Abstract
Diseases of esophageal epithelium (EE) such as reflux esophagitis and cancer are rising in incidence. Despite this, the cellular behaviors underlying EE homeostasis and repair remain controversial. Here we show that in mice, EE is maintained by a single population of cells that divide stochastically to generate proliferating and differentiating daughters with equal probability. In response to challenge with all-trans Retinoic Acid (atRA) the balance of daughter cell fate is unaltered but the rate of cell division increases. However, following wounding, cells reversibly switch to producing an excess of proliferating daughters until the wound has closed. Such fate switching enables a single progenitor population to both maintain and repair tissue without the need for a “reserve” slow-cycling stem cell pool.
Murine EE consists of layers of keratinocytes. These tissue lacks structures such as crypts or glands which form stem cell niches in other epithelia (Figs.1A and B) (1-5). Proliferation is confined to cells in the basal layer (6). On commitment to terminal differentiation, basal cells exit the cell cycle and subsequently migrate to the tissue surface from which they are shed. Early studies suggested all proliferating cells were functionally equivalent, but recent reports propose that a discrete population of slow-cycling stem cells is responsible for both maintenance and wound healing (7-11). This controversy and the importance of EE in disease motivated us to resolve the proliferative cell behavior in homeostatic EE and in tissue challenged by systemic treatment with the vitamin A metabolite all-trans Retinoic Acid (atRA) or acute local wounding (12-13).
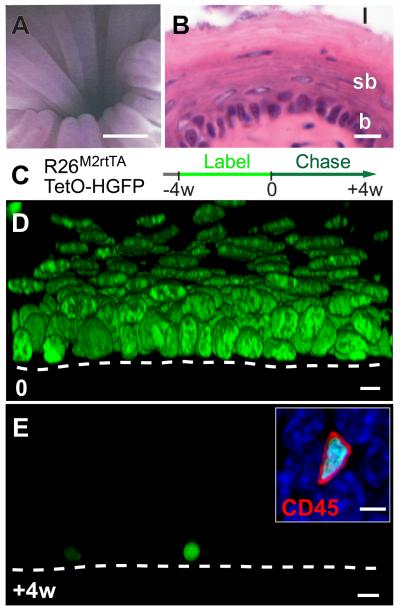
Fig. 1. Esophageal epithelium contains no slow-cycling epithelial cells.
A: Micro-endoscopy showing esophageal lumen, scale bar approx. 500μm.
B: Section of epithelium, basal layer (b), suprabasal layers (sb) and lumen (l), scale bar 10μm.
C: Protocol. Adult Rosa26M2rtTA/TetO-HGFP mice treated with doxycycline (DOX) express HGFP (green). Following DOX withdrawal, HGFP is diluted upon cell division, except in slow-cycling cells.
D, E: Rendered confocal z stacks, showing HGFP (green) at time 0, D, and after 4w chase E (Scale bar 10μm). Dashed line indicates basement membrane. Inset shows CD45 (red) staining in HGFP retaining cell at 4w (DAPI, blue; Scale bar 5μm).
To investigate cell division rates in EE we used a transgenic label retaining cell (LRC) assay (Fig. 1C) (1, 14-15). Doxycycline (DOX) induction of Histone-2B EGFP fusion protein (HGFP) expression in Rosa26M2rtTA/TetO-HGFP mice resulted in nuclear fluorescent labeling throughout the EE (Fig. 1D and fig. S1A). When DOX is withdrawn, HGFP is diluted by cell division, leaving 0.4% basal layer cells (561 out of 140000) retaining label after a 4 week chase (Figs. 1E and S1B). 3D imaging showed these label retaining cells (LRC) had smaller nuclei than the surrounding keratinocytes and did not stain for the basal keratinocyte marker Keratin14 (0 out of 561 LRC, Figs. S1C and D). The stem cell markers CD34 and Lgr5 were also undetectable in LRC or other cells (Figs. S2 and S3) (2, 4, 10, 16). However, 99.9% (2457 out of 2459) of LRC were positive for the pan leukocyte marker CD45 (Fig. 1E inset), comprising of a mixture of Langerhan’s cells and lymphocytes (Figs. S1E and F). These findings lead to the surprising conclusion that, unlike tissues such as the epidermis, there are no slow-cycling or quiescent epithelial stem cells in EE (1, 17). Indeed, HGFP dilution in basal cells was strikingly homogeneous, suggesting that all cells divide at a similar rate of approximately twice per week (Fig. S1G).
Although epithelial cells have the same rate of division, they may still differ in their ability to generate cycling and differentiated progeny. We therefore used inducible cre-lox based genetic marking to investigate if the proliferating cell population is heterogeneous and to quantify cell behavior (18-19). The fate of single cell derived clones was tracked in cohorts of adult AhcreERT R26flEYFP/wt mice at multiple time points over a year post-induction, during which period EE was homeostatic (Figs. 2A and S4). Crucially, analysis of the composition of clones at one year showed they were representative of unlabeled cells (Fig. S5). Over the time course clone number decreased through differentiation, whilst the size of the remaining clones progressively increased (Figs. 2B and C). Although variation in labeling efficiency limits the accuracy with which the proportion of labeled cells can be estimated, within statistical error, this proportion remains constant; consistent with the labeled population being in homeostasis (Fig. 2D).
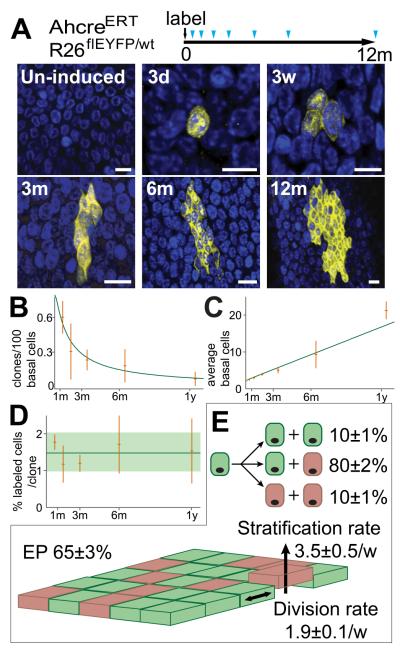
Fig. 2. Proliferating cell fate in esophageal epithelium.
A: Protocol: Clonal labeling was induced in AhcreERTR26flEYFP/wt mice and analyzed at intervals from 3 days to 1 year (triangles). Images are rendered confocal Z stacks of the basal layer showing typical clones at times indicated. EYFP is yellow and DAPI blue. Scale bars 10μm.
B to D: Clone quantification. B, C: clone density and average clone size (basal cells). Observed values (orange) with error bars (s.e.m.), green curves show predictions of model (E). D: average % labeled basal cells at indicated time points (orange), error bars indicate s.e.m. Green line and shading show average and s.e.m. across all time points.
E: Cell Fate in EE. Basal layer comprises 65% functionally equivalent esophageal progenitors (EP, green, dividing at a rate of 1.9/week, (consistent with the rate of dilution of HGFP, Fig. S1G) and 35% post-mitotic cells (pink) which stratify (arrow) at a rate of 3.5/week. 10% of EP divisions generate two EP daughters, 10% two differentiated daughters and 80% one of each fate. Values are optimal fits with 95% plausible intervals.
Significantly, the average size of persisting clones increased linearly with time, while their size distribution acquired long term scaling behavior; a hallmark of a single functionally equivalent population of cells dividing at the same rate (Fig. 2C, and Figs. S6A, B and E) (18-21). Studies of interfollicular epidermis (IFE) revealed that this pattern of clonal evolution was consistent with progenitors dividing stochastically to generate differentiated and cycling daughters with equal probability (18-19). By implementing a Bayesian inference analysis we showed the entire data set conforms to the IFE paradigm (Figs. 2E and S6C to E). We conclude that esophageal progenitors (EP) are functionally equivalent.
The observation of similar progenitor behavior in EE and epidermis, derived from endoderm and ectoderm respectively, argues that squamous epithelia share a common mechanism of homeostasis irrespective of their developmental origin. However EP behavior differs from that of crypt stem cells in the endoderm derived intestinal epithelium, where stochastic fate is a result of competition for limited niche space (16, 22).
Unlike progenitors in other tissues such as the epidermis, EP are not supported by a discrete slow-cycling stem cell population (1). This raises the intriguing question of how the tissue responds to stress or injury. To investigate this issue, we subjected EE to a tissue-wide challenge in the form of atRA treatment and to acute local excisional wounding.
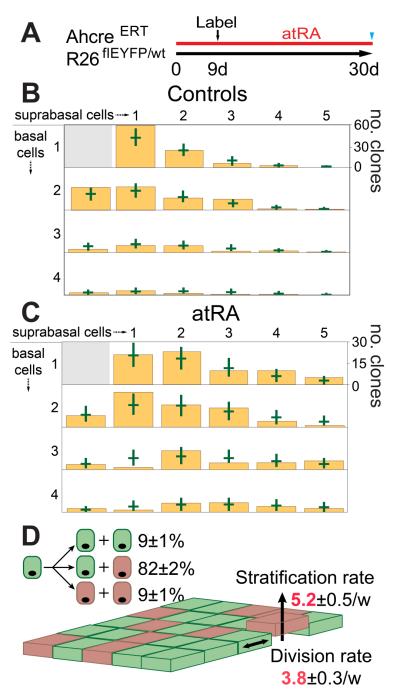
In order to determine the effects of atRA, we selected a dose which induced a “hyper proliferative” response (Fig. S7A) and then used quantitative lineage tracing to define the changes in cell behavior (23-24). Mice were treated for 9 days, clonal labeling induced, and treatment then continued for a further 21 days, when clone size was scored (Fig. 3A). Bayesian analysis revealed the rates of EP proliferation and differentiated cell stratification had approximately doubled. There was a small but statistically significant decrease in the proportion of proliferative cells but, critically, no significant change in the proportions of symmetric and asymmetric divisions indicating the treated tissue was homeostatic (Fig. 3B to D). To test this finding, we used a second experimental schedule in which clonal labeling was induced prior to atRA treatment. The values of parameters determined in the first experiment accurately predicted the number of basal cells per clone on completion of the second protocol (Fig. S7B to D). We conclude that during atRA treatment, EP establish a new homeostatic state.
Fig. 3. alltrans Retinoic Acid (atRA) treatment of EE.
A: Protocol (see text). B and C: Size distribution of multi-cellular clones containing at least one basal cell in control (B, 307 clones), and atRA treated (C, 300 clones) EE. Green bars indicate 95% plausible fit to models in Fig. 2E (control) and 3D (atRA). D: Optimal fit during atRA treatment; proliferation and differentiation rates (red) increase compared with control.
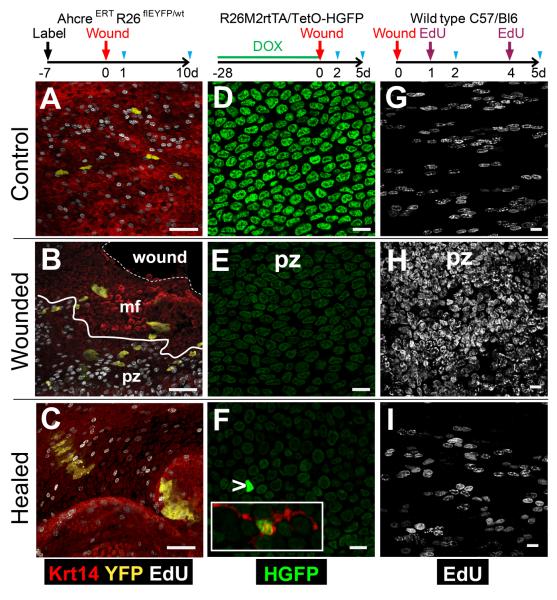
To investigate the repair of EP following wounding, we developed micro-endoscopic biopsy of mouse esophagus (Fig. S8F). Biopsy produced a typical epithelial wound response (25-26). Cells immediately next to the defect formed a migrating front (mf) in which there was minimal proliferation, surrounded by a proliferative zone (pz) in which cell division was dramatically increased (Figs. 4B and S8D). We used three different protocols to analyze cell behavior (Fig. 4). Firstly, we examined clonally labeled EP in AhcreERT R26flEYFP/wt mice induced 1 week before biopsy (Fig. 4 A to C). 24 hours after wounding, fragmented clones of labeled cells were seen, aligned towards the wound and spanning the pz and mf (Figs. 4A ,B and S8D). By 10 days, clones were evident in and around the repaired defect (Fig. 4C and S8 A, E). These findings indicate that EP participate in tissue regeneration post-wounding, a behavior recapitulated in explant culture, suggesting that active recruitment of immune cells is not essential for the switch in EP fate (Fig. S9) (27-28). To investigate the proportion of EP which participate in regeneration we biopsied DOX treated Rosa26M2rtTA/TetO-HGFP mice (Fig. 4D to F)). HGFP was substantially and evenly diluted both within the pz at 2 and 5 days post-biopsy compared with controls, but retained outside the mf (Figs. 4D to F, S8B and S10A, B). We conclude there is widespread mobilization of EP within the pz and that the recruited cells proliferate to similar extent. In a complementary experiment, animals were injected with EdU 24 hours prior to culling, revealing extensive recruitment of cells into cycle in the pz at 2 days, which reverted to control levels at 5 days when the epithelial defect had closed (Fig. 4G to I, S8C and S10C to F). This indicates that the switch in EP fate following wounding is reversible.
Fig. 4. Response of EP to wounding.
Cartoons show protocols, blue triangles indicate sampling.
A to C: Wounding of clonally labeled mice. Confocal z stacks, 1 (B) or 10 (C) days post-biopsy. mf indicates migrating front; pz, proliferative zone; solid line shows pz-mf boundary; dashed line, wound margin. A: day 1 unwounded control. EYFP is yellow, keratin 14 (Krt14) red and EdU grayscale. Scale bars 50μm.
D to F: Dilution of HGFP. Confocal z stacks from unwounded control day 2 (D) and wounded mice at 2 (E) and 5 (F) days post-biopsy, showing HGFP (green). Arrow indicates HGFP bright cell (overexposed to reveal remaining cells), such cells stained for CD45 (red, inset). Scale bars 10μm.
G to I: Cell proliferation. Confocal z stacks from unwounded control at 2 days (G) and experimental mice at 2 (H) and 5 (I) days post-biopsy stained for EdU (grayscale). Scale bars 10μm.
In summary, these results show that EE is both maintained and repaired by a single progenitor cell population capable of reversibly switching between homeostatic and regenerative behavior in response to injury. These findings may be reconciled with the reported proliferative heterogeneity of EE cells in vitro if only some EP cells switch into “wound mode” when placed into culture (10). The widespread participation of progenitors in tissue repair provides a rapid and robust mechanism of wound healing without an underpinning stem cell pool.
Supplementary Material
Acknowledgments
We thank E. Choolun and staff at ARES and CBS Cambridge for technical assistance, D Winton and D. Adams (Cambridge) for mice and M. Gonzalez (London) for the Geminin antibody. We acknowledge the support of the MRC, EPSRC, the NC3Rs, the Wellcome Trust, Sidney Sussex College, Cambridge (DPD), European Union Marie Curie Fellowship PIEF-LIF-2007-220016 (MPA), the Royal College of Surgeons of England (AR) and Cambridge Cancer Centre (AR). This work uses methods included in the patent WO2009010725 (A2), a method of detecting altered behavior in a population of cells, inventors Jones PH, Simons BD, Klein AM.
Footnotes
Supplementary Materials: Materials and Methods Figures S1-S13 Table S1 References (29-30)
References and Notes
- 1.Tumbar T, et al. Science. 2004 Jan 16;303:359. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker N, et al. Cell Stem Cell. 2010 Jan 8;6:25. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 3.Barker N, et al. Nature. 2007 Oct 25;449:1003. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 4.Jaks V, et al. Nat Genet. 2008 Nov;40:1291. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 5.Goetsch E. Am J Anat. 1910;10:1. [Google Scholar]
- 6.Messier B, Leblond CP. Am J Anat. 1960 May;106:247. doi: 10.1002/aja.1001060305. [DOI] [PubMed] [Google Scholar]
- 7.Marques-Pereira JP, Leblond CP. Am J Anat. 1965 Jul;117:73. doi: 10.1002/aja.1001170106. [DOI] [PubMed] [Google Scholar]
- 8.Seery JP, Watt FM. Curr Biol. 2000 Nov 16;10:1447. doi: 10.1016/s0960-9822(00)00803-4. [DOI] [PubMed] [Google Scholar]
- 9.Croagh D, Phillips WA, Redvers R, Thomas RJ, Kaur P. Stem Cells. 2007 Feb;25:313. doi: 10.1634/stemcells.2006-0421. [DOI] [PubMed] [Google Scholar]
- 10.Kalabis J, et al. J Clin Invest. 2008 Dec;118:3860. doi: 10.1172/JCI35012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croagh D, Thomas RJ, Phillips WA, Kaur P. Stem Cell Rev. 2008 Dec;4:261. doi: 10.1007/s12015-008-9031-3. [DOI] [PubMed] [Google Scholar]
- 12.Dent J, El-Serag HB, Wallander MA, Johansson S. Gut. 2005 May;54:710. doi: 10.1136/gut.2004.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jemal A, et al. CA Cancer J Clin. 2011 Mar-Apr;61:69. [Google Scholar]
- 14.Kanda T, Sullivan KF, Wahl GM. Curr Biol. 1998 Mar 26;8:377. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 15.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Cell. 2005 May 6;121:465. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 16.Snippert HJ, et al. Cell. 2010 Oct 1;143:134. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 17.Braun KM, et al. Development. 2003 Nov;130:5241. doi: 10.1242/dev.00703. [DOI] [PubMed] [Google Scholar]
- 18.Clayton E, et al. Nature. 2007 Feb 28;446:185. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 19.Doupe DP, Klein AM, Simons BD, Jones PH. Dev Cell. 2010 Feb 16;18:317. doi: 10.1016/j.devcel.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Klein AM, Doupe DP, Jones PH, Simons BD. Phys Rev E Stat Nonlin Soft Matter Phys. 2007 Aug;76:021910. doi: 10.1103/PhysRevE.76.021910. [DOI] [PubMed] [Google Scholar]
- 21.Klein AM, Simons BD. Development. 2011 Aug;138:3103. doi: 10.1242/dev.060103. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Science. 2010;330:822. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 23.Chapellier B, et al. EMBO J. 2002 Jul 1;21:3402. doi: 10.1093/emboj/cdf331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins CA, Watt FM. Dev Biol. 2008 Dec 1;324:55. doi: 10.1016/j.ydbio.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 25.Potten CS, Allen TD. J Cell Sci. 1975 Mar;17:413. doi: 10.1242/jcs.17.3.413. [DOI] [PubMed] [Google Scholar]
- 26.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Nature. 2008 May 15;453:314. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 27.Martin P, et al. Curr Biol. 2003 Jul 1;13:1122. doi: 10.1016/s0960-9822(03)00396-8. [DOI] [PubMed] [Google Scholar]
- 28.Jacinto A, Martinez-Arias A, Martin P. Nat Cell Biol. 2001 May;3:E117. doi: 10.1038/35074643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.