SUMMARY
Landscape epidemiology and landscape genetics combine advances in molecular techniques, spatial analyses and epidemiological models to generate a more real-world understanding of infectious disease dynamics and provide powerful new tools for the study of RNA viruses. Using dog rabies as a model we have identified how key questions regarding viral spread and persistence can be addressed using a combination of these techniques. In contrast to wildlife rabies, investigations into the landscape epidemiology of domestic dog rabies requires more detailed assessment of the role of humans in disease spread, including the incorporation of anthropogenic landscape features, human movements and socio-cultural factors into spatial models. In particular, identifying and quantifying the influence of anthropogenic features on pathogen spread and measuring the permeability of dispersal barriers are important considerations for planning control strategies, and may differ according to cultural, social and geographical variation across countries or continents. Challenges for dog rabies research include the development of metapopulation models and transmission networks using genetic information to uncover potential source/sink dynamics and identify the main routes of viral dissemination. Information generated from a landscape genetics approach will facilitate spatially strategic control programmes that accommodate for heterogeneities in the landscape and therefore utilise resources in the most cost-effective way. This can include the efficient placement of vaccine barriers, surveillance points and adaptive management for large-scale control programmes.
Key words: Canis familiaris, landscape genetics, phylogeography, spatial heterogeneity, transmission dynamics, vaccination, zoonosis
INTRODUCTION
Landscape epidemiology is the study of the causes and consequences of spatial variation in disease incidence or risk across heterogeneous landscapes (Ostfeld et al. 2005). Landscape structure affects the distribution, abundance and movements of host, vector and pathogen populations and therefore inherently influences localised interactions between infectious and susceptible individuals (Ostfeld et al. 2005; McCallum, 2008). Revealing the landscape factors underlying these interactions calls for an interdisciplinary approach that draws on a range of techniques across different spatial scales (Manel et al. 2003; Ostfeld et al. 2005). Molecular markers provide a basis for this by genetically tracking spatial and temporal dynamics in pathogen and host populations (Biek and Real, 2010). A landscape genetics approach to infectious disease therefore encompasses a range of analytical tools, including geographic information systems, remote sensing, population genetics, phylogenetics and statistical and mathematical modelling techniques (Manel et al. 2003).
RNA viruses represent an ideal group for exploring landscape influences on evolutionary trajectories due to their characteristically high mutation rates and short generation times, which means that epidemiological and population genetic processes occur on a similar timescale (Drummond et al. 2003). The accumulation of mutations over time and space imprints on the structure of viral genomes in a population and can be visualised in data collected over months or years, providing a valuable resource for the elucidation of ecological and evolutionary dynamics. Despite this, the processes that govern phylogeographic patterns in viruses are still poorly understood (Holmes and Grenfell, 2009), pointing to the need for more detailed study into the effect of spatial heterogeneity on viral transmission.
Modern sequence analysis has the power to reveal the historical emergence of pathogen variants, distribution patterns, spillover events, interspecies transmission and changing selection pressures (Archie et al. 2009; Anderson et al. 2010). Advances in sequencing technologies have paved the way for a new generation of approaches to the study of disease dynamics, and next generation sequencing (NGS) techniques are being continually refined, improving their resolution, throughput potential, affordability and accessibility (Holmes and Grenfell, 2009; Metzker, 2010). We envision a paradigm shift to whole genome sequencing as the standard technique for characterizing RNA viral evolution on small spatio-temporal scales, providing greater discrimination between genotypes and finer resolution in population structure (Holmes and Grenfell, 2009). Given the progress in sequencing technology and parallel advances in spatial analytical tools, it is an exciting time to study the landscape epidemiology of RNA viruses from a population genetic perspective.
In this review, we focus on rabies virus (RABV), which presents an excellent model system to illustrate the challenges and prospects of such an approach. RABV is a single stranded, negative-sense RNA virus belonging to the Lyssavirus genus (Family: Rhabdoviridae) (Dietzschold et al. 2005). It is globally distributed and has the ability to infect all mammals, but typically exists in endemic foci as a reservoir host-specific variant with occasional spillover to other species (Rupprecht et al. 2002). Domestic dogs, Canis familiaris, are the principal reservoir of RABV, responsible for 99% of the estimated 55,000 human deaths due to rabies that occur mainly in Asia and Africa every year (Knobel et al. 2005).
Rabies has proven a remarkably valuable system for exploring the effect of landscape processes on host/pathogen interactions. However, most studies have focused on wildlife rabies due to the higher quality of surveillance data and availability of resources in areas with major endemic wildlife foci, e.g. raccoon rabies in eastern North America or fox rabies in Western Europe (Bourhy et al. 1999; Smith et al. 2002; Holmes 2004; Biek et al. 2007; Wheeler and Waller 2008; Szanto et al. 2011). The wealth of research into wildlife rabies provides a basis for comparison with the domestic dog foci that exist in Africa, Asia and parts of Latin America, which, despite their much greater public health burden, have been less well studied.
Domestic dogs are inherently tied to human populations, and various aspects of human ecology, including distribution, habitation and movement patterns, or cultural practices, will directly influence rabies spread in dog populations. Settlements can be considered dog ‘habitat’, and dog densities have been predicted on the basis of human demographics and human geography (Butler, 2000; Knobel et al. 2008). A seemingly ubiquitous feature of countries with persistent dog rabies foci is the free-roaming nature of these populations, often referred to as ‘neighbourhood’ dogs. Dogs in Africa, Asia and parts of Latin America, where canine rabies is endemic, are rarely restricted by leashing or enclosures and their role as domestic animals varies e.g. watch dogs, trade, companion animals. The free movement of dogs would thus be expected to contribute to local rabies transmission, potentially resembling the known features of wildlife rabies and illustrating the complex interplay between anthropogenic and natural drivers of disease spread in this system. A key challenge is to uncover the extent to which natural constraints to rabies flux hold in a host-pathogen system with greater human-mediated dispersal; and what affect this has on phylogeographic signatures.
This review aims to synthesise our current understanding of rabies landscape epidemiology derived from genetic data across different spatio-temporal scales (see Table 1 for a list of relevant studies and associated analytical methods). Using the extensive work on wildlife rabies as a backdrop, we seek to identify commonalities as well as fundamental differences characterizing the dynamics of RABV in domestic dog populations. We will argue that the specific ‘landscape’ supporting the sustained transmission of the virus in dogs is determined by a complex mixture of physical and human geography and that a quantitative understanding of these landscapes will be essential for rabies control and eradication. In addition, control activities are themselves predicted to change molecular epidemiological trajectories, creating interesting opportunities for adaptive management in rabies. Throughout the review we highlight how novel tools and technologies are being used to tackle these problems and identify key areas in which such approaches have future potential.
Table 1.
Studies that have examined sources of spatial heterogeneity in dog rabies dynamics at a landscape scale; Gen = genetic data, Epi = epidemiological data. ML = Maximum Likelihood
| Scale | Sources of spatial heterogeneity | Data | Analytical methods | Key points/Summary | Reference |
|---|---|---|---|---|---|
| Local (∼5 yr period) | Individual host heterogeneity | Epi | Reconstruction of epidemic trees, outbreak simulations, construction of transmission networks based on a spatial infection kernel and generation intervals estimated from epidemiological data using ML | Contact tracing data used to generate robust estimates of epidemiological parameters | Hampson et al. 2009 |
| Local (∼5 yr period) | Spatial configuration of populations | Epi | Patch-occupancy models | Uses dog bite incidence records to explore metapopulation dynamics | Beyer et al. 2011 |
| Regional (20+ yrs) | Socio-economic drivers | Gen | Bayesian inference of phylogeny, molecular clocks and demography | Attributes phylogeographic patterns to economic growth and migration patterns of humans | Carnieli et al. 2011 |
| Regional (10+ yrs) | Translocation, road networks, wildlife hosts | Gen-Epi | Phylogenetic inference | Patterns suggest importance of wildlife hosts in this system. Spread of rabies coincided with major highways, and indicated translocation events. | Coetzee and Nel, 2007 |
| Regional (2 yrs) | Translocation | Gen | Antigenic characterization through monoclonal antibody profiling and molecular sequence comparison | Identified the regional source of a rabid dog case through forensic epidemiological tracing | David et al. 2004 |
| Regional (10 yrs) | Reservoir hosts, cultural drivers | Gen | Bayesian phylogenetics; parsimonious construction of transmission networks | Used statistical parsimony to construct most likely transmission networks. Also discovered the potential influence of social drivers on rabies phylogeographic structure (pastoralist vs. non-pastoralist community structure). | Lembo et al. 2007 |
| Local and Regional (10+ yrs) | Reservoir hosts, transmission clusters in wildlife | Gen-Epi | Bayesian phylogenetics ; generation of epidemic trees based on probabilities of links between possible progenitors and suspected cases weighted by spatio-temporal proximity, ML estimation of spatial infection kernel and generation interval distribution | Most likely reservoir hosts (domestic dogs) inferred. Found evidence for short-lived chains of transmission in wildlife, compared to self-sustaining transmission in domestic dogs | Lembo et al. 2008 |
| Regional (1 yr) | Road networks, population density | Epi | Spatial analysis of reported rabies using GIS. Directional spread of rabies based on mean centre of cases and a standard deviational ellipse weighted by date of cases | Distribution of cases followed road network and towns with high human density and high numbers of free-roaming dogs | Tenzin et al. 2010 |
| Country (3 yrs) | Translocation, cross border incursions | Gen-Epi | Bayesian inference of phylogeny, molecular clock, demography and phylogeographic diffusion based on discrete spatial states | Frequent incursions across country boundary; first molecular evidence for a long distance translocation of a rabies sub-lineage in Africa | Hayman et al. 2011 |
| Between and within country (20+ yrs) | Geopolitical boundaries, translocation, road networks | Gen-Epi | Bayesian inference of phylogeny, molecular clock, demography and phylogeographic diffusion based on discrete spatial states; comparison of different geographic predictors of viral diffusion using model selection; quantification of phylogeographic clustering using association index; spatial simulation to test natural vs. human-mediated dispersal | Demonstrates and quantifies the anthropogenic influence on dog rabies dissemination. Best fit models implicate road networks as important predictors of rabies dispersal | Talbi et al. 2010 |
| Sub-continent (20+ yrs) | Geopolitical boundaries | Gen-Epi | Bayesian inference of phylogeny, molecular clock, demography and phylogeographic diffusion based on discrete spatial states | Strong population subdivision at the country-level according to phylogeographic patterns. | Talbi et al. 2009 |
| Global (20+ yrs) | Mountain ranges, large water bodies, deserts, oceans | Gen-Epi | Bayesian inference of phylogeny, molecular clock, demography; and parsimony-based approach to determine the geographical structure of dog RABV phylogeny | Elucidation of global patterns of dog rabies determined by major natural landscape barriers and historical colonisation events | Bourhy et al. 2008 |
Box 1.
Glossary
Association index (AI): test statistic used as a means of quantifying phylogeny-trait associations i.e. given a discrete character for each tip of a phylogenetic tree, are more closely related taxa likely to share the same trait values than is expected by chance alone. Avoids the issue of lack of independence due to shared ancestry (Wang et al. 2001).
Metapopulation: concept describing the persistence of a species in a spatially heterogeneous environment as a balance between colonisation and extinction in loosely coupled subpopulations or ‘patches’ with different within and between-patch dynamics, which can be applied to infectious diseases (Grenfell and Harwood, 1997)
Most recent common ancestor (MRCA): most recent individual from which all taxa in the group are directly descended.
Oral rabies vaccination (ORV): distribution of oral rabies vaccine baits as a strategy to control the spread of wildlife rabies. Used as a control measure for wildlife rabies in North America, and Europe.
Phylogeography: combines phylogenetics and biogeography to describe the contemporary pattern of an organism's geographic spread according to gene genealogies.
R0(basic reproductive number): the average number of secondary cases derived from a single infectious individual in an entirely susceptible population (Anderson and May, 1991).
Superspreader: an individual causing a disproportionately high number of secondary cases, compared to the mean (represented by R0), depicted in the long tail of a frequency distribution of secondary cases (Lloyd-Smith et al. 2005).
Surfing mutation model: genetic variants at the front of an advancing wave of infection are swept to high frequencies during an epidemic peak, resulting in long-term dominance of colonising genetic lineages (Excoffier and Ray, 2008, Klopfstein et al. 2006).
Box 2.
Key research questions for dog rabies at the landscape scale
-
1.
To what degree do the demography and ecology of dogs, as compared to humans, determine the dynamics of rabies transmission?
As dogs are inherently tied to humans, we might expect the patterns dictated by local dog movements to be confounded by human-mediated long-distance movements or anthropogenic features that facilitate connection of sub-populations.
-
2.
Which aspects of human geography best predict landscape permeability to dog rabies? Are these predictors consistent across different areas and continents?
Although evidence suggests the influence of human geography on dog rabies dispersal, we have yet to uncover the best predictors of this form of spread. Specifically, a quantifiable method of characterizing these landscape features would assist with the creation of guidelines for using dispersal barriers to aid control.
-
3.
How does endemic rabies compare to epidemic spread?
Most studies have focused on outbreak situations in wildlife, but it is not clear how rabies dynamics change in systems where the pathogen has been circulating for centuries, as is the case for dog rabies. How long is phylogeographic structure maintained over time and do initial invasion pathways predict connectivity in the endemic state?
-
4.
Does dog rabies persist in metapopulations?
Dog rabies may best be described as a series of sub-populations with varying inter- and intra-patch dynamics. Exploring the concept of a metapopulation dynamic for dog rabies presents a relatively unexplored area for future research and may uncover the mechanisms that allow pathogen persistence at a local scale.
-
5.
How can information about landscape heterogeneity and genetic structure be incorporated into more efficient control programmes?
Information gathered from spatially explicit landscape models will allow targeted control measures using resources cost-effectively. Moreover, the implementation of vaccine barriers and other forms of control change the complexity of the landscape, potentially altering disease dynamics. Powerful genetic techniques may elucidate the effect of these new landscape heterogeneities, facilitating adaptive management.
RABIES LIFE CYCLE AND PHYLOGEOGRAPHY
Transmission of rabies virus occurs predominantly through the bite of an infected animal, which inoculates virus-laden saliva into the subcutaneous and muscle tissue of a susceptible host (Dietzschold et al. 2005). Once inoculated, the virus enters neurons and migrates to the central nervous system, before spreading to other organs, including the salivary glands where large amounts of infectious virions are shed into the saliva for further transmission (Dietzschold et al. 2005). The incubation period is highly variable with a mean of 22 days in naturally infected dogs (Hampson et al. 2009), but may extend to months or years due in part to localised replication (Hanlon et al. 2007). In contrast, the infectious period is very short, around 3 days, and very rarely exceeds 10 days. The short infectious period severely restricts the spatial scale over which an infectious individual can transmit the virus, while the longer and more variable incubation time may permit the active or passive movement of infected individuals over larger distances, which, as detailed below, helps to explain phylogeographic patterns.
On a global scale, dog RABV forms six major genetic clades, five of which are associated with particular geographic regions: Africa 2, Africa 3, Arctic-related, Asian, and the Indian subcontinent (Bourhy et al. 2008). Grouping of these clades largely reflects major barriers such as oceans, large mountain ranges and deserts, or historical colonisation events. The Arctic-related viruses are relatively well distributed, reflecting the lack of barriers in the far north, and a sixth clade has a cosmopolitan distribution, reflecting historical waves of human migrations and colonisations (Smith et al. 1992; Bourhy et al. 2008). Such efficient dissemination is likely to be a result of the virus's relatively long and variable incubation period occasionally facilitating long-distance transport of infected dogs.
The strong phylogeographic structure exhibited by most dog rabies clades is mirrored in wildlife rabies, where the same pattern is evident even at relatively small spatial scales (Biek et al. 2007) and may be explained by a ‘surfing mutation’ model (Excoffier and Ray, 2008). According to this model, new lineages arising during initial colonisation are able to reach high frequencies driven by an epidemic wave, whereas subsequent lineages do not benefit from such conditions and fail to infiltrate the dominating lineage clusters. Initial invasion events can therefore markedly influence the phylogeographic structure of rabies and, at least in wildlife, this structure can remain intact for decades (Biek et al. 2007; Szanto et al. 2011). However, with ongoing movement of infected individuals and the immigration of new lineages into an area, this structure would be expected to erode over time. How environmental variation affects these processes of emergence and subsequent erosion of phylogeographic structure are questions central to defining the landscape epidemiology of RABV and we will return to them throughout this review.
LANDSCAPE LEVEL EFFECTS ON RABIES DYNAMICS
Since the transmission of rabies is entirely dependent on host movements initiating contact between infectious and susceptible individuals, the landscape that they occupy and disperse within heavily influences the ability of the virus to infect new hosts. Indeed, wildlife rabies epidemics tend to spread as irregular waves that differ in velocity according to heterogeneities in the landscape (Russell et al. 2006). These ‘heterogeneities’ include natural features such as mountain ranges, water bodies and deserts, but also anthropogenic features including roads and vaccine corridors. The influence of spatial heterogeneity on rabies spread can be broken down into three aspects, each of which are discussed in detail in the following sections: (1) host movements: natural versus human-mediated; (2) landscape attributes influencing rabies spread; and (3) population level effects.
The role of human vs. natural dispersal
Throughout history, humans moving animals has repeatedly led to the emergence and spread of rabies in susceptible populations, either through deliberate translocation (e.g. the relocation of animals, some of which may be incubating virus (Nettles et al. 1979)) or inadvertent movement (e.g. raccoons on garbage trucks (Wilson et al. 1997)).
Although human-mediated long-distance movement was found to be a significant feature of raccoon rabies spread (Smith et al. 2002), it was still rare compared to natural dispersal, and stochastic in its occurrence. In contrast, human-mediated dispersal of dog rabies appears more widespread and potentially predictable due to possible links with human activities and migration patterns. The European colonisation of Africa is thought to have led to the introduction and subsequent expansion of canine rabies across the continent (Bourhy et al. 2008; Talbi et al. 2009; Lemey et al. 2010); increasingly frequent records of re-introductions into countries where rabies has been eliminated have been recorded (Zanoni and Breitenmoser, 2003; Weiss et al. 2009; Gautret et al. 2011;), and canine rabies has recently emerged on several previously rabies-free islands in Indonesia via fishermen importing incubating dogs (Windiyaningsih et al. 2004; Susilawathi et al. 2012). Even on smaller scales, phylogeographic patterns frequently imply translocation events that increase the level of mixing between lineages and may obscure patterns of lineage clustering (David et al. 2004; Talbi et al. 2010; Hayman et al. 2011).
Although often inferred, it is difficult to quantify the influence of human-mediated movements on the diffusion of RABV. Though translocations may be frequent, the likelihood of establishment and spread is much lower (Smith et al. 2005). But, unlike for wild animals, where translocation is risky and often unsuccessful, human-mediated displacement from a dog's original home range confers fewer risks. Dog translocations are therefore likely to be more successful in initiating new disease foci, and with potentially higher consequences, introducing disease into previously disease-free areas. Given that long-distance movements instigated by humans are often in response to social drivers, the genetic structure of dog rabies is expected to reflect variation in cultures and socioeconomic factors. For example, rural workers in Thailand often relocate to urban areas where work is more readily available in the off-growing season, taking their dogs, and sometimes rabies, with them (Denduangboripant et al. 2005). Similarly, the resurgence of canine rabies in KwaZulu-Natal in the 1970s was linked to refugee movements from Mozambique (Swanepoel et al. 1993; Cleaveland, 1998) and phylogenetic patterns of RABV in parts of northern Tanzania can be tentatively explained by the movement of nomadic Maasai pastoralists with their dogs (Lembo et al. 2007). Counterproductively, dog-owners have been reported to move their dogs to avoid the threat of culling during attempts to control rabies, which may explain more rapid spread of rabies than would be predicted by dog movement alone. Importantly, many of these aspects of human geography are quantifiable, making it possible in principle to generate testable hypotheses about their role in determining rabies phylogeographic structure.
Talbi et al. (2010) used spatial simulations to show that the observed patterns of spread in North Africa could only be explained by the inclusion of long-distance translocation events as opposed to natural dog movements alone. The wave-like patterns that are a signature of wildlife rabies and determined by natural host movements are less evident in domestic dog rabies, and it is unknown to what extent natural dog movements vs. human-mediated movements determine the observed phylogeographic patterns. A recent study in a Kenyan rangeland found that healthy domestic dogs rarely moved more than 50 metres from their home bases, and the maximum distance recorded was 3·2 km (Woodroffe and Donnelly, 2011). In contrast, dispersal distances in wildlife hosts tend to be at least an order of magnitude higher (Cullingham et al. 2008). However, rabies infection leads to behavioural changes, which may alter movement patterns; in northern Tanzania, most contacts with rabid dogs occurred within a kilometre of an animal's homestead (mean ∼0·88 km), but a small proportion of rabid dogs ran over 15 km while infectious (Hampson et al. 2009). Dog movements also vary depending on the societal context and livelihoods of the communities to which they belong (Woodroffe and Donnelly, 2011). For their dispersal simulations, Talbi et al. (2010) used measures of natural dog movements based on a specific locality in Tanzania (Hampson et al. 2009) but it remains unclear whether such data are transferrable among different geographic localities. Research on domestic dog ecology and movement is lacking and detailed, location-specific data are required in order to reliably test hypotheses for alternative drivers of viral diffusion.
Landscape attributes influencing rabies spread
As a parasite transmitted through direct host-to-host contact, spatially-defined genetic discontinuities in RABV populations may indicate a barrier to host contact (Biek and Real, 2010). We consider a ‘dispersal barrier’ to be any spatial feature of the landscape that impedes the gene flow of a pathogen. Spatially explicit models incorporating landscape features can help uncover barriers to gene flow based on slower progression than expected over a homogeneous area. However, the interpretation of genetic discontinuities requires caution as phylogeographic patterns may arise as a result of historical colonisation events and irrespective of physical barriers (Real and Biek, 2007; Talbi et al. 2009).
Smith et al. (2002) demonstrated a seven-fold reduction in rates of raccoon rabies spread in North America due to the presence of rivers and forest cover. Similarly, the Vistula River separated distinct clusters of a red fox variant and raccoon dog/fox variant in Europe (Bourhy et al. 1999). Clearly, large water bodies impede host movement, but have differential success as barriers according to additional factors such as habitat suitability (Smith et al. 2005; Cullingham et al. 2009), physical geography (Rees et al. 2009), and width and flow rates (Bourhy et al. 1999). In an attempt to assess the differential permeability of barriers, Rees et al. (2008) used genetic simulation modelling to assess the effect of the Niagara River on rabies spread from New York State to Ontario. Comparing genetic population structure derived from field data, with simulated population expansion scenarios they ascertained a 50% barrier effect. In contrast, homogeneous landscapes that lack environmental barriers are particularly vulnerable to the rapid expansion of an introduced pathogen, as demonstrated by the increased speed with which raccoon rabies is predicted to cross central Ohio, which lacks major natural barriers, compared to neighbouring states (Russell et al. 2005). With the exception of major landscape features inferred as barriers to dog rabies on a global scale (Bourhy et al. 2008), we know relatively little about barrier effects at smaller scales. Determining whether natural barriers prevent the dissemination of dog RABV or if ties to human ecology negate their effect is an important research question with significant implications for control.
Political borders are another potential form of barrier that may play a role in the containment of dog rabies, but have no evident impact on wildlife rabies dispersal. For example, distinct and almost monophyletic RABV groups associated with North African countries indicate restricted movement across geopolitical boundaries (Talbi et al. 2010). This well-defined population structure at a regional scale contrasts with a relatively fluid dissemination within countries. On the one hand, this suggests that country level vaccination programmes should have a good chance of eliminating dog rabies even in contiguous landscapes. However, these findings are not generalisable, with epidemiological analyses of RABV in Ghana highlighting frequent cross-border incursions (Hayman et al. 2011). In addition, time-series analyses indicate large-scale synchronous dynamics of rabies across multiple countries in eastern and southern Africa, possibly due to a combination of human or wildlife-mediated long-distance dispersal and a lack of sustained control programmes (Hampson et al. 2007). Understanding the circumstances whereby political boundaries act as dispersal barriers should provide guidance for whether control programmes require regional co-operation or can be sustained at a national level with appropriate border controls. As a first step, it would be useful to compare phylogeographic structure of dog RABV among different parts of the world and across hierarchical spatial scales. Objective measures for such a comparison can be obtained by quantifying the degree to which sequences cluster on a phylogeny according to their geographic location. Several statistics are available for this (Parker et al. 2008), of which the association index (AI) (Wang et al. 2001) has been found to be of particular utility. This measure of phylogeny-trait correlation may provide an initial descriptive analysis of consistency or variability in the hierarchical genetic structure of dog rabies between continents or countries that could determine the generalisation of results across different systems. For example, we might hypothesise that regions with large population densities and a high level of transport infrastructure, e.g. parts of Asia, show less distinctive phylogeographic patterns (a low AI value) compared to more sparsely populated and less developed landscapes like those in sub-Saharan Africa (high AI value).
Quantifying barrier effects, to predict the likelihood of incursions or to exploit them for control programmes, is an important area yet to be tackled for dog rabies. The impact of anthropogenic landscape features on spread, at least in a qualitative sense, has been noted in several countries. For example, Tenzin et al. (2010) mapped the spread of rabies in Bhutan showing a strong visual pattern alongside road networks and towns with high dog-to-human ratios; and phylogeographic patterns in north Mexico and Thailand match the distribution of major migration routes (de Mattos et al. 1999; Denduangboripant et al. 2005). It can be hypothesised that features important to wildlife rabies become less significant to dog rabies as anthropogenic features take over the landscape and mediate the effect of barriers, i.e. bridges, roads and transportation make it possible for dogs to circumvent natural barriers and support a relatively fluid dissemination of rabies across naturally heterogeneous landscapes. These hypotheses remain largely untested and thus present a fruitful area for landscape genetics investigations.
Uncovering those features that facilitate viral spread is an equally important aspect of assessing the landscape. As previously discussed, anthropogenic effects may facilitate the transmission of rabies across a larger scale than natural movements alone allow. The impact of human-mediated dog transport may by explored by phylogenetic analysis, with translocations implicated by the presence of a cluster-specific variant in a distant locality (see David et al. (2004) and Cohen et al. (2007), Table 1), essentially an assignment approach (Paetkau et al. 1995). While this method is not a definitive measure of a translocation, it can identify the most likely dispersal scenario. Talbi et al. (2010) used Bayesian phylogeographic diffusion models (Lemey et al. 2009) in an attempt to quantify the importance of various anthropogenic predictors on the observed spread of rabies cases in North Africa (see Table 1). Model fitting indicated that dispersal patterns among towns were best explained by road distance, consistent with the anticipated role of human movement. Interestingly, further refinements to calculating distance matrices, such as an accessibility index based on road type and travel time, received only limited model support and road distances were only a marginally better predictor than great-circle distances. This may indicate that RABV dispersal follows a rather homogeneous spatial diffusion process, without any particular effect of landscape heterogeneity. More likely however, it means that more useful geographic predictors of human-mediated dispersal have yet to be found. While this was not possible in the Talbi et al. (2010) study, their method can accommodate a wide range of geographical and environmental predictors and thus provides a promising general framework for examining landscape effects in future data sets.
Population level effects and metapopulation dynamics
Infectious disease dynamics are often described in terms of a metapopulation, with host populations divided into smaller, spatially structured sub-populations that exist with different inter- and intra-patch dynamics (Grenfell and Harwood, 1997). Heterogeneities in the spatial configuration of host populations are critical to understanding the persistence of endemic pathogens (Hagenaars et al. 2004) and incorporating this social/spatial structure is particularly important for developing mechanistic models of acute-acting infections like rabies (Cross et al. 2005). In the case of dog rabies, human settlements (village/town/city) can be considered habitat patches that vary in host density and connectivity (Fig. 1). For example, rural areas typically exist as an array of villages connected to larger towns and cities by major roads. Fragmentation and low connectivity, i.e. long distances between settlements, natural barriers and lack of transportation networks, likely restrict the ability of a dog to move across a landscape and hence limit rabies spread. Hypothetically, limited contact between dog populations and correspondingly strong spatial structure of rabies would be expected in the least developed regions where travel is most limited.
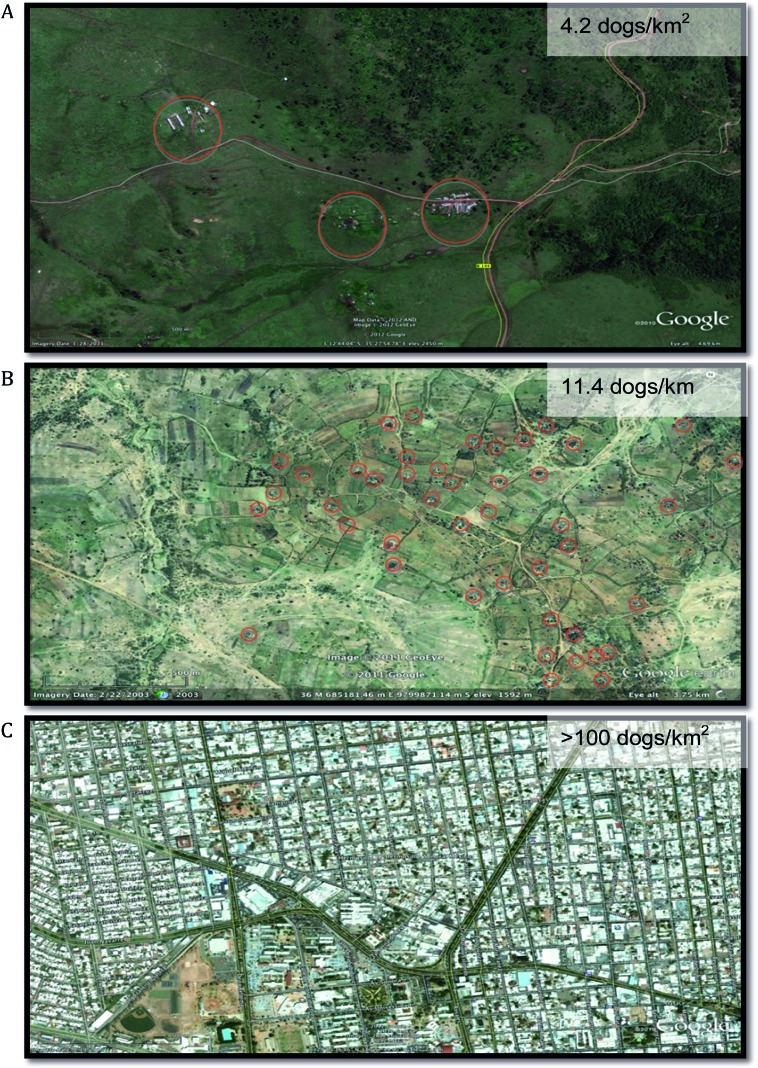
Fig. 1.
Varying spatial complexity in areas with endemic dog rabies as a result of increasing dog population density, A) low density: Ngorongoro District, Tanzania; B) medium density: Serengeti District, Tanzania, C) high density: Hermosillo, Mexico. Red circles highlight settlements in rural areas. Maps obtained using Google Earth (http://earth.google.com).
Local within-patch dynamics are potentially a key component contributing to the persistence and maintenance of disease at larger scales. Therefore, it is important to quantify the relative contribution of different patches to overall disease persistence in a system. This might involve characterizing patches based on dog density, turnover and growth rates, levels of ownership and vaccination, alongside measures of connectivity. For example, dog turnover rates can be extremely high e.g. the Machakos region in Kenya has a dog population estimated to grow by 9% per annum, making rabies control difficult in this area (Kitala et al. 2001). Areas such as these may possibly be identified as hotspots or source patches where rabies has a high chance of being maintained and spread from.
A potentially useful tool for quantifying how rabies dynamics differ among local patches is the basic reproductive number, R0. Theory would suggest that for directly transmitted diseases such as rabies, R0 should increase with host density. However, empirical evidence for density-dependent dynamics is equivocal, with no detectable differences in R0 among dog populations with varying densities around the world (Hampson et al. 2009). While it is unclear why R0 is so insensitive to differences in dog density, e.g. Mexico (∼100 dogs/km2 (Eng et al. 1993)) versus rural Africa (∼10 dogs/km2 (Lembo et al. 2008)), this finding is more consistent with a frequency-dependent mode of rabies transmission. For both density- and frequency-dependent disease dynamics stochastic extinctions are expected once density drops below a certain level, meaning that low-density patches may act as barriers to spread. Indeed, rabies has been observed to spread but appears less able to persist in low density dog populations, e.g. Ngorongoro in Tanzania (average of 4·2 dogs/km2 (Lembo et al. 2008)), suggesting that neighbouring source populations are required for rabies maintenance. Cross et al. (2007) discuss the incorporation of heterogeneities in population structure by expanding on the utility of R0 as a measure of disease transmission. R0 assumes that the population is evenly mixed, but the hierarchical nature of disease invasion requires stochastic models that incorporate both within-patch transmission (R0) and the factors contributing to persistence, i.e. between patch transmission – namely the recruitment of susceptibles, group size and the infectious period. These questions of potential metapopulation structure, patch variability and source-sink dynamics (Pulliam, 1988) represent a particularly pertinent and rich area for future studies of RABV which has yet to be explored using genetic approaches and are vital to inform efficient control strategies.
Host movement rates and connectivity between sub-populations are crucial predictors of disease invasion (Cross et al. 2005; Beyer et al. 2011). Epidemiological data available at a localised scale, e.g. the incidence of dog bites, can provide a simple spatio-temporal measure for metapopulation analysis, as shown by Beyer et al. (2011). Patch-occupancy models identified two metrics of connectivity, the distance between neighbouring villages and the size of villages receiving infection, as significant factors facilitating the transmission of disease. From a population genetics perspective, coalescent analysis of sequence data could provide the means to identify metapopulation dynamics that may not be discernable using epidemiological data alone. Such techniques have been successfully applied, for example, to understand the persistence of influenza A virus, with molecular clock-based estimates of divergence times of most recent common ancestors (MRCA) and demonstrating persistence due to dynamic migration patterns rather than source-sink dynamics (Russell et al. 2008; Bedford et al. 2010).
More generally, the fine-grained spatial genetic structure evident in wildlife rabies that is lacking for dog rabies may be due to differences in time frame. The best-studied wildlife rabies dynamics come from invasions that originated a few decades ago (Bourhy et al. 1999; Real et al. 2005a; Biek et al. 2007), compared to endemic foci in domestic dogs, thought to have persisted for centuries (Bourhy et al. 2008). Factors influencing persistent endemic cycles are likely to differ from those determining epidemic expansions, with viral population turnover, increased mixture of lineages and the greater role of human-mediated movement resulting in less clearly defined fine-grain viral structure. Again, metapopulation models that capture population connectivity at appropriate spatial and temporal scales may shed light on the genetic structure of endemic foci. Characterizing the landscape connectivity between rabies endemic areas (or individual rabies cases, given the resolution of spatial data) may be facilitated using techniques applied in conservation genetics that have yet to be exploited for disease dynamics. For example, the program Fractionnator, (http://www.unil.ch/biomapper/frictionnator/frictionnator.html), provides a ‘strip statistic’ quantifying the effect of landscape features on the relatedness of all possible pairs of individuals sampled. This entails defining grid cells within a strip across the sampled landscape and assessing the abundance of landscape features within each strip. In addition, advances in the development of spatial analysis software such as python-based customised GIS tools (e.g. Etherington, 2001) that allow the visualisation and measurement of genetic relatedness and landscape connectivity based on least cost path analysis, R packages such as Maptools (Lewin-Koh et al. 2012) and alternative open-source software such as PASSaGE 2 (Rosenberg and Anderson, 2011) provide utilities for measuring connectivity between patches, quantifying patterns in spatial data and potentially identifying barriers to dispersal.
Elucidating how heterogeneity at the host level contributes to dynamics at the patch level is an important aspect of understanding the mechanisms underlying rabies persistence and spread. Differential within-patch dynamics resulting from individual heterogeneity including variation in biting frequency, host population size, the structure of a settlement, may confer properties that promote the effectiveness of a patch as a source of infection. Tracing transmission pathways may provide a further means of elucidating source populations that initiate chains of infection, particularly at very fine-scale resolution. Lembo et al. (2007) (Table 1) used parsimonious transmission networks to infer reservoir host dynamics and patterns of interspecific transmission, but this technique could also be used more generally to uncover dispersal patterns, e.g. if a particular patch repeatedly acts as a source of infection. The construction of transmission trees from epidemiological data on the timing and locality of rabies cases is another useful approach for identifying transmission pathways and can be supplemented by contact tracing data (Lembo et al. 2008). Contact tracing of rabies is aided by the memorable nature of rabies bites and therefore presents a unique situation where the collection of contact data is achievable. However, building transmission trees in this way becomes increasingly unreliable as larger numbers of missing links are inferred, and these models may also simplistically predict unlikely transmission events if knowledge of the landscape is not incorporated into algorithms e.g. predictions of transmission across major landscape barriers. Genetic data could be used to quantify mutation rates and build independent time-scaled transmission networks against which epidemiologically constructed transmission trees could be calibrated. Ultimately, transmission trees will be most effectively constructed with the combined use of genetic and epidemiological data, and appropriate datasets are increasingly becoming available for rabies, but further computational and statistical advances are required.
Host transmission is typically heterogeneous, often described by the 80/20 paradigm in which individuals differentially carry and transmit pathogens i.e. 80% of infections are carried by 20% of the population (Woolhouse et al. 1997). In the most extreme case this heterogeneity can exist in the form of superspreaders (Lloyd-Smith et al. 2005). There have been no in-depth studies of superspreading in domestic dogs carrying rabies and the contribution of superspreaders to rabies spread and persistence is controversial (e.g. Hampson et al. 2007 vs. Talbi et al. 2009), but large variations in biting frequency have been observed (Hampson et al. 2009). An important distinction must be made between superspreading in a restricted area, e.g. the same village, and a ‘spatial superspreader’ that has dispersed infection over large distances and multiple settlements (see Fig. 2 for visual example). Although the number of secondary cases caused by such individuals may be equivalent, spatial superspreaders are more important from an epidemiological perspective if they are responsible for connecting sub-populations. Tracing resulting infections to their source through genetic sequence data may identify hotspots where certain patch features promote greater dispersal ability i.e. spatial superspreading events (as suggested by Cross et al. 2007). Ideally, this would utilise retrospective genetic tracing of networks of infection, similar to that used for identifying source infections in foot-and-mouth disease (FMD) (Cottam et al. 2008). Statistically robust methods are yet to be conceived, but accounting for missing links in transmission chains is an important area for future development. As a major advantage compared to FMD, contact that may have lead to transmission is much easier to define for rabies. Contact tracing data, if available, could therefore complement genetic data to offer a more comprehensive view of how individual heterogeneity can impact on dynamics.
Fig. 2.
Dispersal of bites from superspreading dogs resulting in rabies transmission in an area of the Serengeti District in Tanzania. Roads and rivers are shown to highlight the potential influence of landscape features on the dispersal of rabies- tentative observations indicate that superspreader progeny appear to cluster alongside roads and movement may be restricted by the presence of rivers (but other landscape features not shown may also be responsible for influencing dispersal patterns). Two potential types of superspreader are also highlighted in the map: A) a spatial superspreader, which transmits over a large spatial area, potentially connecting sub-populations and may be important from an epidemiological perspective; and B) a superspreader with a limited dispersal range that infects a large number of progeny but remains within a small spatial radius. Inset map shows the location of the Serengeti District within Tanzania.
INTEGRATING LANDSCAPE EPIDEMIOLOGY INTO RABIES CONTROL
Landscape genetics has undoubtedly generated fundamental insights into the dynamics of rabies but has arguably yet to make major contributions to its control. The preceding discussion illustrated potential avenues for exploration and here we aim to more clearly define a landscape genetics research agenda that could directly benefit the planning or implementation of programmes that aim to control or eliminate rabies.
Considerable progress has been made in the development of blueprints and operational toolkits for rabies control (see “Blueprint for rabies prevention and control,” August 2010, http://www.rabiesblueprint.com/, and Lembo et al. 2011). The rabies blueprint presents a major step in the global fight against rabies, providing guidelines and economically feasible strategies to aid policy makers and local communities seeking to embark on rabies intervention and control measures. Landscape genetics research constitutes an important resource within this multidisciplinary approach to help advise and sustain successful control initiatives.
Vaccination is widely deemed the most effective means of rabies control with demonstrated successes in reducing incidence and eliminating disease even in areas with limited resources (Cleaveland et al. 2003; Schneider et al. 2007; Lembo et al. 2010). Molecular genetics has demonstrated that domestic dogs are critical reservoirs for canine rabies, even in parts of Africa with abundant wildlife populations, and therefore indicate that controlling dog rabies through vaccination should eliminate infection from all other species (Lembo et al. 2007, 2008). By changing the susceptibility of populations, mass vaccinations change the landscape in which rabies circulates, and at sufficiently high levels of vaccination coverage, transmission can be interrupted. There are two mechanisms by which vaccination alters the landscape to control rabies. The first is that vaccination itself creates a barrier of susceptible individuals that block the dispersal of the pathogen across the landscape, and the second is via the reinforcement of existing barriers with vaccination. Using landscape genetics to explore the potential impacts of these aspects of landscape control is a logical next step.
Recurrent rabies epidemics occur across large areas where vaccination programmes are patchy and unsustained (Hampson et al. 2007), suggesting that a proactive long-term vaccination programme coordinated across political boundaries is required for success. Indeed, the effectiveness of such a programme has been proven by the intensive vaccinations coordinated by the Pan American Health Organization (PAHO) in Latin America over the past few decades: dog rabies has been eliminated in a large portion of the southern continent, and reported cases from other countries are highly localised due to restrictions on transmission pathways from vaccination barriers (Schneider et al. 2007). Crucially, cases of human rabies have dropped in these areas, reinforcing the importance of effective dog rabies control strategies. Strategically placing a vaccine barrier could maintain freedom from rabies resulting from successful interventions in otherwise landlocked areas.
Environmental or anthropogenic barriers to natural transmission of rabies offer the opportunity to strengthen and smarten vaccination initiatives. Vaccination can be viewed as a form of barrier that impedes rabies spread, and therefore many of the techniques used to draw insights on the permeability of barriers could equally be applied to vaccination programmes. Oral Rabies Vaccination (ORV) campaigns for wildlife, including bait distribution in proximity to a pre-existing natural barrier (the Appalachian Mountains), and utilising existing environmental features to reinforce control campaigns (Wandeler et al. 1988) have successfully contained wildlife rabies. Past experience has shown vulnerability to breaches associated with the differential permeability of barriers and emphasizes the need for ongoing and targeted surveillance to enable early detection and swift responses to incursions (Russell et al. 2005). Yet despite this, few studies have quantified the utility of barriers within a landscape, and there are no guidelines available for the design and implementation of effective cordons sanitaires for dog rabies.
In addition to barrier studies, modern application of spatial data is allowing us to make increasingly accurate measurements of epidemiological parameters that may affect disease dynamics. For example, Bharti et al. (2011) demonstrate the use of remote sensing to test for human predictors of disease. They used anthropogenic light from satellite imagery as a measure of seasonal fluctuations of human populations. The observed fluctuation in light intensity (as a measure of population density) correlated to measles distribution and spread in cities in Niger, and provided an accurate, near real-time representation of short-term population fluctuations that may drive pathogen transmission. This approach demonstrates the use of relatively simple proxies for quantifying migration patterns in poorly resourced regions, often the same regions carrying the highest disease burden from dog rabies.
Large-scale interventions are expensive, and adaptive management is often required alongside intervention. Refinements in resource management will ultimately rely on a combination of knowledge from genetics, landscape and host ecology as part of a reactive programme. Specific landscape elements affecting dog rabies spread are generally not known a priori, so exploration of phylogeographic patterns may identify genetically distinct viral or host populations in areas, which could be targeted for vaccination. However, experience from wildlife rabies suggests that caution is warranted when taking such an approach. Firstly, apparent boundaries between areas dominated by different genetic lineages may have emerged during the initial invasion process and thus do not necessarily reflect areas of low permeability for the virus (Real et al. 2005b; Biek and Real, 2010). Secondly, while the stability of these phylogeographic domains indicates a lack of mixing between them, this simply suggests that immigrating viruses find it difficult to invade areas with an already established focus, but does not signify the absence of viral immigration per se. Such areas with putatively self-contained endemic foci could therefore experience a high risk of rabies re-emergence following successful eradication, unless vaccination effort remains high. Whether the second consideration equally applies to dog rabies is currently not clear as pertinent empirical studies are lacking. As with many research problems in landscape genetics, these types of question may be very productively studied using simulation tools (Epperson et al. 2010). Simulations may indicate the most effective location for vaccine corridors/barriers e.g. vaccination on the far side of a natural barrier (Russell et al. 2006), whereas metapopulation models may elucidate areas with high connectivity that could be the source of persistence in endemic areas. Quantifying the degree of connectivity between sub-populations is of critical importance to understanding how rabies is maintained across landscapes, and is an integral part of designing effective control interventions and cordons sanitaires that limit and contain the virus.
Ongoing surveillance is crucial to the long-term success of rabies control, and considerable value can be added to surveillance initiatives through the incorporation of landscape genetics, with the potential to determine the source of incursions and reveal transmission pathways. Epidemiological surveillance may facilitate active case detection and identification of circulating strains, helping to identify areas missed by vaccination. Using a molecular genetics approach during the 2007 FMD outbreak in the UK, allowed swift and effective containment of the outbreak by directing interventions to the critical areas (Cottam et al. 2008). For countries with limited resources, surveillance in areas with ongoing control programmes and retrospective analysis may be useful for identifying remaining foci of infection to be targeted by vaccination, sources of incursions, or spillover events into or from wildlife. In addition, metapopulation models may be utilized to predict the direction of spread of rabies in naïve populations (Beyer et al. 2011), directing the location of sentinel points and control measures to ‘hotspots’ of infection (Haydon et al. 2006) and putative transmission networks based on sampled cases should indicate how well current levels of surveillance are capturing rabies incidence based on inferred missing links between sampled cases.
Rabies containment and control will require ongoing surveillance and sustained control efforts. In time, when rabies incidence is reduced to low levels (the period when control measures often lapse), genetics can provide a means of directing resources to where they are most needed, maintaining a cost-effective approach. Given the small genome of RABV (12Kb), and advances in NGS sequencing, future exploration of phylogeographic patterns utilizing whole genome sequencing is a realistic prospect. This promises to provide appropriately fine genetic resolution for samples collected on small spatio-temporal scales that might otherwise be uninformative. Application of novel techniques in landscape genetics to other RNA viruses such as FMD and Influenza highlights interesting possibilities for uncovering disease dynamics and aiding control directives (Cottam et al. 2008, Bedford et al. 2010). The technology and analytical power are available but have yet to be fully exploited for dog rabies, and thus offer exciting prospects on what can be achieved with their implementation in the future.
CONCLUSIONS
There is evidently a wide scope for the use of landscape genetics to explore and understand the dynamics of pathogen spread and persistence. Dogs are the principal reservoir of rabies, responsible for the majority of human rabies cases, yet we know little about the dynamics of the pathogen in this host. In order to reduce the many thousands of human rabies deaths that occur annually due to contact with rabid dogs, it is crucial that we uncover the mechanisms governing viral dispersal across the landscape so as to direct successful control interventions. The foundation of knowledge from studies in wildlife populations provides a starting point but aspects of host behaviour, the inherent influence of humans, and the long-term endemic nature of dog rabies foci requires a modified approach be taken for dog rabies. There lies great potential for advancing the effectiveness of control campaigns in areas burdened with disease; specifically landscape genetics has most to contribute to improving surveillance and modifying control strategies based on information gleaned from surveillance. This includes the design and placement of cordons sanitaires, the prioritization of effort towards persistent foci or targeting sources of outbreaks and conduits of transmission. Given the availability of powerful new genetic and spatial techniques, efforts now need to push towards real-world application of landscape genetics to rabies control and elimination.
ACKNOWLEDGEMENTS
We thank Sarah Cleaveland, Tiziana Lembo and an anonymous reviewer for helpful comments on this manuscript.
FINANCIAL SUPPORT
This work was supported by a Medical Research Council doctoral training grant to Glasgow University, the Wellcome Trust and the Research and Policy for Infectious Disease Dynamics (RAPIDD) program of the Science and Technology Directorate, U.S. Department of Homeland Security, and the Fogarty International Centre, NIH.
REFERENCES
- Anderson C. D., Epperson B. K., Fortin M. J., Holderegger R., James P. M. A., Rosenberg M. S., Scribner K. T. and Spear S. (2010). Considering spatial and temporal scale in landscape-genetic studies of gene flow. Molecular Ecology 19, 3565–3575 doi: 10.1111/J.1365-294x.2010.04757.X [DOI] [PubMed] [Google Scholar]
- Anderson R. M. and May R. M. (1991). Infectious Diseases of Humans: Dynamics and Control. Oxford University Press, Oxford: 126, 186–195 [Google Scholar]
- Archie E. A., Luikart G. and Ezenwa V. O. (2009). Infecting epidemiology with genetics: a new frontier in disease ecology. Trends in Ecology and Evolution 24, 21–30 doi: 10.1016/j.tree.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Bedford T., Cobey S., Beerli P. and Pascual M. (2010). Global migration dynamics underlie evolution and persistence of human influenza A (H3N2). PLoS Pathogens 6 doi: 10.1371/journal.ppat.1000918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer H. L., Hampson K., Lembo T., Cleaveland S., Kaare M. and Haydon D. T. (2011). Metapopulation dynamics of rabies and the efficacy of vaccination. Proceedings of the Royal Society of London series B-Biological Sciences 278, 2182–2190 doi: 10.1098/rspb.2010.2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti N., Tatem A. J., Ferrari M. J., Grais R. F., Djibo A. and Grenfell B. T. (2011). Explaining seasonal fluctuations of measles in Niger using nighttime lights imagery. Science 334, 1424–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R., Henderson J. C., Waller L. A., Rupprecht C. E. and Real L. A. (2007). A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proceeding of the National Academy of Sciences, USA 104, 7993–7998 doi:: 10.1073/pnas.0700741104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek R. and Real L. A. (2010). The landscape genetics of infectious disease emergence and spread. Molecular Ecology 19, 3515–3531 doi: 10.1111/j.1365-294X.2010.04679.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy H., Kissi B., Audry L., Smreczak M., Sadkowska-Todys M., Kulonen K., Tordo N., Zmudzinski J. F. and Holmes E. C. (1999). Ecology and evolution of rabies virus in Europe. Journal of General Virology 80, 2545–2557 [DOI] [PubMed] [Google Scholar]
- Bourhy H., Reynes J. M., Dunham E. J., Dacheux L., Larrous F., Huong V. T., Xu G., Yan J., Miranda M. E. and Holmes E. C. (2008). The origin and phylogeography of dog rabies virus. Journal of General Virology 89, 2673–2681 doi: 10.1099/vir.0.2008/003913-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. R. A. (2000). Demography and dog-human relationships of the dog population in Zimbabwean communal lands. Veterinary Record 147, 442–446 [DOI] [PubMed] [Google Scholar]
- Carnieli P. Jr., de Novaes Oliveira R., Macedo C. I. and Castilho J. G. (2011). Phylogeography of rabies virus isolated from dogs in Brazil between 1985 and 2006. Archives of Virology 156, 1007–1012 [DOI] [PubMed] [Google Scholar]
- Cleaveland S. (1998). The growing problem of rabies in Africa. Transactions of the Royal Society of Tropical Medicine and Hygiene 92, 131–134 [DOI] [PubMed] [Google Scholar]
- Cleaveland S., Kaare M., Tiringa P., Mlengeya T. and Barrat J. (2003). A dog rabies vaccination campaign in rural Africa: impact on the incidence of dog rabies and human dog-bite injuries. Vaccine 21, 1965–1973 doi: 10.1016/S0264-410x(02)00778-8 [DOI] [PubMed] [Google Scholar]
- Coetzee P. and Nel L. H. (2007). Emerging epidemic dog rabies in coastal South Africa: a molecular epidemiological analysis, Virus Research 126, 186–195 [DOI] [PubMed] [Google Scholar]
- Cohen C., Sartorius B., Sabeta C., Zulu G., Paweska J., Mogoswane M., Sutton C., Nel L. H., Swanepoel R., Leman P. A., Grobbelaar A. A., Dyason E. and Blumberg L. (2007). Epidemiology and molecular virus characterization of reemerging rabies, South Africa. Emerging Infectious Diseases 13, 1879–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E. M., Wadsworth J., Shaw A. E., Rowlands R. J., Goatley L., Maan S., Maan N. S., Mertens P. P. C., Ebert K., Li Y., Ryan E. D., Juleff N., Ferris N. P., Wilesmith J. W., Haydon D. T., King D. P., Paton D. J. and Knowles N. J. (2008). Transmission pathways of foot-and-mouth disease virus in the United Kingdom in 2007. PLoS Pathogens 4 doi: 10.1371/Journal.Ppat.1000050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross P. C., Lloyd-Smith J. O., Johnson P. L. F. and Getz W. M. (2005). Duelling timescales of host movement and disease recovery determine invasion of disease in structured populations. Ecology Letters 8, 587–595 doi: 10.1111/J.1461-0248.2005.00760.X [DOI] [Google Scholar]
- Cross P. C., Johnson P. L. F, Lloyd-Smith J. O. and Getz W. M. (2007). Utility of R0 as a predictor of disease invasion in structured populations. Journal of the Royal Society 4, 315–324 doi: 10.1098/rsif.2006.0185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullingham C. I., Kyle C. J., Pond B. A., Rees E. E. and White B. N. (2009). Differential permeability of rivers to raccoon gene flow corresponds to rabies incidence in Ontario, Canada. Molecular Ecology 18, 43–53 doi: 10.1111/j.1365-294X.2008.03989.x [DOI] [PubMed] [Google Scholar]
- Cullingham C. I., Pond B. A., Kyle C. J., Rees E. E., Rosatte R. C. and White B. N. (2008). Combining direct and indirect genetic methods to estimate dispersal for informing wildlife disease management decisions. Molecular Ecology 17, 4874–4886 doi: 10.1111/j.1365-294X.2008.03956.x [DOI] [PubMed] [Google Scholar]
- David D., Yakobson B. A., Gershkovich L. and Gayer S. (2004). Tracing the regional source of rabies infection in an Israeli dog by viral analysis. Veterinary Record 155, 496–497 [DOI] [PubMed] [Google Scholar]
- de Mattos C. C., de Mattos C. A., Loza-Rubio E., Aguilar-Setien A., Orciari L. A. and Smith J. S. (1999). Molecular characterization of rabies virus isolates from Mexico: Implications for transmission dynamics and human risk. American Journal of Tropical Medicine and Hygiene 61, 587–597 [DOI] [PubMed] [Google Scholar]
- Denduangboripant J., Wacharapluesadee S., Lumlertdacha B., Ruankaew N., Hoonsuwan W., Puanghat A. and Hemachudha T. (2005). Transmission dynamics of rabies virus in Thailand: implications for disease control. BMC Infectious Diseases 5, 52 doi: 10.1186/1471-2334-5-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzschold B., Schnell M. and Koprowski H. (2005). Pathogenesis of rabies. Current Topics in Microbiology and Immunology 292, 45–56 [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Pybus O. G., Rambaut A., Forsberg R. and Rodrigo A. G. (2003). Measurably evolving populations. Trends in Ecology and Evolution 18, 481–488 doi: 10.1016/S0169-5347(03)00216-7 [DOI] [Google Scholar]
- Eng T. R., Fishbein D. B., Talamante H. E., Hall D. B., Chavez G. F., Dobbins J. G., Muro F. J., Bustos J. L., Ricardy M. D., Munguia A., Carrasco J., Robles A. R. and Baer G. M. (1993). Urban Epizootic of Rabies in Mexico – Epidemiology and Impact of Animal Bite Injuries. Bulletin of the World Health Organization 71, 615–624 [PMC free article] [PubMed] [Google Scholar]
- Epperson B. K., McRae B. H., Scribner K., Cushman S. A., Rosenberg M. S., Fortin M. J., James P. M. A., Murphy M., Manel S., Legendre P. and Dale M. R. T. (2010). Utility of computer simulations in landscape genetics. Molecular Ecology 19, 3549–3564 doi: 10.1111/J.1365-294x.2010.04678.X [DOI] [PubMed] [Google Scholar]
- Etherington T. R. (2011). Python-based GIS tools for landscape genetics: visualising genetic relatedness and measuring landscape connectivity. Methods in Ecology and Evolution 2, 52–55 [Google Scholar]
- Excoffier L. and Ray N. (2008). Surfing during population expansions promotes genetic revolutions and structuration. Trends in Ecology and Evolution 23, 347–351 doi: 10.1016/J.Tree.2008.04.004 [DOI] [PubMed] [Google Scholar]
- Gautret P., Ribadeau-Dumas F., Parola P., Brouqui P, and Bourhy H. (2011), Risk for rabies importation from north Africa. Emerging Infectious Diseases 17, 2187–93. doi: 10.3201/eid1712.110300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenfell B. and Harwood J. (1997). (Meta)population dynamics of infectious diseases. Trends in Ecology and Evolution 12, 395–399 [DOI] [PubMed] [Google Scholar]
- Hagenaars T. J., Donnelly C. A. and Ferguson N. M. (2004). Spatial heterogeneity and the persistence of infectious diseases. Journal of Theoretical Biology 229, 349–359 doi: 10.1016/j.jtbi.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Hampson K., Dushoff J., Bingham J., Bruckner G., Ali Y. H. and Dobson A. (2007). Synchronous cycles of domestic dog rabies in sub-Saharan Africa and the impact of control efforts. Proceedings of the National Academy of Sciences, USA 104, 7717–7722 doi: 10.1073/pnas.0609122104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson K., Dushoff J., Cleaveland S., Haydon D. T., Kaare M., Packer C. and Dobson A. (2009). Transmission Dynamics and Prospects for the Elimination of Canine Rabies. PLoS Biology 7, 462–471 doi: 10.1371/Journal.Pbio.1000053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon C. A., Niezgoda M. and Rupprecht C. E. (2007). Rabies in terrestrial animals In Rabies (ed. Wunner A. C. J. H. W.), pp. 201–258 UK: Elsevier, London [Google Scholar]
- Haydon D. T., Randall D. A., Matthews L., Knobel D. L., Tallents L. A., Gravenor M. B., Williams S. D., Pollinger J. P., Cleaveland S., Woolhouse M. E. J., Sillero-Zubiri C., Marino J., Macdonald D. W. and Laurenson M. K. (2006). Low-coverage vaccination strategies for the conservation of endangered species. Nature 443, 692–695 doi: 10.1038/Nature05177 [DOI] [PubMed] [Google Scholar]
- Hayman D. T. S., Johnson N., Horton D. L., Hedge J., Wakeley P. R., Banyard A. C., Zhang S. F., Alhassan A. and Fooks A. R. (2011). Evolutionary History of Rabies in Ghana. PLoS Neglected Tropical Diseases 5 doi: 10.1371/journal.pntd.0001001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E. C. (2004). The phylogeography of human viruses. Molecular Ecology 13, 745–756 doi: 10.1046/J.1365-294x.2003.02051.X [DOI] [PubMed] [Google Scholar]
- Holmes E. C. and Grenfell B. T. (2009). Discovering the Phylodynamics of RNA Viruses. PLoS Computational Biology 5 doi: 10.1371/Journal.Pcbi.1000505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitala P., McDermott J., Kyule M., Gathuma J., Perry B. and Wandeler A. (2001). Dog ecology and demography information to support the planning of rabies control in Machakos District, Kenya. Acta Tropica 78, 217–230 doi: 10.1016/S0001-706X(01)00082-1 [DOI] [PubMed] [Google Scholar]
- Klopfstein S., Currat M. and Excoffier L. (2006). The fate of mutations surfing on the wave of a range expansion. Molecular Biology and Evolution 23, 482–490 doi: 10.1093/molbev/msj057 [DOI] [PubMed] [Google Scholar]
- Knobel D. L., Cleaveland S., Coleman P. G., Fevre E. M., Meltzer M. I., Miranda M. E. G., Shaw A., Zinsstag J. and Meslin F. X. (2005). Re-evaluating the burden of rabies in Africa and Asia. Bulletin of the World Health Organization 83, 360–368 [PMC free article] [PubMed] [Google Scholar]
- Knobel D. L., Laurenson M. K., Kazwala R. R., Boden L. and Cleaveland S. (2008) A cross-sectional study of factors associated with dog ownership in Tanzania. BMC Veterinary Research 4, 5 doi: 10.1186/1746-6148-4-5.0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo T., Hampson K., Haydon D. T., Craft M., Dobson A., Dushoff J., Ernest E., Hoare R., Kaare M., Mlengeya T., Mentzel C. and Cleaveland S. (2008). Exploring reservoir dynamics: a case study of rabies in the Serengeti ecosystem. Journal of Applied Ecology 45, 1246–1257 doi: 10.1111/j.1365-2664.2008.01468.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo T., Hampson K., Kaare M. T., Ernest E., Knobel D., Kazwala R. R., Haydon D. T. and Cleaveland S. (2010). The Feasibility of Canine Rabies Elimination in Africa: Dispelling Doubts with Data. PLoS Neglected Tropical Diseases 4 doi: e626 doi: 10.1371/journal.pntd.0000626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo T., Haydon D. T., Velasco-Villa A., Rupprecht C. E., Packer C., Brandao P. E., Kuzmin I. V., Fooks A. R., Barrat J. and Cleaveland S. (2007). Molecular epidemiology identifies only a single rabies virus variant circulating in complex carnivore communities of the Serengeti. Proceedings of the Royal Society of London series B-Biological Sciences 274, 2123–2130 doi: 10.1098/rspb.2007.0664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo L., Attlan M., Bourhy H., Cleaveland S., Costa P., de Balogh K., Dodet B., Fooks A. R., Hiby E., Leanes F., Meslin F., Miranda M. E., Müller T., Nel L. H., Rupprecht C. H., Tordo N., Tumpey A., Wandeler A. and Briggs D. J. (2011) Renewed global partnerships and redesigned roadmaps for rabies prevention and control. Veterinary Medicine International, vol. 2011, Article ID 923149, 18 pages, 2011. doi: 10.4061/2011/9231492011:923149 Epub 2011 Jun 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Drummond A. J., Suchard M. A. (2009). Bayesian phylogeography finds its roots. PLoS Computational Biology 5 doi: 10.1371/journal.pcbi.1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Welch J. J. and Suchard M. A. (2010). Phylogeography takes a relaxed random walk in continuous space and time. Molecular Biology and Evolution 27, 1877–1885 doi: 10.1093/molbev/msq067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin-Koh N. J., Bivand R., contributions by Pebesma E. J., Archer E., Baddeley A., Bibiko H-J., Callahan J., Dray S., Forrest D., Friendly M., Giraudoux P., Golicher D., Rubio V. G., Hausmann P., Hufthammer K. O., Jagger T., Luque S. P., MacQueen D., Niccolai A., Short T., Snow G., Stabler B. and Turner R. (2012). Maptools: Tools for reading and handling spatial objects. R package version 0.8–14. http://CRAN.R-project.org/package = maptools)
- Lloyd-Smith J. O., Schreiber S. J., Kopp P. E. and Getz W. M. (2005). Superspreading and the effect of individual variation on disease emergence. Nature 438, 355–359 doi: 10.1038/nature04153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel S., Schwartz M. K., Luikart G. and Taberlet P. (2003). Landscape genetics: combining landscape ecology and population genetics. Trends in Ecology and Evolution 18, 189–197 doi: 10.1016/s0169-5347(03)00008-9 [DOI] [Google Scholar]
- McCallum H. (2008). Landscape structure, Disturbance, and Disease Dynamics In Infectious Disease Ecology: The Effects of Ecosystems on Disease and of Disease on Ecosystems (eds. Ostfeld R. S., Keesing F., and Eviner V. T.), pp. 100–122 Princeton University Press, Princeton, New Jersey [Google Scholar]
- Metzker M. L. (2010). Sequencing technologies – the next generation. Nature Review Genetics 11, 31–46 doi: 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- Nettles V. F., Shaddock J. H., Sikes R. K. and Reyes C. R. (1979). Rabies in Translocated Raccoons. American Journal of Public Health 69, 601–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostfeld R. S., Glass G. E. and Keesing F. (2005). Spatial epidemiology: an emerging (or re-emerging) discipline. Trends in Ecology and Evolution 20, 328–336 doi: 10.1016/j.tree.2005.03.009 [DOI] [PubMed] [Google Scholar]
- Paetkau D., Calvert W., Stirling I. and Strobeck C. (1995). Microsatellite analysis of population structure in Canadian polar bears. Molecular Ecology 4, 347–354 [DOI] [PubMed] [Google Scholar]
- Parker J., Rambaut A. and Pybus O. G. (2008). Correlating viral phenotypes with phylogeny: Accounting for phylogenetic uncertainty. Infection, Genetics and Evolution 8, 239–246 doi: 10.1016/j.meegid.2007.08.001 [DOI] [PubMed] [Google Scholar]
- Pulliam H. R. (1988). Sources, Sinks, and Population Regulation. American Naturalist 132, 652–661 [Google Scholar]
- R Development Core Team (2012). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/ [Google Scholar]
- Real L. A. and Biek R. (2007). Spatial dynamics and genetics of infectious diseases on heterogeneous landscapes. Journal of the Royal Society Interface 4, 935–948 doi: 10.1098/rsif.2007.1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real L. A., Henderson J. C., Biek R., Snaman J., Jack T. L., Childs J. E., Stahl E., Waller L., Tinline R. and Nadin-Davis S. (2005a). Unifying the spatial population dynamics and molecular evolution of epidemic rabies virus. Proceedings of the National Academy of Sciences, USA 102, 12107–12111 doi: 10.1073/pnas.0500057102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real L. A., Russell C., Waller L., Smith D. and Childs J. (2005b). Spatial dynamics and molecular ecology of North American rabies. Journal of Heredity 96, 253–260 doi: 10.1093/jhered/esi031 [DOI] [PubMed] [Google Scholar]
- Rees E. E., Pond B. A., Cullingham C. I., Tinline R., Ball D., Kyle C. J. and White B. N. (2008). Assessing a landscape barrier using genetic simulation modelling: Implications for raccoon rabies management. Preventative Veterinary Medicine 86, 107–123 doi: 10.1016/J.Prevetmed.2008.03.007 [DOI] [PubMed] [Google Scholar]
- Rees E. E., Pond B. A., Cullingham C. I., Tinline R. R., Ball D., Kyle C. J. and White B. N. (2009). Landscape modelling spatial bottlenecks: implications for raccoon rabies disease spread. Biology Letters 5, 387–390 doi: 10.1098/rsbl.2009.0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M. S. and Anderson C. D. (2011). PASSaGE: Pattern Analysis, Spatial Statistics and Geographic Exegesis. Version 2. Methods in Ecology and Evolution 2, 229–232 [Google Scholar]
- Rupprecht C. E., Hanlon C. A. and Hemachudha T. (2002), Rabies Re-examined. The Lancet Infectious Diseases 2, 327–343 [DOI] [PubMed] [Google Scholar]
- Russell C. A., Jones T. C., Barr I. G., Cox N. J., Garten R. J., Gregory V., Gust I. D., Hampson A. W., Hay A. J., Hurt A. C., de Jong J. C., Kelso A., Klimov A. I., Kageyama T., Komadina N., Lapedes A. S., Lin Y. P., Mosterin A., Obuchi M., Odagiri T., Osterhaus A. D., Rimmelzwaan G. F., Shaw M. W., Skepner E., Stohr K., Tashiro M., Fouchier R. A. and Smith D. J. (2008). The global circulation of seasonal influenza A (H3N2) viruses. Science 320, 340–346 doi: 10.1126/science.1154137 [DOI] [PubMed] [Google Scholar]
- Russell C. A., Real L. A. and Smith D. L. (2006). Spatial Control of Rabies on Heterogeneous Landscapes. PLoS One 1 doi: E27 10.1371/Journal.Pone.0000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C. A., Smith D. L., Childs J. E. and Real L. A. (2005). Predictive spatial dynamics and strategic planning for raccoon rabies emergence in Ohio. PLoS Biology 3, 382–388 doi: E88 10.1371/Journal.Pbio.0030088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M. C., Belotto A., Ade M. P., Hendrickx S., Leanes L. F., Rodrigues M. J. D., Medina G. and Correa E. (2007). Current status of human rabies transmitted by dogs in Latin America. Cadernos De Saude Publica 23, 2049–2063 [DOI] [PubMed] [Google Scholar]
- Smith J. S., Orciari L. A., Yager P. A., Seidel H. D. and Warner C. K. (1992). Epidemiologic and historical relationships among 87 rabies virus isolates as determined by limited sequence analysis. Journal of Infectious Diseases 166, 296–307 [DOI] [PubMed] [Google Scholar]
- Smith D. L., Lucey B., Waller L. A., Childs J. E. and Real L. A. (2002). Predicting the spatial dynamics of rabies epidemics on heterogeneous landscapes. Proceedings of the National Academy of Sciences, USA 99, 3668–3672 doi: 10.1073/pnas.042400799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. L., Waller L. A., Russell C. A., Childs J. E. and Real L. A. (2005). Assessing the role of long-distance translocation and spatial heterogeneity in the raccoon rabies epidemic in Connecticut. Preventative Veterinary Medicine 71, 225–240 doi: 10.1016/j.prevetmed.2005.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susilawathi N. M, Darwinata A. E., Dwija I. B. N. P., Budayanti N. S., Wirasandhi G. A. K., Subrata K., Susialarini N. K., Sudewi R. A. A., Wignall F. S. and Mahardika G. N. K. (2012). Epidemiological and Clinical Features of human rabies cases in Bali 2008–2010. BMC Infectious Diseases 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R., Barnard B. J., Meredith C. D., Bishop G. C., Bruckner G. K., Foggin C. M. and Hübschle O. J. (1993). Rabies in southern Africa. Onderstepoort Journal of Veterinary Research 60, 325–46. [PubMed] [Google Scholar]
- Szanto A. G., Nadin-Davis S. A., Rosatte R. C. and White B. N. (2011). Genetic tracking of the raccoon variant of rabies virus in eastern North America. Epidemics, 3, 76–87 doi: 10.1016/j.epidem.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Talbi C., Holmes E. C., de Benedictis P., Faye O., Nakouné E., Gamatié D., Diarra A., Elmamy B. O., Sow A., Adjogoua E. V., Sangare O., Dundon W. G., Capua I., Sall A. A. and Bourhy H. (2009). Evolutionary history and dynamics of dog rabies virus in western and Central Africa. Journal of General Virology 90, 783–91. [DOI] [PubMed] [Google Scholar]
- Talbi C., Lemey P., Suchard M. A., Abdelatif E., Elharrak M., Nourlil J., Faouzi A., Echevarria J. E., Vazquez Moron S., Rambaut A., Campiz N., Tatem A. J., Holmes E. C. and Bourhy H. (2010). Phylodynamics and human-mediated dispersal of a zoonotic virus. PLoS Pathogens 6 doi: 10.1371/journal.ppat.1001166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzin, Sharma B., Dhand N. K., Timsina N. and Ward M. P. (2010). Reemergence of Rabies in Chhukha District, Bhutan, 2008. Emerging Infectious Diseases 16, 1925–1930 doi: 10.3201/Eid1612.100958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandeler A. I., Capt S., Gerber H., Kappeler A. and Kipfer R. (1988). Rabies epidemiology, natural barriers and fox vaccination. Parassitologia 30, 53–57 [PubMed] [Google Scholar]
- Wang T. H., Donaldson Y. K., Brettle R. P., Bell J. E., and Simmonds P. (2001). Identification of shared populations of human immunodeficiency Virus Type 1 infecting microglia and tissue macrophages outside the central nervous system. Journal of Virology 75, 11686–11699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Hoffmann U., Freuling C., Müller T., Fesseler M. and Renner C. (2009). Rabies exposure due to an illegally imported dog in Germany. Rabies Bulletin Europe 33, 5–7 [Google Scholar]
- Wheeler D. C. and Waller L. A. (2008). Mountains, valleys, and rivers: The transmission of raccoon rabies over a heterogeneous landscape. Journal of Agricultural, Biological, and Environmental Statistics 13, 388–406 doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. L., Bretsky P. M., Cooper G. H., Egbertson S. H., VanKruiningen H. J. and Cartter M. L. (1997). Emergence of raccoon rabies in Connecticut, 1991–1994: Spatial and temporal characteristics of animal, infection and human contact. American Journal of Tropical Medicine and Hygiene 57, 457–463 [DOI] [PubMed] [Google Scholar]
- Windiyaningsih C., Wilde H., Meslin F. X., Suroso T. and Widarson H. S. (2004). The Rabies Epidemic on Flores Island, Indonesia (1998–2003). Journal of the Medical Association of Thailand 87, 1389–1393 [PubMed] [Google Scholar]
- Woodroffe R. and Donnelly C. A. (2011). Risk of contact between endangered African wild dogs Lycaon pictus and domestic dogs: opportunities for pathogen transmission. Journal of Applied Ecology 48, 1345–1354 doi: 10.1111/j.1365-2664.2011.02059.x [DOI] [Google Scholar]
- Woolhouse M. E., Dye C., Etard J. F., Smith T., Charlwood J. D., Garnett G. P., Hagan P., Hii J. L., Ndhlovu P. D., Quinnell R. J., Watts C. H., Chandiwana S. K. and Anderson R. M. (1997). Heterogeneities in the transmission of infectious agents: implications for the design of control programs. Proceedings of the National Academy of Sciences, USA 94, 338–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni R. and Breitenmoser U. (2003). Rabies in a puppy in Nyon, Switzerland. Rabies Bulletin Europe 27, 5–7 [Google Scholar]