Abstract
In the course of primary infection with herpes simplex virus 1 (HSV-1), children with inborn errors of TLR3 immunity are prone to HSV-1 encephalitis (HSE) 1–3. We tested the hypothesis that the pathogenesis of HSE involves non hematopoietic central nervous system (CNS)-resident cells. We derived induced pluripotent stem cells (iPSCs) from the dermal fibroblasts of TLR3- and UNC-93B-deficient patients and from controls. These iPSCs were differentiated into highly purified populations of neural stem cells (NSCs), neurons, astrocytes and oligodendrocytes. The induction of IFN-β and/or IFN-γ1 in response to poly(I:C) stimulation was dependent on TLR3 and UNC-93B in all cells tested. However, the induction of IFN-β and IFN-γ1 in response to HSV-1 infection was impaired selectively in UNC-93B-deficient neurons and oligodendrocytes. These cells were also much more susceptible to HSV-1 infection than control cells, whereas UNC-93B-deficient NSCs and astrocytes were not. TLR3-deficient neurons were also found to be susceptible to HSV-1 infection. The rescue of UNC-93B- and TLR3-deficient cells with the corresponding wild-type allele demonstrated that the genetic defect was the cause of the poly(I:C) and HSV-1 phenotypes. The viral infection phenotype was further rescued by treatment with exogenous IFN-α/β, but not IFN-γ1.Thus, impaired TLR3- and UNC-93B-dependent IFN-α/β intrinsic immunity to HSV-1 in the CNS, in neurons and oligodendrocytes in particular, may underlie the pathogenesis of HSE in children with TLR3 pathway deficiencies.
Childhood HSE is a rare, life-threatening, CNS-restricted complication of primary infection with HSV-1, an almost ubiquitous virus that is typically innocuous 4. Children with HSE are not unusually susceptible to other infectious agents, including viruses, or even to HSV-1-related diseases affecting sites other than the CNS 4,5. HSV-1 reaches the CNS from the oral and nasal epithelium, via the cranial nerves 4. We identified autosomal recessive (AR) UNC-93B deficiency as the first genetic etiology of childhood HSE 1. UNC-93B is required for TLR3, TLR7, TLR8 and TLR9 responses 1,6. We then identified AR or autosomal dominant (AD) deficiencies of TLR3 2,3, TRAF3 7, TRIF 8 and TBK19, revealing that childhood HSE can be due to the impairment of TLR3 immunity. HSV-1 produces dsRNA during its replication 10,11 and the dsRNA-sensing TLR3 is expressed and functional in non hematopoietic (neurons, astrocytes, oligodendrocytes) and hematopoietic (microglia) CNS-resident cells, which produce IFN-β and IFN-γ upon TLR3 stimulation 12–15 and can be infected with HSV-1 in vitro 13,16–19. We therefore hypothesized that the pathogenesis of HSE in patients with TLR3 pathway deficiencies involves impaired TLR3-dependent IFN-α/β and/or IFN-γ immunity to HSV-1 in the CNS 1,2.
We tested this hypothesis, by generating iPSCs from control, UNC-93B- and TLR3-deficient dermal fibroblasts (Supplementary Table 1). We first derived and fully characterized iPSC lines from a healthy child, from a HSE child with AR complete UNC-93B deficiency 1, and from a patient with AR complete TLR3 deficiency 2, by reprogramming primary dermal fibroblasts, as previously described 20 (Supplementary Fig. 1 and Methods). A robust stemness and pluripotency profile, karyotypic integrity and patient-specific origin of the iPSCs were confirmed (Supplementary Fig. 1). Whole-exome sequencing for one control, one TLR3-deficient and two UNC-93B-deficient iPSC lines revealed more than 99.9% concordance with the corresponding parental fibroblast lines, in terms of exonic genetic variability (Supplementary Table 2). No synonymous or non synonymous germline and somatic rare variants of any of the 21 known TLR3 pathway genes were found in parental fibroblast and iPSC lines, respectively (Supplementary Table 3). We also used two additional, previously described healthy control iPSC lines 21,22 for subsequent experiments (Supplementary Table 1).
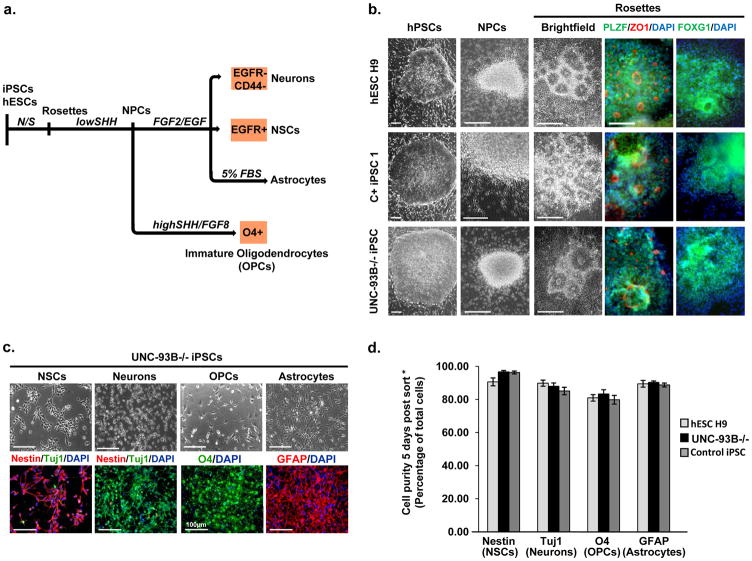
We next induced the differentiation of iPSCs into all major non hematopoietic CNS-resident cells, including neural stem cells (NSCs), neurons, oligodendrocytes and astrocytes 23,24. The selective derivation of each individual neural lineage required a multistep iPSC differentiation approach combined with FACS-mediated cell purification (Fig. 1a). Neural differentiation of UNC-93B-deficient iPSCs, TLR3-deficient iPSCs, control iPSCs and control hESCs (H9 line) was induced by dual SMAD inhibition 21,24 (Fig. 1a and Supplementary Fig. 2a). The resulting polarized columnar neuroepithelial structures expressed PLZF, ZO1 and PAX6, well known markers of neural rosettes 23. Mechanically passaged rosettes retained expression of the forebrain marker FOXG1 (Fig. 1b) and yielded neural precursor cells (NPCs) in the presence of FGF2/EGF (Fig. 1b, Supplementary Fig. 2a). Through differential growth factor treatment and the use of cell type-specific surface markers in NPC-stage cells, we were able to isolate highly enriched populations of NSCs, neurons, astrocytes, or oligodendrocyte lineage cells (Fig. 1c, Supplementary Fig. 2b, c).
Figure 1. Derivation and purification of CNS cells.
a- Schematic diagram of the differentiation and purification protocols used. b- Representative phase-contrast images showing the morphology of the hPSCs, neural rosettes and NPC clusters, from healthy control hESCs, healthy control iPSCs, and UNC-93B−/− iPSCs. Immunocytochemistry analysis revealed the expression of rosette markers (PLZF/ZO1) and a forebrain marker FOXG1. c- Characterization of UNC-93B−/− iPSC-derived CNS cell types: Upper panels: phase-contrast images illustrating the characteristic morphology of each type of neural cell; Lower panels: immunofluorescence analysis for markers of neural stem cells (nestin), neurons (Tuj1), oligodendrocyte progenitor cells (O4) and astrocytes (GFAP). d-Quantification of marker expression for each neural cell type derived from control hESCs, UNC-93B−/− iPSCs, and control iPSCs (error bars = SEM). * Scale bars in b- correspond to 100 μm; those in c- correspond to 50 μm, except for O4 staining (100 μm).
The purified hESC- and iPSC-derived neuronal populations expressed additional lineage-specific markers (Supplementary Fig. 3a) and showed the electrophysiological properties of functional neurons (Supplementary Fig. 3b–e). The identity of the purified GFAP expressing and O4 expressing glial cell populations was confirmed by global gene expression profiling (Supplementary Fig. 4a, b). Immunocytochemistry for stage-specific markers was used to identify the purified O4 “oligodendrocytes” used throughout this study as a mostly immature (premyelinating) oligodendrocyte population (Supplementary Fig. 4c–e). Quantitative analysis demonstrated that our preparations of NSCs, neurons, astrocytes and oligodendrocytes were highly pure (Fig. 1d), and similar gene expression profiles were obtained for neurons and astrocytes derived from disease-specific and control cell lines (Supplementary Fig. 5). Our in vitro CNS cell differentiation system therefore constitutes a reliable platform for the comparative assessment of CNS cell-specific antiviral immunity.
TLR3 expression has been documented in neurons derived in vitro from a human teratocarcinoma cell line 13, and in primary cells, either in human brain tissues in situ (neurons 25) or isolated from human brain ex vivo (oligodendrocytes and astrocytes 12,14,26), but not in human NSCs — self-renewing, multipotent cells responsible for generating neurons, astrocytes, and oligodendrocytes in the CNS 27. We detected mRNA for key genes of the TLR3-responsive pathway, including TLR3 and UNC93B1, in all four cell types tested (Supplementary Fig. 6a–d), whether differentiated from iPSCs or hESCs. We also detected mRNAs for genes encoding key molecules in the IFN-responsive pathways, including the receptors for IFN-α/β and IFN-γ, in these cells (Supplementary Fig. 6e–h). Levels of mRNA for the TLR3- and IFN-responsive pathway genes tested were similar, for each CNS cell type, between cells differentiated from control iPSCs, control hESCs and UNC-93B-deficient iPSCs, with the expected exception of UNC93B1, for which mRNA levels were lower in UNC-93B-deficient cells (Fig. 2a, Supplementary Fig. 6a–d).
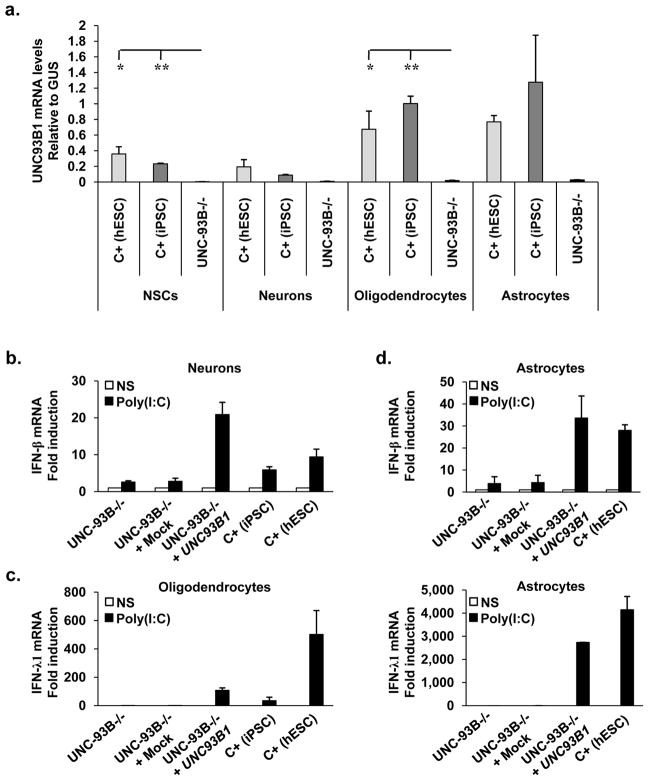
Figure 2. UNC-93B-dependent IFN responses to TLR3 in neurons and glial cells.
a- UNC93B1 mRNA levels in CNS cells differentiated from hESCs from a healthy control (C+ (hESC)) and iPSCs from a healthy control (C+ (iPSC)) and an UNC-93B-deficient patient (UNC-93B−/−). b-, c- IFN-β (b-) or IFN-γ1 (c-) mRNA induction, after six hours of poly(I:C) stimulation, in neurons (b-) or oligodendrocytes (c-) differentiated from one healthy control hESC line, one healthy control iPSC line, and in UNC-93B−/− neurons (b-) or oligodendrocytes (c-), without lentiviral infection, or after infection with a lentivirus containing human WT UNC93B1 (UNC-93B−/− + UNC93B1) or a mock construct (UNC-93B−/− + mock). d- IFN-β (upper panel) or IFN-γ1 (lower panel) mRNA induction, after four (upper panel) or six (lower panel) hours of poly(I:C) stimulation, in astrocytes differentiated from hESCs from a healthy control, in UNC-93B−/− astrocytes, without lentiviral infection, or after infection with a lentivirus containing human WT UNC93B1 (UNC-93B−/− + UNC93B1) or a mock construct (UNC-93B−/− + mock). In a- to d-, Mean values ± SEM were calculated from three (a-) or two (b–d) independent experiments, each carried out in duplicate. ANOVA was performed for data shown in a-. When significant, Dunnett t tests were performed for 2X2 comparisons, significant results are indicated in corresponding panels (* p<0.05, ** p<0.01).
We then studied TLR3 responses in the same cells. IFN-γ1 and IFN-β were induced in a time-dependent manner, by stimulation with the nonspecific TLR3 agonist poly(I:C), a dsRNA analog, in NSCs, neurons, oligodendrocytes and astrocytes differentiated from healthy control iPSCs or hESCs, but not in UNC-93B-deficient iPSC-differentiated CNS cells (Fig. 2b–d, Sup Fig. 6i–l). The induction of NF-κB1, a key IFN-inducing molecule, and that of MX1, a key IFN-inducible molecule, were both impaired in UNC-93B-deficient oligodendrocytes upon poly(I:C) stimulation (Supplementary Fig. 6m, n). The impaired response to poly(I:C) in UNC-93B-deficient CNS cells was consistent with our previous data for UNC-93B-deficient fibroblasts, pointing to a TLR3-dependent response to dsRNA in these cell types 1. Moreover, impaired poly(I:C)-responsiveness in UNC-93B-deficient neurons, oligodendrocytes and astrocytes was rescued by transient expression of the human UNC93B1 gene (Fig. 2b–d). Thus, the UNC-93B-dependent TLR3 pathway is functional in control human iPSC-derived NSCs, neurons, astrocytes and oligodendrocytes, at least for the induction of antiviral IFN-γ1 and IFN-β in response to poly(I:C).
We thus set out to compare the response to HSV-1 in UNC-93B-deficient and control iPSC or hESC-derived CNS cells after infection with HSV-1 and HSV-1-GFP 28. Human NSCs and astrocytes appeared to be more susceptible to HSV-1 infection than neurons and oligodendrocytes, as massive HSV-1-GFP replication was observed earlier (Supplementary Fig. 7 and data not shown). UNC-93B-deficient NSCs and astrocytes derived from two different iPSC lines also showed high levels of HSV-1-GFP replication, similar to those observed in the corresponding cell types derived from two control iPSC lines and the control hESC line (Fig. 3a–c, Supplementary Figs. 8a–c, 9a–c). Treatment of UNC-93B-deficient and control NSCs and astrocytes with recombinant IFN-α2b or IFN-β, but not IFN-γ1, decreased HSV-1-GFP replication levels (Supplementary Figs. 8a, c–f, 9a, c–g). Moreover, treatment with poly(I:C) decreased HSV-1-GFP replication levels in control, but not in UNC-93B-deficient astrocytes (Supplementary Fig. 9f, 10a–d). By contrast, treatment with agonists of TLR9 (CpG-A or CpG-C) did not have such an effect (Supplementary Fig. 10a–d).
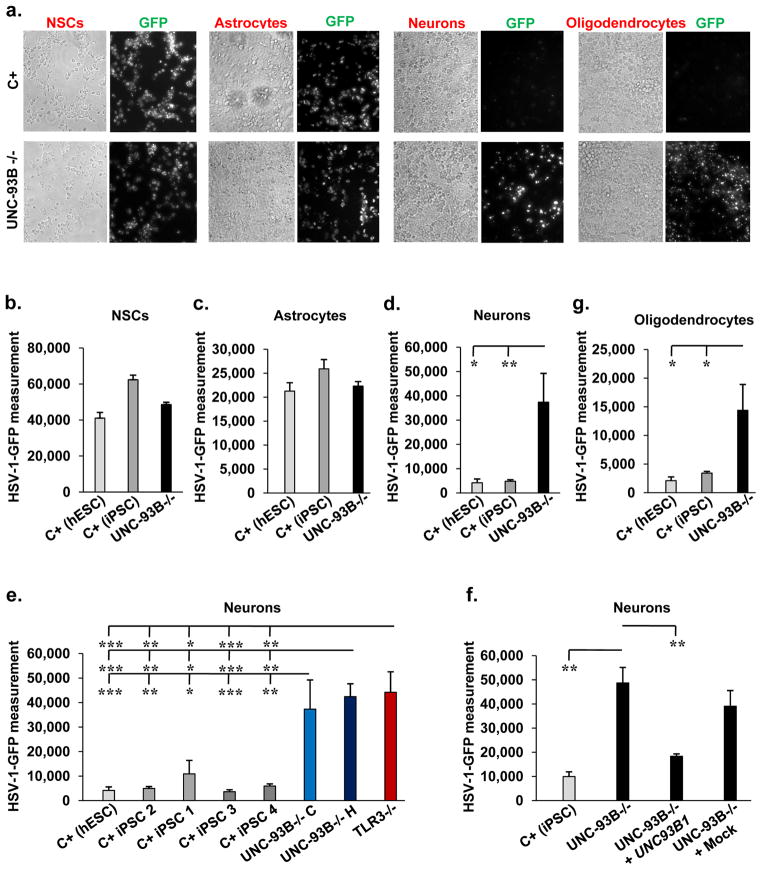
Figure 3. High HSV-1 susceptibility in UNC-93B-deficient neurons and oligodendrocytes.
24 hours of infection with HSV-1-GFP, at a multiplicity of infection of 1, was performed in experiments shown. a- GFP expression in CNS cells differentiated from UNC-93B−/− iPSCs or from hESCs from a healthy control (C+) was visualized by fluorescence microscopy. Phase-contrast photomicrographs from the same view are also shown. b-, c-, d- GFP expression in NSCs (b-), astrocytes (c-) and neurons (d-) differentiated from UNC-93B−/− iPSCs, from hESCs from a healthy control (C+ (hESC)) and one to two iPSC lines each from up to three healthy controls (C+ (iPSC)), was assessed with a fluorescence plate reader. The difference in GFP intensity between HSV-1-GFP-infected cells and uninfected cells is shown. e- GFP expression in neurons differentiated from two lines of UNC-93B−/− iPSCs, one line of TLR3−/− iPSCs, one or two iPSC lines each from three healthy controls, or from one C+ hESCs line. f- GFP expression, in neurons from one C+ iPSC line, in UNC-93B−/− neurons, without lentiviral infection, or after infection with a lentivirus containing human WT UNC93B1 (UNC-93B−/− + UNC93B1) or a mock construct (UNC-93B−/− + Mock). g- GFP expression in oligodendrocytes differentiated from UNC-93B−/− iPSCs, from a C+ hESC line and a C+ iPSC line. Error bars = SEM, calculated from two to three experiments, from the C+ hESC, from the C+ iPSC lines and from the UNC-93B−/− lines, each studied in triplicate (b-, c-, d-, e- and g-); or from one experiment carried out in triplicate, representative of two independent experiments (f-). ANOVA was performed for the data shown in b- to g-. When significant, Dunnett t tests were performed for 2X2 comparisons, significant results are indicated in corresponding panels (* p<0.05, ** p<0.01, *** p<0.001).
When UNC-93B-deficient neurons from the two UNC-93B-deficient iPSC lines were infected with HSV-1-GFP, HSV-1-GFP replication was faster, reaching higher levels than in neurons differentiated from four control iPSC lines or one hESC line (Fig. 3a, 3d, 3e, Supplementary Fig. 11a). The treatment of UNC-93B-deficient neurons with IFN-α2b or IFN-β, but not IFN-γ1, rescued this phenotype (Supplementary Fig. 11a). Similar results were obtained with TLR3-deficient neurons that had been differentiated from TLR3-deficient iPSCs 3,20 (Fig. 3e, Supplementary Fig. 11b). The phenotype of enhanced HSV-1 replication in UNC-93B- and TLR3-deficient neurons was rescued by expression of the wild-type human UNC93B1 and TLR3 genes, respectively (Fig. 3f, Supplementary Fig. 11b). Finally, higher levels and faster replication of HSV-1-GFP were also observed in UNC-93B-deficient oligodendrocytes than in oligodendrocytes differentiated from control iPSCs or hESCs, this phenotype being rescued by treatment with IFN-α2b or IFN-β, but not IFN-γ1 (Fig. 3a, 3g, Supplementary Fig. 11c, d). This is consistent with our previous finding of high susceptibility to HSV-1 and VSV in fibroblasts with TLR3 pathway deficiencies, associated with an impairment of the TLR3-dependent induction of IFN-β and -γ, which can be rescued more effectively by exogenous IFN-α/β than by IFN-γ1 1,2,3.
We further studied the production of IFNs and inflammatory cytokines in UNC-93B-deficient and control CNS cells after infection with HSV-1. UNC-93B-deficient neurons, astrocytes, and oligodendrocytes produced normal amounts of IL-6, as shown by comparison with control iPSC- or hESC-differentiated CNS cell types (Supplementary Fig. 12a–c). UNC-93B-deficient NSCs and astrocytes seemed to produce detectable but lower levels of IFN-γ1 than the control cells tested (Supplementary Fig. 12d, e), suggesting that UNC-93B-independent, TLR3-independent partially compensatory pathways may be involved in triggering IFN responses to HSV-1 in human NSCs and astrocytes. The induction of IFN-β or IFN-γ1 was readily observed in control iPSC- or hESC-differentiated neurons or oligodendrocytes, but was greatly impaired in UNC-93B-deficient neurons and oligodendrocytes, respectively (Supplementary Fig. 12f, g). The induction of MX1 was also impaired in UNC-93B-deficient oligodendrocytes upon HSV-1 infection (Supplementary Fig. 12h). Thus, neurons and oligodendrocytes lacking UNC-93B may be highly vulnerable to HSV-1 infection because of an impairment of the cell-autonomous IFN-α/β or –γ immunity.
Our findings suggest that neurons and oligodendrocytes provide strong, intrinsic protective anti-HSV-1 immunity in the CNS, via an intact TLR3 pathway. Although NSCs and astrocytes lacking UNC-93B are not more susceptible to HSV-1 infection than control cells in vitro, they may play a role in protective anti-HSV-1 immunity in the CNS in vivo. Indeed, mouse astrocytes rely on TLR3 to control HSV-2 29. Hematopoietic cell-derived microglial cells, which also express TLR3 and can be infected with HSV-1 12,14,17, may also contribute to HSE. CNS-intrinsic mechanisms are thus vital for the control of HSV-1 in the course of primary infection in childhood. These new findings, adding to our previous results indicating that most leukocytes and keratinocytes from HSE patients with TLR3 pathway deficiencies respond normally to stimulation with poly(I:C) or HSV-1 2,3,9, are remarkably consistent with the CNS-restricted pattern of lesions during childhood HSE, with no disseminated disease. Human non hematopoietic cells may be the key to survival during the course of primary infection, extending the concepts of host defense beyond innate and adaptive hematopoietic immunity, to non hematopoietic ‘intrinsic’ immunity 30.
Methods
Cell culture
Fibroblasts obtained from an UNC-93B-deficient patient, a TLR3-deficient patint and a healthy control were maintained in DMEM supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine and penicillin (50 U/ml)/streptomycin (50 mg/ml) (hFib media). The patients resided in France, where they were followed up and informed consent was obtained, in accordance with local regulations, with institutional review board (IRB) approval. The experiments described here were conducted in the United States, in accordance with local regulations and with the approval of the IRB of The Rockefeller University, The Harvard Medical School and The Sloan-Kettering Institute for Cancer Research. Induced pluripotent stem cells (iPSCs) and human embryonic stem cells (hESCs; line H9 (WA-09, XX, P40–55) were maintained on CF1-irradiated MEFs (iMEFs, Globalstem Inc) in hESC medium (hESC media) consisting of DMEM/F12 (Invitrogen) supplemented with 20% Knockout Serum (KOSR) (Invitrogen), 10 ng/ml basic fibroblast growth factor (bFGF, Gemini Bio-Products), 1 mM L-glutamine (Invitrogen), 100 μM non essential amino acids, 100 μM 2-β-mercaptoethanol (Sigma-Aldrich) and penicillin (50 U/ml)/streptomycin (50 mg/ml). iPSCs were stimulated to differentiate into embryoid bodies (EB) by culture in bFGF-free hESC medium, without coculture with feeder cells, on non adherent Petri dishes, as previously described 31.
Neural induction and neural subtype specification
MS5 stromal cells were grown in MEM medium supplemented with 10% FBS and 2 mM L-glutamine 32. The neural differentiation of hESCs and iPSCs was induced as previously described 33, but in the presence of Noggin (R&D Systems) and SB431542 (Stemgent) from days 3 to 10 of differentiation (dual-SMAD inhibition 21), to increase the efficiency of rosette formation. Rosettes were harvested mechanically, starting at day 8 of differentiation, and were replated under feeder-free conditions on dishes coated with 10 μg/ml polyornithine (Sigma), 2 μg/ml laminin (Cultrex), and 1 μg/ml fibronectin (Fisher) (Po/Lam/FN), in N2 medium supplemented with Sonic C25 II (20 mg/ml; R&D Systems), ascorbic acid (AA, 0.2 mM; Sigma) and BDNF (20 ng/ml; R&D Systems). Rosettes were allowed to proliferate for a further five days and were then replated, dissociated in Ca2/Mg2-free HBSS and replated again. Emerging clusters of NPCs were harvested for further proliferation or neural subtype specification. For the generation of neural stem cells and neurons, NPCs were maintained in N2 medium supplemented with EGF (20 ng/ml) and FGF2 (20 ng/ml) (R&D Systems) and B-27 supplement without vitamin A (1:100, Invitrogen). For the generation of astrocytes, NPCs were allowed to proliferate and were passaged in the presence of EGF and FGF2 for 40 to 60 days and then exposed to N2 medium containing 5% FBS for an additional 15 to 20 days. For the generation of oligodendrocytes, emerging clusters of NPCs were cultured in N2 medium supplemented with Sonic C25II (125 ng/ml), FGF8 (100 ng/ml; R&D Systems), BDNF (20 ng/ml) and ascorbic acid (AA, 0.2 mM) for 50 to 70 days.
Plasmids and vectors
The polycistronic lentiviral pHAGE–STEMCCA–LoxP vector, carrying the OCT4, SOX2, KLF4, and C-MYC reprogramming factor genes, has been described elsewhere 34. Human UNC93B1 cDNA was amplified from existing cDNA sequences with the following primers: forward 5′-ATAATATGGCCACACATATGGAGGCGGAGCCG-3′ and reverse 5′-GTTGATTAGGATCTATCGTCACTGCTCCTCCGG-3′. The amplified product was inserted downstream from the IRES in a pHAGE2-EF1a-DsRedExpress-IRES-W lentiviral vector (available from the Mostoslavsky laboratory), with the In-Fusion Advantage PCR cloning Kit (Clontech).
Lentivirus production
Lentiviruses containing STEMCCA or pHAGE2-EF1a-DsRedExpress-IRES-UNC93B1 were produced with a five-plasmid transfection system, in 293T packaging cells, by a slightly modified version of a previously described method 35. Briefly, 293T cells were transfected with STEMCCA or pHAGE2-EF1a-DsRedExpress-IRES-UNC93B and four plasmids encoding the packaging proteins Gag-Pol, Rev, Tat and the G-protein of the vesicular stomatitis virus (VSVG), in the presence of the Trans-IT® 293 transfection reagent (Mirus). Viral supernatants were collected every 12 hours, on two consecutive days, starting 48 hours after transfection, and viral particles were concentrated by ultracentrifugation at 49,000 g for 1.5 hours at 4°C.
Lentiviral infection and human iPSC generation
We infected 105 fibroblasts derived from patients or controls with the concentrated polycistronic STEMCCA lentiviral vector and then cultured them at 37°C, under an atmosphere containing 5% CO2, in 2 ml of hFib medium supplemented with 5 μg/ml protamine sulfate, for 24 hours. One day after infection, the viral supernatant was removed and the cells were cultured for 72 hours in hFib medium. They were then transferred onto iMEFs and the medium was replaced with hESC medium. iPSC colonies with an ESC-like morphology were mechanically isolated four to five weeks after infection.
Immunostaining
Cells were fixed by incubation in 4% paraformaldehyde for 30 minutes and permeabilized by incubation with 0.2% Triton X-100 for 30 minutes. Cells were stained in blocking buffer (3% BSA; 5% goat serum) with primary (or conjugated) antibodies at 4°C overnight, washed and stained with secondary antibodies and 1 μg/ml Hoechst 33342 in blocking buffer for 3 hours at 4°C, in the dark. Primary OCT4 and NANOG antibodies (Abcam, Cambridge, MA) were used at a concentration of 0.5 μg/ml, and an Alexa Fluor 555-conjugated anti-rabbit IgG 555 (Invitrogen) was used as the secondary antibody (1:2000). The following conjugated antibodies — TRA-1-60-Alexa Fluor 647, TRA-1-81-Alexa Fluor 488, SSEA-4-Alexa Fluor 647, and SSEA-3-Alexa 488 (Millipore, Billerica, MA) — were used at a dilution of 1:100. FoxG1 antibody was a gift from Dr. S. A. Anderson. Nestin antibody was obtained from Neuromics, Tuj1antibody from Covance, O4 from Millipore, GFAP from Dako, PLZF from Calbiochem, and ZO-1from Zymed. Images were acquired with a Pathway 435 bioimager equipped with a 10x objective (BD Biosciences, San Jose, CA).
Whole-exome sequencing and analysis
DNA (3 μg) was extracted from cells and sheared with a Covaris S2 Ultrasonicator (Covaris). An adapter-ligated library was prepared with the TruSeq DNA Sample Prep Kit (Illumina). Exome capture was performed with the SureSelect Human All Exon 50 Mb kit (Agilent Technologies). Paired-end sequencing was performed on an Illumina HiSeq 2000 (Illumina), generating 100-base reads. The sequences were aligned with the human genome reference sequence (hg19 build), with BWA aligner 36. Downstream processing was carried out with the Genome Analysis Toolkit (GATK) 37, SAM tools 38 and Picard Tools (http://picard.sourceforge.net). Variant calls were made with GATK UnifiedGenotyper. All calls with a Phred-scaled SNP quality ≤20 were filtered out. GATK VariantEval was used to compare the call sets for fibroblasts and iPSCs.
FACS-mediated cell purification
Cells were dissociated with Accutase (Innovative Cell Technologies Inc.) and subjected to FACS with O4 (1:300; Millipore), CD44 FITC (2 μl/106 cells; Abcam), and EGFR PE (10 μl/106 cells; Abcam) antibodies, on a FACS Aria II machine (BD).
Karyotype analysis
Karyotyping and G-banding were performed blind, by Cell Line Genetics.
Mutation analysis
Whole-genome DNA was isolated from fibroblasts and iPSCs with the QiAMP DNA Kit (Qiagen). Exon 8 of the UNC93B1 gene was amplified with the forward primer GCGTGGCTTTGTGCTGAGAG and the reverse primer CAGGAGGGGGATATTTGGGA. Reaction products were purified with the QIAquick PCR purification kit (Qiagen) and sequenced by the DF/HCC DNA Sequencing Facility. The results were analyzed with Sequencher 4.8 software (Gene Codes Corporation).
Microarray analysis
Total RNA was extracted with Trizol reagent (Invitrogen). RNA was collected from astrocytes and from CD44−/EGFR− neurons differentiated from control hESCs or UNC-93B-deficient iPSCs. The RNA was then processed by the MSKCC Genomic core facility and hybridized with Illumina human HT-12 oligonucleotide arrays. Gene expression analysis was carried out with the Partek Genomics Suite: following quantile normalization, all the genes displaying differential expression (FDR of 0.05, fold change of at least ± 2) with respect to hESC (total of 7210 genes) in each population were visualized by clustering. Raw data for the microarray analyses performed in this study are available from the public repository of GEO DataSets (accession no. GSE40593).
Electrophysiology
Whole-cell current clamp recordings were performed at room temperature (23–24°C) in a Multiclamp 700B amplifier (Molecular Devices, Sunnyvale, CA), as previously described 39,40. Neurons were identified under a Nikon microscope equipped with a 4× objective and a 40× water immersion objective. Cells were continuously perfused with freshly prepared extracellular solution containing 126 mM NaCl, 26 mM NaHCO3, 3.6 mM KCl, 1.2 mM NaH2PO4, 1.2 mM MgCl2, 2 mM CaCl2 and 17 mM glucose, and the solution was saturated with 95% O2 – 5% CO2. The intracellular solution contained 135 mM potassium gluconate, 5 mM NaCl, 10 mM HEPES, 0.5 mM EGTA, 3 mM potassium ATP, 0.2 mM sodium GTP and 10 mM sodium phosphocreatine. The pH was adjusted to 7.3 with KOH, and the osmolarity of the solution was ~290 mOsm. Input resistance was calculated from the voltage response elicited by the intracellular injection of a depolarizing (+10 or +20 pA) current pulse. Current steps were applied for 1 s to evoke action potentials. Liquid junction potentials were calculated and corrected off-line. Data were analyzed with Clampfit (Molecular Devices) and SigmaPlot 11 (Systat Software, Chicago, IL) and are presented as means ± SEM.
Reverse transcription-quantitative PCR (RT-qPCR)
For analysis of the expression profiles of key genes involved in stem cell properties and pluripotency, total RNA was extracted with the mirVana™ RNA isolation kit (Ambion). We reverse-transcribed 100 ng of total RNA to generate cDNA, in qScript cDNA Supermix (Quanta). Quantitative real-time PCR was then performed in an AB 7500 Real-Time PCR system (Applied Biosystems), with the PowerSYBR Green PCR Master Mix (Applied Biosystems). The results were analyzed with SDSv1 Software and normalized with respect to β-actin gene expression. Expression levels were determined by the ddCT method and are expressed relative to those in the individual parental cell lines. The primer sequences used have been described elsewhere 41. We assessed the expression levels of genes of the TLR3 and IFN pathways and of the genes for IFN-β, IFN-γ, NF-κB and MX1, by extracting total RNA from NSCs, neurons, oligodendrocytes and astrocytes. RNA was reverse-transcribed directly, with oligo-dT, to determine mRNA levels for TLR3 and IFN pathway genes and for IFN-β, IFN-γ, NF-κB and MX1. RT-qPCR was performed with Applied Biosystems Assays-on-Demand™ probe/primer combinations and 2 x universal reaction mixture, in an ABI 7500 Fast Real-Time PCR System. The β-glucuronidase (GUS) gene was used for normalization. Results are expressed according to the ΔΔCt method, as described by the manufacturer.
TLR3 agonists and viral stimulation
We used a synthetic analog of dsRNA, (polyinosinepolycytidylic acid (poly(I:C); Amersham), a TLR-3 agonist, at a concentration of 25 μg/ml. After incubation with or without poly(I:C) for 2, 4 or 6 hours, cells were collected in Trizol for RNA extraction. For HSV-1 infection, we used the KOS-1 strain, at a multiplicity of infection (MOI) of 1. A GFP-expressing HSV-1 (HSV-1-GFP 281) was used at various MOI to infect fibroblasts, NSCs, neurons, oligodendrocytes and astrocytes.
Cytokine determinations
The production of IFN-α, -β and -γ and of IL-6 was assessed by ELISA. Separate ELISAs were carried out for each of IFN-α (AbCys SA, Paris, France), IFN-β (TFB, Fujirebio, Inc., Tokyo, Japan) and IL-6 (Sanquin, Amsterdam, the Netherlands), according to the kit manufacturers’ instructions. The IFN-γ ELISA was developed in the laboratory, as previously described 42.
HSV-1-GFP infection and quantification
For HSV-1 GFP infection, 104 SV40-transformed fibroblasts, NSCs, neurons, oligodendrocytes or astrocytes were plated in individual wells of 96-well plates and infected with HSV-1-GFP, at various MOI, in a medium appropriate for the cell type concerned. Cells were incubated for two hours, then washed and incubated in 100 μl of culture medium. HSV-1-GFP titers were determined by calculating the 50% end point (TCID50), as described by Reed and Muench, after the inoculation of Vero cell cultures in 96-well plates. The GFP fluorescence of the samples was quantified at the 2 h, 8 h, 18 h and 24 h time points. For assays of cell protection upon viral stimulation, cells were treated with IFN-α2b (104 IU/ml, Schering-Plough, USA), IFN-β (104 IU/ml, PBI Interferonsource, USA), IFN-γ1 (2.5 μg/ml, R&D Systems), poly(I:C) (25 μg/ml), CpG-A (5 μg/ml, Dynavax Technologies, Berkeley, CA) or CpG-C (5 μg/ml, Dynavax Technologies, Berkeley, CA), for 18 hours before infection for fibroblasts and 36 hours before infection for neurons, oligodendrocytes, NSCs and astrocytes, as appropriate.
Statistical tests
Mean values were compared between control cells and cells from the patient, by ANOVA and/or Student’s t-tests, as implemented in the procedures PROC TTEST and PROC ANOVA of SAS version 9.1 (SAS institute, Cary, NC, USA). ANOVA was carried out to compare the means of more than two groups. When significant, ANOVA was followed by t tests for pairwise comparisons (in particular, Dunnett’s t tests comparing the patient with each of the controls).
Supplementary Material
Acknowledgments
We warmly thank our patients, their families and physicians. We would like to thank the members of the three laboratories for helpful discussions and critical reading of this manuscript. The work was funded by grant number 8UL1TR000043 from the National Center for Translational Sciences (NCATS), National Institute of Health (NIH), the Rockefeller University, the St. Giles Foundation, the ANR, INSERM, Paris Descartes University, the March of Dimes, NIH grant 5R01NS072381-02 (to JLC, LS and LDN), NIH grant 1R03AI0883502-01 (to LDN), NIH grant 1R01NS066390 and The Manton Foundation. The Israeli Centers of Research Excellence (I-CORE), Gene Regulation in Complex Human Disease, Center No 41/11 (to IP). FGL is supported by the New York Stem Cell Foundation.
Footnotes
Author contributions
F.G.L., I.M.P., S.Y.Z., J.L.C., L.S. and L.D.N. designed the experiments. F.G.L., I.M.P., S.Y.Z., M.J.C., M.H., A. A., G.M., S.W.Y., S.K., P.A.G, J.O-M., E.J., E.T., Y.E. and T.M.S. performed the experiments. S.A. and M.T. helped to obtain materials from the patients. G.Q.D. and L.A. helped to analyze and describe the data. S.Y.Z. and J.L.C. wrote the manuscript with the aid of F.G.L., I.M.P., L.S. and L.D.N.
Author Information
The authors have no competing financial interests to declare.
References
- 1.Casrouge A, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 2.Zhang SY, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 3.Guo Y, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208:2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitley RJ. Herpes simplex encephalitis: adolescents and adults. Antiviral Res. 2006;71:141–148. doi: 10.1016/j.antiviral.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Abel L, et al. Age-dependent Mendelian predisposition to herpes simplex virus type 1 encephalitis in childhood. J Pediatr. 2010;157:623–629. doi: 10.1016/j.jpeds.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 7.Perez de Diego R, et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33:400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancho-Shimizu V, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121:4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman M, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med. 2012;209:1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquemont B, Roizman B. RNA synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetric transcripts and of double-stranded RNA prepared from them. J Virol. 1975;15:707–713. doi: 10.1128/jvi.15.4.707-713.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bsibsi M, Ravid R, Gveric D, van Noort JM. Broad expression of Toll-like receptors in the human central nervous system. J Neuropathol Exp Neurol. 2002;61:1013–1021. doi: 10.1093/jnen/61.11.1013. [DOI] [PubMed] [Google Scholar]
- 13.Prehaud C, Megret F, Lafage M, Lafon M. Virus infection switches TLR-3-positive human neurons to become strong producers of beta interferon. J Virol. 2005;79:12893–12904. doi: 10.1128/JVI.79.20.12893-12904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jack CS, et al. TLR signaling tailors innate immune responses in human microglia and astrocytes. J Immunol. 2005;175:4320–4330. doi: 10.4049/jimmunol.175.7.4320. [DOI] [PubMed] [Google Scholar]
- 15.Zhou L, et al. Activation of Toll-like receptor-3 induces interferon-lambda expression in human neuronal cells. Neuroscience. 2009;159:629–637. doi: 10.1016/j.neuroscience.2008.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell BM, Bloom DC, Cohrs RJ, Gilden DH, Kennedy PG. Herpes simplex virus-1 and varicella-zoster virus latency in ganglia. J Neurovirol. 2003;9:194–204. doi: 10.1080/13550280390194000. [DOI] [PubMed] [Google Scholar]
- 17.Lokensgard JR, et al. Robust expression of TNF-alpha, IL-1beta, RANTES, and IP-10 by human microglial cells during nonproductive infection with herpes simplex virus. J Neurovirol. 2001;7:208–219. doi: 10.1080/13550280152403254. [DOI] [PubMed] [Google Scholar]
- 18.Bello-Morales R, Fedetz M, Alcina A, Tabares E, Lopez-Guerrero JA. High susceptibility of a human oligodendroglial cell line to herpes simplex type 1 infection. J Neurovirol. 2005;11:190–198. doi: 10.1080/13550280590924179. [DOI] [PubMed] [Google Scholar]
- 19.Marques CP, Hu S, Sheng W, Lokensgard JR. Microglial cells initiate vigorous yet non-protective immune responses during HSV-1 brain infection. Virus Res. 2006;121:1–10. doi: 10.1016/j.virusres.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Pessach IM, et al. Induced pluripotent stem cells: a novel frontier in the study of human primary immunodeficiencies. J Allergy Clin Immunol. 2011;127:1400–1407. doi: 10.1016/j.jaci.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kriks S, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elkabetz Y, et al. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabar V, et al. Migration and differentiation of neural precursors derived from human embryonic stem cells in the rat brain. Nat Biotechnol. 2005;23:601–606. doi: 10.1038/nbt1088. [DOI] [PubMed] [Google Scholar]
- 25.Jackson AC, Rossiter JP, Lafon M. Expression of Toll-like receptor 3 in the human cerebellar cortex in rabies, herpes simplex encephalitis, and other neurological diseases. J Neurovirol. 2006;12:229–234. doi: 10.1080/13550280600848399. [DOI] [PubMed] [Google Scholar]
- 26.Farina C, et al. Preferential expression and function of Toll-like receptor 3 in human astrocytes. J Neuroimmunol. 2005;159:12–19. doi: 10.1016/j.jneuroim.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 27.Taupin P, Gage FH. Adult neurogenesis and neural stem cells of the central nervous system in mammals. J Neurosci Res. 2002;69:745–749. doi: 10.1002/jnr.10378. [DOI] [PubMed] [Google Scholar]
- 28.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinert LS, et al. TLR3 deficiency renders astrocytes permissive to herpes simplex virus infection and facilitates establishment of CNS infection in mice. J Clin Invest. 2012;122:1368–1376. doi: 10.1172/JCI60893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 31.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barberi T, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotechnol. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- 33.Perrier AL, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci U S A. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somers A, et al. Generation of transgene-free lung-disease specific human iPS cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc Natl Acad Sci U S A. 2006;103:16406–16411. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ying SW, Goldstein PA. Propofol-block of SK channels in reticular thalamic neurons enhances GABAergic inhibition in relay neurons. J Neurophysiol. 2005;93:1935–1948. doi: 10.1152/jn.01058.2004. [DOI] [PubMed] [Google Scholar]
- 40.Ying SW, et al. Dendritic HCN2 channels constrain glutamate-driven excitability in reticular thalamic neurons. J Neurosci. 2007;27:8719–8732. doi: 10.1523/JNEUROSCI.1630-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 42.Yang K, et al. Human TLR-7-, -8-, and -9-mediated induction of IFN-alpha/beta and -lambda Is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity. 2005;23:465–478. doi: 10.1016/j.immuni.2005.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.