Abstract
We developed CREST (Clipping REveals STructure), an algorithm that uses next-generation sequencing reads with partial alignments to a reference genome to directly map structural variations at the nucleotide level of resolution. Application of CREST to whole-genome sequencing data from five pediatric T-lineage acute lymphoblastic leukemias (T-ALLs) and a human melanoma cell line, COLO-829, identified 160 somatic structural variations. Experimental validation exceeded 80% demonstrating that CREST had a high predictive accuracy.
Somatically acquired structural variations (SVs) can induce alterations in genes that directly contribute to cellular transformation1. Transcriptome2 and whole genome sequence analysis3, 4 of tumor and matched germ line samples have led to a marked improvement in our ability to identify SVs in cancer. Nevertheless, the accurate identification of SVs using next generation sequencing (NGS) remains challenging. Typically in these analyses, 30–100bp reads from the two ends of a sequence fragment are obtained, mapped to the reference human genome, and discordances in distance, orientation, and/or mapping status (e.g. whether a read is mapped or unmapped to the reference genome) are used to identify structural variations5–9. These approaches only infer the approximate genomic locations of a SV but fail to pinpoint their exact breakpoint at the nucleotide level. Moreover, the available methods tend to generate a high frequency of false positives when applied to experimental data due to the presence of PCR and/or sequencing artifacts and the inherent difficulty of accurately mapping sequences in repetitive regions.
To overcome some of these deficiencies, we explored an alternative approach for SV discovery that is based on directly mapping of SV breakpoints at the nucleotide level without relying on the discordant mapping of paired end reads. By definition a sequence read that spans a bona fide structural variation will have partial alignment to each of the two sides of the junction (Fig. 1a). Current NGS mapping algorithms like BWA10 compute local alignment (that is, partial alignment) for a read either automatically when its mate is globally aligned to the genome or by user request. The unaligned portion is masked by a process termed “soft-clipping” because the unaligned subsequence is retained but not trimmed even though it does not map to the current genomic location (Fig. 1b). With longer NGS read length (≥75bp), these soft-clipped subsequences can be of sufficient length to map unambiguously to a different genomic location, thus, identifying the second breakpoint for a putative structural variation.
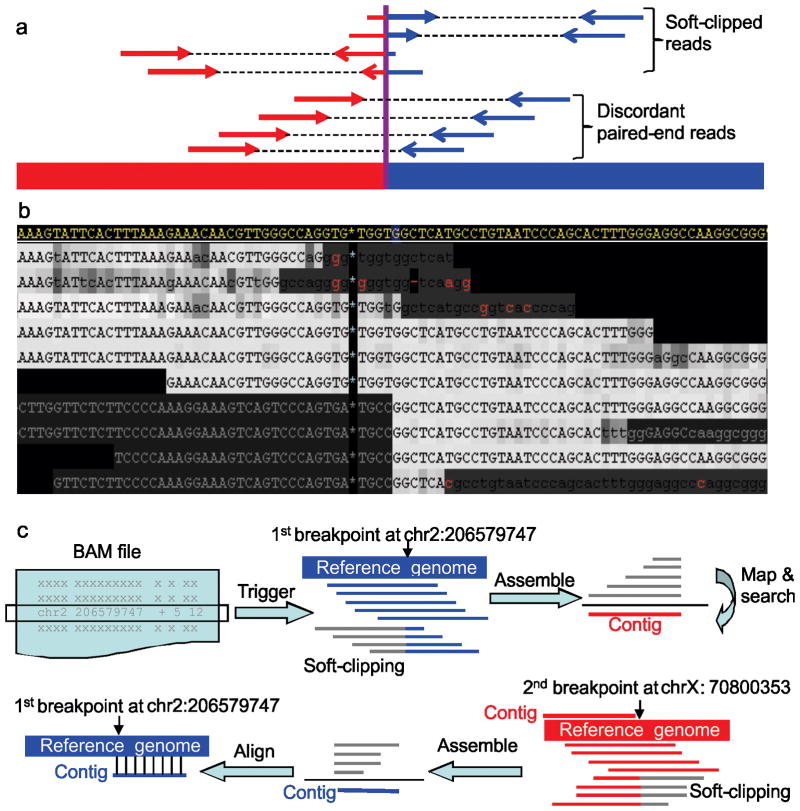
Figure 1.
Mapping SV breakpoints using soft-clipped reads. (a) Illustration of SV analysis using discordantly mapped paired-end reads versus mapping using soft-clipping reads. Red and blue segments represent two discontinuous genomic regions. (b) An example of using soft-clipping signature to identify an interchromosomal translocation. The region in display is 206579705-206579789bp on chromosome 2. The reference genome is shown at the top in yellow while the NGS reads are displayed below. Mismatches to the reference are shown in red letters while the gray letters at lower-left are soft-clipping subsequences not aligned to the reference. Upper-case characters represent high-quality (phred score ≥20) bases and the darkness of shading correlates to lower quality score. In this example, the soft-clipping subsequences map to chromosome X, revealing a chromosome 2 to chromosome X translocation. (c) The five-step CREST algorithm: extraction of soft-clipped reads (gray) in the BAM file; assembly of soft-clipped reads at a putative breakpoint into a contig (red); mapping of the contig against the reference genome to identify candidate partner breakpoints; identification of all possible soft-clipped reads and assembly into a contig (blue); and alignment of the contig derived from the partner (blue) back to the reference genome. A match to the initial breakpoint is considered a SV.
Based on this concept, we developed CREST (Clipping REveals STructure), a software tool that uses the soft-clipping reads to directly map the breakpoints of structural variations. For each SV, the first breakpoint is identified by presence of soft-clipped reads while its partner is found by an assembly-mapping-searching-assembly-alignment procedure (Fig. 1c, Online Methods). The identified SVs are then classified into the following five subtypes based on location and orientation of the breakpoints: (1) inter-chromosomal translocations (CTX), (2) intra-chromosomal translocations (ITX), (3) inversions (INV), (4) deletions (DEL), and (5) insertions (INS) (Supplementary Figs. 1 and 2).
We applied CREST to whole genome DNA sequence data obtained from five cases of childhood T-lineage acute lymphoblastic leukemia (T-ALL) with matched tumor and normal samples that were sequenced as part of the St. Jude Children’s Research Hospital, Washington University Pediatric Cancer Genome Project. This analysis identified a total of 110 SVs (Supplementary Table 1) including 36 CTX, 25 ITX, 1 INV, 26 DEL, 22 INS. PCR primers were designed successfully for 107 (97%) of the predicted SVs and Sanger sequencing of the generated amplicons from the respective tumors confirmed the predicted SV breakpoints in 89 (82% validation rate, representative results are shown in Fig. 2a). Across the five samples, the validated SVs include 31 CTX, 19 ITX, 1 INV, 22 DEL and 16 INS. The validated translocations detected through CREST ranged from balanced translocations to highly complex rearrangements that involved multiple chromosomes. A representative example is shown in Fig. 2b in which a complex rearrangement involving chromosomes 1, 4, 5, and 10 was defined in one sample.
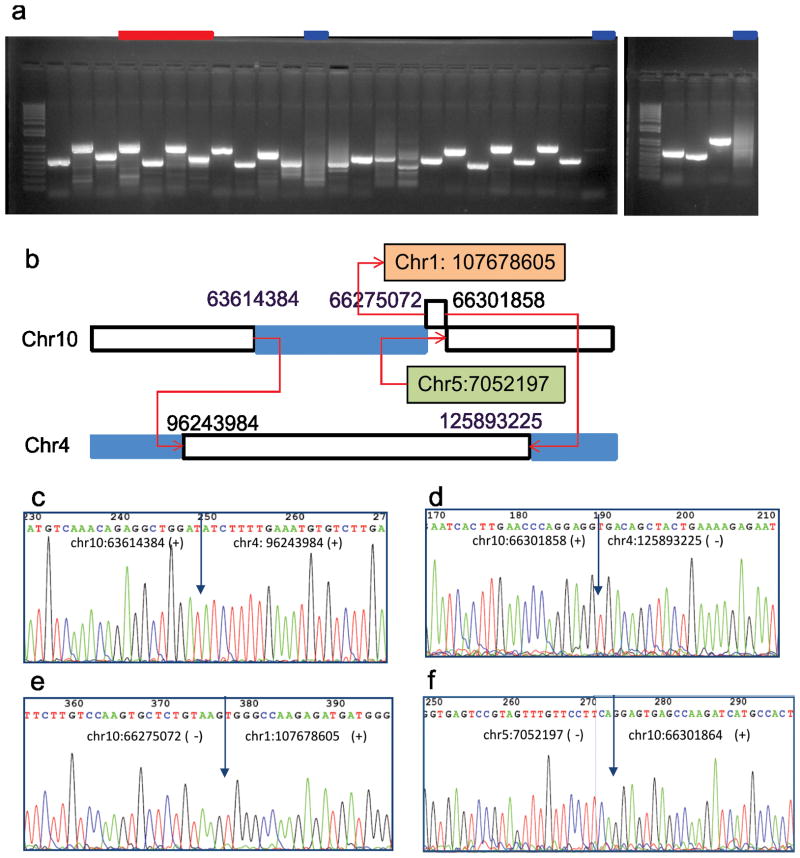
Figure 2.
SV validation result for one T-ALL sample (SJTALL003). (a) PCR amplification of 28 SV breakpoints predicted by CREST. All putative SVs except for those tested in lanes marked in blue were validated by Sanger sequencing. Lanes marked in red point to amplicons listed in figure panels c, e, d and f. (b) A complex inter-chromosomal translocations involving chromosomes 1, 4, 5 and 10. The blue segments on chromosomes 4 and 10 are deletion segments identified by NGS coverage analysis. (c–f) Sanger sequencing data across the 4 breakpoints involved in the complex rearrangement illustrated in panel b.
To compare the performance of CREST to other available algorithms, we first reanalyzed this data set using BreakDancer5, a commonly used tool that implements a paired-end discordance mapping (PEM) algorithm. BreakDancer identified only 27 out of the 89 validated SVs that were defined by CREST. Moreover, although BreakDancer identified another 1,037 putative SVs, none of these survived a post-processing quality check and thus represented false positive predictions. A second PEM algorithm, GSAV, detected 76 (85%) of the validated SVs amongst a total of 5,880,492 predictions, demonstrating that this relatively low false negative rate was achieved with a cost of an extremely high false positive error. Re-analysis using Pindel11, a program that uses unmapped reads across insertion/deletion (indel) breakpoints, detected only five of the 89 validated SVs found by CREST suggesting that different methods are required for finding gross structural variations and indels. Details of the superior performance of CREST compared to these algorithms are provided in Supplementary Data 1.
To further assess the performance of CREST, we applied it to a published whole-genome sequencing dataset from the metastatic melanoma cancer cell line COLO-82912. Using a paired-end discordant mapping method3 the published analysis reported 37 validated SVs12. By comparison, CREST identified 76 SVs (Supplementary Data 2, Supplementary Table 2) including 26 of the 37 reported SVs. Of the 11 reported SVs that were not identified by CREST, 6 were found to have soft-clipped reads in the matching normal sample COLO-829BL, indicating that these six SVs represent germline polymorphisms but not tumor specific somatic SVs. Of the five remaining SVs, three lacked soft-clipped reads, one had soft-clipped reads that mapped to multiple genomic locations and one had low-quality soft-clipped reads across the breakpoints.
CREST identified 50 additional SVs that were not reported previously12. We selected 20 to directly validate by PCR amplification of DNA extracted from the COLO-829 cell line (Supplementary Table 3). 18 of the 20 novel SVs, including 7 CTX, 9 DEL and 2 INS were validated by Sanger sequencing (Supplementary Figs. 3 and 4).
To assess the false negative rate of CREST in identifying germline structural variations, we simulated whole-genome sequencing data for the 887 copy number variations (CNVs) in NA12878, one of the individuals characterized by the 1000 Genomes Project by applying 19 different SV detection methods on high-coverage sequencing data generated by 3 different platforms13. The false negative rate of CREST is 22–27% with 3% false positive calls, demonstrating its superior performance in both sensitivity and accuracy compared with BreakDancer and Pindel (Supplementary Table 4). 52% of the CNVs missed by CREST are in regions of segmental duplications where germline CNVs are frequent (26% of NA12878) but somatically acquired copy number alterations (CNAs) are rare (6% of the 5 T-ALLs) based on the data analyzed in this study, suggesting that the false negative rate of somatic CNAs could be lower than that of germline CNVs. The results of this analysis are presented in more details in Supplementary Data 3
Although the concept of using sequences that span breakpoints has been previously explored for finding chimeric mRNAs2, for mapping viral integration sites by targeted sequencing14 and for identifying indels11, CREST is the first use of this approach for mapping structural variations at the level of the whole genome. CREST is particularly well suited for identifying somatically acquired structural variations in paired tumor-normal samples, where its precision in finding the breakpoints coupled with its integrated ability to subtract common variations present in both germline and tumor samples also allows the removal of false lesions caused by the artifacts generated during library construction and the difficulties inherent in accurately mapping short sequence reads. Although other computational methods for detecting SV have been developed, none outperform CREST in our comparative analysis (see Supplemental Data 1). Moreover, methods specifically designed for the identification of germline deletions15 are not capable of finding inter- and intra-chromosomal rearrangements, which are key mechanisms for creating oncogenic fusion proteins in cancer. The entire CREST package can be downloaded from http://www.stjuderesearch.org/site/lab/zhang with user manual and test data.
Although CREST provides a significant improvement over standard paired-end approaches for identifying SVs, it continues to have difficulty with repetitive DNA sequence regions, rearrangements that occur within or adjacent to germline polymorphic structural variations, and rearrangements that contain non-template DNA sequences that are inserted at the breakpoints and are of similar or longer length than the NGS reads. In addition, CREST, like all mapping methods, demands high quality DNA reads of sufficient coverage to accurately define the DNA sequence (details in Supplementary Discussion). The method provides base-pair level resolution of breakpoints and can therefore be used not only for identifying the number and type of SVs within a tumor genome, but should also allow an analysis of the breakpoint DNA sequence as a way to gain insights into the mechanism responsible for the generation of the structural rearrangement.
ONLINE METHODS
Whole-genome sequencing data
DNA samples were obtained from diagnostic bone marrow aspirates from five pediatric patients with T-ALL and from matched remission bone marrow samples from each patient (as non-tumor controls). The samples were obtained following informed consent given by the patient (if over the age of majority), their parent, or guardian. The study was approved by the Institutional Review Boards of both St Jude Children’s Research Hospital and Washington University. The DNA samples were paired-end sequenced on an Illumina GAIIX using DNA fragments with 300–500bp insert size and DNA sequence reads 100bp, as previously described 4. The mean coverage for the tumor genomes ranged from 32 to 34 fold while that of the matching normal ranged from 24 to 28 fold. All reads were mapped to the human assembly NCBI build 36 using the program BWA10 with the default parameters. In addition to this primary DNA sequence data, paired tumor and normal whole-genome sequencing data from the malignant melanoma cell line COLO-829 were obtained from the Sanger Center in the format of bam files12 which store the read alignments to the human assembly NCBI build 36.
CREST algorithm and SV analysis pipeline
The process flow of the SV analysis pipeline using CREST is shown in Supplementary Figure 2. The input data can either be two bam files representing paired tumor/normal samples or a single bam file. Germline variations are filtered in the paired analysis. The SV detection algorithm first collects all soft-clipped reads, coded as “S” in the CIGAR (Compact Idiosyncratic Gapped Alignment Report) string into a BAM (Binary Alignment/Map format) file. The soft-clipped reads are then used to define putative SV breakpoints that meet the following criteria: (1) the number of soft-clipped reads (c) and the sequence coverage (C) by including reads with >97% sequence identity and a minimum BLAT score of 30 (see methods for details); or (2) the probability of observing at least c soft-clipped reads with the coverage C is greater than 0.05 based on a binomial distribution of observing a heterozygous SV at a user-specified heterogeneity factor. Many putative SV breakpoints have soft-clipped reads as well as wild-type reads because SVs usually occur either in a subset of tumors due to tumor heterogeneity and/or are heterozygous events. Therefore, with the exception for homozygous deletion events, there are usually two groups of reads at a putative breakpoint. Using binomial distribution allows us to evaluate the significance of observing c number of soft-clipped reads (one group) given the coverage (the wild-type group) at the site. Use of the second criterion ensures that statistically significant SVs are retained even if they do not meet the ad-hoc user-defined criteria regions where the NGS coverage is low.
Each putative breakpoint is considered to be the first breakpoint of a potential SV. The corresponding partner breakpoint is identified by applying an assembly-mapping-searching-assembly-alignment procedure consisting of CAP316 for assembly and BLAT17 for genomic search and alignment. If the alignment shows high identity (default set to 90%) between the second contig and the first breakpoint, then the two breakpoints are considered to form a putative structural variation. The identified putative SVs are then filtered to remove false positives due to alignment artifacts by the following process. For each SV, the distance between the second contig to the first breakpoint is required to be within a short distance (user-definable, default set to 15bp) to ensure it does map back to the first breakpoint. For a paired analysis that use both germline and somatic data, presence of SV in the germline sample is further evaluated by extracting germline reads that have soft-clipped bases at the SV breakpoints and re-align them by BLAT to the second contig. This step ensures that a germline SV that may have slightly different soft-clipping position from its somatic counterpart is not classified as a somatic event.
CREST exports three output files: (1) a report file that records the breakpoints of SVs at base-pair resolution, number of soft-clipping reads, and genes located across the breakpoints; (2) a template file for experimental validation with 1000 flanking nucleotides of each breakpoint; and (3) An XML output file that displays the assembled contigs for users who are interested in visually inspecting the predicted SVs. Manual review is optional. The XML file displays alignment of the reads across the breakpoints, quality score of each base and the soft-clipping signature (Supplementary Fig. 5–10). The entire package can be downloaded from http://www.stjuderesearch.org/site/lab/zhang with user manual and test data.
Post-process of structure variations predicted by BreakDancer
For each predicted SV, we first check whether discordant mapping of paired-end reads is caused by repetitive regions in human genome. All supporting reads are extracted in fastq format and each read is re-mapped to the hg18 reference genome using BLAT. If a read-pair is mapped within the library insert range (mean insert size +/− 3 standard deviation), it will NOT be considered a supporting read pair for the SV. All SVs with ≥3 supporting read pairs and BreakDancer score ≥30 after the re-mapping are retained and the tumor-only SVs are considered putative somatic SVs.
The putative somatic SVs are then subjected to an assembly process to evaluate their validity because a valid SV should have at least one cross-junction contig. All reads mapped within 1kb of the two breakpoints along with their unmapped mate pairs are extracted. The mapping information was based on the bam files. We then run phrap to assemble the extracted sequences into contigs by using base call, quality value and paired-end information. Assembly is carried out in two iterations because the first iteration usually generates contigs that represent the wild-type allele unless the alternative allele is a homozygous genomic change. The second iteration starts with reads not assembled in the first iteration, which generates contigs for the heterozygous alternative allele. All contigs are mapped to the reference human genome using BLAT. If a contig has two distinct parts (i.e. two regions with minimum overlapping) mapped to two different genomic regions with high similarity (≥97%) and good read-length (≥30bp), it is considered a cross-junction contig. Once such a contig is identified and there is no germline reads mapped to the breakpoints identified in the blat alignment, the SV is considered an assembly-validated somatic SV.
Estimation of somatic indels across the five T-ALL tumors based on background mutation rate in childhood cancer
Background mutation rate (BMR) for adult glioblastoma multiforme was estimated to be 3.75×10−6 based on synonymous mutations identified in the protein coding regions18. A recent study of childhood cancer medulloblastoma indicates that childhood cancer has somatic mutations fewer by a factor of 5 to 10 than in the adult tumors19, resulting in a background mutation rate of 7.5×10−6 to 3.75×10−7 for childhood cancer. Our analysis of 120 candidate gene sequencing data of 187 childhood leukemia patients shows that the BMR of childhood ALL is within the estimated range but closer to the lower-end of the estimate. Since the estimated BMR was based on substitution variations while somatic indels are approximately one tenth of that of the substitution mutations18, the somatic indel mutation rate in childhood cancer is projected to be 7.5×10−7 to 3.75×10−8. Applying the indel mutation rate to the entire human genome gives an estimated 112 to 225 somatic indels per tumor. Therefore, the total number of somatic indels across the five T-ALL tumors is estimated to be 562 to 1,125.
Preparation of simulated whole-genome sequencing data for NA12878
Simulated whole-genome sequencing data were generated to evaluate the sensitivity of CREST in identifying validated germline structural variations (i.e. deletions, duplications and insertions) compiled as a gold standard data set by the 1000 Genomes Project 13. NA12878 was selected because it was sequenced at high coverage using three sequencing platforms (Illumia/Solexa, Roche/454 and Life Technologies/SOLiD) and analyzed by 19 SV detection methods, 12 of which were evaluated for their sensitivity in detecting deletion polymorphisms. The golden standard data set for NA12878 consists of 642 deletions, 271 duplications and 30 insertions. We were unable to include the 30 insertions for simulation because the inserted sequences were not accessible. Of the 913 deletion/duplication events, 309 at 138 loci are overlapping events with multiple non-reference deletion/duplication alleles. We consider these multi-allele polymorphisms with ≥ 2 non-reference alleles in the population. Two haploid genomes were generated to represent the two non-reference deletion/duplication alleles in these regions. For the 26 loci that have ≥3 overlapping non-reference alleles, two were randomly selected resulting in a loss of 27 events (23 deletions and 4 duplications). We simulated 100-bp paired-end reads with a mean size of 400 bp with a standard deviation (s.d.) of 20bp. using the software MAQ (version 0.7.1)20 and obtained 20-fold coverage to human assembly NCBI build 36 for each haploid genome. Merging the data from the two haploid genomes gives a total of 1,232,167,792 reads for the diploid genome data with a mean coverage of 40. All reads were mapped to the human assembly NCBI build 36 using the program BWA10 with the default parameters.
Two sets of whole-genome simulation data were generated based on the following two quality models. One is a normal quality simulation that derives the sequencing error and quality based on a training data set of 250k empirical reads randomly selected from our T-ALL WGS data while the other is a high quality data set that use only reads with qualities in the range of 32–40 for training. We created the high-quality simulation data because the mapping rate of the normal quality WGS is 10% lower than that of the empirical WGS data for the 10 T-ALL genomes which ranges from 92–95%. On the other hand, high-quality simulation data has a mapping rate of 91% which is close to the empirical mapping rate.
Experimental validation
The COLO-829 cell line was purchased from the American Type Culture Collection (ATCC) (http://www.atcc.org). Oligonucleotide primers for genomic PCR were designed using Primer 3 (http://frodo.wi.mit.edu/primer3). Genomic DNA from T-ALL tumor samples and COLO-829 was PCR amplified using either the Advantage 2 PCR Kit (Clontech, Mountain View, CA) or Phusion High-Fidelity DNA polymerase (New England Biolabs, Ipswich, MA). Thermal cycling conditions for Advantage 2 polymerase were 5 cycles of 94°C for 30 sec and 72°C 3 min, followed by 5 cycles of 94°C for 30 sec, 70°C for 30 sec and 72°C 3 min, followed by 25 cycles of 94°C for 30 sec, 68°C for 30 sec and 72°C 3 min; and for Phusion were 98°C for 30 sec, followed by 35 cycles of 98°C for 15 sec, 66°C for 15 sec and 72°C for 1 min, followed by a final extension step of 72°C for 10 min. PCR products were electrophoresed through 1% agarose gels impregnated with Gel Red, visualized using ultraviolet illumination, and purified directly from PCR mixtures or after gel electrophoresis. Purified PCR products were sequenced using 3730×l capillary sequencers and Big Dye terminator chemistry.
Mapping algorithms for CREST analysis
Since soft-clipping signature which represents a partial (local) alignment to the reference genome is a prerequisite for CREST, output from mapping tools with local alignment function (such as Mosaik (http://bioinformatics.bc.edu/marthlab/Mosaik) and BFAST (http://sourceforge.net/projects/bfast)) can be used after proper reformatting for this analysis. On the other hand, mapping methods that only perform global alignments such as ELAND (Illumina Genome Analyzer Pipeline Software) and bowtie (http://bowtie-bio.sourceforge.net/index.shtml) will not be able to provide the suitable soft-clipping signature.
Supplementary Material
Supplementary Figure 1. Definition of structural variations in CREST analysis
Supplementary Figure 2. Process flow of CREST
Supplementary Figure 3. Validation of 20 putative SVs detected only by CREST but not reported previously in the melanoma cancer cell line COLO-829
Supplementary Figure 4. Sanger sequencing data for the 18 validated novel SVs in COLO-829 cell line
Supplementary Figure 5. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at deletion spanning chr6:51307435-51308086 reported by Pleasance et al.
Supplementary Figure 6. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at deletion spanning chr14:48401118-48403688 reported by Pleasance et al.
Supplementary Figure 7. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at deletion spanning chr14:33583777-33584588 reported by Pleasance et al
Supplementary Figure 8. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at inter-chromosomal alteration spanning chr1:16797227- 145856276 reported by Pleasance et al.
Supplementary Figure 9. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at deletion spanning chr4:131181342-131224000 reported by Pleasance et al.
Supplementary Figure 10. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at deletion spanning chr10:85512695-85513886 reported by Pleasance et al.
Supplementary Figure 11. Illustration of soft-clipped and paired-end discordant mapping signatures across SV breakpoints in short and long NGS reads
Supplementary Figure 12. An example of a validated somatic deletion in a repetitive region detected by CREST
Supplementary Figure 13. An example of a low-frequency 65bp somatic insertion that was co-amplified with wild-type in sample SJTALL012
Supplementary Figure 14. An example of a small somatic deletion detected by CREST. The 26bp somatic deletion (chr1:163486879-163486905) was found in SJTALL003
Supplementary Data 1. Comparison of CREST performance with other SV detection methods
Supplementary Data 2. SV analysis in melanoma cancer cell line COLO-829 by CREST
Supplementary Data 3. Simulation study for assessing CREST false negative rate in finding germline indels
Supplementary Discussion 1. Limitations of CREST and Comparison of CREST with Genome STRiP
Please combine the SI discussion under a single title
Supplementary Table 1. Summary of CREST SV analysis results and validation in five T-ALL samples.
*Supplementary Table 2. Summary of CREST SV analysis results for COLO-829. The sheet labeled SV includes all inter-chromosomal re- arrangements or intra-chromosomal SVs spanning more than 500bp. The second sheet “Small_indels” include all indels less than 500bp. The first four columns (ChrA, PosA, OrtA, #SC) show the chromosome, position, orientation and number of soft-clipped reads observed at the first breakpoint. Column 5–8 (ChrB, PosB, OrtB, #SC) show the chromosome, position, orientation and number of soft-clipped reads observed at the second breakpoint. Column 9 (Type) defines the SV Type. Column 10,11 show GSAV score and predicted SV type. 0 means not predicted by GSAV. Column 12, 13 shows the gene names where the first and the second breakpoint is located, respectively. The entire genomic regions are used to define overlap between gene and a SV.
Supplementary Table 3. Experimental validation results of 20 novel SVs in COLO-829 identified by CREST
Supplementary Table 4. Performance of CREST, BreakDancer GSAV and Pindel on detecting structural variations found in NA12878 using simulated WGS data
Acknowledgments
This study was supported by Cancer Center support grant P30 CA021765 from the National Cancer Institute and the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital. We would like to thank K. Ye for stimulating discussion on the analysis of the COLO-829 cell line. C.G.M. is Pew Scholar in the Biomedical Sciences. Information about the Pediatric Cancer Genome Project can be found at http://www.pediatriccancergenomeproject.org/site.
Footnotes
AUTHOR COTRIBUTIONS
J.Z. conceived and designed the CREST algorithm. J.W. implemented the algorithm. J.R.D. and C.G.W. designed the experiment. J.W., J.Z., S.R., J.M., M.C.R., K.C., C.C.H., L.D., X.F. and L.W. analyzed the data. C.G.W., J.E., S.L.H., L.H. and D.P.-T. performed validation assay. E.R.M., R.K.W. supervised whole genome sequencing data generation. D.Z., J.C.O. and C.N. set up the computing infrastructure. J.R.D. and J.Z. wrote the manuscript.
ACCESSION_CODES
The 5 T-ALL whole-genome sequence data have been submitted to NCBI short read archive (SRA) under the accession SRA029885.1.
REFERENCE LIST
- 1.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 2.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen K, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hormozdiari F, Alkan C, Eichler EE, Sahinalp SC. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19:1270–1278. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sindi S, Helman E, Bashir A, Raphael BJ. A geometric approach for classification and comparison of structural variants. Bioinformatics. 2009;25:i222–230. doi: 10.1093/bioinformatics/btp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills RE, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abel HJ, et al. SLOPE: a quick and accurate method for locating non-SNP structural variation from targeted next-generation sequence data. Bioinformatics. 26:2684–2688. doi: 10.1093/bioinformatics/btq528. [DOI] [PubMed] [Google Scholar]
- 15.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet. 43:269–276. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.TheCancerGenomeAtlasResearchNetwork. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Definition of structural variations in CREST analysis
Supplementary Figure 2. Process flow of CREST
Supplementary Figure 3. Validation of 20 putative SVs detected only by CREST but not reported previously in the melanoma cancer cell line COLO-829
Supplementary Figure 4. Sanger sequencing data for the 18 validated novel SVs in COLO-829 cell line
Supplementary Figure 5. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at deletion spanning chr6:51307435-51308086 reported by Pleasance et al.
Supplementary Figure 6. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at deletion spanning chr14:48401118-48403688 reported by Pleasance et al.
Supplementary Figure 7. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at deletion spanning chr14:33583777-33584588 reported by Pleasance et al
Supplementary Figure 8. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at inter-chromosomal alteration spanning chr1:16797227- 145856276 reported by Pleasance et al.
Supplementary Figure 9. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at deletion spanning chr4:131181342-131224000 reported by Pleasance et al.
Supplementary Figure 10. Alignment of next-gen reads in both tumor (top panel) and normal (bottom panel) of cell line COLO-829 at deletion spanning chr10:85512695-85513886 reported by Pleasance et al.
Supplementary Figure 11. Illustration of soft-clipped and paired-end discordant mapping signatures across SV breakpoints in short and long NGS reads
Supplementary Figure 12. An example of a validated somatic deletion in a repetitive region detected by CREST
Supplementary Figure 13. An example of a low-frequency 65bp somatic insertion that was co-amplified with wild-type in sample SJTALL012
Supplementary Figure 14. An example of a small somatic deletion detected by CREST. The 26bp somatic deletion (chr1:163486879-163486905) was found in SJTALL003
Supplementary Data 1. Comparison of CREST performance with other SV detection methods
Supplementary Data 2. SV analysis in melanoma cancer cell line COLO-829 by CREST
Supplementary Data 3. Simulation study for assessing CREST false negative rate in finding germline indels
Supplementary Discussion 1. Limitations of CREST and Comparison of CREST with Genome STRiP
Please combine the SI discussion under a single title
Supplementary Table 1. Summary of CREST SV analysis results and validation in five T-ALL samples.
*Supplementary Table 2. Summary of CREST SV analysis results for COLO-829. The sheet labeled SV includes all inter-chromosomal re- arrangements or intra-chromosomal SVs spanning more than 500bp. The second sheet “Small_indels” include all indels less than 500bp. The first four columns (ChrA, PosA, OrtA, #SC) show the chromosome, position, orientation and number of soft-clipped reads observed at the first breakpoint. Column 5–8 (ChrB, PosB, OrtB, #SC) show the chromosome, position, orientation and number of soft-clipped reads observed at the second breakpoint. Column 9 (Type) defines the SV Type. Column 10,11 show GSAV score and predicted SV type. 0 means not predicted by GSAV. Column 12, 13 shows the gene names where the first and the second breakpoint is located, respectively. The entire genomic regions are used to define overlap between gene and a SV.
Supplementary Table 3. Experimental validation results of 20 novel SVs in COLO-829 identified by CREST
Supplementary Table 4. Performance of CREST, BreakDancer GSAV and Pindel on detecting structural variations found in NA12878 using simulated WGS data