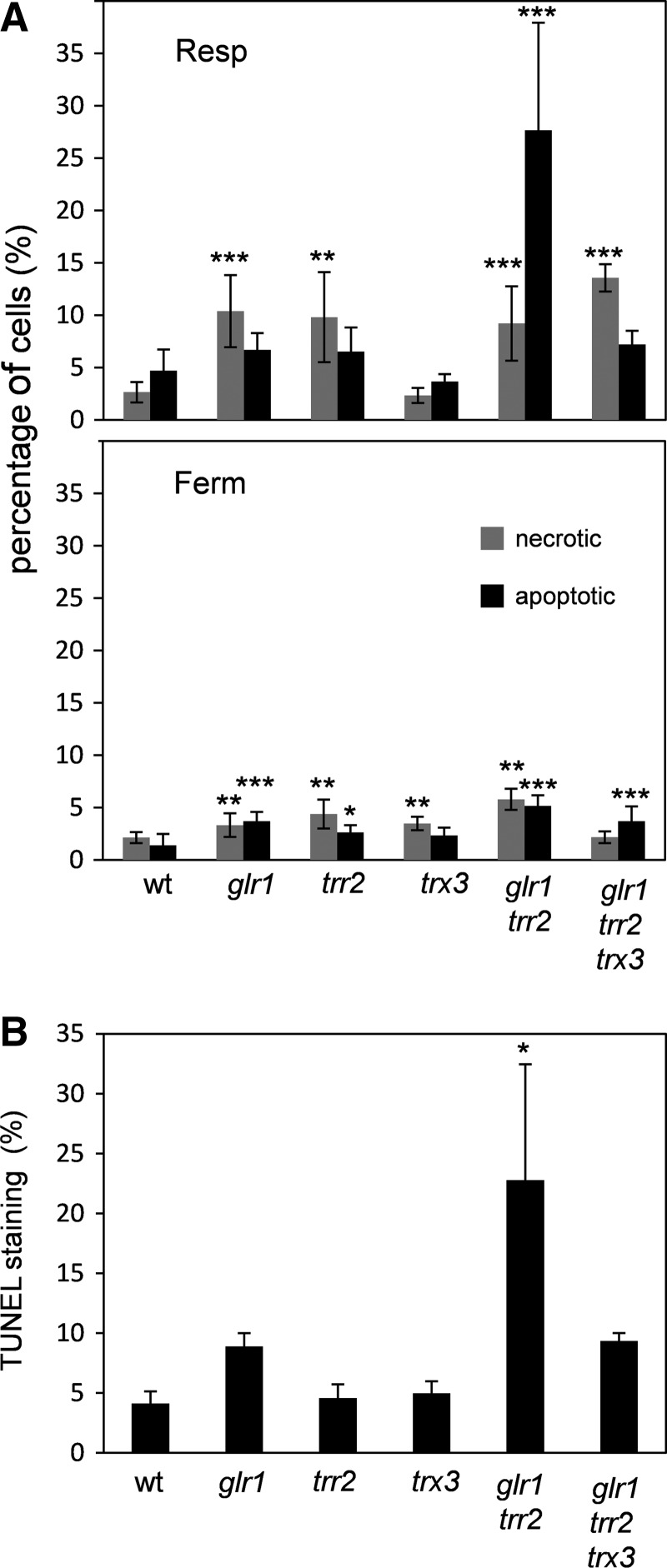
We have previously shown that oxidized Trx3 causes slow growth in a glr1 trr2 mutant (40). We examined whether Trx3 plays a role in yeast cell death in the glr1 trr2 mutant, which might explain this growth defect. Cells were grown under fermentative or respiratory conditions and the percentage of apoptotic cells determined using annexin V staining of phosphatidylserine on the external plasma membrane, which is as an early morphological marker of apoptosis (37). Loss of TRR2 or GLR1 alone, modestly increased the rate of apoptosis compared with a wild-type strain under respiratory conditions (Fig. 1A). In contrast, simultaneous loss of TRR2 and GLR1 caused a significant six-fold increase in apoptosis with approximately 30% of cells displaying annexin V staining. Cell death appears to mainly be mediated by Trx3 in the glr1 trr2 mutant, since deletion of TRX3 significantly reduced annexin V staining (Fig. 1A). Increased apoptosis is mainly a response to respiratory growth conditions, since more modest increases were detected under fermentative growth conditions (Fig. 1A). Apoptosis was significantly increased in all redox mutants examined under fermentative growth conditions, but no Trx3-dependent increase was detected in the glr1 trr2 mutant (Fig. 1A, compare glr1 trr2 and glr1 trr2 trx3). To further confirm these data, DNA fragmentation was detected using the terminal dUTP nick end-labeling (TUNEL) method as another indicator of fungal apoptosis. Significant TUNEL staining was detected in the glr1 trr2 mutant grown under respiratory conditions, which was abrogated by deletion of TRX3 (Fig. 1B).
FIG. 1.
Oxidized Trx3 promotes apoptosis. (A) The wild-type glr1, trr2, trx3, glr1 trr2, and glr1 trr2 trx3 mutant strains were grown to an exponential phase in fermentative (YEPD) or respiratory (YEPGE) media. The percentage of apoptotic (Annexin V staining) and necrotic (propidium iodide staining) cells is indicated. *p<0.05, **p<0.01, ***p<0.001 compared with the wild-type control strain. (B) The proportion of apoptotic cells was determined using the TUNEL assay in the same strains as described above grown under respiratory conditions. *p<0.05 compared with the wild-type control strain. TUNEL, terminal dUTP nick end-labeling; YEPD, yeast extract, peptone dextrose; YEPGE, yeast extract, peptone glycerol, ethanol.
Staining with propidium iodide (PI) was used as a control to differentiate cell death in necrotic cells with disrupted cell membranes. Necrosis was generally elevated under respiratory conditions compared with fermentative growth conditions and this increase was greater in mutants lacking TRR2 or GLR1 (Fig. 1A). However, no Trx3-dependent increase in necrosis was detected in the glr1 trr2 mutant. Since loss of TRX3 did not affect apoptosis in a wild-type background, these data indicate that a gain of Trx3 function, rather than a simple loss of Trx3 activity, most likely promotes apoptosis in the trr2 glr2 mutant.
Trx3 promotes apoptosis in a Yca1-dependent manner
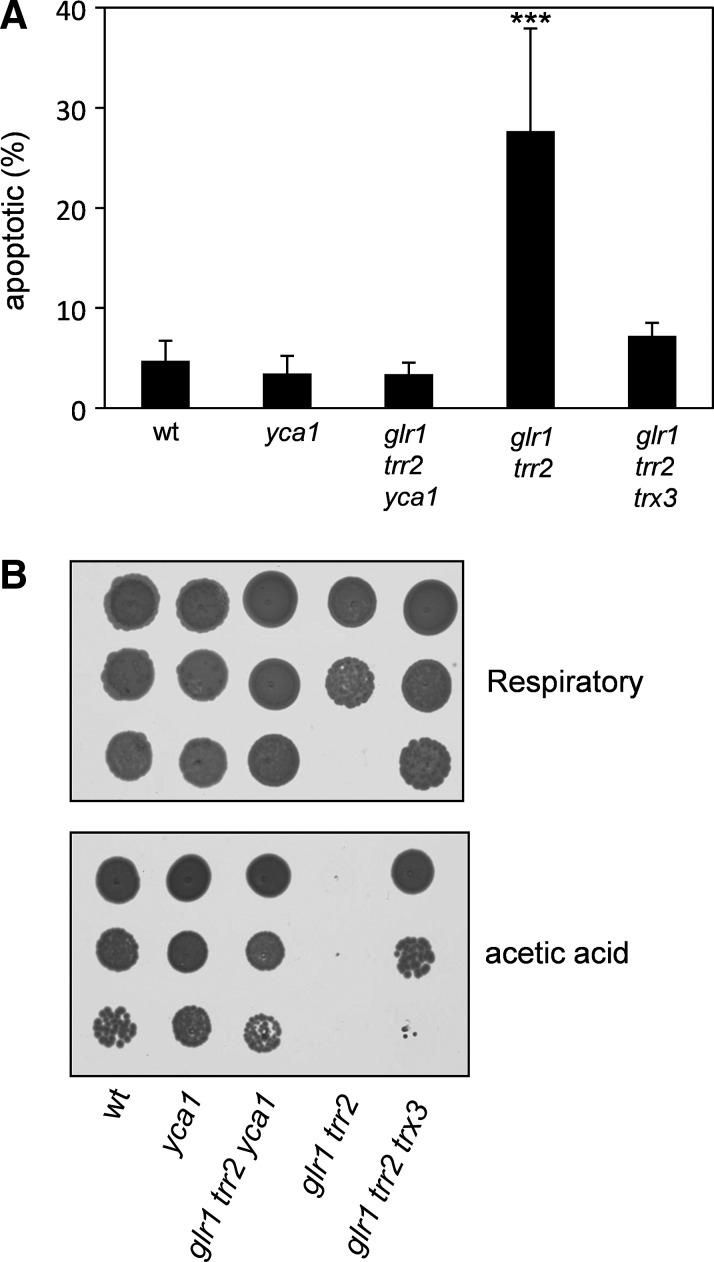
The yeast metacaspase Yca1 is known to mediate cell death in response to various stimuli (19, 25). We therefore tested whether apoptosis in the glr1 trr2 mutant requires Yca1. The proportion of apoptotic cells was determined using annexin V staining and deletion of YCA1 was found to reduce the high rate of apoptosis in the glr1 trr2 mutant to wild-type levels (Fig. 2A). Many different stimuli are known to induce yeast apoptosis, and so we examined whether the glr1 trr2 mutant shows any increased sensitivity to conditions that promote apoptosis. Acetic acid is one such stimulus that is commonly used to induce yeast apoptosis (4, 23). Wild-type and mutant strains were grown to a stationary phase and spotted onto respiratory growth media containing acetic acid (Fig. 2B). Under these conditions, the glr1 trr2 mutant displayed a slow growth, which was rescued by deletion of TRX3 or YCA1 (Fig. 2B). Taken together, these data indicate that oxidation of Trx3 induces PCD in a Yca1-dependent manner.
FIG. 2.
Trx3 promotes apoptosis in a Yca1-dependent manner. (A) The wild-type yca1, glr1 trr2, glr1 trr2 trx3, and glr1 trr2 yca1 mutant strains were grown to an exponential phase in respiratory (YEPGE) media. The percentage of apoptotic cells (Annexin V staining) is indicated. ***p<0.001 compared with the wild-type control strain. (B) Deletion of YCA1 restores the growth of a glr1 trr2 mutant under respiratory and acetic acid stress conditions. The indicated strains were grown to a stationary phase and spotted onto respiratory (YEPGE) media containing acetic acid.
Trx3-mediated apoptosis is not a general response to oxidized mitochondria
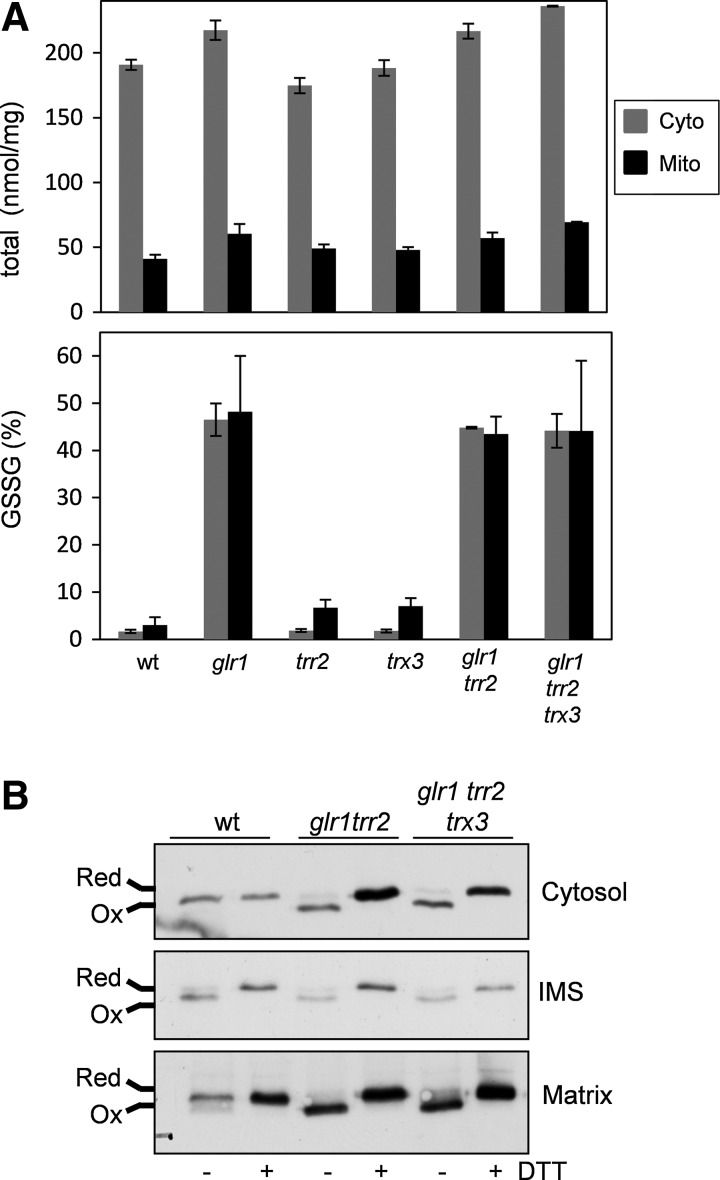
One possible explanation to account for the high rate of apoptosis in glr1 trr2 mutants is that it is a response to the oxidation state of mitochondria in this mutant. We therefore tested whether loss of TRX3 in the glr1 trr2 mutant influences the cytoplasmic or mitochondrial GSH redox couple, which is considered an indicator of the intracellular redox environment. Wild-type and mutant strains were grown under respiratory conditions and total GSH levels were found to be comparable in wild-type, glr1, trr2, trx3, and glr1 trr2 mutants in agreement with previous observations (29, 40). Importantly, deletion of TRX3 in the glr1 trr2 mutant did not affect total GSH levels in cytoplasmic or mitochondrial fractions (Fig. 3A). Oxidized GSSG levels are significantly elevated in both mitochondrial and cytoplasmic fractions from the glr1 mutant consistent with the important role of glutathione reductase in recycling oxidized GSSG to reduced GSH (29). Deletion of TRR2 in the glr1 mutant did not further alter GSSG levels indicating that the glr1 trr2 mutant does not have a more oxidized mitochondrial redox environment compared with a glr1 mutant (Fig. 3A). Additionally, loss of TRX3 did not affect GSSG levels in the glr1 trr2 mutant indicating that the GSH redox state does not correlate with the rates of apoptosis in these mutants.
FIG. 3.
Regulation of GSH metabolism in mitochondrial redox mutants. (A) The wild-type glr1, trr2, trx3, glr1 trr2, and glr1 trr2 trx3 mutant strains were grown to an exponential phase in respiratory media (SGE) and the levels of total GSH and oxidized GSH (GSSG) were determined in cytosolic and mitochondrial fractions. Values shown are the means of three independent determinations and are expressed as an nmol/mg protein. There is no significant difference between the glr1 trr2 and glr1 trr2 trx3 mutants. (B) Redox Western blot analysis. The wild-type, glr1 trr2 and glr1 trr2 trx3 mutant strains expressing cytosol-, IMS- or matrix rxYFP were grown to an exponential phase in respiratory media (SGE). Cell extracts were separated on nonreducing SDS-PAGE gels and Westen blots probed with anti-GFP antibodies. Cells were treated with DTT to fully reduce the redox probes as indicated. Oxidized and reduced proteins are indicated. GSSG, oxidized glutathione; GSH, glutathione; IMS, mitochondrial intermembrane space; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; SGE, synthetic glycerol ethanol.
Accurately measuring GSH levels in organelles, such as mitochondria, can be problematic, since it involves disruptive techniques, including cell breakage and fractionation. We validated our GSH assays using genetically encoded reduction–oxidation-sensitive green fluorescent protein probes, which have been designed to analyze redox states in the cytosol, mitochondrial intermembrane space (IMS), and matrix (17). These sensors equilibrate with the local GSH pool and register thiol redox changes via disulfide bond formation allowing comparisons of the redox state in different intracellular compartments. The cytosolic rxYFP was predominantly reduced in the wild-type strain (Fig. 3B). In comparison, the matrix rxYFP was partially oxidized and the IMS rxYFP was predominantly oxidized in a wild-type strain in agreement with previous observations (17). The IMS rxYFP was unaffected in glr1 trr2 and glr1 trr2 trx3 mutants compared with the wild-type strain in agreement with the idea that the redox state of the IMS is maintained independently of the presence of Glr1 (17). The matrix rxYFP became more oxidized in the glr1 trr2 mutant; however, loss of TRX3 did not alter its redox state in this mutant again suggesting that there is not a correlation between the general mitochondrial redox state and apoptosis in these mutants (Fig. 3B). We therefore examined whether the redox state of Trx3 itself influences the rate of cell death.
Requirement for Trx3 Cys residues to promote cell death
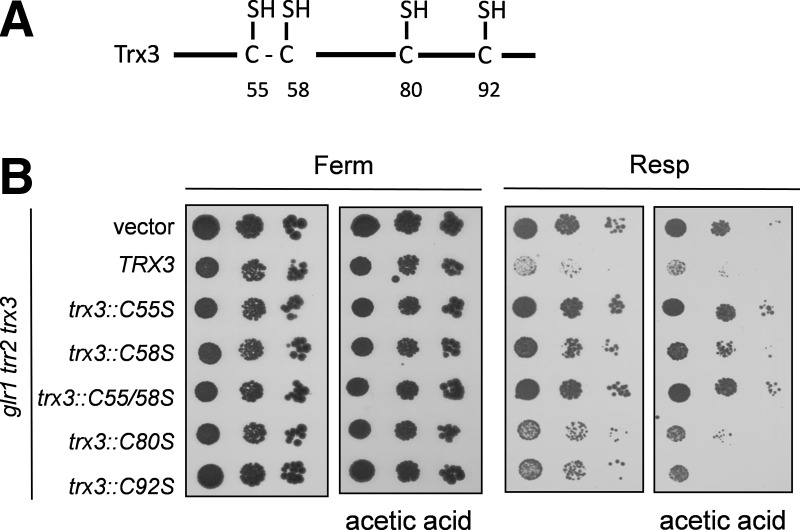
Mitochondrial Trx3 contains two redox active Cys residues (Cys55, Cys58) within the Cys-Gly-Pro-Cys motif that is conserved among thioredoxins from all species (Fig. 4A). In addition, Trx3 contains two extra Cys residues (Cys80, Cys92), which are not present in cytoplasmic Trx1 and Trx2 (30). The requirement for these extra Cys residues is unclear, but mutation of Cys80 or Cys92 has been shown to reduce the catalytic activity of purified Trx3 proteins in an insulin reduction assay by approximately 28% (2).
FIG. 4.
The active site Cys residues of Trx3 are required to promote cell death. (A) Schematic of mitochondrial Trx3 showing the two redox active Cys residues (Cys55, Cys58) within the Cys-Gly-Pro-Cys motif that are conserved among thioredoxins from all species. Trx3 also contains two extra Cys residues (Cys80, Cys92), which are not present in cytoplasmic Trx1 and Trx2. (B) A glr1 trr2 trx3 mutant containing wild-type (TRX3) on a single copy vector (vector) or Cys mutant versions of TRX3 (C55S, C58S, C80S, C92S) were grown to an exponential phase and spotted onto fermentative synthetic dextrose (SD) or respiratory synthetic glycerol ethanol (SGE) media containing acetic acid. Growth was scored after 3 days. Cys, cysteine; SD, synthetic dextrose.
We constructed mutant versions of Trx3 to determine which Cys residues are important for promoting cell death in the glr1 trr2 mutant. For these experiments, mutant versions of Trx3 were expressed in a glr1 trr2 trx3 mutant and the growth was examined using plate assays. The glr1 trr2 trx3 mutant containing an empty vector grew well under respiratory conditions and on media containing acetic acid, whereas, expression of wild-type TRX3 prevented the growth of this mutant in agreement with the finding that Trx3 promotes cell death in the glr1 trr2 mutant (Fig. 4B). Mutations in the Trx3 active site (trx3::C55S, trx3::C58S, trx3::C55S/C58S) rescued growth under both respiratory and acetic acid stress conditions. In contrast, mutations in the additional Cys residues (trx3::C70S, trx3::C92S) modestly improved the growth of the glr1 trr2 mutant under respiratory conditions, but not to the same extent as the complete loss of TRX3 or mutation of its active site Cys residues. These data indicate that the redox activity of Trx3 is important for promoting cell death.
Mitochondrial Prx1 regulates the redox state of Trx3
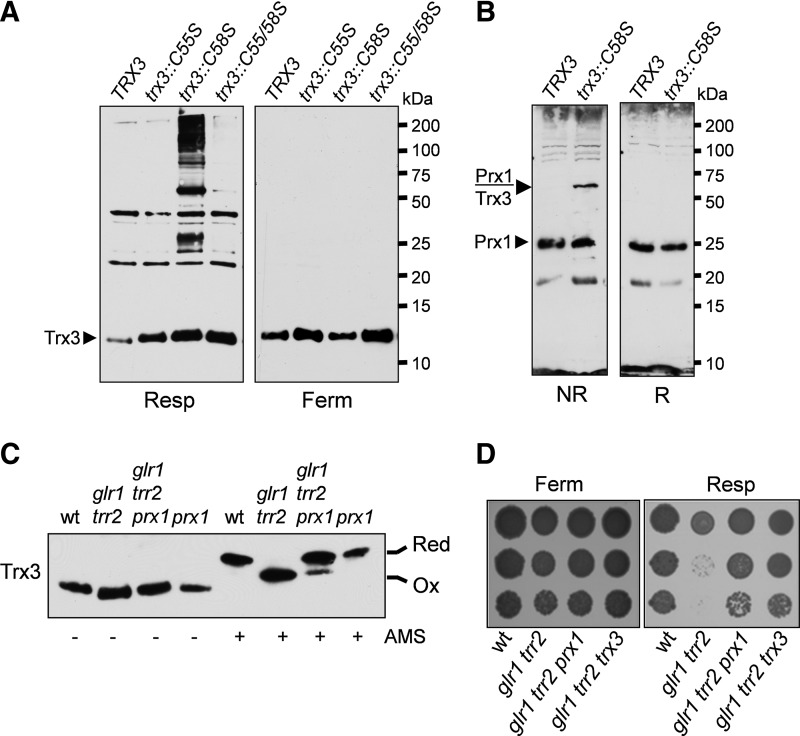
We made use of the Trx3 Cys mutants to search for possible Trx3 disulfide-bonded intermediates. Previous analyses have indicated that disulfide-bonded thioredoxin interactions are quite transient, but can be stabilized in mutants lacking the second Cys residue of the CXXC active site motif (Cys58 in Trx3). Analysis of Trx3 by nonreducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) revealed that the Trx3::C58S mutant forms a number of disulfide-bonded intermediates in a glr1 trr2 mutant (Fig. 5A). Mutation of Cys55 (trx3::C55S/C58S) prevented the formation of these disulfides suggesting that they represent physiological intermediates formed with the first Cys residue of the CXXC active site motif. No disulfide-bonded interactions where stabilized in trx3 mutants lacking the additional Cys80 or Cys92 residues (data not shown). Additionally, no interactions were detected in cells grown under fermentative conditions indicating that their formation correlates with the oxidation of Trx3 under respiratory growth conditions (Fig. 5A).
FIG. 5.
Mitochondrial Prx1 regulates the redox state of Trx3. (A) Trx3::C58S is present in many disulfide-bonded forms in cells grown under respiratory conditions. Western blot analysis of Trx3 is shown for the glr1 trr2 trx3 mutant containing wild-type (TRX3) or Cys mutant versions of TRX3 (C55S, C58S, C55/58S) grown to an exponential phase under fermentative (SD) or respiratory (SGE) growth conditions. Proteins were separated using nonreducing SDS-PAGE. (B) Prx1 can be detected in a disulfide bonded form with Trx3::C58S. Western blot analysis of Prx1 is shown for the glr1 trr2 trx3 mutant containing wild-type (TRX3) or Cys mutant of TRX3 (C58S) grown to an exponential phase under respiratory (SGE) growth conditions. Proteins were separated using nonreducing (NR) or reducing (R) SDS-PAGE. The position of a Prx1-Trx3 dimer is indicated. (C) Prx1 is required to oxidize Trx3 in a glr1 trr2 mutant. The wild-type glr1 trr2, glr1 trr2 prx1, and prx1 mutant strains were grown to an exponential phase in SGE media. Proteins were precipitated with TCA and free thiols modified by reaction with AMS. Samples were separated using SDS-PAGE and Trx3 detected by Western blot analysis. Oxidized and reduced proteins are indicated. (D) Deletion of PRX1 restores the growth of a glr1 trr2 mutant under respiratory conditions. The wild-type, glr1 trr2, glr1 trr2 prx1, and glr1 trr2 trx3 mutant strains were grown to a stationary phase and spotted onto fermentative (YEPD) or respiratory (YEPGE) media. AMS, 4-acetamido-4′maleimidyldystilbene-2,2′-disulfonic acid; TCA, trichloracetic acid.
We have previously shown that the mitochondrial thioredoxin system can support Prx1 activity in an in vitro peroxidase assay and suggested that Trx3 can directly reduce the sulfenic acid form of the Prx1 peroxidatic Cys residue (13). We therefore tested whether Prx1 accounts for one of the Trx3 disulfide-bonded intermediates detected in the glr1 trr2 mutant. Western blot analysis of Prx1 indicated that a significant proportion of Prx1 forms an intermediate with the trx3::C58S mutant (Fig. 5B). This interaction was confirmed to be a Trx3-Prx1 intermediate, since it is not detected in a prx1 mutant (data not shown). This intermediate was not detected under reducing conditions confirming that it is mediated by a disulfide bond.
Given the well-characterized roles of Prx's in redox signaling, we tested whether mitochondrial Prx1 might regulate the redox state of Trx3. The cellular oxidation state was preserved by rapidly treating cells with trichloracetic acid (TCA), which protonates free thiol groups. Extracts were reacted with the thiol-specific probe 4-acetamido-4′maleimidyldystilbene-2,2′-disulfonic acid (AMS). AMS alkylates Cys residues in a free-SH, but not in an oxidized state, increasing their relative molecular mass which can be detected by SDS-PAGE and Western blot analysis (39). The migration of all Trx3 from the wild-type strain grown under respiratory conditions was decreased following treatment with AMS indicating that the vast majority is present in the reduced form (Fig. 5C). Simultaneous loss of TRR2 and GLR1 shifted the Trx3 redox balance to an oxidized state in agreement with our previous observations (40). Interestingly, deletion of PRX1 in the trr2 gl1 mutant restored the Trx3 redox balance to a more reduced state indicating that Prx1 promotes Trx3 oxidation in a glr1 trr2 mutant (Fig. 5C). We reasoned that if Prx1 is responsible for Trx3 oxidation in a glr1 trr2 mutant, then deletion of PRX1 should restore the growth of a glr1 trr2 mutant under respiratory conditions comparable to deletion of TRX3. This was indeed found to be the case indicating that Prx1 appears to act as a signaling molecule that oxidizes Trx3 under respiratory conditions, promoting cell death (Fig. 5D).
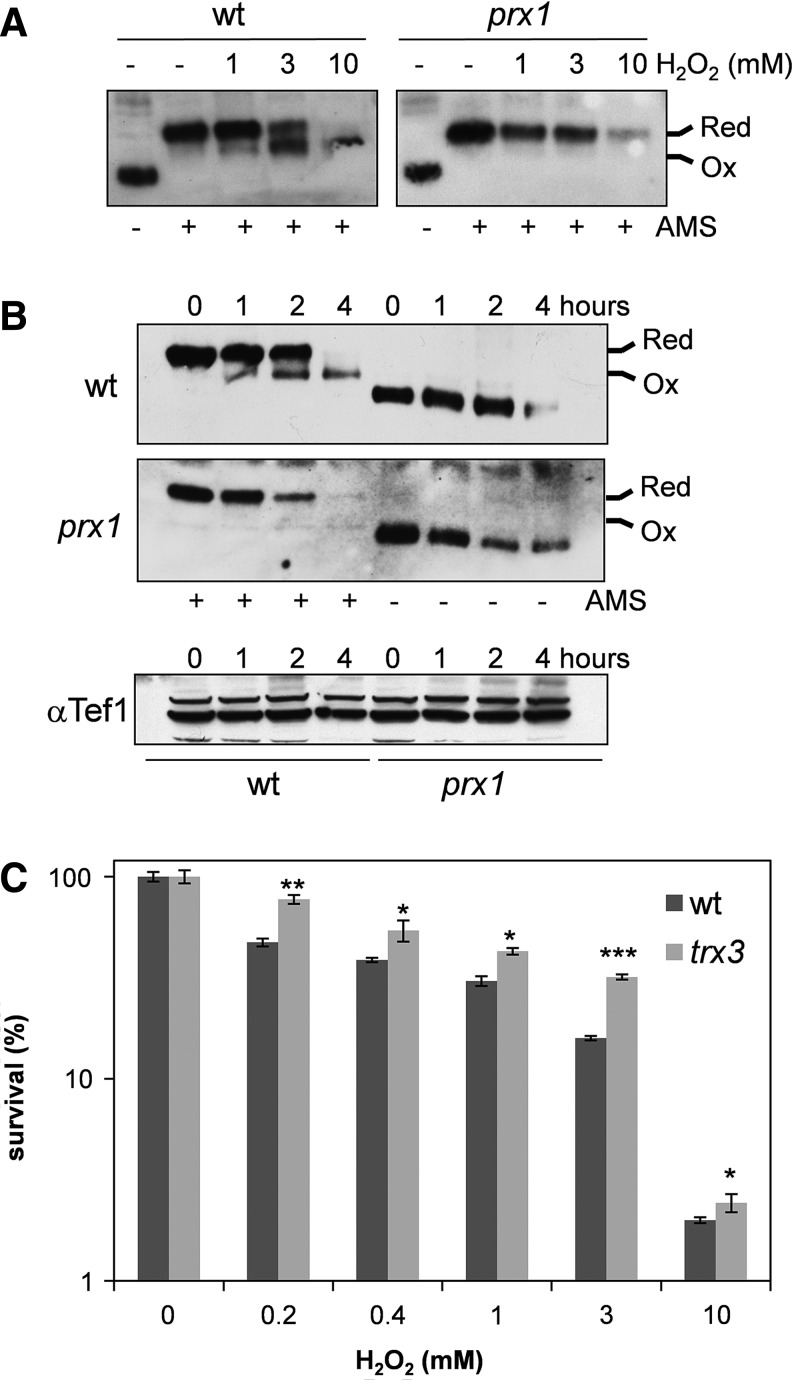
Loss of TRX3 increases resistance to hydrogen peroxide (H2O2) stress. Our previous analysis of mitochondrial Trx3 showed that it is relatively resistant to oxidation by H2O2 compared with cytoplasmic thioredoxins (40). We therefore examined the redox state of Trx3 following a longer term exposure and higher concentrations of peroxide in cells grown under respiratory conditions. Analysis of the Trx3 redox state revealed that it becomes progressively more oxidized following treatments with increasing concentrations of H2O2 for 4 h (Fig. 6A). Oxidation is detectable following exposure to 1 mM H2O2, and Trx3 appears predominantly oxidized following a treatment with 10 mM H2O2 for 4 h. Interestingly, this oxidation was found to require Prx1, since Trx3 remained in a reduced state following H2O2 treatments in a prx1 mutant (Fig. 6A). Less Trx3 was detected in both the wild-type and prx1 mutant strains following the 10 mM treatment, suggesting that Trx3 may be subject to degradation following exposures to higher concentrations of H2O2. We examined the time course of oxidation following the 10 mM treatment and found that Trx3 was partially oxidized within 1 h and fully oxidized by 4 h (Fig. 6B). Oxidation of Trx3 depends on the presence of Trx3, since no oxidation was detected in the trx3 mutant. A loading control was probed with an antibody that recognizes an elongation factor 1A (Tef1) and showed no differences in Tef1 proteins levels following exposure of wild-type and trx3 mutant cells to 10 mM H2O2 (Fig. 6B).
FIG. 6.
Prx1 oxidizes Trx3 in response to H2O2 stress. (A) The wild-type and prx1 mutant strains were grown to an exponential phase in YEPGE media and treated with 1 mM, 3 mM, or 10 mM H2O2 for 4 h. Proteins were precipitated with TCA and free thiols modified by reaction with AMS. Oxidized and reduced Trx3 are indicated. (B) The wild-type and prx1 mutant strains were grown to an exponential phase in YEPGE media, and the Trx3 redox state analyzed following exposure to 10 mM H2O2 for 1, 2, and 4 h. Oxidized and reduced Trx3 are indicated. A Western blot is shown probed with antibodies against the elongation factor 1A (Tef1) as a loading control following the same treatments. (C) A trx3 mutant shows increased resistance to H2O2 stress. The wild-type and trx3 mutant strains were grown to an exponential phase in YEPGE media and treated with the indicated concentrations of H2O2 for 4 h. Percent survival is expressed relative to that of untreated cultures. Experiments were repeated in triplicates and values shown are means. *p<0.05, **p<0.01, ***p<0.001 for comparisons between the trx3 mutant and the wild-type strain. H2O2, hydrogen peroxide.
If the oxidation state of mitochondrial Trx3 is an important trigger for apoptosis, then one prediction would be that mutants lacking TRX3 may be resistant to agents that oxidize Trx3. We tested this idea by examining the resistance of a trx3 mutant to various concentrations of H2O2. Loss of TRX3 was found to increase cell viability during H2O2 exposure over a concentration range up to 10 mM (Fig. 6C).
Discussion
Yeast mitochondrial thioredoxin reductase and the GSH system play an overlapping role in maintaining the redox state of mitochondrial Trx3 (40). During normal fermentative growth conditions, deletion of both TRR2 and GLR1 leads to partial oxidation of Trx3, which is even more pronounced during oxidative stress or respiratory growth conditions. In this current study, we have shown that the accumulation of oxidized Trx3 in the glr1 trr2 mutant during respiratory growth conditions promotes apoptosis. ROS exposure and the resulting oxidative stress have been well characterized as an insult that induces a form of PCD in yeast (25). PCD is thought to provide a mechanism for the selective death of a subset of the yeast population, which can be beneficial in a unicellular organism under certain stress conditions (10). Many stress conditions that produce ROS, including H2O2 exposure and depletion of GSH, have been shown to promote yeast PCD (24, 33–34). Saccharomyces cerevisiae contains a metacaspase (Yca1) that mediates cell death in response to various stimuli, including oxidative stress, caused by H2O2 exposure and yca1 mutants are resistant to H2O2 consistent with apoptosis accounting for cell death during ROS exposure (19, 25). Similarly, our current data indicate that Yca1 is required to induce PCD mediated by oxidized Trx3 in a glr1 trr2 mutant and trx3 mutants are more resistant to H2O2.
Mitochondria are a main source of ROS in eukaryotic cells and play well-established roles in cell death pathways (1). In mammalian cells, the intrinsic cell death pathway is stimulated by ROS exposure causing a complex cascade of events, including permeabilization of the outer mitochondrial membrane and release of cytochrome C (1, 7). Increasing evidence suggests that redox regulatory mechanisms are important in these processes. For example, GSH levels and redox state are decreased in response to ROS-mediated apoptosis (7). Mammalian mitochondrial Trx2 is required for normal development of the mouse embryo, and lack of Trx2 results in increased rates of apoptosis presumably due to ROS accumulation (27). Furthermore, mitochondrial Trx2 is oxidized in mammalian cells undergoing apoptosis and oxidized thioredoxin has been proposed to sensitize cells to ROS-induced apoptotic cell death (6, 43). Mitochondrial Trx2 has also been shown to prevent mitochondrial ROS-induced apoptosis following tumor necrosis factor -alpha exposure (15). One key mechanism by which Trx2 influences apoptosis is through modulation of the apoptosis signal-regulating kinase 1 (ASK1) (42). Inhibition and oxidation of mitochondrial thioredoxin systems may therefore represent common events in the activation of PCD in eukaryotic cells.
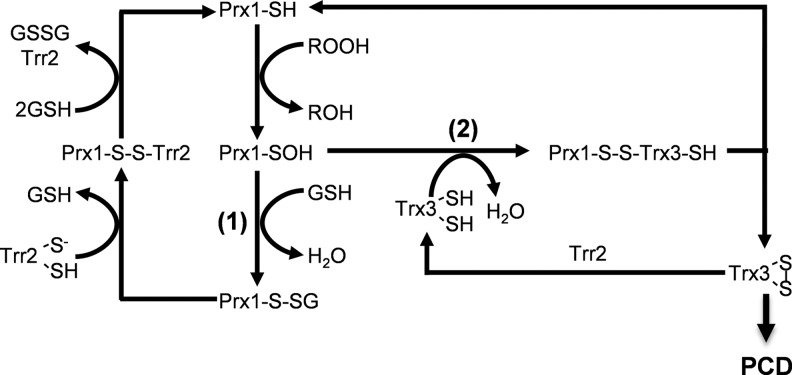
It was originally reported that Trx3 is the physiological electron donor for Prx1 (31). This is unexpected since 1-Cys Prx's are not thought to form a disulfide that can act as a substrate for thioredoxins. Our previous in vitro assays confirmed that mitochondrial Trx3 and Trr2 can support Prx1 activity with H2O2 as a substrate (13). Since Trx3 therefore appears to be able to reduce the sulfenic acid form of the Prx1 peroxidatic Cys residue, it is unclear why the mitochondrial thioredoxin does not support Prx1 activity in vivo. Mammalian Prx's (Prx3, Prx5) have been proposed to function in mitochondrial redox signaling (8). Kinetic analyses also suggests that Prx's are major targets of oxidation by mitochondrial H2O2, which means that they will play a major role in determining the levels of mitochondrial H2O2 available for signaling redox sensitive proteins, such as thioredoxins. Additionally, Prx's can act as direct signal transducers by oxidizing thiol proteins. We were able to trap Prx1 in a mixed disulfide with the Trx3::C58S mutant. Based on the formation of a Prx1-Trx3 reaction intermediate, we propose the mechanism shown in Figure 7. Under oxidative stress conditions, the peroxidatic Cys residue of Prx1 is oxidized to the sulfenic acid form. Reduced Prx1 can be regenerated in a reaction mechanism requiring Trr2 and GSH as previously described (13). Alternatively, oxidized Prx1 may react with Trx3 resulting in the generation of oxidized Trx3, which can promote PCD. This means that Prx1 normally functions as an antioxidant that detoxifies hydroperoxides. However, during an oxidative stress, Prx1 can function as a signaling molecule that oxidizes Trx3 and promotes apoptosis. This would mean that under conditions where yeast cells cannot detoxify mitochondrial ROS, they can induce PCD presumably to remove the affected cells.
FIG. 7.
Proposed reaction mechanism for regulation of PCD by Trx3. Reduction of H2O2 by Prx1 results in oxidation of the peroxidatic Cys residue (Cys91) of Prx1 to the sulfenic acid form (Prx1-SOH). Reaction with GSH generates glutathionylated Prx1 (Prx1-S-SG) (1), which is a substrate for reduction by Trr2 in a reaction that requires GSH to regenerate reduced Prx1. Alternatively, Prx1-SOH reacts with the catalytic Cys residue (Cys55) of Trx3 to form a disulfide-bonded intermediate (2). This intermediate can be reduced by the second Cys residue in Trx3 (Cys58) generating oxidized Trx3 and reduced Prx1. Oxidized Trx3 can be reduced by Trr2 to prevent activation of PCD. PCD, programmed cell death.
The finding that the peroxidatic Cys residue of Trx3 (Cys55) is required to activate PCD indicates that Trx3 may directly oxidize another substrate protein that forms part of the cell death pathway. Thioredoxins are well-known oxidoreductases, which may therefore directly catalyze thiol/disulfide exchange with an, as yet, unidentified protein. Mutation of Cys55 would prevent oxidation of this unidentified protein under conditions, where Trx3 normally becomes oxidized. Similarly, mutation of the second Cys residue (Cys58) in Trx3 also prevented the activation of PCD. Presumably, this mutant may trap substrate proteins in a mixed disulfide preventing them from functioning to activate PCD. Alternatively, oxidation and reduction of the Trx3 catalytic residues (Cys55 and Cys58) may cause some conformational change in Trx3, which triggers PCD. This could be analogous to the activation of apoptosis by ASK1 in mammalian cells, which can be regulated by redox-dependent binding of Trx2 (42). Thus, direct oxidation–reduction of thioredoxin can alter its noncovalent interaction with other proteins. Further work will be required to elucidate the exact pathway by which Trx3 induces PCD.
Materials and Methods
Yeast strains and plasmids
The S. cerevisiae strains used in this study were isogenic derivatives of W303 (MATa ura3-52 leu2-3 leu2-112 trp1-1 ade2-1 his3-11 can1-100). Strains deleted for the mitochondrial thioredoxin (trx3::kanMX4), mitochondrial thioredoxin reductase (trr2::HIS3), mitochondrial peroxiredoxin (prx1::kanMX4), and glutathione reductase (glr1::TRP1) have been described previously (11, 13, 40). Strains deleted for YCA1 were constructed using a one-step polymerase chain reaction amplification protocol that replaced its entire open reading frame with the LEU2 gene (3). Standard yeast genetic techniques were used to construct mutants lacking multiple genes (glr1 trr2, glr1 trr2 trx3, glr1 trr2 yca1, and glr1 trr2 prx1). Mutant versions of Trx3 (trx3::C55S, trx3::C58S, trx3::C80S, and trx3::92S) were made in TRX3 contained on plasmid pRS416 using the QuikChange method (Stratagene).
Growth conditions
Strains were grown in a rich yeast extract, peptone dextrose (YEPD) medium (2% w/v glucose, 2% w/v bactopeptone, 1% w/v yeast extract) or a minimal synthetic dextrose (SD) medium (0.17% w/v yeast nitrogen base without amino acids, 5% w/v ammonium sulfate, 2% w/v glucose supplemented with appropriate amino acids and bases). For growth on nonfermentable carbon sources, synthetic glycerol ethanol (SGE) and yeast extract, peptone glycerol, ethanol (YEPGE) contained 3% (v/v) glycerol and 1% (v/v) ethanol. Media were solidified by the addition of 2% (w/v) agar. Acetic acid sensitivity was determined by growing cells to a stationary phase, diluting and spotting onto agar plates containing 20 mM acetic acid.
Redox measurements
Protein extracts were electrophoresed under reducing or nonreducing conditions on SDS-PAGE minigels and electroblotted onto polyvinylidene fluoride membrane (Amersham Pharmacia Biotech). The bound antibody (αTrx3, αPrx1) was visualized by chemiluminescence (ECL; Amersham Pharmacia Biotech). The redox state of Trx3 was measured by covalent modification with the thiol-reactive probe AMS (Molecular Probes) as described previously (39). Glutathione (GSH and GSSG) levels were determined as described previously (12). Intracellular redox states were measured using reduction–oxidation-sensitive green fluorescent protein probes (cytosolic rxYFP, matrix rxYFP, IMS rxYFP) as described previously (17).
Cell death assays
Cell viability was determined by growing cells to an exponential phase and treating with H2O2 for 4 h. Aliquots of cells were diluted into fresh YEPD media and plated in triplicate on YEPD plates to obtain viable counts after 3 days growth. Apoptotic (Annexin V staining) versus necrotic (PI staining) cells were detected using the ApoAlert® Annexin V Kit (Clontech). DNA fragmentation was measured using the TUNEL method (FragEL™ DNA Fragmentation Kit; Calbiochem). For scoring fluorescent cells, three or more independent views of at least 50 cells from biological replicates were visualized using an Olympus widefield microscope and MetaVue software (Bioimaging Facility, Faculty of Life Science, University of Manchester). Results are expressed as means±standard error. Significant differences were analyzed using the Student's t-test. Values of p<0.05 were considered statistically significant.
Abbreviations Used
- AMS
4-acetamido-4′maleimidyldystilbene-2,2′-disulphonic acid
- ASK1
apoptosis signal-regulating kinase 1
- Cys
cysteine
- GSH
glutathione
- GSSG
oxidized glutathione
- H2O2
hydrogen peroxide
- IMS
mitochondrial intermembrane space
- PCD
programmed cell death
- PI
propidium iodide
- Prx
peroxiredoxin
- ROS
reactive oxygen species
- SD
synthetic dextrose
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SGE
synthetic glycerol ethanol
- TCA
trichloracetic acid
- TUNEL
terminal dUTP nick end-labeling
- YEPD
yeast extract, peptone dextrose
- YEPGE
yeast extract, peptone glycerol, ethanol
Acknowledgments
We thank Caryn Outten (University of South Carolina) for the gift of the rxYFP plasmids used in this study. This work was supported by funding from the Wellcome Trust and the Biotechnology and Biological Sciences Research Council.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Abdelwahid E. Rolland S. Teng X. Conradt B. Hardwick JM. White K. Mitochondrial involvement in cell death of non-mammalian eukaryotes. Biochim Biophys Acta. 2011;1813:597–607. doi: 10.1016/j.bbamcr.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao R. Zhang Y. Zhou CZ. Chen Y. Structural and mechanistic analyses of yeast mitochondrial thioredoxin Trx3 reveal putative function of its additional cysteine residues. Biochim Biophys Acta. 2009;1794:716–721. doi: 10.1016/j.bbapap.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 3.Baudin A. Ozier-Kalogeropoulos O. Danouel A. Lacroute F. Cullin O. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucl Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmona-Gutierrez D. Eisenberg T. Buttner S. Meisinger O. Kroemer G. Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- 5.Chance B. Sies H. Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y. Cai J. Jones DP. Mitochondrial thioredoxin in regulation of oxidant-induced cell death. FEBS Lett. 2006;580:6596–6602. doi: 10.1016/j.febslet.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Circu ML. Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic Biol Med. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox AG. Winterbourn CC. Hampton MB. Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem J. 2010;425:313–325. doi: 10.1042/BJ20091541. [DOI] [PubMed] [Google Scholar]
- 9.Day AM. Brown JD. Taylor SR. Rand JD. Morgan BA. Veal EA. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol Cell. 2012;45:398–408. doi: 10.1016/j.molcel.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 10.Gourlay CW. Du W. Ayscough KR. Apoptosis in yeast—mechanisms and benefits to a unicellular organism. Mol Microbiol. 2006;62:1515–1521. doi: 10.1111/j.1365-2958.2006.05486.x. [DOI] [PubMed] [Google Scholar]
- 11.Grant CM. Collinson LP. Roe J-H. Dawes IW. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol Microbiol. 1996;21:171–179. doi: 10.1046/j.1365-2958.1996.6351340.x. [DOI] [PubMed] [Google Scholar]
- 12.Grant CM. Perrone G. Dawes IW. Glutathione and catalase provide overlapping defenses for protection against hydrogen peroxide in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;253:893–898. doi: 10.1006/bbrc.1998.9864. [DOI] [PubMed] [Google Scholar]
- 13.Greetham D. Grant CM. Antioxidant activity of the yeast mitochondrial 1-Cys peroxiredoxin is dependent on thioredoxin reductase and glutathione in vivo. Mol Cell Biol. 2009;29:3229–3240. doi: 10.1128/MCB.01918-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith OW. Meister A. Origin and turnover of mitochondrial glutathione. Proc Natl Acad Sci U S A. 1985;82:4668–4672. doi: 10.1073/pnas.82.14.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen JM. Zhang H. Jones DP. Mitochondrial thioredoxin-2 has a key role in determining tumor necrosis factor-alpha-induced reactive oxygen species generation, NF-kappaB activation, and apoptosis. Toxicol Sci. 2006;91:643–650. doi: 10.1093/toxsci/kfj175. [DOI] [PubMed] [Google Scholar]
- 16.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 17.Hu J. Dong L. Outten CE. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J Biol Chem. 2008;283:29126–29134. doi: 10.1074/jbc.M803028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang SW. Baines IC. Rhee SG. Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J Biol Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- 19.Khan MA. Chock PB. Stadtman ER. Knockout of caspase-like gene, YCA1, abrogates apoptosis and elevates oxidized proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:17326–17331. doi: 10.1073/pnas.0508120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HY. Kim JR. Thioredoxin as a reducing agent for mammalian methionine sulfoxide reductases B lacking resolving cysteine. Biochem Biophys Res Commun. 2008;371:490–494. doi: 10.1016/j.bbrc.2008.04.101. [DOI] [PubMed] [Google Scholar]
- 21.Kumar C. Igbaria A. D'Autreaux B. Planson AG. Junot C. Godat E. Bachhawat AK. Delaunay-Moisan A. Toledano MB. Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J. 2011;30:2044–2056. doi: 10.1038/emboj.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J-C. Straffon MJ. Jang T-Y. Grant CM. Dawes IW. The essential and ancillary role of glutathione in Saccharomyces cerevisiae: studies with a grande gsh1 disruptant strain. FEMS Yeast Res. 2001;1:57–65. doi: 10.1111/j.1567-1364.2001.tb00013.x. [DOI] [PubMed] [Google Scholar]
- 23.Ludovico P. Sousa MJ. Silva MT. Leao C. Corte-Real M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology. 2001;147:2409–2415. doi: 10.1099/00221287-147-9-2409. [DOI] [PubMed] [Google Scholar]
- 24.Madeo F. Frohlich E. Ligr M. Grey M. Sigrist SJ. Wolf DH. Frohlich KU. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madeo F. Herker E. Maldener C. Wissing S. Lachelt S. Herlan M. Fehr M. Lauber K. Sigrist SJ. Wesselborg S. Frohlich KU. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- 26.Meister A. Mitochondrial changes associated with glutathione deficiency. Biochem Biophys Acta. 1995;1271:35–42. doi: 10.1016/0925-4439(95)00007-q. [DOI] [PubMed] [Google Scholar]
- 27.Nonn L. Williams RR. Erickson RP. Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol Cell Biol. 2003;232:916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olafsdottir K. Reed DJ. Retention of oxidized glutathione by isolated rat liver mitochondria during hydroperoxide treatment. Biochim Biophys Acta. 1988;964:377–382. doi: 10.1016/0304-4165(88)90038-4. [DOI] [PubMed] [Google Scholar]
- 29.Outten CE. Culotta VC. Alternative start sites in the S. cerevisiae GLR1 gene are responsible for mitochondrial and cytosolic isoforms of glutathione reductase. J Biol Chem. 2004;279:7785–7791. doi: 10.1074/jbc.M312421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedrajas JR. Kosmidou E. Miranda-Vizuete A. Gustafsson J-A. Wright APH. Spyrou G. Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J Biol Chem. 1999;274:6366–6373. doi: 10.1074/jbc.274.10.6366. [DOI] [PubMed] [Google Scholar]
- 31.Pedrajas JR. Miranda-Vizuete A. Javanmardy N. Gustafsson J-A. Spyrou G. Mitochondria of Saccharomyces cerevisiae contain one-conserved cysteine type peroxiredoxin with thioredoxin peroxidase activity. J Biol Chem. 2000;275:16296–16301. doi: 10.1074/jbc.275.21.16296. [DOI] [PubMed] [Google Scholar]
- 32.Pedrajas JR. Padilla CA. McDonagh BN. Bárcena JA. Glutaredoxin participates in the reduction of peroxides by the mitochondrial 1-CYS peroxiredoxin in Saccharomyces cerevisiae. Antioxid Redox Signal. 2010;13:249–258. doi: 10.1089/ars.2009.2950. [DOI] [PubMed] [Google Scholar]
- 33.Pereira C. Silva RD. Saraiva L. Johansson B. Sousa MJ. Corte-Real M. Mitochondria-dependent apoptosis in yeast. Biochim Biophys Acta. 2008;1783:1286–1302. doi: 10.1016/j.bbamcr.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Perrone GG. Tan SX. Dawes IW. Reactive oxygen species and yeast apoptosis. Biochim Biophys Acta. 2008;1783:1354–1368. doi: 10.1016/j.bbamcr.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 35.Porras P. Padilla CA. Krayl M. Voos W. Barcena JA. One single in frame AUG codon is responsible for a diversity of subcellular localizations of glutaredoxin 2 in Saccharomyces cerevisiae. J Biol Chem. 2006;281:16551–16562. doi: 10.1074/jbc.M600790200. [DOI] [PubMed] [Google Scholar]
- 36.Rhee SG. Chae HZ. Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 37.Sharon A. Finkelstein A. Shlezinger N. Hatam I. Fungal apoptosis: function, genes and gene function. FEMS Microbiol Rev. 2009:1–22. doi: 10.1111/j.1574-6976.2009.00180.x. [DOI] [PubMed] [Google Scholar]
- 38.Trotter EW. Grant CM. Thioredoxins are required for protection againts a reductive stress in the yeast Saccharomyces cerevisiae. Mol Microbiol. 2002;46:869–878. doi: 10.1046/j.1365-2958.2002.03216.x. [DOI] [PubMed] [Google Scholar]
- 39.Trotter EW. Grant CM. Non-reciprocal regulation of the redox state of the glutathione/glutaredoxin and thioredoxin systems. EMBO Rep. 2003;4:184–189. doi: 10.1038/sj.embor.embor729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trotter EW. Grant CM. Overlapping roles of the cytoplasmic and mitochondrial redoc regulatory systems in the yeast Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:392–400. doi: 10.1128/EC.4.2.392-400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wood ZA. Schroder E. Harris JR. Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 42.Zhang R. Al-Lamki R. Bai L. Streb JW. Miano JM. Bradley J. Min W. Thioredoxin-2 inhibits mitochondria-located ASK1-mediated apoptosis in a JNK-independent manner. Circ Res. 2004;94:1483–1491. doi: 10.1161/01.RES.0000130525.37646.a7. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X. Zheng Y. Fried LE. Du Y. Montano SJ. Sohn A. Lefkove B. Holmgren L. Arbiser JL. Holmgren A. Lu J. Disruption of the mitochondrial thioredoxin system as a cell death mechanism of cationic triphenylmethanes. Free Radic Biol Med. 2011;50:811–820. doi: 10.1016/j.freeradbiomed.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]