Abstract
Successful completion of diverse cellular functions, such as mitosis, positioning organelles, and assembling cilia, depends on the proper assembly of microtubule-based structures. While essentially all of the proteins needed to assemble these structures are now known, we cannot explain how even basic features such as size and shape are determined. As steps towards filling this knowledge gap, there have been several recent efforts towards reconstituting, with purified proteins, the basic structural motifs that recur in diverse cytoskeletal arrays. We discuss these studies and highlight how they shed light on the self-organized assembly of complex and dynamic cytoskeleton-based cellular structures.
Introduction
Microtubules are polar polymers of αβ-tubulin heterodimers required for directional transport and micro-mechanical functions in eukaryotic cells. In dividing cells, a complex and dynamic array of microtubules, called the bipolar spindle, is needed to segregate chromosomes and position the cell division plane (Glotzer, 2009; Wittmann et al., 2001). In non-dividing cells, microtubules are needed for a wide range of processes, including positioning organelles (Carazo-Salas and Nurse, 2006), polarized growth (Wasteneys and Ambrose, 2009), migration (Kaverina and Straube, 2011; Watanabe et al., 2005), and the assembly of flagella and cilia (Ishikawa and Marshall, 2011). In differentiated cells, such as neurons, organized microtubule bundles provide tracks for the intracellular transport needed for axonal growth (Conde and Caceres, 2009; Stiess and Bradke, 2011). Microtubules also contribute to the polarized secretion needed for cell-cell interactions at the immunological synapse (Rey et al., 2007). In all these contexts, whether the microtubules provide tracks for transport, cues to organize the cytoplasm, or to generate force, it is crucial that these dynamic polymers are precisely organized.
Essentially all the different microtubule associated proteins (MAPs) needed to assemble microtubule-based structures have now been identified and can be divided into four groups. The first group consists of motor proteins (e.g. kinesins and dynein) that use the energy from ATP hydrolysis to step along microtubule tracks to transport cargo (Vale, 2003). The second group includes crosslinking proteins that align filaments with a specific geometry and stabilize structures (Bratman and Chang, 2008; Peterman and Scholey, 2009). The third set comprises proteins that modulate microtubule number. These include regulators of nucleation (Kollman et al., 2011; Luders and Stearns, 2007) and enzymes that sever pre-existing filaments (Roll-Mecak and McNally, 2010; Sharp and Ross, 2012). The fourth group consists of regulators of dynamic instability, a characteristic property of microtubules that involves abrupt switch-like transitions between periods of assembly and disassembly from filament ends (Desai and Mitchison, 1997; Howard and Hyman, 2009). These proteins can regulate different parameters of dynamic instability, such as the rates of tubulin assembly, or the frequencies of catastrophe (i.e. the switching from assembly to disassembly) and rescue (i.e. the transitions from disassembly to growth). Different combinations of MAPs from these four groups somehow work together to assemble microtubule arrays involved in diverse cellular processes.
While the discovery of these MAPs represents major advances in the field, we still cannot explain how the size and shape of different microtubule-based architectures, whose dimensions are orders of magnitude greater than that of the molecules involved, are determined. It is also becoming clear that the rules that explain the assembly of large multi-component well-ordered structures, such as the ribosome, may not directly apply to the assembly of microtubule arrays, such as the bipolar spindle, as these structures can be highly dynamic and, irregular in composition and molecular contacts. Therefore, while understanding the stereospecific interactions between the key molecular components is important, it is not sufficient to shed light on how the size or the shape of micron-scale microtubule arrays is determined. To answer these questions, we believe that an important step is to determine and characterize the smallest set of components needed to build key structural motifs, such as aligned microtubule bundles or asters, recurring in the diverse microtubule arrays needed for cellular function (Figure 1). A useful framework for the assembly of these structures is self-organization on the micron length scale (Mitchison, 1992; Nedelec et al., 2003). In such models, these dynamic architectures are believed to emerge from the activities of numerous proteins that follow simple rules and respond to local cues, which could be chemical or mechanical. Self-organization does not depend on a blueprint or a master organizer and is distinct from self-assembly as it requires energy input.
Figure 1. Schematic representation of the microtubule arrangements observed in different filament arrays.
(A) Microtubule organization in a neuron. Insets show magnified views of the arrangement of aligned microtubules in the axon and dendrites. (B) Bipolar metaphase spindle in mitotic cells. Inset shows a magnified view of microtubule arrangement in an aster at the spindle pole. (C) Anaphase spindle formed in mitotic cells. (D) Interphase microtubule assemblies in fission yeast. (E) Cortical microtubule bundles in the cell wall of plants. (Schematics are not drawn to scale)
A structural motif comprised of two aligned microtubules that overlap is found in several different arrays and contributes to the dynamic organization of the cytoplasm in at least two ways. First, it can generate forces to drive intracellular movement. For example, overlapping pairs of microtubules with antiparallel orientations can push centrosomes apart at the start of cell division (Tanenbaum and Medema, 2010; Wittmann et al., 2001). Second, these arrays can encode intracellular position. For example, the overlap region in an antiparallel array can recruit proteins to ‘mark’ the midpoint between segregating anaphase chromosomes (Glotzer, 2009). Such ‘marks’ are proposed to help position the site of cell cleavage. To carry out cellular functions with fidelity the overall length and the overlap length of aligned microtubule bundles must be precisely controlled.
In this review, we highlight recent advances in our understanding of how size and shape of microtubule-based structures are determined. To frame our discussion of important recent findings relating to motor and non-motor MAPs, we focus on the self-organized assembly of aligned bundles of two overlapping microtubules. The parameters that describe this motif are the overall end-to-end distance of the aligned microtubules and the overlap length between the two filaments. We discuss reconstitution studies with purified proteins and structural analyses that reveal how motor and non-motor proteins set these parameters by regulating the relative position of two microtubules, the lengths of filaments, and the extent of filament overlap. We also discuss how these findings inform on the organization of the microtubule aster, another basic building block. Finally, we highlight some of the similarities and differences between prokaryotic and eukaryotic cytoskeletons.
Controlling the relative position of two microtubules
Can a two-component system, composed of microtubules and a crosslinking motor protein, align two microtubules such that the size of this simple motif is controlled? Turns out, this is not the case. When two anti-parallel microtubules are crosslinked by a motor protein, the relative displacement of the filaments will continue until the motor protein reaches the end of the track, and the overlap between the two microtubules will be lost (Figure 2A). This is exactly what is observed in TIRF microscopy-based assays examining relative microtubule sliding by widely conserved kinesins required for eukaryotic cell division. Kinesin-5, the plus-end directed motor needed for bipolar spindle assembly, and kinesin-14, the minus-end directed motor needed to properly organize the two ends (called ‘poles’) of the bipolar spindle, have been shown to slide two antiparallel microtubules apart (Braun et al., 2009; Fink et al., 2009; Kapitein et al., 2005). However, the action of these proteins alone cannot keep microtubules aligned with different overlap lengths.
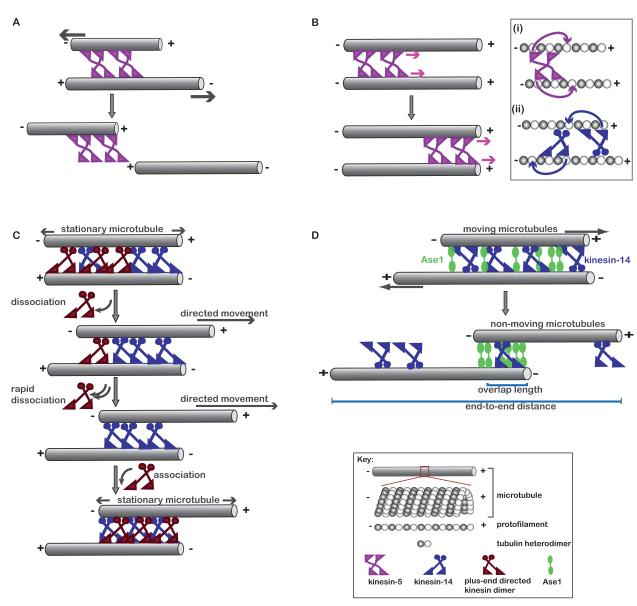
Figure 2. Formation of aligned microtubule bundles by motor and non-motor crosslinking proteins.
(A) Schematic shows how crosslinking and sliding of anti-parallel microtubules (grey) by kinesin-5 (pink) can completely separate the two filaments. (B) Movement of kinesin-5 between two parallel microtubules does not result in relative displacement of the filaments. Inset shows the stepping of (i) kinesin-5 (pink) and (ii) kinesin-14 (blue) motor proteins on two parallel microtubules. (C) A model for bi-directional oscillations of anti-parallel microtubules crosslinked by two antagonistic motor proteins (plus-end directed motor protein, red; minus-end directed motor protein, blue). (D) Proposed mechanism by which the non-motor crosslinking protein, Ase1 (green), and the motor protein, kinesin-14 (blue), generate anti-parallel microtubule overlap of fixed length.
Analysis of two parallel microtubules crosslinked by a motor protein reveals that the filaments do not move relative to each other, and therefore, the size of this simple array cannot be modulated by protein activity (Figure 2B). Why is this? For kinesin-5, which is a homotetramer that crosslinks two microtubules by binding each filament with a pair of motor domains, the stepping of the motor domains towards the plus-ends of each of the parallel filaments results in the motion of the kinesin itself, but no relative movement of the microtubules (Figure 2B, inset (i)). Parallel filaments do not move when crosslinked by kinesin-14, which uses a pair of motor domains to walk on one microtubule and a non-motor domain to interact with the second filament. To crosslink two microtubules, kinesin-14 orients stochastically and its motor domains are equally likely to bind either of the two filaments. Therefore, as shown in inset (ii) of Figure 2B, the stepping of one kinesin-14 molecule towards the minus-end of lower filament opposes another kinesin-14 molecule stepping on the top filament, thereby preventing relative filament motion. While the ability of other motor proteins to slide apart parallel filaments still needs to be examined, the findings thus far indicate that another molecular component is needed to stably align microtubule bundles and build a structure of fixed length.
In principle, the additional component needed to control the size of an aligned microtubule pair could be another motor protein. This minimal system would involve one motor protein that walks towards the microtubule plus-end and another that walks towards the minus-end. The antagonizing activities of these two different motor proteins could be balanced by tuning relative concentrations. Thus far, thorough experimental tests of this hypothesis have been carried out with the plus-end motor protein, kinesin-5, and the minus-end motor protein, kinesin-14 (Hentrich and Surrey, 2010; Tao et al., 2006). These experiments, as well as computer simulations, have led to the conclusion that a persistent balance of forces is not achieved by these two motors. Instead, bi-directional oscillations are observed in which microtubules moves back and forth, frequently pausing between reversals of direction (Civelekoglu-Scholey et al., 2010; Hentrich and Surrey, 2010; Tao et al., 2006). The same behavior has also observed with another pair of antagonistic motor proteins, kinesin-1 and dynein (Vale et al., 1992). To understand this phenomenon, one has to consider both the stochastic fluctuations in motor protein numbers during sliding and the force-dependent detachment of motors from microtubules. Briefly, when a balance in opposing activities is achieved the numbers of motor proteins will fluctuate due to binding and unbinding kinetics (Figure 2C). If the number of plus-end directed kinesins decreases, the force balance will tip and increase the dissociation of additional molecules of the same kinesin due to higher opposing force. The filaments will thus move relative to each other and eventually, re-association of the plus-end directed kinesin will be favored and the force balance will be restored (Figure 2C). However, this is only transient, as the same events will disrupt the balance and result in oscillations. Our current understanding of these systems indicates that a three-component system comprising of microtubules and opposing motor proteins is also not sufficient to generate aligned microtubules that have fixed overlap and overall length.
Another possible three component system that can regulate the size of these basic building blocks is one comprised of microtubules, a motor protein, and a non-motor crosslinking protein. Compared to motor proteins, non-motor crosslinking proteins are a poorly understood class of MAPs. Recently, important roles for these proteins in the organization of overlapping microtubule arrays have been suggested from examining the PRC1/Ase1/MAP65 family of microtubule crosslinking proteins. These proteins play key roles in organizing microtubules in a variety of different cellular contexts, such as directing cell growth in plants, nuclear positioning in yeast, and cell division in all eukaryotes. Members of this protein family are characterized by their ability to selectively crosslink microtubules in an anti-parallel orientation (Bieling et al., 2010; Janson et al., 2007; Kapitein et al., 2008; Subramanian et al., 2010). Cellular studies of their function suggest that these proteins participate in the organization of anti-parallel microtubules in conjunction with a number of different motor proteins. In particular, organization of interphase microtubule arrays in fission yeast has suggested a model in which Ase1, the fission yeast PRC1, counteracts kinesin-14’s activity to control microtubule overlap (Janson et al., 2007). Importantly, in vitro studies of this 3-component system, comprising of microtubules, recombinant Ase1, and kinesin-14, are consistent with this model (Braun et al., 2011). Ase1, like other PRC1 homologs, binds with high affinity to anti-parallel microtubule overlap regions. TIRF-imaging assays revealed that when kinesin-14 slides apart a pair of anti-parallel microtubules crosslinked by Ase1, the density of Ase1 in the overlap increases as overlap length decreases. The accumulated Ase1 opposes motor protein-driven sliding and stalls the relative sliding of the two microtubules (Figure-2d). Under the experimental conditions of this study, stalling occurs when the ratio of Ase1:kinesin-14 in the anti-parallel overlap exceeds 4:1 (Braun et al., 2011). These findings indicate that a 3-component system can align two microtubules such the overlap length can be controlled by relative amounts of Ase1 and kinesin-14 in the overlap region. Once the overlap length is set, the end-to-end distance of the aligned filament array is simply determined by the lengths of the two microtubules and the overlap between them.
Is this a general mechanism for the organization of microtubule arrays by the PRC1 family of MAPs? The answer to this question appears to be no. In contrast to Ase1, when human PRC1-crosslinked anti-parallel microtubules are moved apart by kinesin-5, PRC1 density in the overlap region does not increase as overlap length reduces. Instead, the two microtubules completely separate, as is observed with the motor protein alone (Subramanian et al., 2010). Why these differences are observed between the human and yeast homolog of PRC1 is an open question. It is possible that this is due to the multimerization of Ase1 under some conditions, a property not observed for PRC1 (Kapitein et al., 2008). Alternatively, Ase1/PRC1 crosslinks may respond differently when filaments are moved by the two different motor proteins, kinesin-5 and kinesin-14. While this needs to be examined further, it is possible that these differences are adaptations for Ase1/PRC1’s cellular functions. In budding yeast, the sliding of Ase1-crosslinked microtubules by kinesin-5 motors is required for the elongation of the anti-parallel microtubules of the spindle at anaphase. Therefore, it may be advantageous for Ase1 to not act as a ‘brake’ against kinesin-5 (Khmelinskii et al., 2009). However, in the formation of overlapped bundles in the interphase cells of fission yeast, the same non-motor crosslinker is paired with kinesin-14, which it can effectively stall (Carazo-Salas and Nurse, 2006; Janson et al., 2007). PRC1/Ase1/MAP65 proteins also function together with other motor proteins, such as kinesin-6 (or Mklp1) (Fu et al., 2009; Gruneberg et al., 2006), kinesin-7 (or Cenp-E) (Kurasawa et al., 2004), kinesin-4 (or Kif4A) (Kurasawa et al., 2004; Zhu and Jiang, 2005). Interestingly, analysis of how PRC1 and kinesin-4 regulate the assembly of anti-parallel arrays (see below) has revealed a very different mechanism than what has been proposed for Ase1 and kinesin-14. Additional studies will help unravel the different ways by which motor and non-motor proteins can control the size of a basic structural motif comprised of two microtubules.
It is helpful to consider the biophysical basis of how a combination of motor and non-motor proteins may control the relative sliding of microtubules to determine size. The magnitude of friction forces due to a crosslinking non-motor protein, as would be needed to oppose a kinesin that slides two microtubules apart, is proportional to: (1) its frictional coefficient, (2) filament velocity, and (3) number of molecules. The frictional coefficient is inversely proportional to the coefficient for 1-D diffusion of the MAP on the microtubule lattice. For human PRC1, estimates of frictional force based on measured diffusion coefficients suggest that over 100 PRC1 molecules are needed to counteract the force generated by kinesin-5 moving at ~20 nm/s. As the frictional force is proportional to velocity, its magnitude will decrease as the relative filament sliding velocity reduces (Bormuth et al., 2009). Therefore, to further slow-down the sliding of crosslinked filaments, additional non-motor molecules must somehow accumulate. Further, the extent to which a motor protein will slow down due to opposing force will depend on the motor protein’s ‘force-velocity’ relationship. For example, the velocity of kinesin-5 is not greatly reduced over a wide range of opposing loads, while other kinesins, such as kinesin-1, exhibit steep dependencies in the same range (Svoboda and Block, 1994; Valentine et al., 2006). Consequently, all else being similar, kinesin-5 is less likely to stall compared to kinesin-1. While force generation by motor proteins has been carefully examined, the frictional forces generated by non-motor proteins remain poorly characterized and additional work is needed to understand how these proteins may function as ‘brakes’ in the organization of microtubules into aligned bundles of fixed length
Regulating microtubules length
Are the ‘slide and stall’ mechanisms sufficient to control the length of simple microtubule-based motifs in cells? The in vitro experiments discussed so far are typically performed with taxol-stabilized microtubules and do not take into consideration microtubule dynamics. Microtubules exhibit dynamic instability and individual polymers continuously grow or shrink in the presence of GTP (Figure 3A) (Desai and Mitchison, 1997). Therefore, even when a stable overlap is achieved by crosslinking proteins, microtubule length will continuously fluctuate. This may also result in the separation of the two microtubules. It is therefore not surprising that the length of microtubule-based structures depend on proteins that regulate microtubule polymerization.
Figure 3. Regulation of microtubule length.
(A) Schematic of microtubule dynamic instability. (B-E) Proposed models for the regulation of microtubule dynamics by motor and non-motor proteins. Microtubule destabilization by the motor proteins kinesin-13 (B) and kinesin-8 (C). Enhancement of microtubule growth by the TOG-domain containing proteins XMAP215 (D) and CLASP (E).
Microtubule destabilization by motor proteins
While kinesins are best known for their function in transporting cargoes along microtubules, widely conserved members of this superfamily directly regulate the depolymerization of the microtubule tracks, harnessing the energy from ATP hydrolysis to disassemble microtubules. Here we discuss insights into the functions of these atypical motor proteins that have come from studies of MCAK, a kinesin-13, and Kip3, the yeast kinesin-8. Both of these proteins have important functions during cell division. Kinesin-13 plays a role in determining microtubule lengths in the mitotic spindle and chromosome-microtubule attachment (Wordeman, 2005). Kinesin-8 is required for proper chromosome alignment to the spindle equator during metaphase (Gardner et al., 2008; Su et al., 2011).
TIRF-microscopy analysis with recombinant proteins and dynamic microtubules shows that both kinesin-13 (Helenius et al., 2006; Hunter et al., 2003) and kinesin-8 (Gupta et al., 2006; Varga et al., 2006) accumulate at the ends of dynamic microtubules, where they increase catastrophe frequency (Figures 3B and 3C). How do these proteins find microtubule ends? It turns out that these motor proteins use two different mechanisms. Kinesin-8 walks to the plus-end of a microtubule and specifically destabilizes this end (Figure 3C) (Gupta et al., 2006; Varga et al., 2006). In contrast, kinesin-13 finds microtubule ends by 1-D diffusion along the filament lattice and can destabilize either end (Figure 3B) (Helenius et al., 2006; Hunter et al., 2003). Recently, Patronin and Microspherule protein 1 (MCRS1) as have been identified as putative suppressors of kinesin-13 microtubule depolymerization activity at the microtubule minus-end, thereby allowing control over kinesin-13’s microtubule-end specificity (Goodwin and Vale, 2010; Meunier and Vernos, 2011).
Interestingly, kinesin-8 can destabilize longer microtubules faster than shorter microtubules. How is this achieved? It has been shown that kinesin-8 is a processive motor protein with a low dissociation rate from microtubules (Varga et al., 2006). As a consequence, most of the kinesin-8 molecules that land on a microtubule have a high probability of getting to the end of that filament. Longer microtubules have more binding sites and will accumulate more kinesin-8 than shorter microtubules. Therefore, the plus-ends of longer filaments will accumulate more kinesin-8. In current models, co-operative interactions between kinesin-8 molecules at microtubule ends physically ‘bump-off’ a kinesin-8 bound to a tubulin dimer to disassemble the filament (Varga et al., 2009) (Figure-3c). Therefore, more kinesin-8 molecules at the ends of longer filaments lead to faster disassembly of filaments.
What are the advantages of these different length regulation mechanisms for the biological function of these destabilizing kinesins? Indiscriminate and fast depolymerization, as seen with kinesin-13, may be useful under conditions where extensive microtubule depletion is desired. This may be advantageous during mitosis for efficient depolymerization of microtubules that make improper attachments to chromosomes. Selective depolymerization of long microtubules, as seen with kinesin-8, is better suited for processes that require fine control over microtubule length, such as during the alignment of chromosomes at equator of the metaphase spindle during mitosis.
Further insight into the function of these motor proteins has come from structural studies of kinesin-13. Electron micrographs of kinesin-13-bound microtubules show that in the presence of ATP analogs, the tight binding of these kinesins at microtubule ends results in a distortion of the microtubule protofilaments such that they curl and unravel (Desai et al., 1999; Moores et al., 2002). Cryo electron microscopy studies have shown that depolymerizing microtubule ends are associated with increased curvature, which disrupts the lateral interaction between protofilaments (Hyman et al., 1995). Together, these structural studies suggest that disassembly of microtubules is induced by kinesin-13-dependent stabilization of curved protofilaments. A similar mechanism has been proposed for stathmin, a non-motor microtubule destabilizing protein (Belmont et al., 1996; Cassimeris, 2002). The crystal structure of the stathmin bound to tubulin shows that the tubulin dimer adopts a curved conformation in this complex (Gigant et al., 2000; Ravelli et al., 2004). Though kinesin-8 is also speculated to have a similar effect on microtubule ends as kinesin-13, the structural basis of microtubule depolymerization by kinesin-8 is poorly understood (Peters et al., 2010). Currently, it is unclear how the kinesin-8 motor domain distinguishes between tubulin in the middle of the microtubule, where it walks processively, and at filament ends, where it promotes destabilization. How cooperative interactions between kinesin-8 molecules at microtubule ends trigger destabilization also requires structural studies.
Microtubule growth by non-motor microtubule associated proteins
The activities of microtubule destabilizing motor proteins in cells are antagonized by non-motor MAPs that increase microtubule length by promoting polymerization. The best understood among these are the evolutionarily conserved proteins, XMAP215 and CLASP(Orbit/Stu1), which promote microtubule growth in a variety of biological contexts, such as directional growth in plants, cell polarization in neurons and cell division (Al-Bassam and Chang, 2011;Howard and Hyman, 2009).
To promote the addition of tubulin at microtubule ends, XMAP215 and CLASP must (i) bind unpolymerized tubulin, (ii) recruit it to the end of a microtubule, and (iii) unbind only after transferring the tubulin dimer to the growing microtubule. In these structurally related proteins, a series of two to five conserved TOG domains are used to bind unpolymerized tubulin (Al-Bassam and Chang, 2011; Slep and Vale, 2007; Widlund et al., 2011). A basic amino acid-rich domain in these proteins mediates interaction with microtubule lattice to increase the probability of finding filament ends. The final step of tubulin transfer from the TOG domain to the microtubule end requires that these domains discriminate between the subtle structural features in polymerized and unpolymerized tubulin.
How are microtubule ends found by XMAP251 and CLASP? Analysis of the microtubule interactions of XMAP215 at the single molecule level shows that it diffuses in 1-D along the microtubule lattice (Brouhard et al., 2008). At the microtubule plus end, XMAP215 binds with high affinity and tracks the tip of the growing microtubule (Figure 3D). In this end-bound state, it catalyzes multiple (~25) rounds of tubulin addition to the microtubule end (Brouhard et al., 2008). In contrast to XMAP215, CLASP does not autonomously target to microtubule ends. In vitro, it binds uniformly along the microtubule lattice (Al-Bassam et al., 2010). In cells, CLASP piggy-backs on other microtubule binding proteins to localize to distinct subsets of microtubules (Akhmanova et al., 2001; Bratman and Chang, 2008; Mimori-Kiyosue et al., 2005). When a depolymerizing microtubule end encounters a CLASP-bound region within the lattice, the probability that the filament undergoes a rescue event increases (Figure 3E).
How do these TOG-domain containing proteins transfer tubulin to microtubule ends? Insights into this question have come from a recent crystal structure of one of the TOG domains (TOG1) from the yeast XMAP215 homolog, Stu2p, bound to αβ-tubulin (Ayaz et al., 2012). The structure shows that the TOG-domain bound tubulin adopts a ‘curved’ conformation in this complex. This structural form is adopted by tubulin heterodimers in solution or at filament ends but not in the middle of microtubule lattice (Hyman et al., 1995; Lowe et al., 2001; Ravelli et al., 2004). This suggests a model in which the basic amino-acid rich domain aids the interaction of XMAP215 with the microtubule lattice. When this molecule encounters a microtubule end, one of the multiple TOG domains will bind the curved tubulin-heterodimers in this region. This will place the other TOG-domain that is carrying the unpolymerized tubulin close to the growing end. Microtubule assembly will occur by ‘hand-off’ of the TOG-domain-bound soluble tubulin to the filament tip (Figure 3D). Concomitant straightening of the lattice will promote the dissociation of XMAP215 for another round of tubulin addition. While the ‘hand-off’ model is appealing, additional work is needed to obtain direct evidence for tubulin transfer.
While TOG domains are the major regulators of microtubule growth in many cell types, other proteins that promote this reaction have also been identified. Among these is CRMP2, which has important roles in axonal growth in neurons but unlike XMAP215 and CLASP, lacks TOG domains. In CRMP-2, the tubulin binding domain adopts a fold seen in the metabolic enzyme dihydropyrimidinase (Stenmark et al., 2007). The tubulin-CRMP-2 complex is likely transported to filament plus-end by kinesin-1, where tubulin is transferred to the growing microtubule ends (Fukata et al., 2002; Kimura et al., 2005). Whether it does so by discriminating different tubulin structural forms remains to be seen. It is possible that an active transport process ensures a more efficient accumulation of CRMP2 and tubulin to microtubule ends compared to the diffusion-based processes that target the TOG-domain containing proteins to microtubules.
Microtubules undergoing dynamic instability can have broad length distributions that are modulated by MAPs (Gardner et al., 2011; Kinoshita et al., 2001). Thus far, experiments with combinations of growth and catastrophe factors cannot yield microtubules with a narrow length distribution (Kinoshita et al., 2001). Variability in microtubule lengths is advantageous when filaments are utilized for exploring the cellular space, as would be needed for chromosome capture during cell division. However, a more precise control over microtubule length is required when encoding intracellular location, as would be needed to recruit proteins to the midpoint between segregating chromosomes during anaphase. It is becoming clear that this higher precision in controlling microtubule organization requires regulators of microtubule polymerization interacting with MAPs with additional function.
Regulating microtubule overlap length
There have been important advances in our understanding of how the overlap length of two aligned microtubules can be precisely determined, independent of the end-to-end distance of the filaments. In particular, insights into this mechanism have come from analyzing anti-parallel microtubule arrays involving the crosslinking protein, PRC1. Cell biological studies have indicated that PRC1 and kinesin-4 (kif4A/Xklp2) work together to maintain the overlap length of the central spindle, the microtubule array that emerges after chromosome segregation starts. This array contributes to the proper positioning of the site of cell cleavage and keeps the segregated chromosomes apart. A recent study show that PRC1 and kinesin-4 are sufficient to generate such anti-parallel arrays of fixed overlap length in vitro (Bieling et al., 2010). Interestingly, this regulation does not depend on frictional forces due to PRC1 stalling kinesin-drive filament sliding. Instead, PRC1 acts as a ‘tag’ for the microtubule overlap region and recruits kinesin-4, a motor protein that regulates polymerization dynamics.
How is the regulation of microtubule overlap length achieved by these two MAPs? In vitro analysis of microtubule dynamics in the presence of kinesin-4 shows that it acts as a suppressor of growth and catastrophe. However, unlike the other regulators of microtubule dynamics such as kinesin-8 or XMAP215 that act in single microtubules, the activity of kinesin-4 is confined by PRC1 to filament plus-ends that are part of an anti-parallel array. Kinesin-4 alone has weak microtubule binding affinity and is recruited by PRC1 to regions of overlapping anti-parallel microtubules (Figure 4) (Bieling et al., 2010). The motor protein then walks processively to microtubule ends in the overlap region and acts as a suppressor of microtubule polymerization dynamics (Figure 4) (Bieling et al., 2010). The final steady-state length of anti-parallel microtubule overlap in this system is dependent on kinesin-4 concentration and initial length of the overlap (Bieling et al., 2010). Longer overlap regions recruit more kinesin-4. This increases the probability with which the motor protein will arrive at the plus-ends of longer microtubules, for faster inhibition of filament dynamics. The overlap length of microtubules in this aligned array therefore acts as an antenna to regulate its own length. This is analogous to the length-dependent microtubule destabilization by kinesin-8 (Varga et al., 2006). The reconstitution of microtubule organization by PRC1 and kinesin-4 nicely demonstrates a biochemical mechanism by which lengths of anti-parallel overlaps can be controlled by suppression of microtubule dynamics. However, in these experiments, relative microtubule sliding is restricted as the distal ends of the filament are attached to the surface of the coverslip. It will be of interest to examine how the length of anti-parallel overlaps established by PRC1 and kinesin-4 is altered by relative sliding of the two microtubules by another motor protein. This is relevant in dividing cells, in which motor proteins (e.g., kinesin-6) are recruited to the anti-parallel microtubule array generated by PRC1 and kinesin-4.
Figure 4. Formation of anti-parallel microtubule overlaps of fixed length by PRC1 and kinesin-4.
Schematic depicts the proposed mechanism by which the non-motor crosslinking protein PRC1 (green) and an inhibitor of microtubule dynamics kinesin-4 (orange) form an anti-parallel overlap of fixed length between two elongating microtubules.
A crucial feature of this model is that PRC1 selectively tags the overlap region of pairs of aligned antiparallel microtubules. Recent structural studies have begun to explain how this selective microtubule crosslinking can be achieved. PRC1 is a modular protein with a helical N-terminus that is responsible for dimerization, a central spectrin domain that mediates microtubule binding, and a C-terminus ‘Lys-Arg’-rich predicted unstructured domain that enhances the microtubule binding affinity (Subramanian et al., 2010). Tomograms of pairs of microtubules crosslinked by PRC1 shows that the protein forms well-ordered rod-like striations connecting two microtubules that are 35±2 nm apart. In contrast, PRC1 appears more flexible on single microtubules, with only the spectrin domain adopting a well-defined conformation (Subramanian et al., 2010). These observations lead to the current working model in which the microtubule-interacting spectrin domains of PRC1 decode filament polarity. While it is inherently a flexible molecule, PRC1 adopts a relatively rigid conformation when crosslinking two anti-parallel microtubules. In this configuration, the relative orientation of the two spectrin domains is restricted such that anti-parallel crosslinks are favored. Further insights into this crosslinking mechanism are likely to come from crystal structures of PRC1 dimers. These studies should also shed light on how PRC1 binds kinesin-4 so that the motor protein can step along microtubules within the overlap region.
A less appreciated aspect of how motor and non-motor MAPs crosslink microtubules is the role of inter-filament spacing. Clues that this parameter is important come from studies of acto-myosin networks in the vertebrate muscle showing that force generation during contraction is impacted by inter-filament spacing (Matsubara et al., 1984). Measurements of uniform inter-filament spacing indicate this parameter can vary significantly between different microtubule arrays. For example, insect cells expressing the dendritic crosslinking protein, MAP2, shows an inter-microtubule spacing of 61.7±9.0 nm while this distance in the axonal crosslinking protein, Tau, is 19.8±4.1 (Chen et al., 1992). It is unclear how different motor proteins, whose lengths can also vary, slide these crosslinked arrays. For example, kinesin-5 is a 90 nm long dumbbell shaped tetramer and kinesin-4 is a 120 nm long dimer (Kashina et al., 1996; Sekine et al., 1994). Both of these proteins are able to slide the microtubules crosslinked by PRC1, which are spaced ~35 nm apart.
How are proteins of widely different molecular sizes accommodated in a microtubule array? Given the high persistence length of microtubules, the probability of local filament deformations is low (Gittes et al., 1993). We favor the model in which one crosslinker dictates the inter-microtubule spacing. The network then act as a ‘molecular-sieve’ to selectively exclude crosslinkers that cannot be accommodated based on size. Motor proteins and MAPs that do bind may need to adopt a particular crosslinking conformation. This could alter their association kinetics or force generation. Thus, by controlling the localization and activity of different proteins, the structure of the crosslinked filament network may regulate its organization to form structures of defined size and shape.
Implications for aster formation
Another recurring motif in microtubule-based structures is an aster. Similar to the anti-parallel arrays, asters act as force generators and provide spatial cues in the cytoplasm. For example, astral microtubules play important roles in processes such as directed cell migration (Watanabe et al., 2005) and embryonic nuclear positioning (Morris, 2003; Wuhr et al., 2009). An aster is formed when microtubules intersect at their ends rather than align parallel to each other. This structural motif can be defined by the length of the two filaments, the relative angle between them, and filament density. This raises the question: what is the minimum number of components needed to build an aster?
As it turns out, asters are among the easiest of structures to assemble in vitro. A two-component system comprised of a motor crosslinking protein and microtubules can generate asters (Hentrich and Surrey, 2010; Nedelec et al., 1997; Surrey et al., 2001). The formation of asters requires that a motor protein crosslinks and moves two microtubules relative to each other until their ends come together. At the ends of microtubules, the motor must stay bound so that the filaments remain connected (Figure-5a). One of the best examples of aster formation is an in vitro reconstitution study demonstrating that asters of steady state size could spontaneously assemble in mixtures of dynamic microtubules and the minus-end directed motor protein, kinesin-14 (Hentrich and Surrey, 2010; Surrey et al., 2001). Not all crosslinking motor proteins form asters. In the same experiment as above, it was observed that the plus-end directed crosslinking motor protein, kinesin-5, does not form asters (Hentrich and Surrey, 2010). One reason for this observation may be the low microtubule crosslinking efficiency of kinesin-5 (Hentrich and Surrey, 2010). Another possibility is the low velocity of kinesin-5 that could make it difficult for the protein to reach the end of a rapidly growing microtubule (Hentrich and Surrey, 2010). The average length of filaments in an aster will be determined by microtubule dynamics. What determines the relative orientation of two filaments in an aster? Though this is not yet experimentally verified, filament orientation in asters is likely to be determined by the properties of the crosslinking molecule as in the case of aligned microtubules.
Figure 5.
Organization of microtubules into an aster.
Model for aster formation by: (A) the minus-end directed crosslinking motor protein, kinesin-14, and (B) the combined activity of the non-motor protein NuMA (green), the plus-end directed motor protein kinesin-5, and the minus-end directed motor proteins dynein (red) and kinesin-14 (blue).
How do asters form in cells? Studies in cell-free mitotic extracts (Gaglio et al., 1996; Gaglio et al., 1995) have led to current models for how asters assemble in cells. Briefly, the minus end directed motor proteins, Kinesin-14 and dynein, carry out the minus-end directed transport to coalesce microtubule ends. The plus-end directed motor protein, Kinesin-5, partially opposes the minus end directed forces to prevent filament separation. The non-motor protein NuMA, crosslinks microtubules minus-ends and acts as a brake against filament separation. The crosslinking activity of NuMA is restricted to the vicinity of microtubule minus-ends by interaction with dynein (Figure-5b) (Merdes et al., 2000; Merdes et al., 1996). Thus far, the biochemical studies of this multicomponent system are incomplete and we lack in vitro reconstitutions with purified components. In addition, the structural basis of microtubule crosslinking by NuMA and the geometry it imposes on microtubules that comprise the aster remain to be elucidated. However, studies so far suggest that the net minus-end directed forces generated by motor proteins and the preferential minus-end crosslinking by a non-motor MAP are important factors that help determine the shape of this structural motif, and keep the microtubules from becoming aligned as a bundle.
Filament organization in prokaryotic cytoskeletal networks
Cytoskeletal filaments, once considered to be hallmarks of eukaryotic cells, are also present in prokaryotes. These prokaryotic filaments play important roles in determining cell shape (Margolin, 2009; Shaevitz and Gitai, 2010), accurate segregation of plasmid DNA (Gerdes et al., 2010; Salje et al., 2010), and positioning the cell-division plane (Cabeen and Jacobs-Wagner, 2010; Erickson et al., 2010). As in the case of microtubule-based architectures, these functions require precise organization of these filaments into structures of specific shapes and lengths. A major difference between the two systems is that motor proteins have not yet been identified in any prokaryotic cells. Thus filament organization must be driven entirely by crosslinking and regulation of filament length.
Similar to microtubules, crosslinking of prokaryotic filaments is likely to be important for their organization into different structures. A prominent example is the FtsZ ring. FtsZ is a structural homolog of tubulin which polymerizes to form filaments (Adams and Errington, 2009; Erickson et al., 2010). During cell division, FtsZ filaments are bundled to form a ring in the middle of dividing rod-shaped bacterial cells. This structure has been proposed to provide positional cues and generate forces during cell fission (Erickson et al., 2010). While FtsZ filaments appear to be capable of organizing into bundles on their own, a group of proteins, referred to as the Z-associated proteins (ZapA-D), have been identified as stabilizers of FtsZ bundles in cells (Adams and Errington, 2009; Kirkpatrick and Viollier). At least one of these proteins, ZapA, has been proposed to stabilize the Z-ring by crosslinking FtsZ filaments (Gueiros-Filho and Losick, 2002). It is postulated that positively charged regions in oligomeric ZapA could interact with the acidic C-terminus tail of FtsZ to form crosslinks in a manner reminiscent of microtubule interactions of non-motor proteins (Low et al., 2004). Therefore, the ZAP proteins may have functions that are analogous to those of microtubule crosslinking proteins. How the length of FtsZ filaments and their relative placement in the ring is determined remains unknown.
Like microtubules, the formation of structures of precise length will require the regulation of prokaryotic filament dynamics. Remarkably, in vitro reconstitution shows that filaments of the actin homolog, ParM undergo dynamic instability similar to microtubules (Garner et al., 2004). These dynamics are harnessed for DNA capture and force generation for plasmid segregation in a manner reminiscent of the mitotic spindle. This process was recapitulated in an impressive in vitro reconstitution experiment with 3 components: dynamic ParM filaments, DNA containing the centromere-like site, parC, and ParR, which is a parC binding protein. In current models, the ParR/parC nucleoprotein complex acts as a regulator of parM dynamic. When the dynamically unstable ParM filament is captured by the ParR/parC complex on plasmid DNA, the filaments are stabilized against catastrophe. This promotes ParM elongation, which in turn provides the pushing force for plasmid segregation (Garner et al., 2007). Therefore, as seen in eukaryotes, precise length control of the ParM filament can be achieved by tuning its intrinsic dynamics by a regulatory protein.
There have also been important advances in examining the components of the prokaryotic cytoskeleton at the structural level. The crystal structure of the ParR-DNA nucleo-protein complex and electron micrographs of ParM bound to ParR-ParC have been obtained (Salje et al., 2010). These structural studies suggest a model in which both ends of ParM are effectively capped by the large solenoid shaped ParR to reduce the rate of catastrophe, while still allowing for monomer addition at the filament ends. The organization of FtsZ bundles is much less understood. Electron cryotomographic reconstructions of dividing Caulobacter cells suggest that the Z-ring is made of short curved filaments (Erickson et al., 2010). How these filaments are structurally interconnected by the Zap-proteins is not known. Biophysical analysis of the interactions of these proteins with the FtsZ filament as well as high resolution structural analysis of the crosslinks formed will shed light on how the Z-ring is organized.
Future outlook
Elucidating the mechanisms of the self-organized assembly of microtubule-based structures requires an understanding of the properties of proteins involved, the reactions they participate in, and the final structure that emerges. As summarized in this Review, significant progress has been made in understanding the formation of some of the basic building blocks, such as a pair of antiparallel microtubules or an aster, that recur in wide-range of microtubule-based structures needed for cellular function. The next step is to extend these analyses to less understood structural motifs, such as the parallel microtubule bundles in axons or the mixed-polarity arrays in dendrites. Another major gap in our understanding of the formation of microtubule structures is how the filament number in these different arrays is controlled. We believe that visualization of microtubule nucleation in real-time is the breakthrough needed. Together, these studies will reveal how not only the shape and length, but also the number of filaments are controlled in the formation of microtubule-based structures.
During development and cell division, the formation of microtubule-based structures is tightly regulated by signaling molecules such as kinases and phosphatases. While significant advances have been made in the biochemical and structural analysis of motor proteins and MAPs, an understanding of the roles of signaling molecules in the formation of microtubule-based structures has largely been restricted to cell biological analysis. Yet even in the formation of a simple anti-parallel bundle, kinases play critical roles. For example, during cell division, the mitotic kinase, Plk1 (Polo Like Kinase-1), is recruited to the anti-parallel microtubule bundles. On these microtubule arrays, the interaction between PRC1 and Plk1 alters both the kinase activity of Plk1 and the microtubule binding of PRC1 (Hu et al., 2012; Neef et al., 2007). How such cross-talk between signaling and mechanical processes impacts the formation of microtubule-based structures of precise length needs to be addressed by in vitro reconstitution and structural methods.
In addition to motor proteins, non-motor MAPs, and signaling molecules, the size of microtubule assemblies are also likely to be controlled by tubulin post-translational modifications. A diverse set of modifications such as acetylation, detyrosination, polyglutamylation and poly-glycylation of αβ-tubulin, have been proposed to act as a ‘code’ for the recruitment and activity of microtubule binding proteins (Janke and Bulinski, 2011; Verhey and Gaertig, 2007). For example, it is suggested that detyrosination decreases the depolymerization activity of kinesin-13 (Peris et al., 2009). However, for the majority of modifications, their role in microtubule organization remains unclear due to the difficulty in obtaining homogeneously modified tubulin for biochemical and biophysical studies. With recent advances in both the expression of recombinant tubulin (Drummond et al., 2011; Johnson et al., 2011) and strategies for incorporating modified amino-acids into proteins (Davis and Chin, 2012; Foley and Burkart, 2007; Vila-Perello and Muir, 2010), the stage is now set for elucidating the role of tubulin post-translational modification on the formation of microtubule arrays of specific size and shape.
Conclusion
The elaborate micron-scale microtubule-based architectures that assemble in cells function with remarkable fidelity. We have just begun to understand the molecular mechanisms that set the size and shape of the elementary motifs frequently found in diverse cytoskeletal structures. This is an important step towards deciphering how these arrays assemble and actually function. Complex nanoscale DNA-based motifs can now be built using relatively simple rules and components (Torring et al., 2011). We are optimistic that like DNA origami, microtubule origami will soon be possible to allow us to synthesize dynamic structures that can carry out complex cellular functions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, et al. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM. A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L, Mitchison T, Deacon HW. Catastrophic revelations about Op18/stathmin. Trends Biochem Sci. 1996;21:197–198. [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Bormuth V, Varga V, Howard J, Schaffer E. Protein friction limits diffusive and directed movements of kinesin motors on microtubules. Science. 2009;325:870–873. doi: 10.1126/science.1174923. [DOI] [PubMed] [Google Scholar]
- Bratman SV, Chang F. Mechanisms for maintaining microtubule bundles. Trends Cell Biol. 2008;18:580–586. doi: 10.1016/j.tcb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Braun M, Drummond DR, Cross RA, McAinsh AD. The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat Cell Biol. 2009;11:724–730. doi: 10.1038/ncb1878. [DOI] [PubMed] [Google Scholar]
- Braun M, Lansky Z, Fink G, Ruhnow F, Diez S, Janson ME. Adaptive braking by Ase1 prevents overlapping microtubules from sliding completely apart. Nat Cell Biol. 2011;13:1259–1264. doi: 10.1038/ncb2323. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. The bacterial cytoskeleton. Annu Rev Genet. 2010;44:365–392. doi: 10.1146/annurev-genet-102108-134845. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Nurse P. Self-organization of interphase microtubule arrays in fission yeast. Nat Cell Biol. 2006;8:1102–1107. doi: 10.1038/ncb1479. [DOI] [PubMed] [Google Scholar]
- Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol. 2002;14:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- Chen J, Kanai Y, Cowan NJ, Hirokawa N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature. 1992;360:674–677. doi: 10.1038/360674a0. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Tao L, Brust-Mascher I, Wollman R, Scholey JM. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J Cell Biol. 2010;188:49–68. doi: 10.1083/jcb.200908150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C, Caceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Drummond DR, Kain S, Newcombe A, Hoey C, Katsuki M, Cross RA. Purification of tubulin from the fission yeast Schizosaccharomyces pombe. Methods Mol Biol. 2011;777:29–55. doi: 10.1007/978-1-61779-252-6_3. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol. 2009;11:717–723. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- Foley TL, Burkart MD. Site-specific protein modification: advances and applications. Curr Opin Chem Biol. 2007;11:12–19. doi: 10.1016/j.cbpa.2006.11.036. [DOI] [PubMed] [Google Scholar]
- Fu C, Ward JJ, Loiodice I, Velve-Casquillas G, Nedelec FJ, Tran PT. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev Cell. 2009;17:257–267. doi: 10.1016/j.devcel.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, Menager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, et al. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Compton DA. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Odde DJ, Bloom K. Kinesin-8 molecular motors: putting the brakes on chromosome oscillations. Trends Cell Biol. 2008;18:307–310. doi: 10.1016/j.tcb.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Mullins RD. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 2004;306:1021–1025. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- Gigant B, Curmi PA, Martin-Barbey C, Charbaut E, Lachkar S, Lebeau L, Siavoshian S, Sobel A, Knossow M. The 4 A X-ray structure of a tubulin:stathmin-like domain complex. Cell. 2000;102:809–816. doi: 10.1016/s0092-8674(00)00069-6. [DOI] [PubMed] [Google Scholar]
- Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SS, Vale RD. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143:263–274. doi: 10.1016/j.cell.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Li X, Chan EH, Chalamalasetty RB, Nigg EA, Barr FA. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol. 2006;172:363–372. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 2002;16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr., Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- Hentrich C, Surrey T. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J Cell Biol. 2010;189:465–480. doi: 10.1083/jcb.200910125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- Hu CK, Ozlu N, Coughlin M, Steen JJ, Mitchison TJ. Plk1 negatively regulates PRC1 to prevent premature midzone formation before cytokinesis. Mol Biol Cell. 2012;23:2702–2711. doi: 10.1091/mbc.E12-01-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Chretien D, Arnal I, Wade RH. Structural changes accompanying GTP hydrolysis in microtubules: information from a slowly hydrolyzable analogue guanylyl-(alpha,beta)-methylene-diphosphonate. J Cell Biol. 1995;128:117–125. doi: 10.1083/jcb.128.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec FJ, Tran PT. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–368. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Johnson V, Ayaz P, Huddleston P, Rice LM. Design, overexpression, and purification of polymerization-blocked yeast alphabeta-tubulin mutants. Biochemistry. 2011;50:8636–8644. doi: 10.1021/bi2005174. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Janson ME, van den Wildenberg SM, Hoogenraad CC, Schmidt CF, Peterman EJ. Microtubule-driven multimerization recruits ase1p onto overlapping microtubules. Curr Biol. 2008;18:1713–1717. doi: 10.1016/j.cub.2008.09.046. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I, Straube A. Regulation of cell migration by dynamic microtubules. Semin Cell Dev Biol. 2011;22:968–974. doi: 10.1016/j.semcdb.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Roostalu J, Roque H, Antony C, Schiebel E. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev Cell. 2009;17:244–256. doi: 10.1016/j.devcel.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Kimura T, Watanabe H, Iwamatsu A, Kaibuchi K. Tubulin and CRMP-2 complex is transported via Kinesin-1. J Neurochem. 2005;93:1371–1382. doi: 10.1111/j.1471-4159.2005.03063.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman AA. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294:1340–1343. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CL, Viollier PH. New(s) to the (Z-)ring. Curr Opin Microbiol. 14:691–697. doi: 10.1016/j.mib.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol. 2011;12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Moncrieffe MC, Lowe J. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J Mol Biol. 2004;341:839–852. doi: 10.1016/j.jmb.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Lowe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara I, Goldman YE, Simmons RM. Changes in the lateral filament spacing of skinned muscle fibres when cross-bridges attach. J Mol Biol. 1984;173:15–33. doi: 10.1016/0022-2836(84)90401-7. [DOI] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Meunier S, Vernos I. K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat Cell Biol. 2011;13:1406–1414. doi: 10.1038/ncb2372. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus- end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ. Self-organization of polymer-motor systems in the cytoskeleton. Philos Trans R Soc Lond B Biol Sci. 1992;336:99–106. doi: 10.1098/rstb.1992.0049. [DOI] [PubMed] [Google Scholar]
- Moores CA, Yu M, Guo J, Beraud C, Sakowicz R, Milligan RA. A mechanism for microtubule depolymerization by KinI kinesins. Mol Cell. 2002;9:903–909. doi: 10.1016/s1097-2765(02)00503-8. [DOI] [PubMed] [Google Scholar]
- Morris NR. Nuclear positioning: the means is at the ends. Curr Opin Cell Biol. 2003;15:54–59. doi: 10.1016/s0955-0674(02)00004-2. [DOI] [PubMed] [Google Scholar]
- Nedelec F, Surrey T, Karsenti E. Self-organisation and forces in the microtubule cytoskeleton. Curr Opin Cell Biol. 2003;15:118–124. doi: 10.1016/s0955-0674(02)00014-5. [DOI] [PubMed] [Google Scholar]
- Nedelec FJ, Surrey T, Maggs AC, Leibler S. Self-organization of microtubules and motors. Nature. 1997;389:305–308. doi: 10.1038/38532. [DOI] [PubMed] [Google Scholar]
- Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, Barr FA. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol. 2007;9:436–444. doi: 10.1038/ncb1557. [DOI] [PubMed] [Google Scholar]
- Peters C, Brejc K, Belmont L, Bodey AJ, Lee Y, Yu M, Guo J, Sakowicz R, Hartman J, Moores CA. Insight into the molecular mechanism of the multitasking kinesin-8 motor. EMBO J. 2010;29:3437–3447. doi: 10.1038/emboj.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris L, Wagenbach M, Lafanechere L, Brocard J, Moore AT, Kozielski F, Job D, Wordeman L, Andrieux A. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J Cell Biol. 2009;185:1159–1166. doi: 10.1083/jcb.200902142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman EJ, Scholey JM. Mitotic microtubule crosslinkers: insights from mechanistic studies. Curr Biol. 2009;19:R1089–1094. doi: 10.1016/j.cub.2009.10.047. [DOI] [PubMed] [Google Scholar]
- Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- Rey M, Sanchez-Madrid F, Valenzuela-Fernandez A. The role of actomyosin and the microtubular network in both the immunological synapse and T cell activation. Front Biosci. 2007;12:437–447. doi: 10.2741/2073. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A, McNally FJ. Microtubule-severing enzymes. Curr Opin Cell Biol. 2010;22:96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J, Gayathri P, Lowe J. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol. 2010;8:683–692. doi: 10.1038/nrmicro2425. [DOI] [PubMed] [Google Scholar]
- Sekine Y, Okada Y, Noda Y, Kondo S, Aizawa H, Takemura R, Hirokawa N. A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J Cell Biol. 1994;127:187–201. doi: 10.1083/jcb.127.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaevitz JW, Gitai Z. The structure and function of bacterial actin homologs. Cold Spring Harb Perspect Biol. 2010;2:a000364. doi: 10.1101/cshperspect.a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Ross JL. Microtubule-severing enzymes at the cutting edge. J Cell Sci. 2012;125:2561–2569. doi: 10.1242/jcs.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark P, Ogg D, Flodin S, Flores A, Kotenyova T, Nyman T, Nordlund P, Kursula P. The structure of human collapsin response mediator protein 2, a regulator of axonal growth. J Neurochem. 2007;101:906–917. doi: 10.1111/j.1471-4159.2006.04401.x. [DOI] [PubMed] [Google Scholar]
- Stiess M, Bradke F. Neuronal polarization: the cytoskeleton leads the way. Dev Neurobiol. 2011;71:430–444. doi: 10.1002/dneu.20849. [DOI] [PubMed] [Google Scholar]
- Su X, Qiu W, Gupta ML, Jr., Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 2010;142:433–443. doi: 10.1016/j.cell.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surrey T, Nedelec F, Leibler S, Karsenti E. Physical properties determining self-organization of motors and microtubules. Science. 2001;292:1167–1171. doi: 10.1126/science.1059758. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol. 2006;16:2293–2302. doi: 10.1016/j.cub.2006.09.064. [DOI] [PubMed] [Google Scholar]
- Torring T, Voigt NV, Nangreave J, Yan H, Gothelf KV. DNA origami: a quantum leap for self-assembly of complex structures. Chem Soc Rev. 2011;40:5636–5646. doi: 10.1039/c1cs15057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Vale RD, Malik F, Brown D. Directional instability of microtubule transport in the presence of kinesin and dynein, two opposite polarity motor proteins. J Cell Biol. 1992;119:1589–1596. doi: 10.1083/jcb.119.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol. 2006;8:470–476. doi: 10.1038/ncb1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6:2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- Vila-Perello M, Muir TW. Biological applications of protein splicing. Cell. 2010;143:191–200. doi: 10.1016/j.cell.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys GO, Ambrose JC. Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol. 2009;19:62–71. doi: 10.1016/j.tcb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Noritake J, Kaibuchi K. Regulation of microtubules in cell migration. Trends Cell Biol. 2005;15:76–83. doi: 10.1016/j.tcb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, Howard J. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci U S A. 2011;108:2741–2746. doi: 10.1073/pnas.1016498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Hyman A, Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol. 2001;3:E28–34. doi: 10.1038/35050669. [DOI] [PubMed] [Google Scholar]
- Wordeman L. Microtubule-depolymerizing kinesins. Curr Opin Cell Biol. 2005;17:82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Wuhr M, Dumont S, Groen AC, Needleman DJ, Mitchison TJ. How does a millimeter-sized cell find its center? Cell Cycle. 2009;8:1115–1121. doi: 10.4161/cc.8.8.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Jiang W. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci U S A. 2005;102:343–348. doi: 10.1073/pnas.0408438102. [DOI] [PMC free article] [PubMed] [Google Scholar]