Abstract
Polycomb group of proteins (PcG), by controlling gene silencing transcriptional programs through cell cycle, lock cell identity and memory. Recent chromatin genome-wide studies indicate that PcG targets sites are bivalent domains with overlapping repressive H3K27me3 and active H3K4me3 mark domains. During S phase, the stability of epigenetic signatures is challenged by the replication fork passage. Hence, specific mechanisms of epigenetic inheritance might be provided to preserve epigenome structures. Recently, we have identified a critical time window before replication, during which high levels of PcG binding and histone marks on BX-C PRE target sites set the stage for subsequent dilution of epigenomic components, allowing proper transmission of epigenetic signatures to the next generation. Here, we extended this analysis to promoter elements, showing the same mechanism of inheritance. Furthermore, to gain insight into the inheritance of PREs bivalent marks, we analyzed dynamics of H3K4me3 deposition, a mark that correlates with transcriptionally active chromatin. Likewise, we found an early S-phase enrichment of H3K4me3 mark preceding the replication-dependent dilution. This evidence suggests that all epigenetic marks are inherited simultaneously to ensure their correct propagation through replication and to protect the “bivalency” of PREs.
Keywords: polycomb, chromatin, histone marks, epigenetic inheritance, DNA replication, bivalent domain, S phase, PRE
Introduction
Polycomb group of proteins (PcG) are critical regulators of development and cell differentiation that act through repression of gene transcription.1 They reside in two main complexes, termed Polycomb repressive complexes 1 and 2 (PRC1 and PRC2). PRC2 contains a histone methyl transferase activity (HMTase), which catalyzes histone H3 lysine 27 tri-methylation (H3K27me3).2-5 This chromatin mark is specifically recognized by PRC1 complex,2 which exerts several catalytic functions believed to be important for transcriptional repression.6,7 In Drosophila, PcG function is mediated by specialized epigenetic DNA modules called Polycomb response elements (PREs).8 Similarly, a few cis-elements with “PRE properties” were recently reported in mammals,9,10 although their identification on a large scale in that system remains to be completed. While PcG functions in transcriptional repression via histone methylation, and higher order structures formation1,11-14 has been extensively demonstrated, the mechanisms by which PcG-mediated signatures are inherited during cell cycle remain elusive. Studies in mammalian cells suggest that all three core components of PRC2 form a combined binding surface that can insert and recognize the H3K27me3 modification, thus generating a positive feedback loop that helps to propagate H3K27me3 mark through DNA replication.15,16 Taking into consideration that in Drosophila, chromatin histone proteins may be loaded on DNA not only during S phase,17 the ability of PcG proteins to bind their own mark, occurring during all phases of cell cycle and reinforcing the epigenetic repressed status of target genes, could partially explain the stability of epigenetic signatures despite their high turnover in the cell. However, during replication, stability of epigenetic marks is challenged by the replication fork passage. Hence, specific mechanisms of epigenetic inheritance in S-phase must be provided in order to preserve PcG-dependent silencing program. We have recently reported that, during S phase, PcG engagement and characteristic H3K27me3 histone mark deposition on repressed late replicating PREs are restricted to a brief interval in early S phase occurring before DNA replication of the same regions.18 This suggested a model in which the correct transmission of epigenetic information is achieved by preventing the replication-dependent dilution of epigenetic signatures on daughter strands. Interestingly, in such a model the PcG-dependent H3K27me3 mark would be inherited by dilution and not by de novo methylation occurring at the time of replication. Here, we performed cell cycle-dependent analysis of epigenetic signatures at the BX-C PcG-bound promoter, extending our model to another class of PcG targets. In line with our previous data, we found that PcG proteins and H3K27me3 mark are enriched at the repressed and late-replicating abdA promoter during early S phase and subsequently diluted, suggesting a common mechanism of inheritance for all PcG binding sites.
Further, increasing evidence suggests that Polycomb (PcG) and trithorax-group (TrxG) proteins with their associated histone modifications are critical for the plasticity of the pluripotent state, for the dynamic changes in gene expression that accompany cell differentiation and for subsequent maintenance of lineage-specific gene expression programs.19 Indeed, a feature of pluripotent cells is a high representation of genomic regions consisting of overlapping PcG-dependent repressive H3K27me3 and TrxG-dependent active H3K4me3 marks, termed bivalent domains. These play a key role in keeping developmental regulators “poised” for alternate fates.20,21 Upon cell differentiation, most bivalent promoters resolve to a “univalent” state. Induced genes become further enriched for H3K4me3 and lose H3K27me3, while many non-induced genes retain H3K27me3 but lose H3K4me3. Recent studies suggest that Polycomb binding sites, like bivalent domains, carry not only the repressive H3K27me3 modifications, but are also enriched for the activating, trxG-associated H3K4me3 mark.20,22 Studies in Drosophila confirm these findings, showing that repressive and active mark can co-exist on PcG target genes. Moreover, PcG and txG complexes colocalize and are dynamically bound to their target sites during embryogenesis.23-26
Although accumulated evidences clarified some aspects of epigenome inheritance during replication,15,18,27 other features, such as the inheritance of bivalent domains during S phase, remain unexplored. To address this issue, we followed the H3K4me3 active mark at PREs through replication. We found that the low levels of H3K4me3 present at PREs show the same dynamics of enrichment before replication, indicating that all epigenetic signatures controlling the current PRE transcriptional state and its potential are inherited simultaneously.
Results and Discussion
PcG proteins and repressive mark H3K27me3 are enriched at abdA promoter before replication
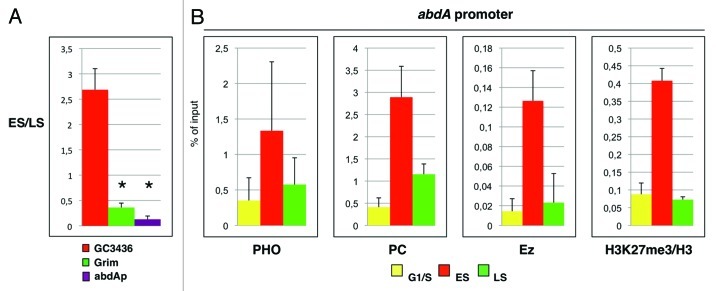
In Drosophila, PcG complexes exert their function both at PREs and transcription start sites of their target genes.8,24 Combining data from replication timing analysis and ChIP assays, we recently reported that PcG complex’s engagement and histone repressive mark deposition is uncoupled from and precedes PREs replication.18 Here, we asked if this S-phase dynamic of epigenetic signatures also takes place on PcG-bound promoters. To this aim, we performed epigenetic analysis on the BX-C-repressed abdA promoter. To measure the replication timing, asynchronous S2 cells were labeled with bromodeoxyuridine triphosphate (BrdU) and FACS (fluorescence-activated cell sorting) sorted into two S-phase fractions representative of early and late S phase according to DNA content.18 BrdU-labeled DNA was immunoprecipitated from these S-phase fractions to enrich for genomic sequences that replicate during the labeling period. Quantitative real-time PCR (qRT-PCR) with primers specific for abdA promoter and control regions was performed to measure the relative amount of analyzed regions. Ratios between the amounts of amplified products in early and late S phase showed that abdA promoter replicates during late S phase (Fig. 1A). We confirmed this result on early and late S-phase fractions of S2 cells collected after HU synchronization (data not shown). We then performed chromatin immunoprecipitation (ChIP) in HU synchronized S2 cells to measure the occupancy of PcG proteins on the AbdA promoter during S phase. Chromatin collected from G1/S, early and late S phase (ES and LS, respectively) was immunoprecipitated with antibodies against PHO, PC and Ez (Fig. 1B), which are members of PhoRC, PRC1 and PRC2 complexes, respectively. As observed for BX-C PREs,18 we found that the amount of PcG proteins bound on AbdA promoter varied over S-phase progression, following the same dynamics. In particular, we observed an increase in early S phase followed by a drop in PcG binding in late S phase, returning to G1/S basal levels. To analyze PcG-dependent HMTase function on chromatin, we measured the levels of histone lysine methylation during S phase with antibodies that recognize total H3 and H3K27me3. As expected, the ratio between H3K27me3 and H3 peaked in early S phase (Fig. 1B, reviewed in ref. 18) following PcG protein loading onto PREs, ensuring the correct epigenetic signatures propagation through replication.
Figure 1. PcG proteins and repressive mark H3K27me3 are enriched at abdA promoter before replication. (A) Replication timing of abdA promoter as measured by quantitative real-time PCR (qRTPCR) in BrdU immunoprecipitated DNA. Ratios between the amplified products in early and late S phase are shown. We amplified positive controls for the early and late S phase in red and green, respectively. Gene names correspond to their entries in FlyBase. All data points were generated from an average of at least three independent experiments. Standard error of the mean is indicated. Two-tailed t-test was applied for statistical analysis. Asterisks indicate statistically relevant differences: 〈 = 0.05. p-values: CG3436/Grim, p = 0.0006; CG3436/abdA promoter, p = 0.0005. (B) ChIP analyses of abdA promoter using indicated antibodies on synchronized cells are presented as percentage of input chromatin. ChIP analysis with antibodies against H3K27me3 were normalized to histone H3 density. Mock enrichment is below 0.003% of the input. Data obtained in HU-treated cells (G1/S) are shown in yellow. Data obtained in cells collected 1 h and 2 h from HU block release (ES and LS) are in red and green, respectively. Each graph shows the result from at least four independent immunoprecipitation reactions done on different chromatin preparations. Standard error of the mean is indicated.
Inheritance of bivalent domains through replication
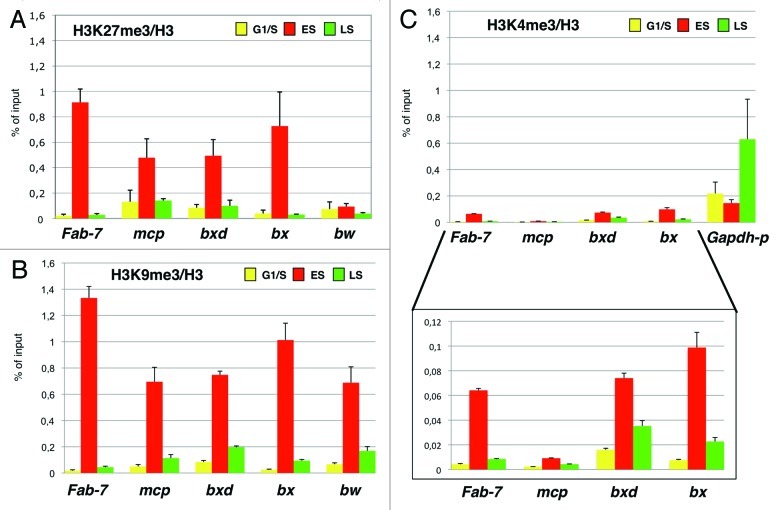
In Drosophila, genome-wide ChIP-seq and transcriptional analysis, in parallel with the detection of transcription start sites (TSS), revealed new features of Polycomb distribution along the Drosophila genome.24 One particularly clear feature is that Polycomb often targets TSSs with a stalled RNAPolII. These sites are also enriched in H3K4me1/me2, and these specific signatures at TSSs might serve transcriptional pausing of key developmental genes. Although the H3K27me3 and H3K4me3 marks do not generally coexist in Drosophila,8 these transcriptionally paused promoters could be functionally considered as the fly analogs of the “bivalent domains” in mammals, which represent poised states for lineage-specific activation of developmental genes.24 This was further confirmed in S2 cells, where repressed PcG targets show occupancy of repressive marks in combination with active marks, general transcription factors and RNA PolII.23 We have previously shown that the inheritance of distinct repressive marks at BX-C PREs during replication share a common timeframe.18 In particular, we found that the H3K9me3 repressive mark, present on PREs,23,26 shows a trend similar to H3K27me3 (Fig. 2A and B), being enriched in early S phase and subsequently diluted in late S phase, when PRE replication takes place. This result is in agreement with the evidence that H3K9me3 histone mark is controlled by PcG proteins26 and suggests that repressive epigenetic signatures are simultaneously inherited during replication. Here, we analyzed the S-phase dynamic of H3K4me3 deposition, a mark that correlates with transcriptionally active chromatin. As expected, we confirmed the presence of H3K4me3 mark at PREs, although at lower levels compared with H3K27me3 (Fig. 2C). Surprisingly, quantification of H3K4me3 enrichment in different S-phase fractions revealed that the active mark deposition follows a similar tendency compared with H3K27me3 and H3K9me3 marks. This dynamic is specific for PREs, because H3K4me3 levels on the transcriptionally active and early replicating Gapdh promoter show a different trend, being diluted from G1 to early S phase (Fig. 2C). Altogether, these results indicate that all three epigenetic marks responsible for the bivalent transcriptional potential of PREs are inherited at the same time to preserve their epigenetic state.
Figure 2. Active and repressive histone marks are inherited simultaneously during replication. ChIP analysis are presented as percentage of input chromatin precipitated for each region normalized to histone H3 density. Mock enrichment is below 0.003% of the input. Data obtained in HU-treated cells (G1/S) are shown in yellow. Data obtained in cells collected 1 h and 2 h from HU block release (ES and LS) are in red and green, respectively. (A–C) ChIP analysis on PREs using antibodies against H3K27me3, H3K9me3 and H3K4me3, respectively, on synchronized cells. As a negative control, we used the promoter region of brown (bw) that is repressed in S2 but it is not under the control of PcG proteins.28 Each graph shows the result from at least four independent immunoprecipitation reactions done on different chromatin preparations. Standard error of the mean is indicated. Magnification with adjusted scale is presented below (C) to highlight H3K4me3 mark dynamics during replication.
Conclusion
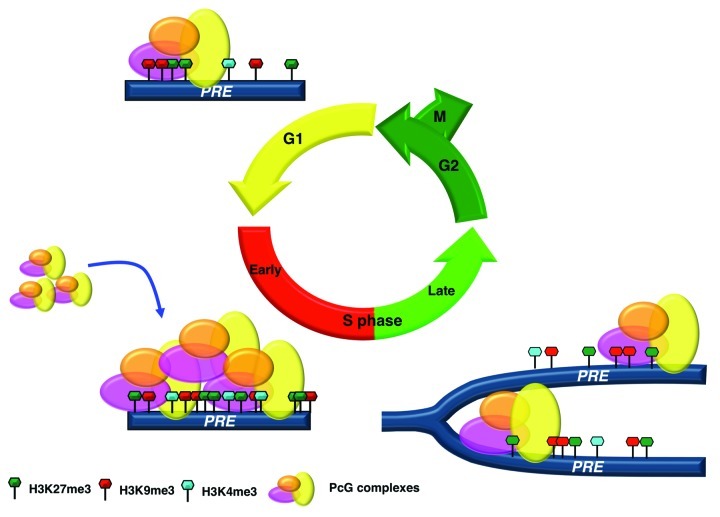
Our findings determine the early S phase as the critical time point for the Polycomb cell memory system integrating recent observations.18 We suggest that PcG complex’s binding and enrichment for all repressive and active histone marks that determine the “epigenetic bivalency” of PcG bound elements are uncoupled from and precede PcG targets replication, when epigenetic signatures are redistributed on daughter strands (Fig. 3). This time-dependent dynamics would allow the local conservation of the epigenetic structures through DNA replication and is necessary for the inheritance of the epigenome.
Figure 3. Schematic representation for trasmission of epigenetic signatures at PREs. The scheme shows a PcG bound bivalent domain. In G1 phase minimal amounts of PcG complexes and H3K27me3 and H3K9me3 marks, sufficient for transcriptional repression, are present at PREs. Low levels H3K4me3 mark are also present. During the early S phase PcG complexes are uploaded on their targets and all repressive and active histone marks are enriched. After PREs replication the redistribution on daughter strands reinstates epigenetic marks to the previous steady-state.
Materials and Methods
Culture cell growth
Drosophila embryonic S2 cells were grown at 25°C in serum-free insect culture medium (HyQ SFX; Hyclone).
Replication timing analysis and chromatin immunoprecipitation
The replication timing analysis and ChIP were performed as previously described in reference 18. Primer sequences: CG3436-f 5′-ATC GCT AAC AGC CAT GTC GG-3′, CG3436-r 5′-CTT ACC GAT TCA AGG AGC GC-3′; Grim-f 5′-TTC CCG AGT CTC TCA CCG C-3′, Grim-r 5′-ACA GGA ACC CAC ACC ACT GAC-3′; abdApr-f 5′-TTG AGT CAG GGA GTG AGC C-3′, abdApr-r 5′-CGC TTT GAG TCG TTG GAG AC-3′.
Antibodies
Antibodies against PC were kindly provided by R. Paro and antibodies against Pho and E(z) by J. Muller. Commercial rabbit polyclonal antibodies against methylated Lysine 27 of histone H3 (Upstate, 07–449), methylated Lysine 9 of histone H3 (Abcam, ab8898), methylated Lysine 4 of histone H3 (Abcam, ab8580) and histone H3 (Abcam, ab1791) were used.
Acknowledgments
We thank J. Muller and R. Paro for kindly providing antibodies that have been essential for this study. We are grateful to Gennaro Oliva for help in graphic design. We thank Maria Vivo, Beatrice Bodega and all members of the lab for stimulating discussions and constructive criticisms. This work was supported by Telethon (grant S00094TELA) and Associazione Italiana Ricerca sul Cancro (AIRC) to V.O.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/19710
References
- 1.Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–26. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 2.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 3.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96. doi: 10.1016/S0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 4.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/S0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 6.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–7. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–5. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 9.Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, et al. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–97. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 10.Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–20. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cléard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet. 2006;38:931–5. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- 13.Comet I, Savitskaya E, Schuettengruber B, Nègre N, Lavrov S, Parshikov A, et al. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev Cell. 2006;11:117–24. doi: 10.1016/j.devcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–74. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 15.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 16.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–7. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–4. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanzuolo C, Lo Sardo F, Diamantini A, Orlando V. PcG complexes set the stage for epigenetic inheritance of gene silencing in early S phase before replication. PLoS Genet. 2011;7:e1002370. doi: 10.1371/journal.pgen.1002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prezioso C, Orlando V. Polycomb proteins in mammalian cell differentiation and plasticity. FEBS Lett. 2011;585:2067–77. doi: 10.1016/j.febslet.2011.04.062. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 21.Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–8. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 22.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiling A, O’Neill LP, D’Eliseo D, Turner BM, Orlando V. Epigenome changes in active and inactive polycomb-group-controlled regions. EMBO Rep. 2004;5:976–82. doi: 10.1038/sj.embor.7400260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res. 2011;21:216–26. doi: 10.1101/gr.114348.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orlando V, Jane EP, Chinwalla V, Harte PJ, Paro R. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 1998;17:5141–50. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papp B, Müller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–54. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–22. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paro R, Zink B. The Polycomb gene is differentially regulated during oogenesis and embryogenesis of Drosophila melanogaster. Mech Dev. 1993;40:37–46. doi: 10.1016/0925-4773(93)90086-D. [DOI] [PubMed] [Google Scholar]