Abstract
Hidden genetic variations harbor potential for the evolution of new traits. Molecular chaperones, that assist protein folding, may conceal genetic variations in protein coding regions. Here, we investigate if the chemical milieu of cells has the potential to alleviate intracellular protein folding; potentially implicating a role of osmolytes in concealing genetic variations. Using the model osmolyte TMAO, we uncover that it can buffer mutations that impose kinetic traps in the folding pathways of two model proteins. Using this information, we rationally designed TMAO-dependent mutants in vivo, starting from a TMAO-independent protein. Strikingly, we delineate different osmolytes to have a unique spectrum of buffered-mutations. Consequently, the chemical milieu of cells may alter the folding pathways of unique mutant variants in polymorphic populations and lead to unanticipated spectra of genetic buffering.
Keywords: Protein folding, chaperone, chemical chaperone, Proteostasis, TMAO, genetic buffering, canalization
Alterations in gene sequences are a major source of variation in protein structure and function leading to phenotypic variation 1,2. Since native structures of proteins are only marginally stable, majority of non-conserved changes in the protein sequences lead to the formation of non-functional or metastable proteins 3,4. Metastable proteins are functional under permissive conditions but are rendered inactive under special circumstances, which could be a major source of hidden genetic diversity, a phenomenon referred to as genetic-canalization 5. Metastability, through canalization, may allow robust protein structures to adopt alternate conformations without losing in vivo activity; a significant step towards evolution of new protein function6-12. To establish this link we need to understand if protein folding modulators assist intracellular folding of metastable mutants.
Since intracellular milieu is enriched with osmolytes that act as chemical chaperones, we asked if these could favor genetic canalization. Although there are preliminary reports of osmolyte dependent folding in vivo 13 ,14 ,15, it is unknown if osmolytes affect canalization by assisting the folding of metastable proteins. Since osmolytes may also affect chaperone structure and function 16,17, demonstration of direct action of osmolytes in buffering mutations is a prerequisite to underline the importance of chemical milieu in shaping sequence evolution. Mechanistically, chemical chaperones are believed to act through solvents 18 and this finding is based on in vitro studies performed under solution abundant conditions. It remains to be seen if osmolytes are active in the crowded cellular environment 19-21, where the solvent volume is heavily diminished.
It is important to understand the mechanism of folding assistance afforded by specific osmolytes in vivo, which would determine if specific mutations are preferentially buffered by osmolytes. Furthermore, it is highly likely that different chemical chaperones, depending on their mode of action, may have differential buffering potential and substrate specificity like molecular chaperones 22,23.
To understand osmolyte dependent modulation of intracellular folding and specific mutational buffering we chose to use TMAO as a model chemical chaperone. It is physiologically abundant in certain class of organisms which accumulate large concentrations of Urea24. As a first step towards reconstituting osmolyte-dependence, we investigated the molecular basis of TMAO-assisted protein folding. We used a slow folding mutant of Maltose Binding Protein (DM-MBP), to demonstrate a direct correlation between TMAO assisted folding in vitro and in vivo. Using an unrelated test protein (GCN5 related acetyl transferase, Serratia marescens, aminoglycoside 3-N-acetyltransferase Gm-R), we reconstituted TMAO-dependence, confirming that TMAO assists proteins that encode flexibility driven traps in folding pathway. This mode of action was found to be unique to TMAO and Trehalose, with chemical chaperones like proline and glycerol being minimally-functional in buffering these mutations. Distinct spectrum of mutational buffering in the presence of chemical chaperones was observed with mutant libraries of two different proteins. We suggest that naturally occurring osmolytes have distinct proteostatic potential to alter metastability in vivo depending on mode of frustration in the folding pathway, and thus have a distinct spectrum of buffered-genotypes.
RESULTS
Chemical chaperone assists folding kinetics in vivo
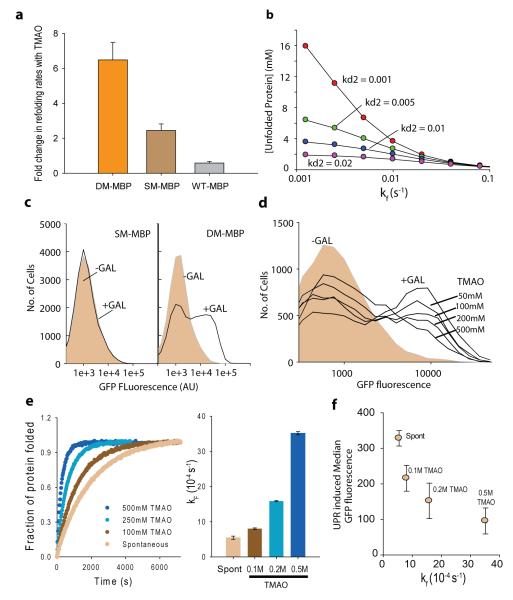
To investigate whether osmolytes buffer genetic variations by chaperoning protein folding, we chose TMAO as a model osmolyte. DM-MBP was chosen as an ideal candidate protein. It is a slow folding variant of Maltose Binding Protein (MBP) with two mutations Y283D and V8G (Supplemental results, Supplementary Figure 1) and is known to be a TMAO-dependent folder 25. We asked if refolding of the parent proteins MBP and SM-MBP (MBP(Y283D)) are also assisted by TMAO. Although TMAO accelerates the refolding rate of DM-MBP by ~7 fold (Fig 1a and Supplementary Figure 2), it affords no acceleration to the refolding rate of MBP. SM-MBP refolding is accelerated only modestly by a factor of ~2.4 (Fig 1a and Supplementary Figure 2). Thus, Y283D mutation in SM-MBP imposes a modest TMAO dependence whereas the additional V8G mutation renders the DM-MBP significantly more dependent on TMAO. This implicates that mutations may render the folding of a protein TMAO-dependent even when the folding of wild-type variant is not assisted by TMAO. These proteins differ by a maximum of two residues and have similar native states indicating that TMAO-assistance may be primarily determined by the folding pathway of the protein. Thus, DM-MBP provided an ideal platform to investigate osmolyte dependent folding and mutational buffering.
Figure 1.
Chemical chaperone assists rapid attainment of native structure. (a) Single amino acid substitution renders MBP TMAO dependent. Fold change in refolding rates of DM-MBP, SM-MBP and Wt-MBP, in presence of TMAO as compared to spontaneous refolding. The rates were obtained by fitting the curves to single exponential equations. (b) Kinetic simulation output showing steady state level of unfolded protein increasing with decreasing folding rates. Accumulation of non-native protein with varying folding rates is shown for different rates of degradation of the non-native protein. Degradation rate was varied from 0.001 to 0.02 s−1. (c) Induction of UPR as observed by increase in GFP fluorescence on Galactose induction of DM-MBP as compared to SM-MBP. GFP fluorescence was recorded using FACS. (d) UPR Induction observed by increase in GFP fluorescence on inducing DM-MBP expression in presence of different concentrations (50mM, 100mM, 200mM and 500mM) of TMAO. (e) Refolding rates of DM-MBP as a function of TMAO concentration. Effect of TMAO was measured by carrying out refolding of DM-MBP in buffer containing 100mM to 500mM TMAO. Goodness of fit is provided in Supplementary Figure 8. (f) In vitro refolding rate obtained at different concentrations of TMAO are plotted against the median UPR-induced GFP fluorescence obtained in S. cerevisiae, at the same concentrations of TMAO. All averages shown in panel a, e and f are averages of 3 independent experiments. (See also Supplementary Figure 1-10)
Subsequently we tested if V8G mutation renders folding of MBP TMAO-dependent in vivo. We chose to use quantitative reporter of Unfolded Protein Response (UPR) as a surrogate for the accumulation of misfolded protein. To test if difference in folding rates could be sensed by UPR, we performed kinetic simulations assuming constant transcription and translation rate for SM-MBP and DM-MBP with only their folding rates being different (Supplementary Figure 3 for details of the simulation). This model predicted the steady state level of unfolded protein in the ER to increase with decreasing folding rates (Fig 1b). Since UPR activation is dependent on accumulation of misfolded protein in ER26, in principle, UPR could be used for investigating difference in folding rates in vivo. Towards this, we generated a Saccharomyces cerevisiae BY4742 derivative that harbors a UPR inducible GFP27 and genome-integrated and galactose-inducible SM-MBP (strain ynk001) or DM-MBP (strain ynk002) that is targeted to ER along with C-terminal ER-retention signal. UPR induced upon the overexpression of SM-MBP or DM-MBP was read out from the increase in GFP fluorescence. Consistent with the simulation results, DM-MBP induced a marked UPR whereas SM-MBP was unable to mount a significant response (Fig 1c). This was confirmed not be due to reduced expression levels of SM-MBP compared to DM-MBP (Supplementary Figure 4). We generated an Ire1 deletion strain over the background of ynk002. GFP induction was absent in the deletion strain confirming that signaling through Ire1 was essential for sensing the accumulation of DM-MBP in ER (Supplementary Figure 5). Thus difference in folding rates is quantitatively sensed by UPR in vivo.
Using this reporter strain (yNK002) as the platform, we sought to address if TMAO may have an effect on intracellular folding environment thereby establishing a correlation between in vitro and in vivo folding assistance provided by TMAO. Firstly, we investigated if TMAO is able to penetrate S. cerevisiae when added in the external medium, by measuring the intracellular concentration of TMAO upon growth of yeast in YPD media containing TMAO. There was a linear correlation between the externally added TMAO and the measured intracellular concentrations (Supplementary Figure 6), allowing us to investigate the perturbation of folding rate as a function of intracellular TMAO concentration. When yNK002 was grown in presence of different concentrations of TMAO, we observed a concentration dependent decrease in the UPR induced upon over-expression of DM-MBP (Fig 1d) without any discernible changes in its expression levels (Supplementary Figure 7). Notably, the decrease in UPR correlated well with the increase in the in vitro refolding-rates of DM-MBP with increasing concentration of TMAO (Fig 1e and 1f, Supplementary Figure 8). To exclude the possibility that TMAO interferes with UPR, independent of the expression of DM-MBP, we checked that DTT-induced UPR is not affected by TMAO (Supplementary Figure 9). Additionally, as concentrations of ER chaperones are controlled through UPR, absence of UPR induction in cells grown in presence of TMAO indicated that TMAO action was not routed through the induction of molecular chaperones (Supplementary Figure 9). To check for specificity of TMAO in reducing DM-MBP mediated UPR we asked if Proline, an osmolyte that accelerated refolding of DM-MBP only marginally (Supplementary Figure 10), may act to decrease DM-MBP-induced UPR. Consistent with our hypothesis, DM-MBP-induced UPR was only marginally alleviated by Proline (Supplementary Figure 10). The decrease was well-correlated to the slight folding assistance afforded by Proline in vitro, further supporting our claim that UPR is able to provide a semi-quantitative read-out of intracellular refolding rate of DM-MBP. Correlation between in vitro and in vivo folding assistance suggests that TMAO action in vivo, even in the presence of molecular chaperones, is routed through similar mechanisms as in conditions that is devoid of molecular chaperones in vitro. Thus osmolytes may alter intracellular folding environment even in presence of specialized folding machinery. This, taken together with the finding that SM-MBP folding is independent of TMAO, indicates that single site mutations may convert an osmolyte-independent folder to an osmolyte-dependent folder in vivo, suggesting that chemical-chaperones may buffer genetic variations.
TMAO eliminates kinetic traps in folding landscape in vivo
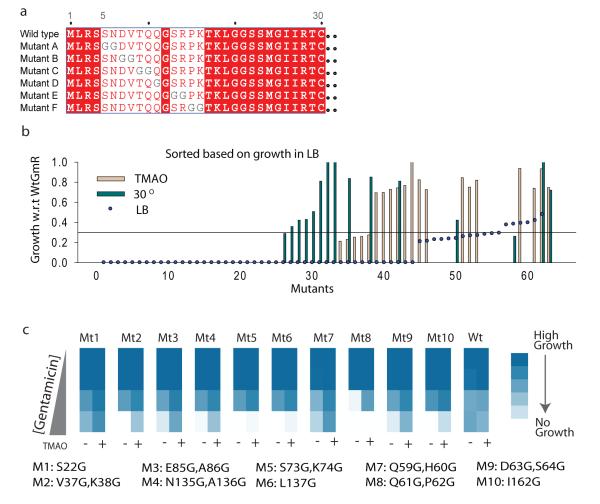
Correlation between in vitro and in vivo TMAO-assisted folding prompted us to investigate whether certain mutations are more likely to be buffered by TMAO than others. Since it has been hypothesized that osmolytes primarily induce compaction of unfolded states by inducing structure formation 28,29, we anticipate that engineering flexibility-driven kinetic traps in a TMAO-independent protein should make it dependent on the chemical chaperone. Towards testing the generality of our hypothesis, we chose a model protein unrelated to MBP: a GCN5 related acetyl transferase, Serratia marescens aminoglycoside 3-N-acetyltransferase (Gm-R), that transfers an acetyl group from Acetyl-CoA to gentamicin30. The active Gm-R protein confers gentamicin resistance to E. coli cells comprising a simple model for in vivo activity assays. In vivo folding of GmR is independent of TMAO as Gm-R expressed from an Arabinose inducible promoter is able to confer gentamycin resistance to E. coli at 0.1% Arabinose in absence of TMAO in the medium. Flexibility driven traps in folding pathway may be engineered by disrupting crucial nucleating interactions that lead to loss of structure formation thereby imparting flexibility to unfolded chains. Similar traps may also be designed by incorporation of glycines that populate larger conformational space than other amino acids 31. Substitution with glycines increase main-chain flexibility and hence should result in entropic-trap for intermediates or unfolded states that lack strong native-like interactions. We took the later approach and replaced each two consecutive residues with two glycines hence creating a comprehensive library of mutants of Gm-R containing double glycine substitutions at most of the positions (Fig 2a). We let the different mutants grow in media containing gentamicin and 0.1% Arabinose in absence and in the presence of 200mM TMAO (Supplementary Figure 11, for detailed method). Most of the glycine-duplet substitution mutants were either inactive or partially active and did not grow well in presence of Gentamicin (Supplementary Table 1). There were also a few partially active mutants that were rendered inactive in TMAO. Interestingly, 15 out of a total of 63 mutants exhibited significantly higher gentamicin resistance in 200mM TMAO than in its absence (Fig 2b, Supplementary Figure 12), whereas wild-type Gm-R transformed cells did not exhibit any difference in growth (Supplementary Figure 13). Thus, presence of mutants that are more active in TMAO suggests that some of these mutants may be buffered by this osmolyte.
Figure 2.
Rationally designing TMAO-dependent conditional activity. (a) Schematic of Glycine-duplet substitution library showing representative mutations and the pattern of mutations. Two consecutive residues were substituted by Glycine-Glycine residues to obtain a comprehensive library of Glycine-duplet substitutions. (b) The mutants were grown with gentamicin selection in absence of TMAO at 37°C (LB), in presence of TMAO at 37°C (TMAO), and in absence of TMAO at 30°C (30D). Growth normalized with respect to cells transformed with wtGmr, that grew equally well in these three conditions. Mutants are sorted according to their growth in LB and growth in the other two conditions is plotted as bars for each mutant. (c) Gentamicin resistance as obtained by growing each mutant in 5 different concentrations of gentamicin (2 μg/ml, 4 μg/ml, 8 μg/ml, 16 μg/ml and 32μg/ml) in presence and absence of TMAO. Growth is shown as a colormap with increasing color density representing increase in growth. Details of GmR Glycine-duplet mutants used for this analysis are mentioned at the bottom of the panel. (See also supplemental Figure 11-17)
To validate the buffering phenomenon we asked if TMAO-dependent growth may result from reduced bactericidal activity of gentamicin in presence of TMAO. If gentamicin efficacy is reduced in presence of TMAO we expect to see a general decrease in gentamicin sensitivity in presence of TMAO independent of the presence of GmR gene. Cells transformed with inactive GmR did not grow even in the presence of TMAO excluding the possibility that TMAO reduces gentamicin efficacy (Supplementary Figure 14). To exclude the possibility that certain mutants form toxic aggregates in absence of TMAO, thereby exhibiting TMAO-assisted growth, we investigated the growth of E. coli, expressing these mutant proteins, in the absence of gentamicin selection (Supplementary Figure 15). Growth was similar irrespective of the presence or absence of TMAO eliminating the possibility of growth difference due to misfolding-induced toxicity. To confirm that differences in growth seen in presence of gentamicin reflect TMAO-dependent enhancement of gentamicin-resistance, we chose ten most significant hits. We checked for gentamicin resistance of these mutants in presence and absence of TMAO by monitoring growth as a function of gentamicin concentration (Fig 2c). Consistent with the previous growth data, all the TMAO-sensitive mutants exhibited augmented gentamicin resistance in presence of TMAO, while gentamicin sensitivity of WtGmR was not altered. Enhanced gentamicin resistance in presence of TMAO is a convoluted function of the amount of active protein in cytosol and specific activity of the native protein. Specific activity of the mutants may change either due to alterations in kcat or KM, in presence of TMAO. To investigate this, we purified two of the TMAO-sensitive mutants, GmR(N135G,A136G) and GmR(Q59G,H60G), and the wild-type protein and monitored gentamicin acetyl transferase activity in vitro in presence and absence of TMAO (Supplementary Figure 16). Specific activity of the proteins were not enhanced by TMAO, on the contrary, there was a marginal decrease in activity of GmR (N135G, A136G) (Supplementary Figure 16). This excludes the possibility of enhanced specific activity in presence of TMAO suggesting that TMAO buffer these mutations by increasing the concentration of active protein in the cytosol. Next, we investigated if TMAO acts by increasing solubility of mutant proteins. Though GmR(N135G,A136G) was significantly more prone to aggregation than wtGmR (Supplementary Figure 17), partitioning of the protein in supernatant and pellet fraction was only marginally affected by TMAO; this could not explain the large change in gentamicin resistance in presence of TMAO. This indicated that TMAO acts to increase the pool of active proteins rather than the total soluble fraction of the protein. The total amount of GmR declined in presence of TMAO thereby excluding the possibility that TMAO increases activity by generally increasing expression level of proteins. However, this raises the possibility that TMAO may allow better folding by decreasing rate of protein synthesis in vivo. Decrease in total WtGmR expression in presence of TMAO does not increase gentamicin resistance suggesting that TMAO does not allow better folding/solubility by decreasing protein expression. To further confirm this, we sought to decrease expression level by growing E. coli at 30°C. The library when grown at 37°C and 30°C revealed seven temperature sensitive (TS) mutants that were more active at 30°C than at 37°C (Fig 2b). We found that of these seven TS mutants, only two were also buffered by TMAO at 37°C (Supplementary Figure 12). This reveals that not all temperature sensitive mutants can benefit from the chaperoning effect of TMAO. Interestingly, of the fifteen TMAO-buffered mutants only two were rescued at lower temperature. This suggests that TMAO-dependent mutations are different from the spectrum of mutations that are rendered active at lower temperatures.
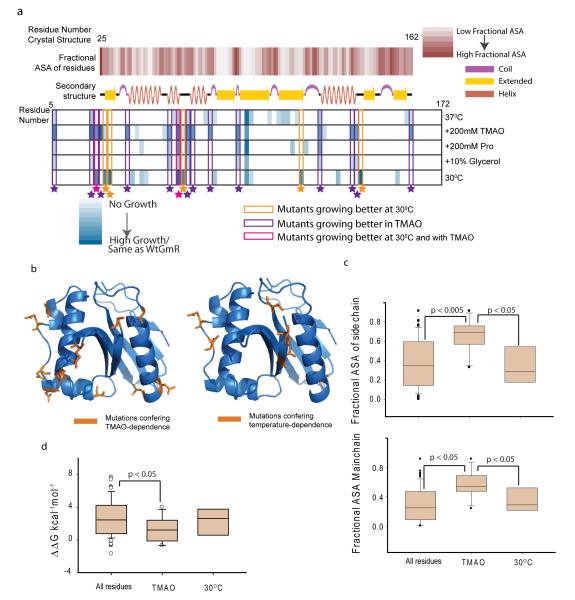
This provided the first evidence that chemical chaperones are able to buffer specific de novo mutations in vivo. Mutations that result in TMAO-dependent or temperature-dependent Gm-R activity, were mapped on the crystal structure of Gm-R (PDB-id: 1BO430) (Fig 3a & 3b). It was found that most of the TMAO-dependent mutations lie on the surface of the protein (Fig 3c) either on the exposed loop regions or on helices’ termini (Fig 3a, second row and fourth row). While the TMAO-dependent mutations were exposed, the low temperature mutants were significantly more buried as indicated both by main chain and side chain accessibility (Fig 3c, lower and upper panel), suggesting differences in mechanism by which TMAO and low temperature may act to assist intracellular protein folding. Re-constitution of TMAO-dependence suggests that mutants with enhanced flexibility of unfolded states have a general tendency to benefit from the presence of TMAO.
Figure3.
Analysis of TMAO assisted buffering. (a) Structural features of osmolyte and temperature sensitive mutants. First row: fractional Accessible Surface Area (ASA) of the main chain atoms of the residues calculated from the crystal structure 1bo430. Second row: secondary structure cartoon of 1bo4. Third to Seventh row: Color scale showing growth of different mutants at different conditions. Growth at 37°C(third row), in presence of TMAO at 37°C(fourth row), at 37°C in 200mM Proline (fifth row), at 37°C in Glycerol (sixth row), at 30°C (seventh row). Mutants exhibiting differential growth are enclosed in colored boxes. (b). Residues in GmR structure (1bo4) colored in orange, which when mutated to glycine-duplets lead to TMAO-dependent activity (left panel) or temperature-dependent activity (right panel). (c). Box plot showing side chain fractional ASA (top panel) and main chain fractional ASA (bottom panel) of all the residues that were mutated to obtain the library, mutants that are more active in presence of TMAO and mutants that are more active at 30°C. The significance is based on two tailed Students-t test. (d). Box plot for predicted ΔΔG (kcal−1mol−1 FU ) of all the residues mutated to obtain the library, mutants that are more active in presence of TMAO and -mutants that are more active at 30°C. Since ΔΔGFU values do not follow a normal distribution, significance was calculated using non-parametric Mann-Whitney test (see also Supplementary Figure 18,19).
To gain further insight into stability perturbations that are tolerated by TMAO, we calculated the energy of each of the Glycine-duplet substitution mutations using FoldX32. Even though this measure is only semi-quantitative, it provides us with an estimate of the perturbation tolerated by mutational buffering. Almost all the mutations that were generated had a positive ΔΔGF-U compared to the wild-type protein, indicating destabilization, as expected (Supplementary Figure 18). Interestingly there was no correlation between in vivo activity of the mutants and the predicted destabilization (Supplementary Figure 19) indicating that in vivo activity may be a convoluted function of folding thermodynamics and kinetics 2. ΔΔGF-U values of the TMAO-dependent mutants were marginally lower than the ΔΔGF-U of all the mutants whereas there was no apparent difference between TMAO and temperature-dependent sets (Fig 3d). However, even with prediction of marginal energetic perturbation, majority of the TMAO-sensitive mutants show extremely low activity in vivo in the absence of TMAO.
We tested gentamicin resistance of the glycine-duplet substitution library of Gm-R in presence of chemical chaperones like proline and glycerol. Only four of the fifteen TMAO-buffered mutants exhibited differential growth in presence and absence of these osmolytes (Fig 3a, fifth and sixth row). Proline buffered four and Glycerol buffered one of the TMAO-dependent mutants, albeit to much lesser extent than TMAO. Proline also buffered three other mutants, though the growth was significantly less than that of the TMAO- or temperature buffered mutants. Thus mutations that lead to flexibility-driven traps in the folding pathway were uniquely buffered by TMAO. The overlap between temperature dependent and proline- or glycerol- dependent mutations were also negligible suggesting that mutational buffering of the tested osmolytes are not routed through decrease in growth rate. This also suggests that different osmolytes may drive mutational buffering in a unique manner that is dependent on the mechanism of their action on protein-folding pathways in vivo.
Chemical chaperones buffer distinct spectrum of mutations
Since glycine-duplet mutations provide only a limited view of mutational buffering, we generated a single-site random mutant library for each codon of gmr gene to understand the breadth of buffering affected by the osmolytes(Supplementary Figure 20 and supplemental methods). We obtained approximately 5000 mutants, of which twenty mutants were sequenced and found to be mutated for single residues. The theoretically possible size for a single-site substitution library of a 170 aa protein is 3230 in terms of amino acid substitutions. By screening the activity of 5000 mutants we aimed to achieve at least 50% saturation, i.e. we could screen approximately ten amino acid substitutions per site. Using replica plating and image analysis we were able to pick up approximately 500 inactive or partially active mutants (Supplementary Figure 21). Thirty of the five hundred clones tested were temperature sensitive (Supplementary Figure 22, middle panel), while ten clones turned out to be TMAO-dependent (Supplementary Figure 22, left panel). Only one of the ten TMAO-buffered clones exhibited temperature dependence (Supplementary Figure 22). Similarly, only one of the temperature dependent clones was rescued by TMAO (Supplementary Figure 23). This validated the results from glycine-duplet substitution library and confirmed that not all temperature sensitive mutants would benefit from TMAO-assisted folding. Since E. coli reacts to heat shock by upregulating chaperones, we asked if excess chaperone concentration can buffer TMAO-dependent mutants. These clones did not regain activity when grown without TMAO at 42°C (Supplementary Figure 22) indicating that increased concentration of molecular chaperones at higher temperature is not able to correct the folding of TMAO-sensitive mutants. Thus corroborating the previous result that TMAO action is not routed through molecular chaperones.
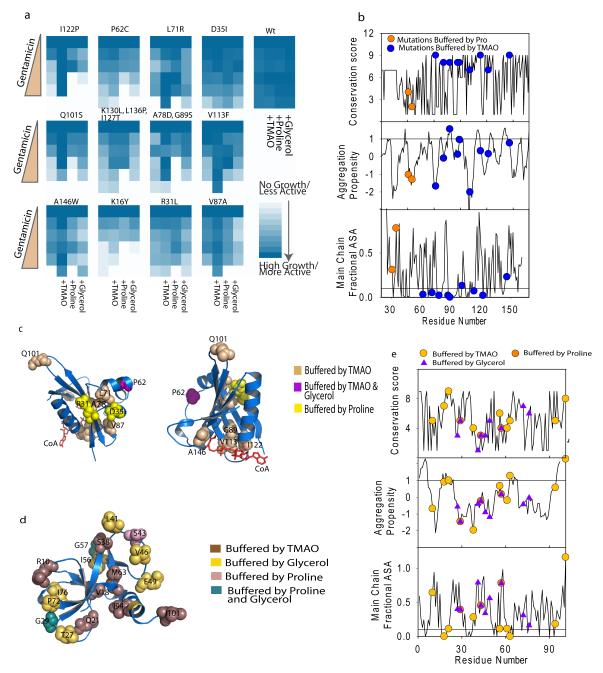
Using this library we screened for clones that exhibited enhanced gentamicin resistance in presence of three different osmolytes. Initial screen was performed with one concentration of gentamicin and clones that exhibited differential growth in presence of osmolytes (90) were chosen for detailed analysis. We grew all these clones in four different media conditions containing different osmolytes and five different gentamicin concentrations to obtain a quantitative indication of gentamicin resistance. Forty of these mutants show reproducible changes in gentamicin resistance in presence of different osmolytes. Sequencing results provided satisfactory sequences for thirteen clones; the other clones were not analyzed due to lack of quality sequence information. While TMAO was most effective in buffering a large number of mutations, Proline and Glycerol were also able to buffer certain mutations (Fig 4a). A few of the mutations were buffered uniquely by Proline. The mutations were not preferentially located in aggregation-prone segments of the protein as predicted by Zyggregator 33 (Fig 4b). Summarily, TMAO-sensitive mutations tend to lie in the buried regions (Fig 4b), whereas the ones buffered by Proline, either in combination with TMAO or singularly, tend to lie on the surface. This was distinct from the results obtained from the glycine-duplet substitution library, where majority of TMAO sensitive mutations mapped to the protein surface. This difference may be justified by the fact that property of the substituting amino acid plays an important role in determining perturbation to folding pathway, which may in turn determine the buffering capability of the osmolyte. Majority of the changes that were buffered by TMAO consisted of distinct non-conservative changes like A78D, V113F or A151W, that either substituted a buried non-polar residue with negatively charged one or replaced smaller amino acids with larger residues(Fig 4a,c). These mutations alter core packing and hence may have important implications in subtle alterations of protein structures. TMAO allowed substitutions in regions of proteins that are highly conserved whereas the two mutations exclusively buffered by Proline were at less conserved positions (Fig 4b). TMAO-buffered mutations majorly clustered at two hydrophobic cores of GmR (Fig 4c). The first cluster consisted of mutations at V113, G89 and I122, which line the acetyl-CoA binding pocket, indicating that folding as well as ligand-binding may be compromised in these mutants while the second group of mutants, L71 and A78, lie in a core that is far removed from the substrate binding pocket. Overlapping but non-identical set of mutations buffered by the different osmolytes suggest that buffering capability would depend on their unique mechanisms of folding-assistance.
Figure4.
Chemical chaperones have specific spectrum of mutational buffering. (a). Gentamicin resistance of 12 random mutants in presence of osmolytes is shown as colormap. WtGmR transformed cells were taken as control. (b) Properties of mutated residues are plotted. Conservation (top panel), aggregation propensity (middle panel) and main chain fractional accessible surface area (bottom panel) are shown along with the residues that were buffered by the osmolytes. Line in the middle panel at aggregation propensity value of 1 denotes the generally accepted cutoff value; peptide segments above this value are considered to be aggregation prone. The line in the lower panel signifies the ASA of 0.1. Residues lying below this line are considered to be completely buried. (c) Residues buffered by osmolytes are mapped on the crystal structure of GmR, 1bo4. Color codes are listed in the legend. (d) Residues involved in mutational buffering is mapped to the crystal structure of Ccdb, 3vub47 (e) Properties of CcdB mutants were analyzed as described in (b) (see also Supplementary Figure 20-27).
To further underline implications of chemical chaperones in vivo, we used a mutant library of an unrelated protein CcdB (cell death and differentiation protein). This library consisted of single site substitution by lysine/aspartate/glutamate randomly spanning the entire CcdB sequence. Active CcdB kills E. coli by interacting with and inhibiting the activity of DNA-Gyrase 34. In vivo functionality of this protein is easy to assay (See Supplementary Figure 24 and supplemental methods), and has been used extensively for mutagenesis based definition of structural and functional parameters 4,35. We grew 288 mutants of CcdB (Supplementary Figure 25), in presence and absence of the different chemical chaperones. A substantial number of mutants were buffered in the presence of TMAO, glycerol or proline (Supplementary Figure 26 and Supplementary Table 2). Consistent with GmR data, TMAO was able to buffer negative charge substitutions at buried sites, like glutamate substitution at the residue M63 (Fig 4d, Supplementary Figure 27). I94D and V18K also exhibited TMAO-dependence. Notably, I94 and V18 interact with M63 to form a tightly packed hydrophobic core. This corroborates our finding from GmR single-site random mutant library, that TMAO is able to buffer alterations in core residues. Like in GmR, majority of the CcdB mutations buffered by TMAO are buried except for I101D and R10K (Fig 4d). I101 has been implicated in the interaction of CcdB with gyrase 36; thus a mutation that may potentially disrupt interaction between CcdB and gyrase is buffered by TMAO suggesting the role of osmolytes in buffering mutations that perturb protein-protein interactions. Consistent with GmR results, Proline and Glycerol buffered mutations that lie on the surface of the protein and were predominantly positive charge substitutions (Fig 4d & e, Supplementary Figure 27). While TMAO-dependence was conferred by substitutions at moderately conserved residues in CcdB (Fig 4e) mutations buffered by the osmolyte were not preferentially located on aggregation prone segments as predicted by Zyggregator (Fig 4e). Interestingly, while I56K, a partially active mutant is buffered by glycerol and not by TMAO, I56E, another substitution at the same position that renders the protein completely inactive, is buffered by TMAO but not by glycerol. This indicates that different osmolytes and chemical environment may indeed assist protein evolution in distinct manner that is dependent on their mechanism of action.
Osmolyte action not routed through stress response pathways
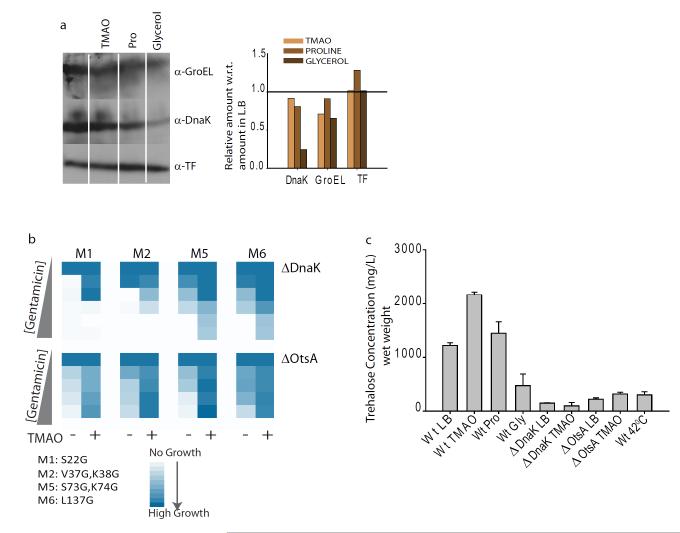
Since molecular chaperones are up-regulated by stress response pathways, we asked if sustained growth in osmolytes is associated with chronic induction of stress response pathways. To have an unbiased view of the transcriptional pathways upregulated upon growth in osmolytes, we performed microarray based gene expression profiling of E. coli (Supplementary Figure 28, 29). Importantly, most of the genes that are upregulated upon heat shock (Supplementary Figure 28,left panel) or osmotic shock (Supplementary Figure 29, Right Panel) are involved in maintaining proteostasis 37. These genes were not upregulated when E. coli cells were grown in TMAO, Proline and Glycerol. It was also validated at the level of gene product using western blots, that the canonical markers of misfolding stress, DnaK, GroEL or Trigger Factor, are not upregulated upon sustained growth of E. coli in different osmolytes (Fig 5a, Supplementary Figure 30). In fact, growth in glycerol leads to a marked decrease in DnaK, suggesting that buffering action of glycerol is unlikely through its effect on DnaK. Microarray data revealed that osmolyte-upregulated pathways were different for different osomolytes, indicating the absence of a ubiquitous stress. Since the in vivo activity assay for GmR was based on sustained gentamicin resistance activity over the entire growth period, transient upregulation of stress response genes upon osmotic shock(approx 30 minutes)38, would be incapable of affecting buffering activity as shown by the osmolytes.
Figure5.
Stress response pathways do not mediate osmolyte mediated buffering. (a) Expression levels of GroEL, DnaK and Trigger Factor in presence of osmolytes like TMAO, Proline and glycerol, measured using western-blotting (left panel)quantified and is denoted as bar graph (right panel). (b). color map showing growth of four mutants of glycine-duplet substitution library, transformed in ΔdnaK and ΔotsA strains, at different gentamicin concentrations.(c). Intracellular trehalose concentrations estimated in cells grown at different conditions. Triplicate measurements were made for each growth conditions and the experiments were repeated twice to average the values. (See also Supplementary Figure 28-34).
To further investigate if TMAO-buffering may have contributions from osmotic-stress response, we compared the activity of ~500 GmR mutants grown in presence of TMAO or in presence of 0.7M NaCl (Supplementary Figure 31). Even though salt-stress is able to increase the growth of certain mutants, only three of the ten TMAO-dependent mutants exhibited any buffering in high-salt condition (Supplementary Figure 31). Furthermore, we investigated if TMAO, may have its action routed through DnaK 17, the most abundant chaperone of prokaryotic cytosol. Towards this we transformed four of the TMAO-dependent glycine-duplet substitution mutants, in E. coli ΔdnaK strain (Fig 5b, upper panel). Assuringly, we found that TMAO buffered these mutations indicating that TMAO action is not dependent on the presence of DnaK. This is also substantiated by the fact that DM-MBP, which is not assisted by any molecular chaperone of ER for folding, is chaperoned by TMAO in vivo. Mutational buffering activity of TMAO in the cytosol of E. coli and the ER of S. cerevisiae (Fig 1d), two unrelated compartments in terms of chaperone composition, indicated that TMAO action is likel to be direct.
Since E. coli cytosol contains an endogenous osmolyte, trehalose, which is believed to prevent protein aggregation 39, we asked whether trehalose level increases upon osmolyte-treatments. When treated with TMAO we observed an increase in the intracellular concentration of trehalose (Fig 5c). The increase was only ~2 fold, whereas osmotic insults increase trehalose concentration over ~10 fold 40. Notably, TMAO-treated E. coli ΔdnaK strain exhibited Trehalose concentrations significantly lower than the basal level present in untreated cells(Fig 5c). This taken together with the fact that TMAO is able to buffer mutations even in ΔdnaK strain indicates that Trehalose may play only a minor role, in TMAO assisted buffering. To investigate if TMAO action is Trehalose-dependent, we checked for folding of four of the TMAO-dependent glycine-duplet mutants in E. coli ΔotsA strain deficient in synthesizing Trehalose. We checked that background concentration of the sugar was extremely low in this strain and did not increase in presence of TMAO (Fig 5c). The glycine-duplet mutants exhibited TMAO-dependent folding even in this strain lacking trehalose, indicating that TMAO-assisted buffering is independent of enhanced trehalose accumulation (Fig 5b, lower panel). Since Trehalose concentration was not altered by Proline or Glycerol (Fig 5c), it is unlikely that their action is dependent on Trehalose. Taken together this suggests that though the endogenous osmolyte, Trehalose, is induced by TMAO, it is not essential for TMAO-assisted buffering. Thus osmolytes added in the medium had a direct effect on intracellular protein folding.
Since Trehalose concentration was upregulated by TMAO-treatment, we investigated Trehalose assisted folding in vitro using DM-MBP as the model protein. Interestingly, we found that trehalose, like TMAO, acts prominently on DM-MBP folding (Supplementary Figure 32). If Trehalose, like TMAO, also assists by increasing rigidity of flexible folding intermediates, we posited, that synthetically decreasing the flexibility of non-native states would decrease the effect of these osmolytes on folding. To decrease the flexibility of folding intermediates we used previously reported disulphide bonded variants of DM-MBP, DM-MBP(18,296) and DM- MBP(184,362) 25 (Supplementary Figure 33). Action of Trehalose and TMAO on the refolding of disulfide-bonded protein was equally reduced indicating that Trehalose, like TMAO, assists folding of DM-MBP by decreasing the flexibility of non-native states (Supplementary Figure 32 and 34).Thus in different organisms, depending on whether they accumulate TMAO or Trehalose under certain conditions, mutational buffering may be promoted in similar direction. Interestingly Trehalose concentration was found to decrease in presence of sustained heat shock, when E. coli was grown overnight at 42°C (Fig 5c), and in strains that harbour a deletion of DnaK, indicating that Trehalose-dependent mutants may be rendered inactive at higher temperatures or under conditions of chaperone depletion.
DISCUSSION
Protein folding has been implicated in chaperone mediated mutational buffering12. Here, using small-molecule modulators of in vivo protein-folding, we demonstrate that modifiers of protein folding may generally buffer mutational variations in vivo. Furthermore, our work uncovers differential mutational buffering by different osmolytes.
We uncover that each of the chemical chaperones tested has a subset of mutations that it can buffer in vivo, possibly due to difference in the mechanism of action of the different osmolytes. While TMAO (and possibly Trehalose) may buffer mutations that destabilize the core significantly or lead to a drastic increase in flexibility of non-native conformations, other osmolytes like Proline and Glycerol may buffer mutations that are on protein surface. Thus, complex milieu of physiological osmolytes in the cell may be associated with an unappreciated spectrum of genetic buffering. This source of canalization adds to the processes already known to be involved in imparting robustness to cellular phenotypes including processes linked to protein folding.
We find that trehalose has similar mechanism of folding assistance as TMAO. In light of evidence that certain pathogenic prokaryotes, like Mycobacterium tuberculosis, accumulate large concentration of the osmolyte 41, we speculate that it may promote access to a larger amino acid sequence space thereby increasing the probability of generating drug-resistant variations. Trehalose also extends lifespan of nematodes and is more predominant in mutants that exhibit extended lifespan42. Since ageing is linked to collapse of proteostasis43, it will be interesting to uncover if trehalose decelerates aging by assisting protein folding . It is also important to note that, in E. coli, chronic heat shock or chaperone depletion was able to decrease the cellular pool of trehalose; we posit that a small but significant fraction of the mutants that have been reported to be heat sensitive could have been trehalose-sensitive mutants. Further work on the role of metabolites in protein folding and mutational buffering will be needed to understand if metabolic shifts may lead to altered spectrum of mutational buffering and alter proteostasis. Thus, understanding small-molecule mediated modulation of proteostatic potential, either directly or through molecular chaperones44,45, will be crucial for appreciating the principles of proteostasis-driven protein evolution as well as for therapeutic interventions in misfolding disorders. More immediately, applications of these chemical chaperones for in vitro protein evolution should enhance the sequence space accessible to folded structures thereby allowing the evolution of enzymes with novel or enhanced functions.
Materials and Methods
Protein expression and purification
pBAD-MBP, pCH-SM-MBP (Y283D), pCH-DM-MBP (V8G, Y283D) and double cysteine mutant pCH-DM-MBP (18C, 296C) clone constructs were kind gifts from Prof. F. Ulrich Hartl. MBP and its mutant proteins were purified as described previously46. Protein concentration was spectrophotometrically determined using the molar extinction coefficient. at 280nm.
PCR-based mutagenesis
Site directed and random mutagenesis for Gm-R was performed using modified Quick Change protocol of Stratagene using megaprimer based methods as discussed elaborately in the supplemental experimental procedures.
Genetic screens, activity assays, yeast work
Detailed experimental procedures are outlined in the supplemental experimental procedures.
Supplementary Material
Acknowledgment
KS is a UGC(India) Junior Research Fellow. KC is a CSIR research fellow and an Intermediate Fellow of the Wellcome Trust-DBT India alliance. The work was funded by the CSIR-EMPOWER programme and in part by grants from Wellcome Trust-DBT India alliance. KC acknowledges CSIR for funds to IGIB and infrastructural support. We thank Raghavan Varadarajan, Jonathan Weissman, Ulrich Hartl and Manajit Hayer-Hartl for generous gift of reagents. We acknowledge NBRP-E.coli at NIG for providing strains from the Keio Collection.
Footnotes
Author Contribution : KC,AB and KS wrote the manuscript. AB, KS and NK performed the experiments. AB, KS, NK,VD, NB, AR, and SM generated the reagents. KC and SS supervised the work.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–87. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 2.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Camps M, Herman A, Loh E, Loeb LA. Genetic constraints on protein evolution. Crit Rev Biochem Mol Biol. 2007;42:313–26. doi: 10.1080/10409230701597642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajaj K, Chakrabarti P, Varadarajan R. Mutagenesis-based definitions and probes of residue burial in proteins. Proc Natl Acad Sci U S A. 2005;102:16221–6. doi: 10.1073/pnas.0505089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waddington C. Canalization of Development and the Inheritance of Acquired Characters. Nature. 1942;150:563–565. [Google Scholar]
- 6.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLoS Comput Biol. 2008;4:e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 8.Moczek AP. On the origins of novelty in development and evolution. Bioessays. 2008;30:432–47. doi: 10.1002/bies.20754. [DOI] [PubMed] [Google Scholar]
- 9.Povolotskaya IS, Kondrashov FA. Sequence space and the ongoing expansion of the protein universe. Nature. 2010;465:922–927. doi: 10.1038/nature09105. [DOI] [PubMed] [Google Scholar]
- 10.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–42. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 11.Lindquist S. Protein Folding Sculpting Evolutionary Change. Cold Spring Harb Symp Quant Biol. 2010 doi: 10.1101/sqb.2009.74.043. [DOI] [PubMed] [Google Scholar]
- 12.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–24. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 13.Ignatova Z, Gierasch LM. Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc Natl Acad Sci U S A. 2006;103:13357–61. doi: 10.1073/pnas.0603772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nascimento C, Leandro J, Tavares de Almeida I, Leandro P. Modulation of the activity of newly synthesized human phenylalanine hydroxylase mutant proteins by low-molecular-weight compounds. Protein J. 2008;27:392–400. doi: 10.1007/s10930-008-9149-9. [DOI] [PubMed] [Google Scholar]
- 15.Schultz T, Liu J, Capasso P, de Marco A. The solubility of recombinant proteins expressed in Escherichia coli is increased by otsA and otsB co-transformation. Biochem Biophys Res Commun. 2007;355:234–9. doi: 10.1016/j.bbrc.2007.01.149. [DOI] [PubMed] [Google Scholar]
- 16.Street TO, Krukenberg KA, Rosgen J, Bolen DW, Agard DA. Osmolyte-induced conformational changes in the Hsp90 molecular chaperone. Protein Sci. 2010;19:57–65. doi: 10.1002/pro.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diamant S, Eliahu N, Rosenthal D, Goloubinoff P. Chemical chaperones regulate molecular chaperones in vitro and in cells under combined salt and heat stresses. J Biol Chem. 2001;276:39586–91. doi: 10.1074/jbc.M103081200. [DOI] [PubMed] [Google Scholar]
- 18.Bolen DW, Baskakov IV. The osmophobic effect: natural selection of a thermodynamic force in protein folding. J Mol Biol. 2001;310:955–63. doi: 10.1006/jmbi.2001.4819. [DOI] [PubMed] [Google Scholar]
- 19.Konopka MC, Shkel IA, Cayley S, Record MT, Weisshaar JC. Crowding and confinement effects on protein diffusion in vivo. J Bacteriol. 2006;188:6115–23. doi: 10.1128/JB.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eggers DK, Valentine JS. Crowding and hydration effects on protein conformation: a study with sol-gel encapsulated proteins. J Mol Biol. 2001;314:911–22. doi: 10.1006/jmbi.2001.5166. [DOI] [PubMed] [Google Scholar]
- 21.Ando T, Skolnick J. Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion. Proc Natl Acad Sci U S A. 2010;107:18457–62. doi: 10.1073/pnas.1011354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerner MJ, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–20. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 23.Wang JD, Michelitsch MD, Weissman JS. GroEL-GroES-mediated protein folding requires an intact central cavity. Proc Natl Acad Sci U S A. 1998;95:12163–8. doi: 10.1073/pnas.95.21.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–22. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 25.Chakraborty K, et al. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell. 2010 doi: 10.1016/j.cell.2010.05.027. (Accepted) [DOI] [PubMed] [Google Scholar]
- 26.Kimata Y, Kohno K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr Opin Cell Biol. 2010;23:135–42. doi: 10.1016/j.ceb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–7. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong J, Gierasch LM. Macromolecular crowding remodels the energy landscape of a protein by favoring a more compact unfolded state. J Am Chem Soc. 2010;132:10445–52. doi: 10.1021/ja103166y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holthauzen LM, Rosgen J, Bolen DW. Hydrogen bonding progressively strengthens upon transfer of the protein urea-denatured state to water and protecting osmolytes. Biochemistry. 2010;49:1310–8. doi: 10.1021/bi9015499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolf E, et al. Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase. Cell. 1998;94:439–49. doi: 10.1016/s0092-8674(00)81585-8. [DOI] [PubMed] [Google Scholar]
- 31.Ramakrishnan C, Ramachandran GN. Stereochemical criteria for polypeptide and protein chain conformations. II. Allowed conformations for a pair of peptide units. Biophys J. 1965;5:909–33. doi: 10.1016/S0006-3495(65)86759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schymkowitz J, et al. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tartaglia GG, Vendruscolo M. The Zyggregator method for predicting protein aggregation propensities. Chem Soc Rev. 2008;37:1395–401. doi: 10.1039/b706784b. [DOI] [PubMed] [Google Scholar]
- 34.Bernard P, et al. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J Mol Biol. 1993;234:534–41. doi: 10.1006/jmbi.1993.1609. [DOI] [PubMed] [Google Scholar]
- 35.Bajaj K, et al. Structural correlates of the temperature sensitive phenotype derived from saturation mutagenesis studies of CcdB. Biochemistry. 2008;47:12964–73. doi: 10.1021/bi8014345. [DOI] [PubMed] [Google Scholar]
- 36.Dao-Thi MH, et al. Molecular basis of gyrase poisoning by the addiction toxin CcdB. J Mol Biol. 2005;348:1091–102. doi: 10.1016/j.jmb.2005.03.049. [DOI] [PubMed] [Google Scholar]
- 37.Gunasekera TS, Csonka LN, Paliy O. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol. 2008;190:3712–20. doi: 10.1128/JB.01990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meury J, Kohiyama M. Role of heat shock protein DnaK in osmotic adaptation of Escherichia coli. J Bacteriol. 1991;173:4404–10. doi: 10.1128/jb.173.14.4404-4410.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singer MA, Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell. 1998;1:639–48. doi: 10.1016/s1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- 40.Rod ML, Alam KY, Cunningham PR, Clark DP. Accumulation of trehalose by Escherichia coli K-12 at high osmotic pressure depends on the presence of amber suppressors. J Bacteriol. 1988;170:3601–10. doi: 10.1128/jb.170.8.3601-3610.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodruff PJ, et al. Trehalose is required for growth of Mycobacterium smegmatis. J Biol Chem. 2004;279:28835–43. doi: 10.1074/jbc.M313103200. [DOI] [PubMed] [Google Scholar]
- 42.Honda Y, Tanaka M, Honda S. Trehalose extends longevity in the nematode Caenorhabditis elegans. Aging Cell. 2010;9:558–69. doi: 10.1111/j.1474-9726.2010.00582.x. [DOI] [PubMed] [Google Scholar]
- 43.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106:14914–9. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mu TW, et al. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–81. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ong DS, Mu TW, Palmer AE, Kelly JW. Endoplasmic reticulum Ca2+ increases enhance mutant glucocerebrosidase proteostasis. Nat Chem Biol. 2010;6:424–32. doi: 10.1038/nchembio.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang YC, et al. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125:903–14. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 47.Loris R, et al. Crystal structure of CcdB, a topoisomerase poison from E. coli. J Mol Biol. 1999;285:1667–77. doi: 10.1006/jmbi.1998.2395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.