Abstract
Sir2 regulates lifespan in model organisms, which has stimulated interest in understanding human Sir2 homolog functions. The human Sir2 gene family comprises seven members (SIRT1–SIRT7). SIRT1, the human ortholog of the yeast Sir2 by closest sequence similarity, is a nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase with enzymatic properties indistinguishable from the yeast enzyme. We studied the involvement of SIRT1 in normal human keratinocyte physiology by a transcriptional microarray analysis of primary keratinocytes either overexpressing or underexpressing SIRT1. Using a systems biology analytical approach, we predicted that SIRT1 induces keratinocyte differentiation through a pathway integral to or overlapping with that of calcium-induced differentiation. We experimentally assayed this prediction and found that the SIRT1 inhibitor nicotinamide inhibited expression of keratinocyte differentiation markers, whereas a SIRT1 activator, resveratrol, enhanced expression of keratinocyte differentiation markers. Similar results were obtained in keratinocytes manipulated to overexpress or underexpress SIRT1, and modulating SIRT1 significantly affected keratinocyte proliferation rates. We conclude that SIRT1 functions in normal human keratinocytes to inhibit proliferation and to promote differentiation.
INTRODUCTION
Human SIRT1 (silent mating type information regulation 2 homolog 1) is a nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase controlling gene expression, cellular metabolism, and cellular stress responses (Haigis and Guarente, 2006). SIRT1 regulates adipocyte, muscle, liver, and endocrine pancreas physiology (Fulco et al., 2003; Picard et al., 2004; Rodgers et al., 2005; Bordone et al., 2006). Modulation of SIRT1 activity in those tissues impacts signaling networks, including insulin signals, controlling cell metabolism, and stress responses (Rodgers et al., 2005; Bordone et al., 2006). Additionally, SIRT1 has been shown to regulate cell differentiation in both myocytes and white adipocytes. Overexpressing SIRT1 negatively regulates their differentiation, whereas SIRT1 RNA interference (RNAi) enhances differentiation (Fulco et al., 2003; Picard et al., 2004). This study expands on these findings by investigating the role of human SIRT1 in skin cells using human primary keratinocytes as a model system for skin biology. Computational systems biology methods are combined with laboratory validation assays to develop a causal network model for SIRT1 function in primary keratinocytes.
In model organisms including yeast, flies, and worms, the SIRT1 ortholog Sir2 regulates lifespan (Kaeberlein et al., 1999; Tissenbaum and Guarente, 2001; Rogina and Helfand, 2004; for review see Blander and Guarente, 2004). In yeast, sir2 null mutants have shorter replicative lifespan and overexpressing yeast sir2 extends their lifespan (Kaeberlein et al., 1999). In worms and flies, overexpressing sir2 also increases their lifespan, and in worms null sir2 mutations decreases the lifespan (Tissenbaum and Guarente, 2001; Rogina and Helfand, 2004; Viswanathan et al., 2005). An ongoing aging-related research in mice indicates that SIRT1 is required for normal physiology, but specific effects on lifespan remain to be determined (McBurney et al., 2003). Given that SIRT1 homologs impact aging patterns, it is important to determine the cell and molecular events that SIRT1 controls in human skin cells.
Small molecule activators of SIRT1 have been identified (Howitz et al., 2003). Of these, the most potent activator is resveratrol, which is implicated in a number of health benefits. Resveratrol increases lifespan in yeast, worms, and flies (Howitz et al., 2003; Wood et al., 2004). In isolated human cells, resveratrol increases cell survival after DNA damage (Howitz et al., 2003). Also, resveratrol has recently been shown to inhibit pig preadipocyte differentiation, whereas nicotinamide, a SIRT1 inhibitor, greatly stimulated the proliferation and differentiation of pig preadipocytes (Bai et al., 2008). Less is known about resveratrol effects in other tissue contexts such as the skin.
To understand the function of SIRT1 in the skin, primary human keratinocytes either overexpressing or underexpressing SIRT1 were subjected to gene expression microarray analysis. Using a systems biology approach, we determined that SIRT1 overexpression recapitulates a molecular signature, which overlaps with epidermal differentiation in vivo, whereas SIRT1 underexpression recapitulates a molecular signature, which overlaps with epidermal proliferation in vivo. Hypotheses developed in the systems level analysis of microarray data were then assayed in vitro. In support of these hypotheses, we found that inhibiting SIRT1 by either RNAi or the chemical inhibitor nicotinamide repressed keratinocyte differentiation while overexpressing or activating SIRT1-induced differentiation. Finally, when measuring the replication capacity of these cells, we found that the cells overexpressing SIRT1 replicated fewer times than their controls whereas cells underexpressing SIRT1 replicated more than their controls. These data strongly suggest that SIRT1 is an important regulator of the keratinocyte differentiation pathway and is a potential regulator of skin aging.
RESULTS
To investigate the role of SIRT1 in growth and differentiation of normal human primary keratinocytes (NHEK), a microarray analysis of SIRT1-overexpressing and SIRT1 knockdown NHEK cells was conducted. For this purpose, four transgenic keratinocyte populations were generated and characterized as follows: SIRT1 overexpressing cells, pBABE-vector-negative controls, SIRT1 RNAi-treated cells, and pSUPER-RNAi-negative controls. SIRT1 protein overexpression and knockdown is evident by western blot in the respective NHEK transgenic cells, using actin and tubulin abundance as a loading control (Figure 1a). These genetically modified NHEK cells provide an in vitro context for studying SIRT1 function in skin cells.
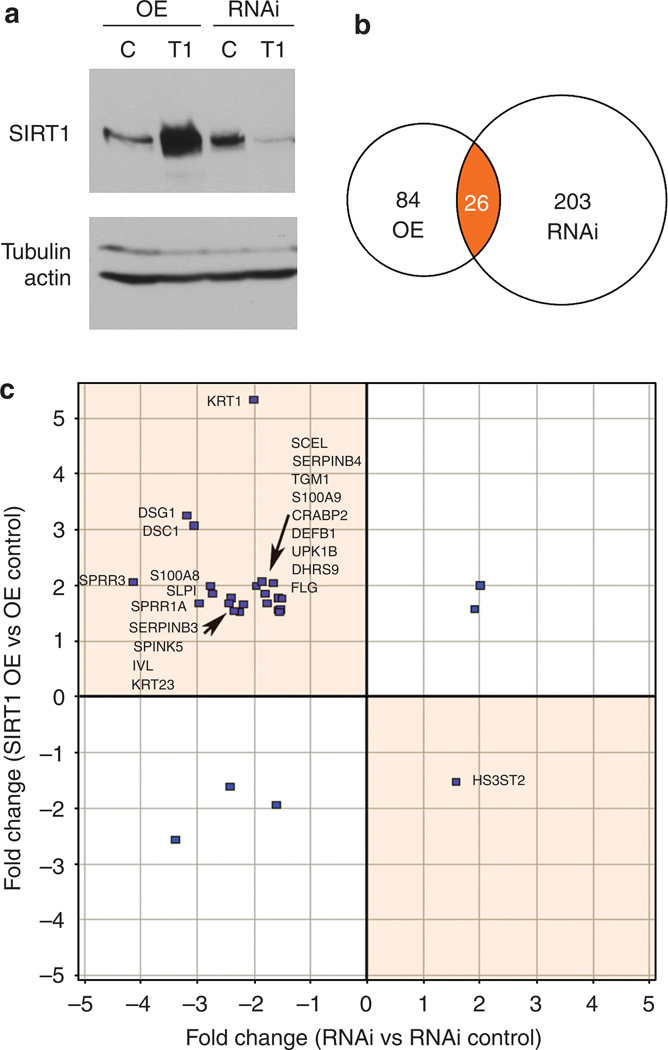
Figure 1. Gene expression changes induced by either overexpressing or underexpressing SIRT1 in primary keratinocytes identify a subset of expression changes oppositely affected by the two treatments.
(a) NHEK cells were infected with one of the following: SIRT1 overexpression virus (OE, T1) or its control virus (OE, C), SIRT1 RNAi virus (RNAi, T1) or its control virus (RNAi, C). A western blot analysis of SIRT1 demonstrates overexpression and reduced expression of SIRT1 relative to the tubulin and actin loading controls. (b) Venn diagram of the SIRT1 overexpression (OE) and the SIRT1 RNAi (RNAi) state changes. The OE and the RNAi experiments include 109 and 228 RNA expression state changes, respectively. Orange-colored region indicates the overlapping OE and the RNAi state changes. (c) Scatter plot analysis of genes affected by both treatments indicates that 21 of 26 are oppositely modulated (orange sectors).
SIRT1-overexpressing and SIRT1 knockdown NHEK cells were subject to a gene expression microarray analysis using the Affymetrix U133a GeneChip technology. The microarray data were collected (each of the four keratinocyte preparations was run in triplicate) for a total of 12 arrays measuring 22,278 probe sets in each microarray. The signals of the SIRT1-overexpressing and SIRT1 knockdown cells were compared with their respective controls using RMA (Robust Multichip Average) analysis to determine which probe sets were significantly different between experimental and negative control groups. Significantly changed probe sets had an adjusted P-value of <0.05 and a fold change of > 1.3. A total of 109 gene expression changes in the SIRT1-overexpressing cells and 228 gene expression changes in the SIRT1 knockdown cells were identified by these criteria (Figure 1b). The observed transcriptional changes caused by SIRT1 overexpression or knockdown in primary keratinocytes form the basis for the systems-level causal network modeling.
Because both perturbations were performed on the same cell type, an initial Venn analysis was performed comparing the two microarray data sets to identify overlapping transcripts changed in both experiments (Figure 1b). A total of 26 transcripts were modulated in both the SIRT1-overexpressing and the SIRT1 knockdown NHEK cells (Figure 1b, orange). Of the 26 transcripts, the expressions of 21 transcripts were modulated in opposite directions (Figure 1c; Table 1). Twenty genes were increased and one was decreased in the SIRT1-overexpressing cells. A survey of this list of genes changed in both experiments identifies the high number of keratinocyte differentiation markers (SPRR3, SPRR1A, KRT1, KRT23, DSC1, DSG1, S100A8, S100A9, FLG, IVL, and so on) (Table 1), suggesting increased cell differentiation in the SIRT1-overexpressing cells. However, a causal network analysis of all the data (109 gene expression changes in SIRT1-overexpressing cells and 228 gene expression changes in SIRT1 knockdown cells) is required to better understand the molecular network controlling these observed gene expression changes.
Table 1.
Gene expressions oppositely affected by SIRT1 OE and knockdown (RNAi)
| Gene symbol | Gene title | SIRT1 versus OE control | RNAi versus RNAi control |
|---|---|---|---|
| SPRR3 | Small proline-rich protein 3 | 2.06003 | −4.14393 |
| SPRR1A | Small proline-rich protein 1A | 1.67404 | −2.97523 |
| SERPINB3 | Serpin peptidase inhibitor, clade B | 1.78097 | −2.4144 |
| SERPINB4 | Serpin peptidase inhibitor, clade B | 1.67404 | −1.77892 |
| SPINK5 | Serine peptidase inhibitor, Kazal type 5 | 1.52344 | −2.26524 |
| SLPI | Secretory leukocyte peptidase inhibitor | 1.86176 | −2.74473 |
| KRT1 | Keratin 1 (epidermolytic hyperkeratosis) | 5.32704 | −2.01391 |
| KRT23 | Keratin 23 | 1.54114 | −2.34947 |
| DSC1 | Desmocollin 1 | 3.06524 | −3.06312 |
| DSG1 | Desmoglein 1 | 3.25427 | −3.19393 |
| S100A8 | S100 calcium binding protein A8 | 1.98069 | −2.77599 |
| S100A9 | S100 calcium binding protein A9 | 1.84634 | −1.81252 |
| FLG | Filaggrin | 1.52028 | −1.5533 |
| IVL | Involucrin | 1.66094 | −2.20228 |
| TGM1 | Transglutaminase 1 | 1.54757 | −1.56663 |
| CRABP2 | Cellular retinoic acid binding protein 2 | 1.76826 | −1.57207 |
| DEFB1 | Defensin, beta 1 | 2.06146 | −1.87341 |
| UPK1B | Uroplakin 1B | 1.75564 | −1.5049 |
| DHRS9 | Dehydrogenase/reductase | 1.56085 | −1.54757 |
| SCEL | Sciellin | 2.04202 | −1.66056 |
| HS3ST2 | Heparan sulfate 3-O-sulfotransferase 2 | −1.5351 | 1.56699 |
OE, overexpression.
Causal network modeling is a systematic analysis of an entire ‘omic’ data set. To identify the cellular processes and the molecular factors modulated in the SIRT1-overexpressing and SIRT1 RNAi cells, the Genstruct Causal Modeling platform was used to computationally derive a mechanistic model for the observed gene expression changes (see the Materials and Methods section for details). Automated causal reasoning on the SIRT1 overexpression and SIRT1 knockdown data yielded 15 and 74 statistically significant hypotheses, respectively. We focused on those computationally derived hypotheses that are evidenced in both experiments and proposed to change in opposite directions, as these hypotheses suggest core SIRT1-dependent molecular networks.
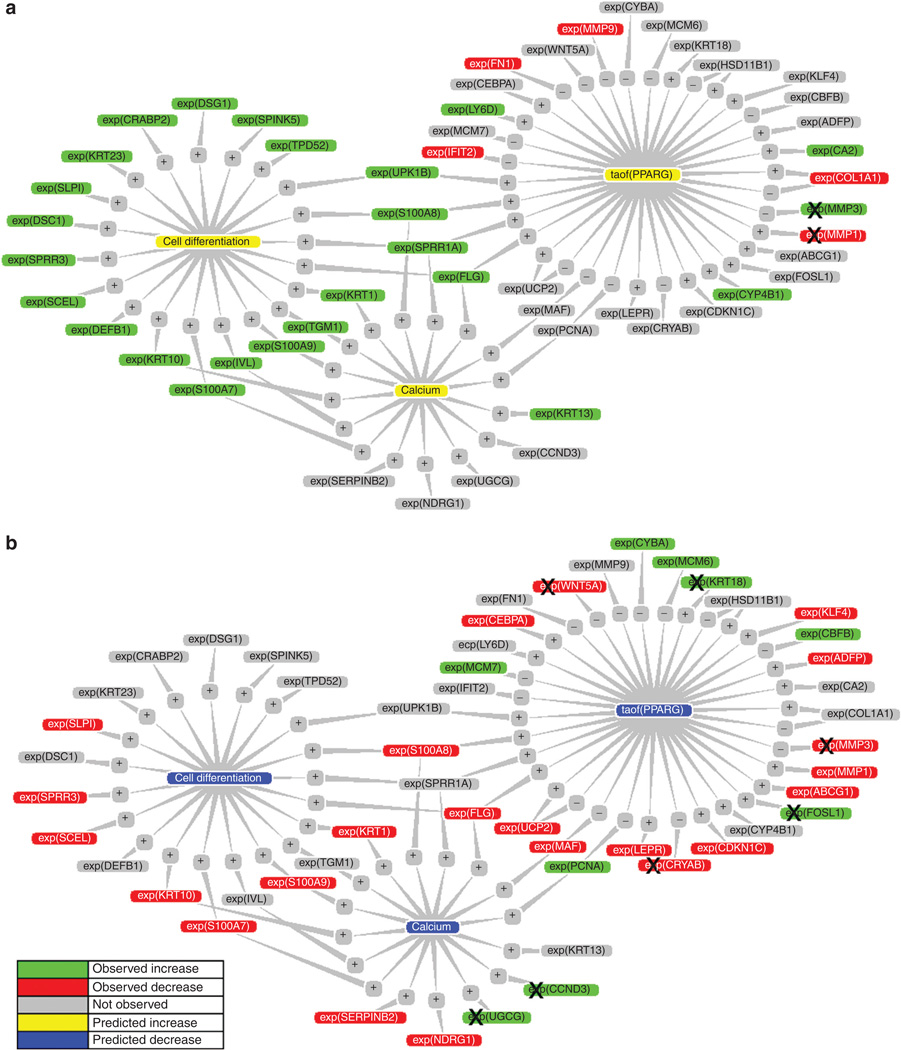
Causal modeling identifies cell differentiation, calcium-dependent processes, and the transcriptional activity of peroxisome proliferator-activated receptor gamma (PPARG) as cellular processes or molecular components mechanistically relevant to both experiments. All three hypotheses are proposed upregulated in the SIRT1 overexpression and downregulated in the SIRT1 knockdown experiment. These three hypotheses and their supporting and conflicting microarray observations are displayed in Figure 2a for the SIRT1 overexpression and Figure 2b for the SIRT1 RNAi experiment. These three hypotheses become the framework for understanding the molecular networks affected by SIRT1.
Figure 2. Causal network modeling of microarray observations predicts that increasing and decreasing SIRT1 levels in NHEK cells have opposite effects on cell differentiation, calcium-mediated signaling, and the transcriptional activity of PPARG.
(a) In the SIRT1 overexpressing cells. (b) In the SIRT1 RNAi cells. Diagram of RNA state changes consistent with or contrary to the indicated hypothesis. Green—observed increase in RNA expression of a given gene; red—observed decrease in RNA expression of a given gene; yellow—hypothesized increase in biological processes or protein activity; blue—hypothesized decrease in biological processes or protein activity. “+” symbolizes causal activation; “−” symbolizes casual inhibition; “+” and “−” nodes are supported by published findings supporting the causal assertion between the hypothesis and the RNA state change. An “X” over an observed RNA expression change indicates a contradiction (the direction of the observed RNA expression change is inconsistent with the hypothesis it is connected to). General note: In figures where gene expression is depicted, expression is noted by “exp” and the NCBI gene symbol is in parentheses. For example, exp(KRT10) indicates a change in keratin 10 expression. In addition, placement within a particular color indicates whether the change in expression was observed to increase (green) or observed to decrease (red). Also predicted increase (yellow) or predicted decrease (blue) may also be indicated.
The network of hypotheses supporting increased cell differentiation in the SIRT1-overexpressing cells are displayed in Figure S1. In addition to the three core hypotheses, other computationally derived hypotheses unique to each experiment further delineate the molecular networks proposed by causal modeling. Gene expression data support an increased transcriptional activity of CCAAT/enhancer-binding protein alpha (CEBPA) in the SIRT1-overexpressing primary keratinocytes (Figure S1). CEBPA is a known PPARG-binding protein that functions as a transcriptional coactivator to PPARG. Further support for CEBPA activity in SIRT1 signaling is suggested by the SIRT1 knockdown experiment, in which CEBPA expression is observed decreased by SIRT1 knockdown, supporting the proposed decreased transcriptional activity of PPARG in SIRT1 knockdown cells (Figure 2b). Even though there was no corresponding increase in CEBPA transcription in the SIRT1-overexpressing cells, causal modeling, by identifying known CEBPA target genes observed to change in a manner consistent with increased CEBPA transcriptional activity, predicts increased CEBPA activity.
The network of hypotheses supporting reduced cell differentiation in the SIRT1 knockdown cells are displayed in Figure S2. A causal analysis of the SIRT1 knockdown experiment identified functionally relevant hypotheses unique to that experiment. In addition to the three core hypotheses, decreased retinoate signaling and increased cell proliferation were inferred in that experiment (Figure S2). Decreased retinoate signaling is consistent with reduced cell differentiation, and because retinoate is a known activator of PPARG activity, decreased retinoate signaling is also consistent with decreased PPARG activity proposed in the causal network model.
Causal modeling further predicts that cell cycle regulators, particularly E2F1 and CCND1 (cyclin D1), are proposed to increase their activity by SIRT1 knockdown (Figure S3). The predicted increases in E2F1 and CCND1 activity are supported by gene expression changes of known cell cycle components, including MCM6 (mini chromosome maintenance complex component 6), TTK (TTK protein kinase), and PCNA (proliferating cell nuclear antigen) (Figure S3). In summary, causal network modeling proposes that for SIRT1 knockdown in NHEK cells, decreased calcium, retinoate, and PPARG activities, as well as increased cell cycle regulators function in a molecular network that inhibits primary keratinocyte differentiation and promotes cell proliferation.
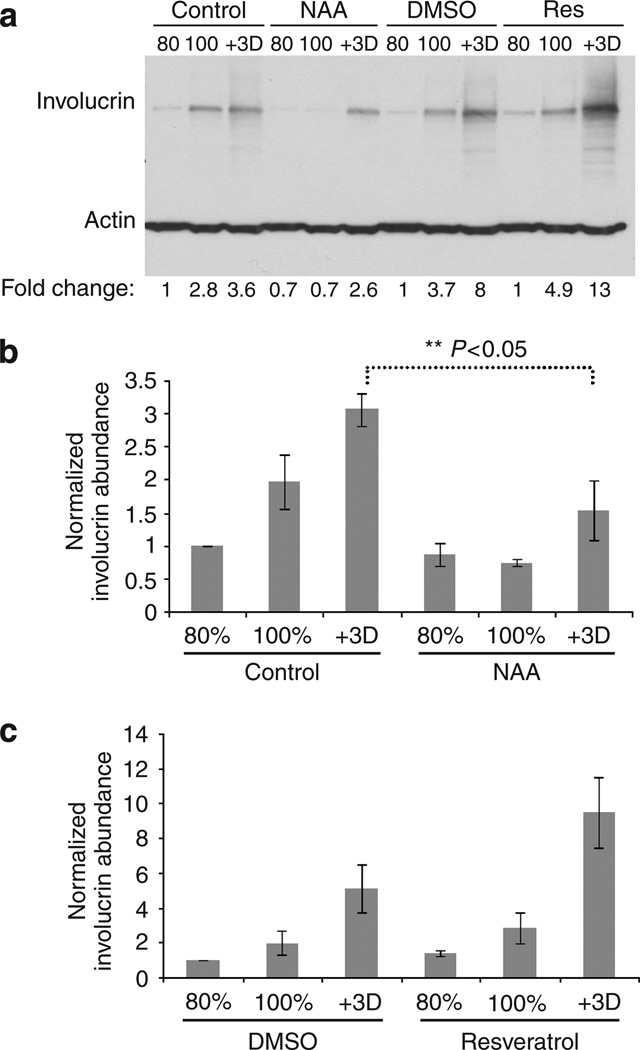
Causal network modeling was used to develop a mechanistic model of SIRT1 function in NHEK cells that provides a rationale for further experimentation. To test the predicted effects of SIRT1 modulation on NHEK cell differentiation, NHEK cells were treated with either SIRT1 agonists or antagonists. NHEK cells were incubated with nicotinamide (NAA, a known SIRT1 inhibitor), H2O (a vehicle control for nicotinamide treatment), resveratrol (a SIRT1 activator), or DMSO (a vehicle control for resveratrol treatment), and the NHEK cells were assayed for cell differentiation by measurement of known keratinocyte differentiation markers. Western blot analysis using anti-involucrin (and anti-actin as a total protein loading control) indicates that the SIRT1 antagonist NAA inhibits confluence-induced differentiation and that the SIRT1 agonist resveratrol promotes cell differentiation (Figure 3a). The quantification of the NAA-treated cells revealed between 1.5- and 4-fold reductions in involucrin abundance in the 100% confluent and between 1.5- and 3.5-fold in +3 days time points (Figure 3a and b). The quantification of the resveratrol-treated cells revealed between 1.5- and 2-fold increase in the involucrin intensity in the +3-day time point (Figure 3a and c). Although not meeting the statistical significance level (t-test, P<0.05), resveratrol treatment trended toward increasing normalized involucrin abundance (t-test, P = 0.058). A valid possibility is that the resveratrol differentiation effect is due to inhibition or activation of its other targets (Baur and Sinclair, 2006). Thus, as predicted by causal modeling, SIRT1 pharmacological antagonist inhibits NHEK cell differentiation whereas SIRT1 pharmacological agonist trend toward promoting NHEK cell differentiation.
Figure 3. SIRT1 activation and SIRT1 inhibition have opposite effects on keratinocyte differentiation.
(a) NHEK cells treated with growth media (Control); nicotinamide (NAA) (8 mm), a SIRT1 inhibitor; DMSO, a control for resveratrol; resveratrol (RES) (3 µm), a SIRT1 activator. Cells were harvested at 80% confluence (80), 100% confluence (100), or 3 days after 100% confluence (+ 3D). A western blot was performed to determine the levels of involucrin and actin. (b and c) Statistical analysis of three biological replicates for NAA- and Res-treated NHEK cells shown in panel a. Panels b and c mean ± SEM for each treatment. NAA treatment reduces normalized involucrin abundance (t-test, P<0.05), and although not meeting the same level of statistical significance, resveratrol treatment trended toward increasing normalized involucrin abundance (t-test, P = 0.058).
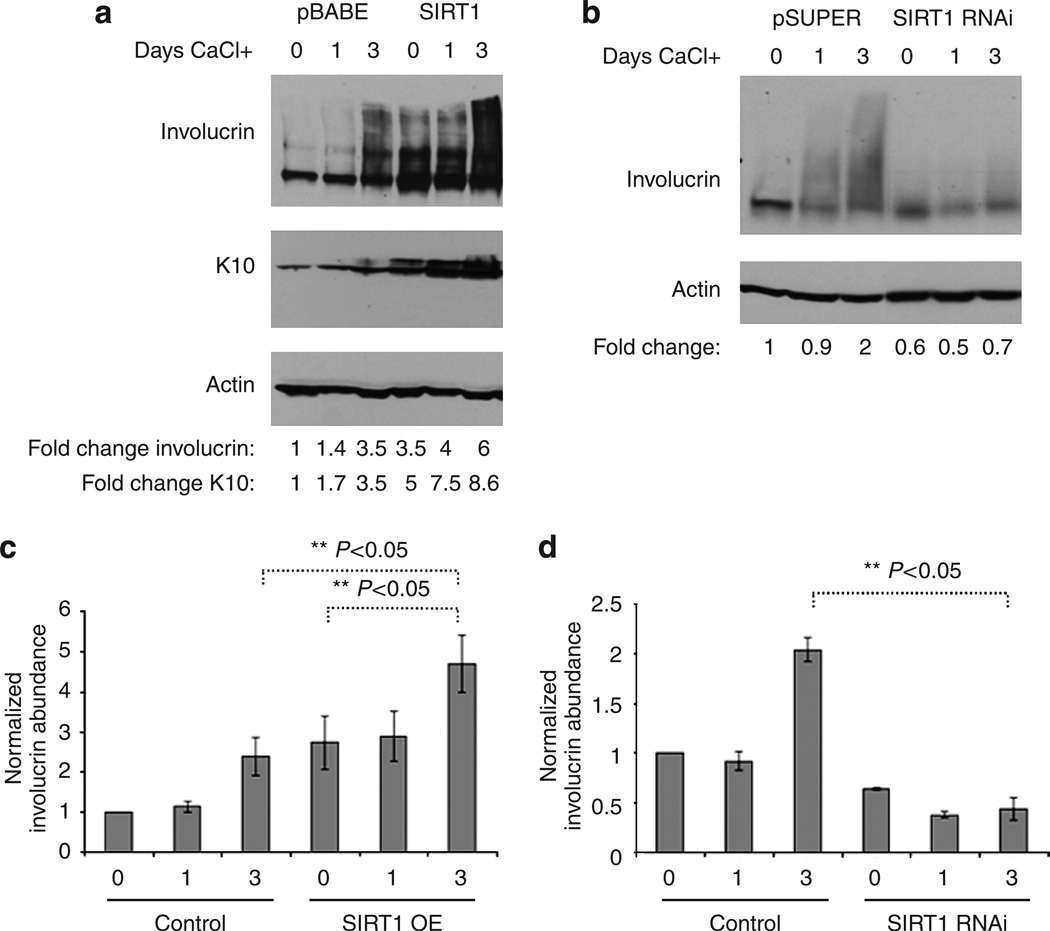
Another cellular process predicted to increase in the SIRT1-overexpressing cells and to decrease in the SIRT1 knockdown cells is calcium-mediated signal transduction, a potent inducer of keratinocyte differentiation (Sharpe et al., 1989). To test the role of SIRT1 in calcium (Ca2+)-dependent NHEK cell differentiation, SIRT1-overexpressing and SIRT1 knockdown cells were treated with calcium and assayed by measuring keratinocyte differentiation markers. Western blots of these cells before Ca2+ addition, 1 day after Ca2+ addition, and 3 days after Ca2+ addition revealed that the cells overexpressing SIRT1 expressed higher levels of crosslinked involucrin than did their controls (Figure 4a). The quantification of involucrin in the SIRT1-overexpressing cells revealed between 1-and 3.5-fold increase at day 0, 1.5- to 3-fold increase at day 1, and 3- to 5-fold increase at day 3 (Figure 4a and c). Additionally, SIRT1 overexpression had an additive effect with calcium in this experiment (Figure 4a, compare SIRT1 overexpression (OE) day 0 with day 3). Conversely, the SIRT1 knockdown (SIRT1 RNAi-infected) cells had less cross-linked involucrin compared with the RNAi controls, which indicates less cell differentiation (Figure 4b). The quantification of involucrin levels in the SIRT1 RNAi cells revealed a 2-fold decrease at day 0, a 1.5- to 3-fold decrease at day 1, and a 2.5- to 7-fold decrease at day 3 (Figure 4b and d). Thus, these biochemical findings confirmed that calcium-dependent signaling is related to SIRT1 function, and that reducing SIRT1 abundance inhibits Ca2+-mediated signals controlling cell differentiation.
Figure 4. SIRT1 upregulation induces and SIRT1 downregulation inhibits calcium-dependant keratinocyte differentiation.
(a) NHEK cells infected with control virus (pBABE) or SIRT1 overexpression virus (SIRT1) were harvested before the CaCl2 addition (0), 1 day after the addition (1), and 3 days after addition (3). Proteins were extracted, and a western blot was run to detect involucrin, K10, and actin. A representative blot from three different experiments is shown. (b) NHEK cells infected with control virus (pSUPER) or SIRT1 RNAi virus (SIRT1 RNAi) were harvested before the CaCl addition (0), 1 day after the addition (1), and 3 days after the addition of 1.6 mm Ca2+ (3). Proteins were extracted, and a western blot was run to detect involucrin and actin. A representative blot from three different NHEK experiments is shown. (c and d) Statistical analysis of three biological replicates for SIRT1 overexpression and RNAi, respectively. Panels c and d show mean ± SEM for each treatment. SIRT1 overexpression increases normalized involucrin abundance (t-test, P<0.05). The combination of calcium and SIRT1 further increases the level of normalized involucrin abundance (t-test, P<0.05). Panel d shows that SIRT1 RNAi has the opposite effect (t-test, P<0.05).
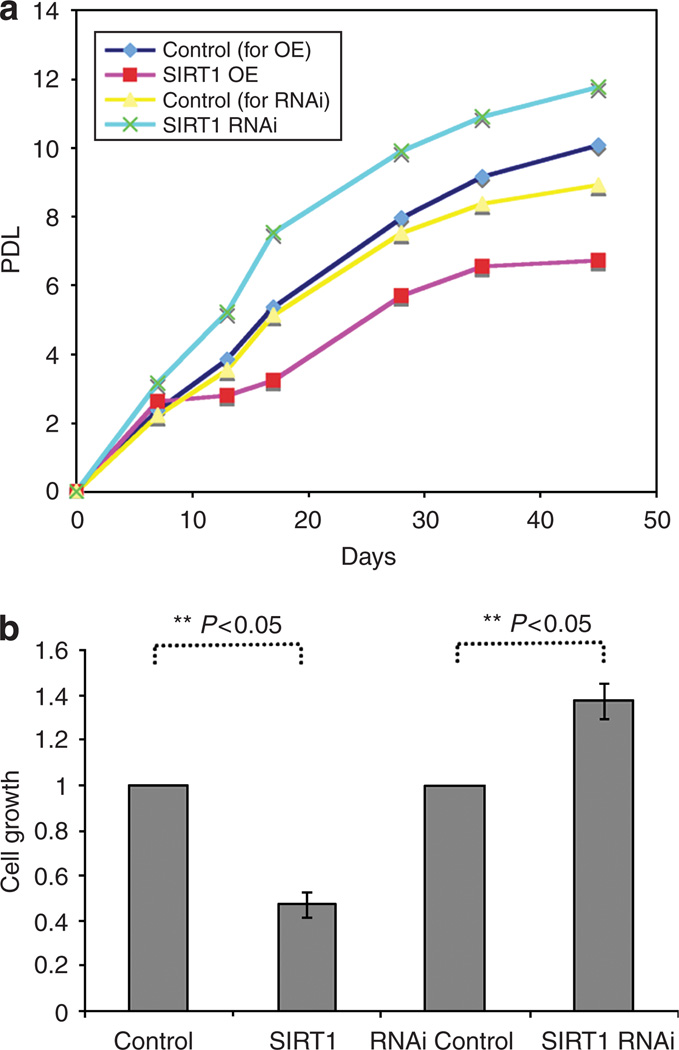
Finally, the role of SIRT1 in cell proliferation was examined. The proliferation rates of NHEK cells overexpressing or underexpressing SIRT1 were compared with negative control NHEK cells. As shown in Figure 5a, in a span of 45 days, the SIRT1-overexpressing NHEK cells went through only 7.2 population doublings (PDs) whereas their control cells went through 10.1 PDs; in the same length of time, the NHEK cells infected with the SIRT1 RNAi underwent 12.4 PDs whereas their control cells went through 9.3 PDs. Statistical analysis of five replicates at day 5 supports this conclusion (Figure 5b). Thus, as inferred by causal network modeling, SIRT1 limits the rate of human keratinocyte proliferation.
Figure 5. SIRT1 downregulation increases and SIRT1 upregulation decreases the replicative lifespan of human keratinocytes in culture.
(a) Example lifespan of NHEK cells overexpressing or underexpressing SIRT1 is shown. After infection and selection, cells were counted and were seeded for the lifespan experiment. PD (population doubling) was determined by the formula: PD = (log Nf−log Ni)/log2, where Nf is the number of cells counted and Ni the number of cells seeded. (b) Statistical analysis of five biological replicates (mean ± SEM) for SIRT1 overexpression and RNAi, respectively, indicates that the two treatments have opposite effects on cell growth (t-test, P<0.05 for each).
DISCUSSION
Sir2 homologs are known regulators of aging, structurally conserved from yeast to humans (for review see Blander and Guarente, 2004). Although it is known that SIRT1, the human Sir2 ortholog, controls differentiation in adult mammalian tissues such as muscle and white adipose, it has been less clear how SIRT1 functions in the maintenance of skin. In the present study, we characterize by gene expression microarray the molecular signaling networks occurring in primary keratinocytes experimentally manipulated to either overexpress or underexpress SIRT1. Such an approach generates a large gene expression change data set calling for a systems level analysis. Causal modeling uses artificial intelligence algorithms to generate hypotheses relevant to the observed microarray data. In this study, we followed-up these hypotheses by testing them in cultured human epidermal keratinocytes. We demonstrated in vitro that SIRT1 controls keratinocyte proliferation and differentiation, and that calcium-induced keratinocyte differentiation requires SIRT1.
Using causal modeling of the gene expression microarray data, we inferred that SIRT1 inhibits keratinocyte proliferation. To test this cell proliferation hypothesis, we measured the replication capacity of the SIRT1 overexpression and SIRT1 knockdown keratinocytes. Reducing SIRT1 expression in keratinocytes increased their proliferation rate, whereas overexpressing SIRT1 inhibited proliferation (Figure 5a and b). Consistent with the causal model for SIRT1 function developed in this study, SIRT1 has recently been shown to bind E2F1 and to inhibit E2F1 transactivation activity (Wang et al., 2006). In summary, the SIRT1 effect on keratinocyte proliferation might work via the inhibition of E2F1 activity.
Causal network modeling of the microarray data proposes that SIRT1 overexpression promotes cell differentiation and SIRT1 knockdown inhibits cell differentiation. To test the cell differentiation hypothesis, we have measured the differentiation dynamics of keratinocytes overexpressing or underexpressing SIRT1 by assaying for keratinocyte differentiation markers. Indeed, SIRT1 overexpression promotes keratinocyte differentiation and SIRT1 knockdown blocks keratinocyte differentiation. Similar effects are seen in keratinocytes treated with nicotinamide, a SIRT1 antagonist, and polyphenol resveratrol, a putative SIRT1 agonist. Interestingly, other polyphenols such as epigallocatechin-3-gallate also appear to increase human keratinocyte differentiation (Hsu et al., 2003; Eckert et al., 2006). In conclusion, SIRT1 induces keratinocyte differentiation, as predicted by causal network modeling.
The effect of SIRT1 on keratinocytes differentiation is the opposite of what is observed in muscle and fat cells. SIRT1 overexpression in muscle and fat cells actually inhibits differentiation (Fulco et al., 2003; Picard et al., 2004). Again, this may reflect fundamental differences in the physiology of these tissues. For example, SIRT1 in keratinocytes may regulate a different set of transcription factors or cofactors in the skin compared with muscle and fat cells. SIRT1 inhibits adipocyte differentiation by repressing PPARG by interacting with NCOR1 (nuclear receptor co-repressor 1) (Picard et al., 2004). One possibility is that adipocytes and keratinocytes express different PPARG cofactors. In the keratinocytes, SIRT1 might activate PPARG by binding and activating CEBPA, a PPARG agonist. Consistent with this, SIRT1 binds and regulates CEBPA function (Qiao and Shao, 2006). Another possibility is that in the keratinocytes SIRT1 might activate PPARG by direct deacetylation.
Increased intracellular Ca2+ is a known promoter of keratinocyte differentiation both in vivo and in vitro (Tu et al., 2004). Causal network modeling identified increased Ca2+ as a hypothesis in the SIRT1-overexpressing cells and decreased Ca2+ as a hypothesis in the SIRT1 knockdown cells. To test the Ca2+ hypothesis, we measured the differentiation capacity of cells with higher and lower SIRT1 protein levels and found that calcium-induced keratinocyte differentiation was all but entirely blocked in SIRT1 knockdown cells and activated in SIRT1-overexpressing cells. In fact, SIRT1 overexpression itself was sufficient to trigger keratinocyte differentiation. This suggests that SIRT1 is an important effector of intracellular Ca2+mediated signals controlling differentiation. However, it cannot be excluded based on this experimental evidence that Ca2+ and SIRT1 may also have some independent regulatory functions controlling the differentiation of primary keratinocytes.
In conclusion, causal network modeling of gene expression microarray data followed by experimental validation provides evidence that SIRT1 promotes keratinocyte differentiation acting downstream of Ca2+. Additionally, SIRT1 negatively regulates human keratinocyte proliferation, which might work via E2F1 inhibition. Our findings have implications for understanding the maintenance of the skin in healthy individuals, and may be important to the understanding of skin diseases characterized by hyperproliferation and under-differentiation, such as psoriasis.
MATERIALS AND METHODS
Cells
NHEK cells obtained from Cascade Biologics (Portland, OR), and maintained under standard conditions in Epilife Medium with Ca2+ (Cascade Biologics) and supplemented with human keratinocyte growth supplement (Cascade Biologics).
Plasmids
SIRT1 RNAi plasmids constructed using pSUPER-RETRO (Oligo Engine, Seattle, WA) using the SIRT1 siRNA sequence GGAAATATA TCCTGGACAA. SIRT1 was overexpressed using pBabePuro.
Infection
NHEK cells were infected for 3–6 hours and selected with puromycin (0.5 µm) for 3 days.
Differentiation assay (Ca2+)
Control cells were incubated without additional Ca2+ (0.00065 mm supplied in the media), whereas the treated cells were incubated with 1.6 mm Ca2+, samples were harvested at days 0, 1, and 3. The experiment was run in triplicate.
Differentiation assay (confluency) and treatment with SIRT1 inhibitor and activator
The concentration of nicotinamide in the medium was 8 mm and the concentration of resveratrol in the medium was 3 µm (DMSO used as a solvent control). For each treatment, one plate was harvested at 80% confluency, another at 100% confluency, and the last 3 days after 100% confluency. The experiment was run in triplicate.
Western blot analysis
The harvested NHEK cells were lysed in protein sample buffer and separated by SDS-PAGE, transferred to nitrocellulose, and incubated as indicated (SIRT1 polyclonal antibody (custom-made in laboratory), involucrin mAb (Neomarkers, Fremont, CA), actin C-4 antibody (Sigma, St Louis, MO) or a KRT10 antibody (Labvision, Fremont, CA)). The densitometric analysis was performed on scanned images of western blots using ImageJ software (NIH Image analysis website http://rsb.info.nih.gov/ij/). Quantification provided as the ratio to actin abundance as a normalization control.
Cellular growth
For each condition tested, NHEK cells were seeded at a 200,000 cells per 6 cm plate. Upon reaching 80% confluency, these cells were counted and then split. PD was determined by the formula: PD = (log Nf−log Ni)/log2, where Nf is the number of cells counted and Ni the number of cells seeded. Two long-term trials and four short-term trials were conducted.
GeneChip hybridizations
Three replicates of each sample (SIRT1 overexpression, vector control, SIRT1 RNAi, and RNAi control) were hybridized to GeneChip arrays (HG-U133A_2, Affymetrix, Santa Clara, CA). Hybridizations and scanning were carried out at the MIT BioMicro Center.
Image and data analysis
To enable direct GeneChip comparisons, data were normalized using RMA (Irizarry et al., 2003). Fold changes were calculated by dividing the average intensity values of the experimental samples (either RNAi or overexpression) by the average of the reference control samples.
Causal network modeling (automated hypothesis generation using reverse causal analysis)
Reverse causal reasoning interrogates the human knowledge assembly model to identify upstream controllers (hypotheses) for the transcriptional changes observed by microarray analysis (RNA state changes). Each hypothesis is scored according to two probabilistic scoring metrics, richness, and concordance, which examine distinct aspects of the probability of a hypothetical cause explaining a given number of RNA state changes. Richness is the probability that the number of observed RNA state changes connected to a given hypothesis could have occurred by chance alone. Concordance is the probability that the number of observed RNA state changes that match the directionality of the hypothesis (e.g., increased or decreased kinase activity for a kinase and increased or decreased transcriptional activity for a transcription factor) could have occurred by chance alone. A hypothesis is considered to be statistically significant if it meets richness and concordance cutoffs of P<0.05. Automated reverse causal reasoning on the SIRT1 overexpression and SIRT1 knockdown data yielded 15 and 74 statistically significant hypotheses, respectively, which were further investigated and prioritized by evaluation of their biological relevance to the experimental context. Causal links between biologically relevant hypotheses were identified and integrated into a causal network model for the role of SIRT1 in primary keratinocytes.
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (L.G) and an Estee Lauder Fellowship (G.B). We acknowledge Dr Rebecca Fry for assistance and advice on analytical processing for the microarray data. We also thank Danica Chen, Alina Berdichevsky, and Angeliki Chalkiadaki for stimulating discussions and helpful advice and Adair Swain for graphic art support.
Abbreviations
- Ca2+
calcium
- CEBPA
CCAAT/enhancer-binding protein alpha
- NAA
nicotinamide
- NHEK
normal human primary keratinocytes
- PD
population doubling
- PPARG
peroxisome proliferator-activated receptor gamma
- RNAi
RNA interference
- SIRT1
Silent mating type information regulation 2 homolog 1
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest. Mary S. Matsui, Daniel Maes and Thomas Mammone are full-time employees of the Estee Lauder Company.
SUPPLEMENTARY MATERIAL
Figure S1. Causal model for SIRT1-induced keratinocyte differentiation.
Figure S2. Causal model for SIRT1-induced keratinocyte differentiation.
Figure S3. Cell cycle hypothesis in the SIRT1 RNAi cells. As in Figure 2.
REFERENCES
- Bai L, Pang WJ, Yang YJ, Yang GS. Modulation of Sirt1 by resveratrol and nicotinamide alters proliferation and differentiation of pig preadipocytes. Mol Cell Biochem. 2008;307:129–140. doi: 10.1007/s11010-007-9592-5. [DOI] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;6:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert RL, Crish JF, Efimova T, Balasubramanian S. Opposing action of curcumin and green tea polyphenol in human keratinocytes. Mol Nutr Food Res. 2006;50:123–129. doi: 10.1002/mnfr.200500125. [DOI] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Hsu S, Bollag WB, Lewis J, Huang Q, Singh B, Sharaway M, et al. Green tea polyphenols induce differentiation and proliferation in epidermal keratinocytes. J Pharm Exp Ther. 2003;306:29–34. doi: 10.1124/jpet.103.049734. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L, Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein alpha transcriptional complex. J Biol Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe GR, Gillespie JI, Greenwell JR. An increase in intracellular free calcium is an early event during differentiation of cultured human keratinocytes. FEBS Lett. 1989;254:25–28. doi: 10.1016/0014-5793(89)81002-6. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tu CL, Oda Y, Komuves L, Bikle DD. The role of the calcium-ensing receptor in epidermal differentiation. Cell Calcium. 2004;35:265–273. doi: 10.1016/j.ceca.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C.elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]