Abstract
Using a method in which DNA adducts are discovered based on their conversion in a nucleotide form to phosphorimidazolides with isotopologue benzoylhistamines (or p-bromobenzoylhistamine) prior to detection by MALDI-TOF-MS, we have profiled the adducts that form when calf thymus DNA is reacted in vitro with p-benzoquinone (BQ). We find, as relative values normalized to 100% of adducts observed, 79% BQ-dCMP, 21% BQ-methyl-dCMP (a new DNA adduct), and trace amounts of BQ-dAMP and BQ-dGMP. Since mC is 5% of C in this DNA, the reaction of BQ with DNA in vitro is about five times faster at methyl-C than C. When equal amounts of dCMP and methyl-dCMP are reacted with BQ, equal amounts of the corresponding adducts are observed. Thus, the microenvironment of methyl-C in DNA enhances its reactivity relative to C with BQ. In a prior, similar study, but based on analysis by 32P-postlabeling, the second most abundant adduct was assigned to BQ-A, apparently because of co-migration of the BQ-A and BQ-methyl-C adducts (as bisphosphates) in the chromatographic step. Since the calf thymus DNA (used as received) was contaminated with RNA, we also detected the ribonucleotide adduct, BQ-CMP.
INTRODUCTION
Humans are exposed to quinones as endogenous biomolecules, dietary natural products, environmental contaminants, and as metabolites of natural and synthetic aryl and phenolic compounds. Some of this exposure is a concern since quinones, such as p-benzoquinone (BQ), a metabolite of benzene and phenol, can be genotoxic.1,2 Much attention has been given to the genotoxicity of benzene since it is a product of common combustion processes such as smoking and operation of vehicles that burn gasoline or diesel.3 Polychlorinated biphenyls are rodent carcinogens, and can be metabolized to quinones that form DNA adducts.4 The genotoxicity of the related molecule, hydroquinone, has been studied extensively since it is mutagenic and has a wide range of commercial applications.5 Maternal benzene exposure was associated with decreases in birth weight and head circumference,6 and prenatal exposure to a hydroxylated polychlorinated biphenyls was associated with decreased mental development.7
Towards a goal of learning more about the genotoxicity of environmental mixtures, we have developed a method that broadly discovers and provides some characterization of DNA adducts by mass spectrometry.8 As pointed out before,8 the method, via its greater ability (based on isotopologue mass tagging) to discover unknown DNA adducts, is complementary to detecting adducts by isotope dilution HPLC-MS/MS. The principle of the method is the same as that of 32P-postlabeling, except that our method: (1) involves chemical labeling (DNA adducts as nucleotides are converted to phosphorimidazolides with benzoylhistamine, BH, in an ordinary or isotopologue form, in the presence of a carbodiimide); (2) detects adducts specifically (accurate masses of adducts are detected by MALDI-TOF-MS); (3) provides semi-quantitation (labeling yield of 60% along with normalization of response with a factor of less than three); and (4) provides structural information by MALDI-TOF/TOF-MS including differentiation of deoxyribonucleotides from ribonucleotides based on formation and detection of a BH-phosphate-sugar fragment ion.8,9
As a first step in using our method to build up a further understanding of the genotoxicity of quinones, we elected to study the ability of BQ to form DNA adducts. While such reactivity by BQ has been investigated before, relying on analysis by 32P-postlabeling (sometimes in conjunction with synthesis and NMR), we noticed some discrepancies and incomplete interpretation of results even for the in vitro reaction of BQ with DNA. Chenna and co-workers reported three adducts (and relative adduct levels) as follows: 70.3% BQ-dCMP; 25.1% BQ-dAMP, and 1.7% BQ-dGMP.10 Bodell and coworkers similarly reported the same three major adducts in similar relative amounts: BQ-dCMP (74.2%), BQ-dAMP (22.4%), and BQ-dGMP (1.09%).11 However, Jiang and co-workers reported four adducts: 71.07% BQ-dCMP, minor BQ-dGMP, minor maybe BQ-dAMP, and minor unknown.12 In this latter study, the relative abundances of the three minor adducts were 11.03, 9.46, and 8.44%, but these values were not connected by the authors to any of the minor species just cited.
EXPERIMENTAL
Calf thymus DNA, 2´-deoxycytidine-5´-monophosphate, disodium salt (dCMP), hydroquinone, p-benzoquinone, nuclease P1, ferric chloride, triethylammonium acetate buffer (TEAA, 1M), 2-N-(morpholino)ethanesulfonic acid (MES), 1-ethyl-3-[3-(dimethylamino)propyl]carbodiimide (EDC), α-cyano-4-hydroxy-cinnamic acid (CCA), and ammonium citrate (dibasic) were from Sigma-Aldrich (St. Louis, MO). 5-Methyl-2´-deoxycytidine-5´-monophosphate, disodium salt (mdCMP) was a gift from Affymetrix (Cleveland, Ohio). Phosphodiesterase I was from Worthington (Lakewood, NJ). BIOMAX-5 Ultrafree MC centrifugal filter devices were from Millipore (Billerica, MA). Propanesulfonic acid silica was from J. T. Baker (Phillipsburg, NJ). Microcentrifuge tubes, pipetter tips, and HPLC grade acetonitrile (ACN) were from Fisher Scientific (Pittsburgh, PA). The OASIS columns were from Waters (Milford, MA). All materials were used as received. Benzoylhistamine (BH), benzoylhistamine-d4 (BH(d4), where the ethylene moiety is tetradeuteriated), and p-bromo-benzoylhistamine (Br-BH) were synthesized as described.9
Hydroquinone was reacted in the presence of ferric chloride with calf thymus DNA as described.10 Our method for measuring DNA adducts, described before,8,9 is summarized here. The digestion of the modified DNA to nucleotides was done with nuclease P1 at pH 5.5 followed with phosphodiesterase I at pH 9, 3 hours at 45 °C for both. The sample was purified with an OASIS column (186000383, Waters) and dried. The labeling reaction was performed by adding 3.5 μL of 12 mM BH (6 mM BH and 6 mM BH(d4)), or 12 mM Br-BH, and 3.5 μL of 80 mM EDC in 0.01 M MES buffer at pH 6, to the dried OASIS collection vial, and then, after mixing, the sample was kept at room temperature in the dark for 3 hours. The sample was separated on a capillary HPLC column (PepMap 180 μm I.D×150 mm, Dionex, Sunnyvale, CA) using a gradient flow from 3% to 60 % ACN in 40 min at 2.2 μL/min. A droplet was collected onto a MALDI plate every 20 sec with a Probot Fraction Collector (Dionex, Sunnyvale, CA). CCA matrix (0.5 μL, 5 mg/mL in ACN:water, 50:50, v/v, with 2.5 mM of ammonium citrate, dibasic) was deposited onto each dried spot, followed by air-drying for 5 min. Analysis was done on a Voyager DE STR MALDI-TOF-MS or a Model 5800 MALDI-TOF/TOF-MS (AB SCIEX, Foster City, CA) in a negative ion mode with a delay time of 150 ns. Each sample well was surveyed to find a “sweet spot”, and then 400 laser pulses were averaged to generate a spectrum. MS/MS was performed with a medium pressure of air and a mass resolution window of 400 with the metastable-ion suppressor on.
p-Benzoquinone was reacted with dCMP or mdCMP as described.13 The nucleotide (8 mg of dCMP or 8.3 mg of mdCMP, 0.023 mmol) and p-benzoquinone (16 mg, 0.15 mmol) were allowed to react in 1 mL of 0.1 M sodium acetate buffer (pH 5) at 37 °C. After 17 h, the diluted reaction mixture (1:100 in water) was mixed with CCA matrix in a 1:5 ratio and subjected to MALDI-TOF-MS in a negative ion mode. Also, MS/MS in a positive ion mode was performed on the protonated modified nucleobase formed in MALDI ion source. Further, a combined sample (1 μL of each diluted reaction mixture) was labeled with Br-BH and subjected to MALDI analysis directly.
RESULTS AND DISCUSSION
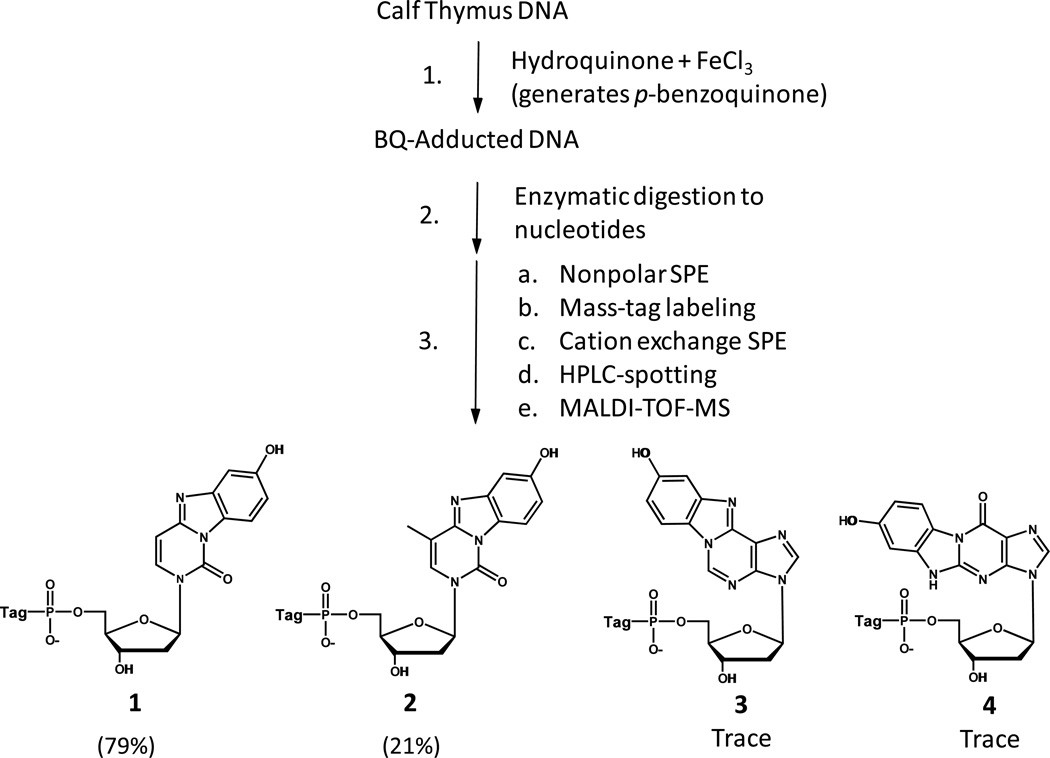
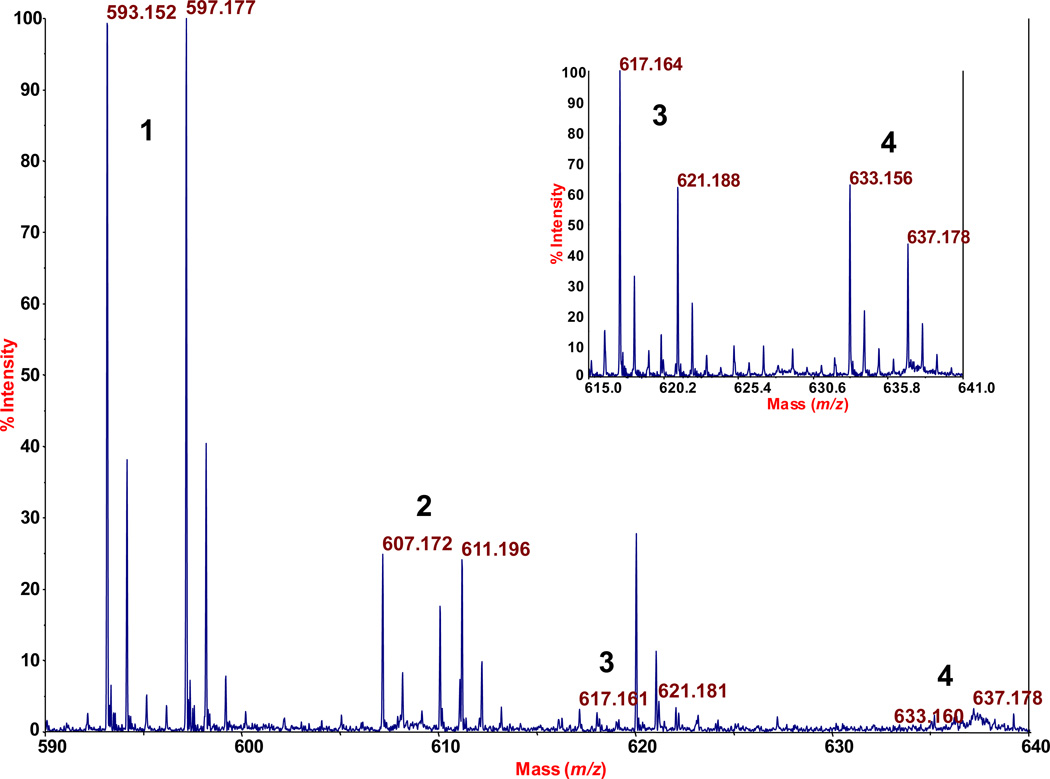
We reacted calf thymus DNA with hydroquinone in the presence of ferric chloride (which generates p-benzoquinone) as described by Chenna and coworkers,10 and subjected the DNA, after precipitation with ethanol and washing with ethanol, to our method for detecting nonpolar DNA adducts by mass-tag MS.9 (A version of our assay that discovers polar DNA adducts also has been reported.8) The reaction scheme, and the four DNA adducts of benzoquinone that we detected, including their relative amounts, are presented in Figure 1. As seen, the relative amounts were as follows: 1, BQ-dCMP (79%); 2, BQ-mdCMP (21%); 3, BQ-dAMP (trace); and 4, BQ-dGMP (trace), taking the sum of these peak heights to be 100%. The values were based on the relative peak heights of the adducts by MALDI-TOF-MS, where each adduct was detected in an isotopologue (do,d4) mass-tagged from. The latter data is shown in Figure 2. For each adduct, the average height of the two isotopologue peaks was used to calculate the relative amounts of the adducts.
Figure 1.
Reaction scheme for the formation and detection of p-benzoquinone (BQ) DNA adducts. SPE: solid phase extraction on an OASIS cartridge. (Also observed, as product 5, was a BQ-cytosine RNA adduct, since the DNA was used as received: see Supplemental Figure 1.)
Figure 2.
Analysis by mass tagging-MALDI-TOF-MS of DNA adducts formed when p-benzoquinone (BQ) is reacted with calf thymus DNA. Assignments: 1 (m/z 593.152 and 597.177): BH- and BH(d4)-conjugates of BQ-dCMP; 2 (607.172 and 611.196), corresponding conjugates of BQ-mdCMP; 3 (617.164 and 621.188, in inset), corresponding conjugates of BQ-dAMP; and 4 (633.156 and 637.178 in inset), corresponding conjugates of BQ-dGMP.
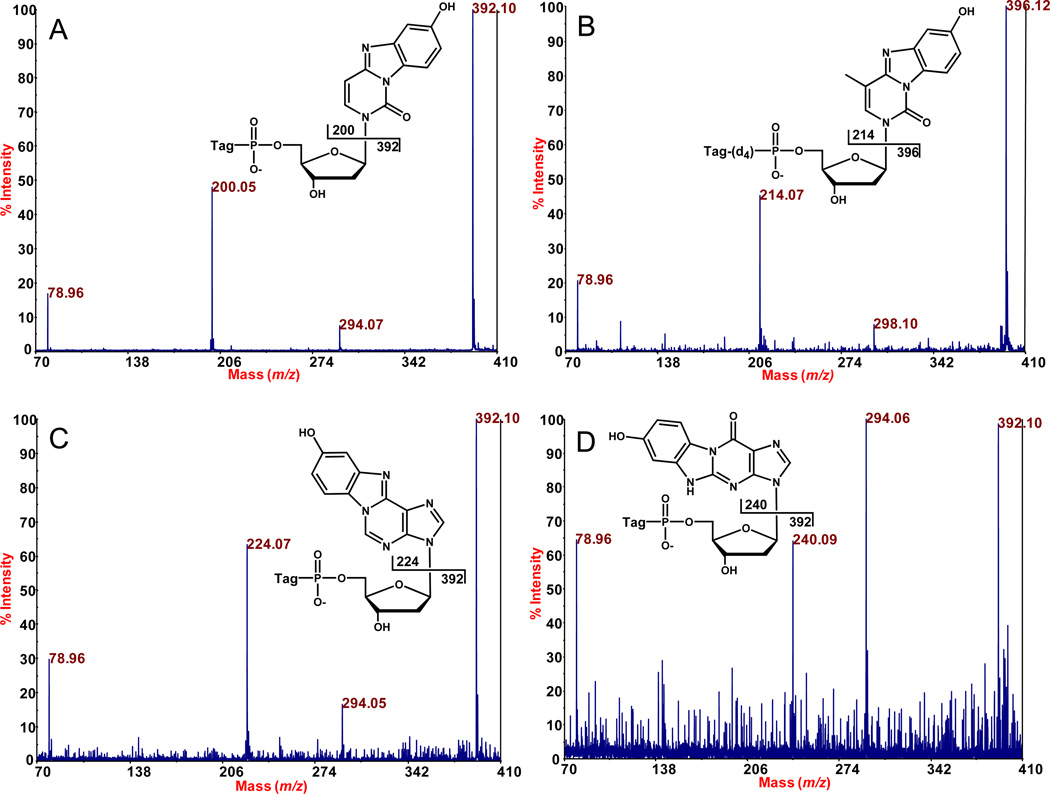
The assignments in Figure 2 were supported by the collision-induced dissociation data shown in Figure 3. Subjecting the parent ion for BH-BQ-dCMP at m/z 593.152 to MALDI-TOF/TOF-MS gave the spectrum presented in Figure 3A. In this latter spectrum, m/z 78.96 is phosphate, m/z 200.05 is BQ-C, m/z 294.07 is BH-phosphate, and m/z 392.10 is BH-phosphate-deoxyribose. Figure 3B shows the MS/MS spectrum assigned to BH(d4)-BQ-mdCMP (parent ion at m/z 611.196). While we also tested the corresponding nondeuteriated compound (parent mass at m/z 607.178), its nucleobase fragment ion was observed at m/z 214.07, an ambiguous mass in this study (data not shown). This is because the BH anion has an exact mass of 214.098, and the exact mass of mC as an anion is 214.062. These two masses are difficult to discriminate in a definitive way at a trace level on our instrument. (Further study of this aspect is presented below.) MS/MS on the ion at m/z 611.196 for BH(d4)-mdCMP (Figure 3B) eliminated this uncertainty. Figures 3C and 3D show fragmentation data for BH-BQ-dAMP and BH-BQ-dGMP, respectively.
Figure 3.
Fragmentation analysis by MALDI-TOF/TOF-MS of parent ions of the DNA adducts detected in Figure 2. A, BH-BQ-dCMP; B, BH(d4)-BQ-mdCMP; C, BH-BQ-dAMP; and D, BH-BQ-dGMP. The peaks at m/z 78.96 and 294.06 are from phosphate and deoxyribose phosphate respectively.
Since the calf thymus DNA that we reacted with BQ was contaminated with RNA (a common occurrence in commercial DNA), it is not surprising that some BQ-CMP was observed as well, as seen in Supplemental Figure 1. Part A shows the detection of the (M - H)− ion of this adduct (BH-labeled) in the two isotopic forms (m/z 609.152 and 613.175, respectively). Part B shows the major fragments that form from metastable m/z 609.152. As seen, the observation of m/z 408.10 (vs m/z 392.10 in parts A, C and D of Figure 3) makes it clear that this adduct is a ribonucleotide.
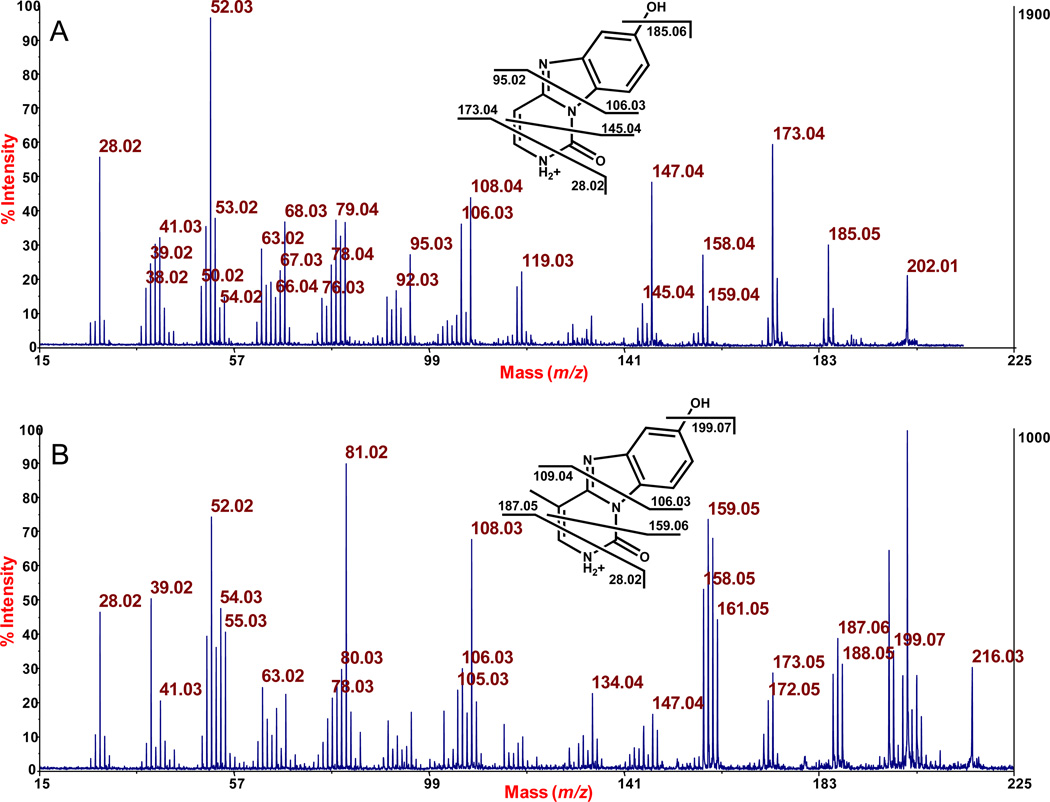
At this point, to overcome the ambiguity of the m/z 214 ion in the negative ion mode, we changed our tag to p-bromobenzoylhistamine (Br-BH), and conducted some additional experiments. First, we repeated the analysis of the DNA adducts from the reaction of DNA with hydroquinone/FeCl3, and observed the same ratio (4:1) of BQ-dCMP to BQ-mdCMP (Supplemental Figure 2). Trace, equivalent amounts of BrBH-BQ-dAMP and BrBH-BQ-dGMP were also observed, similar to the results with the BH tag, but the compounds were in a different MALDI spot (and a different laser intensity was needed to boost their signals) so the data (not shown) provided no quantitative information about the adducts of A and C relative to those of C and mC. We then switched to the positive ion mode, and subjected the protonated nucleobase from BrBH-BQ-dCMP and BrBH-BQ-mdCMP (that forms in the ion source) to collision-induced dissociation after the ion source. This gave equivalent fragmentation data from each (aside from the presence of the methyl group in mdCMP), although the baseline noise was somewhat high, as seen in this figure. Nevertheless, major peaks could be assigned, as shown in Figure 4. Noteworthy, as structurally-informative peaks, is that both adducts share m/z 106.03 from cleavage through the middle (upper fragment), whereas the corresponding, bottom fragments are at m/z 95.02 and 109.64 for C and mC, respectively. (Fragmentation analysis of these products from nucleotide reactions is presented next). We relied on the known structure of the BQ-C adduct16 in making these assignments.
Figure 4.
Fragmentation analysis by MALDI-TOF/TOF-MS of the protonated nucleobase from BQ-dCMP, m/z 202.06 (A), and from BQ-mdCMP, m/z 216.08 (B) formed by reacting BQ with calf thymus DNA.
Second, we reacted equal amounts of dCMP and mdCMP (separately as authentic nucleotides) with BQ, and observed equal amounts of the corresponding BQ adducts (Supplemental Figure 3). Because of the absence of mass tagging, the relative heights of the assigned peaks in each spectrum do not define the relative amounts of corresponding reacted and nonreacted compounds. The two reactions were conducted under identical conditions, and it is clear from the essentially equivalent peak heights for BQ-dCMP and BQ-mdCMP (including comparison with matrix ions) that essentially equal amounts of these two products were formed.
We also conducted MALDI-TOF-MS analysis of these latter BQ adducts of dCMP and mdCMP after labeling with BrBH. Although the spectrum is noisy, once again we observed essentially equal amounts of these adducts, as shown in Supplemental Figure 4. Acetyl products were observed as well (in essentially the same equal ratio for C relative to mC) since acetate was not removed prior to conducting the labeling reaction.
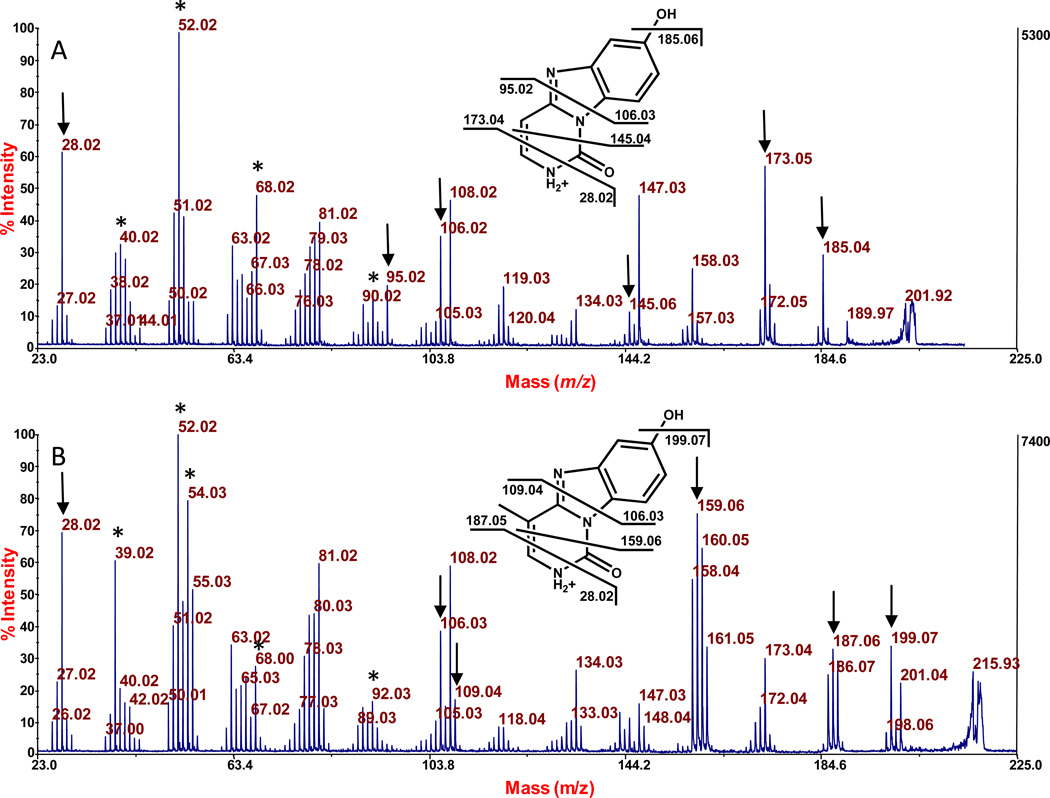
Fragmentation of the protonated nucleobases of the nucleotides of BH-dCMP and BH-mdCMP is shown in Figure 5, where the baseline noise is lower than for the corresponding analysis shown in Figure 4. As seen, the same fragmentation patterns are present in both of these Figures. The lower baseline in Figure 5 permitted the assignment of additional peaks, and this information is presented in Supplemental Figure 5. Further, in regard to Figure 5, a fragment ion corresponding to loss of ammonia from the protonated molecules is not seen, although it is seen from protonated cytosine and 5-methylcytosine (data not shown), consistent with fusion of the exocyclic amine of C and mC with BQ.
Figure 5.
Fragmentation analysis by MALDI-TOF/TOF-MS of the protonated nucleobase from BQ-dCMP, m/z 202.06 (A); and from BQ-mdCMP, m/z 216.08 (B). These compounds were formed by reacting BQ with dCMP and mdCMP, respectively. The peaks that are interpreted in this figure are marked with an arrow, with theoretical values on the structures. The peaks marked with an asteric are interpreted in Supplemental Figure 5.
While co-testing of authentic standards is necessary in general to provide definitive quantitative results by MALDI-TOF-MS, it is clear that BQ-mdCMP is the second most abundant adduct. For adducts studied to date, BH-labeling normalizes response even for diverse adducts to within a factor of less than 3, and more so for similar adducts.8
Apparently BQ-mdCMP was misinterpreted as BQ-dAMP previously in two of the prior studies10,11 because it co-migrated with authentic BQ-dAMP under the conditions employed for 32P-postlabeling. Our ratio of BQ-dCMP to BQ-mdCMP + BQ-dAMP (4:1) is similar to the ratios of 2.8:1 and 3.4:1 reported before for the ratio of BQ-dCMP to BQ-dAMP, supporting our hypothesis that BQ-mdCMP and BQ-dAMP co-migrated in the prior studies. Our data also agree qualitatively with the report by Jiang and coworkers12 of four adducts by 32P-postlabeling from the reaction of calf thymus DNA with BQ.
In calf thymus DNA, 5-methylcytosine (mC) is 5% of cystosine (C), so the expected ratio of BQ-dCMP to BQ-mdCMP is 19:1 if BQ reacts equally with C and mC. However, we observe a ratio of about 4:1. Thus our data indicates that BQ preferentially targets mC over C by about 5-fold when reacted in vitro with DNA. The basis for this enhanced reactivity at mC apparently is its microenvironment in DNA, based on the data presented above (the free nucleotides react equally with BQ), but we did not study the potential role of ss-DNA vs ds-DNA for this reactivity (calf thymus DNA was used as received).
Our in vitro results are not informative for the reaction of BQ with DNA in vivo, and more studies are needed in this area. Snyder and coworkers14 incubated rat liver mitoplasts first with[3H]dGTP to form DNA tritiated in this way, and then exposed the mitoplasts to [14C]benzene, which yielded several DNA adducts of guanine. They similarly incubated mitoplasts with [3H]dATP before adding [14C]benzene, which yielded a single, major adduct of adenine. They also reacted BQ in vitro with dG and characterized spectroscopically the major guanine adduct formed from the latter reaction. There was no study of whether this adduct corresponded to any of the in vivo adducts. Bodell and coworkers reported (based on 32P-postlabeling) that reaction of HL-60 and bone marrow cells with HQ (in the presence of FeCl3) or BQ gave the same single DNA adduct in each case (unidentified), and that this adduct did not correspond to any of the three adducts formed when HQ (in the presence of FeCl3), or BQ, is reacted with calf thymus DNA.11
The heightened reactivity of mC in DNA in vitro towards BQ raises concern (if it happens in vivo) that this exposure (and perhaps some other quinone exposures) may have an epigenetic effect, especially via triggering (we assume) nucleotide excision repair in a region of DNA where mC residues are clustered. This might help to explain the hypomethylation of LINE-1 by exposure of humans to benzene,15 although the overall situation may be much more complicated since hypermethylation was observed in p15 simultaneously, and benzene gives rise to multiple reactive metabolites.16 Some of the higher mutability of mC relative to C17,18 might be due to reaction with quinones. Evidence against benzoquinone as a carcinogen has been summarized.16
5-Methylcytosine in DNA also is more prone than cytosine to deaminate hydrolytically, and to form a photodimer.19 Oxidation of the methyl moiety of mC has been reviewed.19 Whether the G of a hemimethylated or fully methylated CpG sequence in DNA is more or less reactive than an unmethylated CpG depends on the G-reactive chemical as has been summarized.19–22 For example, (+)-trans-benzo[a]pyrene diolepoxide is more reactive with cytosine-methylated sequences.20 This enhanced reactivity has been attributed to intercalation19 or a hydrophobic interaction.22 Perhaps such interactions are an important part of the microenvironment of 5-mC in DNA that enhances its reactivity with BQ.
It is well known that 32P-postlabeling is limited in its ability to provide qualitative and quantitative analysis of DNA adducts. In addition to deoxynucleotides, ribonucleotides are labeled23 and non-nucleotides can be labeled.24 Some nonbulky DNA adducts have been misinterpreted for many years as bulky aryl adducts.25 Labeling yield can depend markedly on adduct structure.26,27 While mass tag labeling provides more specificity including structural information, and gives a consistent labeling yield, it is less sensitive in general at the present time than 32P-postlabeling, as we have discussed previously.8
CONCLUSION
Isotopologue mass tag labeling with benzoylhistamine is a useful technique for discovery of DNA adducts, especially because it provides exact peak pairs to help discriminate signals from noise. Here this technique was used to discover a benzoquinone adduct of 5-methylcytosine, and to learn that BQ-mdCMP (rather than BQ-dAMP as reported before based on 32P-postlabeling) is the second most abundant adduct when DNA is reacted in vitro with p-benzoquinone. This encourages additional work in this area, such as similar analysis of DNA adducts after an in vivo exposure to p-benzoquinone, and similar study of other quinones to which humans may be exposed.
Supplementary Material
Acknowledgments
Funding
This work was supported by NIH Grant P42ES017198 from NIEHS. Contribution number ___ from the Barnett Institute.
ABBREVIATIONS
- BH
benzoylhistamine
- BQ
p-benzoquinone
- BrBH
p-bromobenzoylhistamine
- C
cytosine
- BH(d4)
benzoylhistamine-d4, where the ethylene moiety is tetradeuteriated
- G
guanine
- HQ
hydroquinone
- MALDI-TOF-MS
matrix assisted laser desorption ionization time-of-flight mass spectrometry
- mC
5-methylcytosine
Footnotes
The authors declare no competing financial interest.
Contributor Information
Poguang Wang, Email: p.wang@neu.edu.
Jianxin Gao, Email: gjianxin@hotmail.com.
Guodong Li, Email: gli54@hotmail.com.
Olga Shimelis, Email: oshimelis@sial.com.
Roger W. Giese, Email: r.giese@neu.edu.
REFERENCES
- 1.Gaskell M, McLuckie EIE, Farmer PB. Comparison of the mutagenic activity of the benzene metabolites, hydroquinone and para-benzoquinone in the supF forward mutation assay: a role for minor DNA adducts formed from hydroquinone in benzene mutagenicity. Mutation Res. 2004;554:387–398. doi: 10.1016/j.mrfmmm.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 2.Snyder R, Witz G, Goldstein BD. The toxicology of benzene. Environ. Health Perspect. 1993;100:293–306. doi: 10.1289/ehp.93100293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hook GER, Lucier GW. Environmental Perspectives Supplements: Benzene Toxicity, Carcinogenesis, and Epidemiology. Environ. Health Perspect. 1996;104(Supplement 6) [Google Scholar]
- 4.Zhao S, Narang A, Ding X, Eadon G. Characterization and Quantitative Analysis of DNA Adducts Formed from Lower Chlorinated PCB-Derived Quinones. Chem. Res. Toxicol. 2004;17:502–511. doi: 10.1021/tx034245b. [DOI] [PubMed] [Google Scholar]
- 5.McGregor D. Hydroquinone: An evaluation of the human risks from its carcinogenic and mutagenic properties. Crit. Rev. Toxicol. 2007;37:887–914. doi: 10.1080/10408440701638970. [DOI] [PubMed] [Google Scholar]
- 6.Siama R, Thiebaugeorges O, Goua V, Aussel L, Sacco P, Bohet A, Forhan A, Ducot B, Annesi-Maesano I, Heinrich J, Magnin G, Schweitzer M, Kaminski M, Charles MA Eden Mother-Child Cohort Study Group. Maternal personal exposure to airborne benzene and intrauterine growth. Environ. Health Perspect. 2009;117:1313–1321. doi: 10.1289/ehp.0800465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park H-Y, Park J-S, Sovcikova E, Kocan A, Linderholm L, Bergman A, Trnovec T, Hertz-Picciotto I-H. Exposure to hydroxylated polychlorinated biphenyls (OH-PCBs) in the prenatal period and subsequent neurodevelopment in eastern Slovakia. Environ. Health Perspect. 2009;117:1600–1606. doi: 10.1289/ehp.0900611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, Fisher D, Rao A, Giese RW. Nontargeted nucleotide analysis based on benzoylhistamine labeling-MALDI-TOF/TOF-MS: discovery of putative 6-oxo-thymine in DNA. Anal. Chem. 2012;84:3811–3819. doi: 10.1021/ac300532z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P, Li G, Giese RW. Non-signal imidazole reagents for mass spectrometry analysis of phosphomonoesters. 8,003,395. U.S. Patent. 2011;B2
- 10.Chenna A, Hang B, Rydberg B, Kim E, Pongracz K, Bodell WJ, Singer B. The benzene metabolite p-benzoquinone forms adducts with DNA bases that are excised by a repair activity from human cells that differs from an ethenoadenine glycosylase. Proc. Natl. Acad. Sci.: USA. 1995;92:5890–5894. doi: 10.1073/pnas.92.13.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodell WJ, Levay G, Pongracz K. Investigation of benzene-DNA adducts and their detection in human bone marrow. Environ. Health Perspect. 1993;99:241–244. doi: 10.1289/ehp.9399241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang X, Liu S, Xu X. DNA-benzoquinone adducts analyzed by nuclease P1 mediated 32P-postlabeling method. J. Environ. Sci (China) 1992;4:97–105. [Google Scholar]
- 13.Chenna A, Singer B. Large Scale Synthesis of p-Benzoquinone-2´-Deoxycytidine and p-Benzoquinone-2´-Deoxyadenosine Adducts and Their Site-Specific Incorporation in DNA Oligonucleotides. Chem. Res. Toxicol. 1995;8:865–874. doi: 10.1021/tx00048a007. [DOI] [PubMed] [Google Scholar]
- 14.Snyder R, Jowa L, Kalf G, Rushmore T. Formation of reactive metabolites from benzene. Arch. Toxicol. 1987;60:61–64. doi: 10.1007/BF00296948. [DOI] [PubMed] [Google Scholar]
- 15.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun H-M, Jiang J, Marinelli B, Pesatori AC, Bertazzi PA, Yang AS. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 16.Harris CM, Stec DF, Christov PP, Kozekov ID, Rizzo CJ, Harris TM. Deoxyguanosine Forms a Bis-Adduct with E,E-Muconaldehyde, an Oxidative Metabolite of Benzene: Implications for the Carcinogenicity of Benzene. Chem. Res. Toxicol. 2011;24:1944–1956. doi: 10.1021/tx2002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laird PW, Jaenisch R. DNA methylation and cancer. Human Molecular Genetics. 1994;3:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA, Rideout WM, Shen J-C, Spruck CH, Tsai YC. Methylation, mutation and cancer. BioEssays. 1992;14:33–36. doi: 10.1002/bies.950140107. [DOI] [PubMed] [Google Scholar]
- 19.Darwanto A, Van Ornam JD, Lao VV, Sowers LC. Chemical Carcinogenesis and Epigenetics in Penning, T.M. (ed) Chemical Carcinogenesis, Current Cancer Research, Springer. Science. 2011;2011:245–266. [Google Scholar]
- 20.Weisenberger DJ, Romano LJ. Cytosine Methylation in a CpG Sequence Leads to Enhanced Reactivity with Benzo[a]pyrene Diol Epoxide that Correlates with a Conformational Change. J. Biol. Chem. 1999;274:23948–23955. doi: 10.1074/jbc.274.34.23948. [DOI] [PubMed] [Google Scholar]
- 21.Glick J, Xiong W, Lin Y, Noronha AM, Wildes CJ, Vouros P. The influence of cytosine methylation on the chemoselectivity of benzo[a]pyrene diol epoxide-oligonucleotide adducts determined using nanoLC/MS/MS. J. Mass Spectrom. 2009;44:1241–10248. doi: 10.1002/jms.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matter B, Wang G, Jones R, Tretyakova N. Formation of Diastereomeric Benzo[a]pyrene Diol Epoxide-Guanine Adducts in p53 Gene-Derived DNA Sequences. Chem. Res. Toxicol. 2004;17:731–741. doi: 10.1021/tx049974l. [DOI] [PubMed] [Google Scholar]
- 23.Godschalk RW, Maas LM, Kleinjans JC, Van Schooten FJ. Influences of DNA isolation and RNA contamination on carcinogen-DNA adduct analysis by 32P-postlabeling. Environ. Mol. Mutagen. 1998;32:344–350. [PubMed] [Google Scholar]
- 24.Masento MS, Hewer A, Grover PL, Phillips DH. Enzyme-mediated phosphorylation of polycyclic hydrocarbon metabolites: detection of non-adduct compounds in the 32P-postlabeling assay. Carcinogenesis. 1989;10:1557–1559. doi: 10.1093/carcin/10.8.1557. [DOI] [PubMed] [Google Scholar]
- 25.Arif JM, Dresler C, Clapper ML, Gairola CG, Srinivasan C, Lubet RA, Gupta RC. Lung DNA Adducts Detected in Human Smokers are Unrelated to Typical Polyaromatic Carcinogens. Chem. Res. Toxicol. 2006;19:295–299. doi: 10.1021/tx0502443. [DOI] [PubMed] [Google Scholar]
- 26.Shields PG, Harris CC, Petruzzelli S, Bowman ED, Weston A. Standardization of the 32P-postlabeling assay for polycyclic aromatic hydrocarbon-DNA adducts. Mutagenesis. 1993;8:121–126. doi: 10.1093/mutage/8.2.121. [DOI] [PubMed] [Google Scholar]
- 27.Goncalves LL, Beland FA, Marques MM. Synthesis, characterization, and comparative 32P-postlabeling efficiencies of 2,6-dimethylanilline-DNA adducts. Chem. Res. Toxicol. 2001;14:165–174. doi: 10.1021/tx0002031. [DOI] [PubMed] [Google Scholar]
- 28.Hook GER, Lucier GW. Benzene Toxicity, carcinogenesis, and Epidemiology, Environ. Health Perspect. 1996;104 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.