Abstract
Context
Neuroblastoma is diagnosed over a wide age range from birth through young adulthood, and older age at diagnosis is associated with a decline in survivability.
Objective
To identify genetic mutations that are associated with age at diagnosis in patients with metastatic neuroblastoma.
Design, Setting and Patients
We performed whole genome sequencing of DNA from diagnostic tumors and their matched germlines from 40 patients with metastatic neuroblastoma obtained between 1987 and 2009. Age groups at diagnosis included infants (0-<18 months), children (18 months-<12 years), and adolescents and young adults (≥12 years). To confirm the findings from this discovery cohort, validation testing using tumors from an additional 64 patients obtained between 1985 and 2009 was also performed. Formalin-fixed paraffin-embedded tumor tissue was used for immunohistochemistry and fluorescent in situ hybridization. Telomere lengths were analyzed using the whole genome sequencing data, quantitative polymerase chain reaction and fluorescent in situ hybridization.
Main Outcome Measure
Somatic recurrent mutations in tumors from patients with neuroblastoma correlated with the age at diagnosis and telomere length.
Results
We identified mutations in the ATRX gene in 100% (5/5) (95% CI, 50% – 100%) of tumors from patients in the adolescent and young adult group, 17% (5/29) (95% CI, 7% – 36%) of tumors from children, and 0% (0/6) (95% CI, 0% – 40%) of tumors from infants in the discovery cohort (n=40). In the validation cohort (n=64), we identified mutations in the ATRX gene in 33% (9/27) (95% CI, 17% – 54%) of tumors from patients in the adolescent and young adult group, 16% (4/25) (95% CI, 6% – 35%) of tumors from children, and 0% (0/12) (95% CI, 0% – 24%) of tumors from infants. We identified mutations in the ATRX gene in 44% (14/32) (95% CI, 28% – 62%) of tumors from patients in the adolescent and young adult group, 17% (9/54) (95% CI, 9% – 29%) of tumors from children, and 0% (0/18) (95% CI, 0% – 17%) of tumors from infants in the combined cohort (n=104). ATRX mutations were associated with an absence of ATRX protein in the nucleus and with long telomeres.
Conclusions
ATRX mutations were associated with age at diagnosis in children and young adults with stage 4 neuroblastoma.
Clinical Protocol
“Molecular Characterization of Neuroblastic Tumor: Correlation with Clinical Outcome” (clinical trials.gov: NCT00588068).
INTRODUCTION
Neuroblastoma, a tumor of the sympathetic nervous system, is the most common extracranial solid tumor of childhood and accounts for 15% of all cancer-related deaths in children. Through the 1980s, deaths from neuroblastoma were usually an early event: less than 5% occurred beyond 3 years from diagnosis and less than 1% occurred after 5 years. 1,2 Survival for more than 30 months after recurrence of stage 4 neuroblastoma was the exception, even with myeloablative retrieval therapy. 3 During the 1990s, several studies began to describe late recurrences of neuroblastoma at 4–13 years from diagnosis, with some patients continuing to survive for up to 19 years from diagnosis. 4 Time to death after tumor recurrence among adolescents was significantly longer than among children. This difference was most evident for patients with advanced stage neuroblastoma.
Today, approximately 70% of patients with neuroblastoma present with metastatic disease, and with current treatment approaches the age at diagnosis has proven to be one of the most powerful predictors of outcome.5 The probability of overall survival is 88% in infants (<18 months at diagnosis), 49% in children (18 months-<12 years), and only 10% in adolescents or young adults (≥12 years).6,7 A majority of neuroblastoma occurring in adolescents and young adults as well as in some older children have a protracted course, with death occurring many years after diagnosis.8,9 This clinical subtype is now referred to as indolent or chronic neuroblastoma.4
Genetic mutations associated with neuroblastoma and its clinical course are incompletely understood. To define the mutational landscape of metastatic neuroblastoma, the St. Jude Children’s Research Hospital - Washington University Pediatric Cancer Genome Project performed whole genome sequence analysis of DNA from tumors and matched germ line samples from 40 pediatric patients with metastatic neuroblastoma diagnosed and/or treated at Memorial Sloan-Kettering Cancer Center. Tumors from patients diagnosed as infants, children, and adolescents and young adults were included.
METHODS
Patients and Tissues
Use of human tissues for genetic studies was approved by the institutional review boards of Memorial Sloan-Kettering Cancer Center (MSKCC), St. Jude Children’s Research Hospital, and Washington University in St Louis. Written informed consent was obtained from patients or legal guardians at the time of surgical resection or bone marrow procedure. All patients diagnosed with metastatic neuroblastoma at MSKCC had tumor samples stored in the tumor inventory. For this study, tumor samples were selected at random from the tumor registry to represent each of the 3 patient diagnostic age groups for both a discovery (n=40) and validation (n=64) cohort. The proportion of samples from each of the 3 age groups in this study is not representative of the relative frequency of metastatic neuroblastoma in those age groups. Only neuroblastomas from patients with metastatic disease were chosen. They were selected from a pool of 158 available tumors diagnosed across the age spectrum, i.e. infant, children, adolescent and adult. As these tumors passed pathology review for adequate tumor content (>50%), and adequate DNA quality, they were entered into the discovery set (n=40), and subsequently into the validation set (n=64). No preference was given to gender, ethnicity, or clinical outcome. In addition, 1 patient from another institution had tissue submitted for evaluation in the validation cohort and approved for molecular studies under a waiver from the institutional review board. Patients were diagnosed between 1987 and 2009 for the discovery cohort and 1985 and 2009 for the validation cohort. Information on age at diagnosis, sex, race/ethnicity, tumor stage, and survival were taken from the clinical database. Information on race/ethnicity was provided by the participant or parent and was included because some single nucleotide polymorphisms are differentially distributed across different populations. Fresh tumor samples were cryopreserved at the time of surgery. Remission bone marrow or blood samples were cryopreserved to serve as a matched germline reference for each tumor. Formalin-fixed paraffin-embedded (FFPE) tumor samples were used for immunohistochemistry (IHC) and fluorescent in situ hybridization (FISH) studies. Whole genome sequencing (WGS) was done on the discovery cohort to identify genetic lesions. Polymerase chain reaction (PCR) and Sanger Sequencing were then carried out on both the discovery and validation cohorts to validate somatic lesions. The association between age group and identified mutations was examined. Telomere analysis was performed in the discovery cohort because telomere length has been associated with outcome in neuroblastoma patients. Mutations of ATRX were recently identified in pancreatic neuroendocrine tumors and shown to be associated with lengthened telomeres generated by a mechanism know as alternative lengthening of telomeres (ALT).10 We also determined if neuroblastoma tumors with ATRX mutations had longer telomeres.
Whole-genome sequencing
Using a paired-end sequencing approach, we sequenced DNA from the tumors and matching germline controls in the discovery cohort with an average of 35.1× haploid coverage per genome. Single nucleotide variations (SNVs) and insertions/deletions (indels) were identified as previously described. 11,12 Structural variations were detected using the CREST algorithm. 13 The WGS data are deposited at the European Bioinformatics Institute (EBI) with accession number: EGAS00001000213.
Sequence Validation
PCR and Sanger sequencing were performed to validate somatic lesions in the discovery and validation cohorts. PCR amplification was performed for 35 cycles using Advantage 2 polymerase (Clontech) with an annealing temperature of 68 °C. A 5-μL sample of the resulting PCR product was purified using 2 μL ExoSAP-IT (Affymetrix) prior to Sanger sequencing. PCR primer sequences are available upon request.
MYCN Copy Number Analysis
MYCN amplification was based on three independent methods: (1) review of the clinical database where amplification was determined by southern blotting or by FISH, (2) quantitative PCR using previously published methods, 14 and (3) WGS. All three methods gave identical results. For samples where the MYCN copy number were >10, the number of genes in the amplicon was derived from WGS. For samples with ≤ 10 copies of MYCN, the size of the amplified region was determined as focal (≤ 5 genes), large segment (> 5 genes) or chromosomal (whole chromosome 2 gain).
Telomere Analysis
Telomeres were analyzed in the discovery cohort using 3 different methods. The whole genome sequencing (WGS) data was analyzed for telomere length for all tumors and germline DNA in the discovery cohort. Quantitative PCR was performed to validate the results from WGS analysis for all 10 tumors in the discovery cohort with ATRX mutations and an additional 4 samples with wild type ATRX to serve as controls. Telomere FISH was performed on all tissue samples in the discovery cohort that had available FFPE samples. All of the samples that were analyzed by telomere FISH were also analyzed for ATRX protein expression by immunohistochemistry.
Telomere length was predicted in silico by counting the number of next-generation sequencing reads containing the telomeric-repeat sequence TTAGGG. 15 The resulting number of reads was normalized to the average genomic coverage, and the difference in diagnostic and germline telomeric sizes was calculated. Telomere length was validated in vitro in neuroblastomas expressing an ATRX aberration as described previously. 16,17 Briefly 15–20ng of diagnostic and germline WGA amplified DNA was subject to qPCR using 2 sets of primers in separate reactions, 1 to amplify telomeric sequence and 1 to amplify a common gene; 36B4 (RPLP0). Ct values obtained were compared to those of 2 standard curves, a telomeric standard curve performed on known quantities of a telomeric 84mer and 1 using an oligomer of 36B4 (RPLP0). All reactions were performed in triplicate with both tumor and germline DNA and both assays on the same plate. All reactions were carried out using Brilliant III Ultra-Fast SYBR Green master mix (Agilent) on a Stratagene Mx3000 thermal cycler using a melting temperature of 60°C. This allowed us to determine the telomere length in Kb per diploid genome. The forward primer for telomere analysis was:
5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′
The reverse primer for telomere analysis was:
5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCC-3′
The forward primer for the internal control 36B4 (RPL0) gene was:
5′-CAGCAAGTGGGAAGGTGTAATCC-3′
The reverse primer for the internal control 36B4 (RPL0) gene was:
5′-CCCATTCTATCATCAACGGGTACAA-3′
The standard used to generate the standard curve for telomeres was:
5′-(TTAGGG)14-3′
The standard used to generate the standard curve for the internal control 36B4 (RPL0) was:
5′-CAGCAAGTGGGAAGGTGTAATCCGTCTCCACAGACAAGGCCAGGACTCG TTTGTACCCGTTGATGATAGAATGGG-3′
ATRX Immunohistochemistry
Formalin-fixed, paraffin-embedded tissues were cut into 4-μm-thick sections and immunostained with a polyclonal antibody against ATRX (1:600; Sigma-Aldrich) by using heat-induced epitope retrieval and Leica Polymer Refine Detection Kit (Leica Microsystems) on a Leica Bond system after 15-minute antibody incubation.
Telomere FISH
Interphase FISH was performed on 4-μm-thick, formalin-fixed, paraffin-embedded tissue sections. The Cy3-labeled TelG probe (PNAbio) was co-denatured with the target cells on a hotplate at 90 °C for 12 minutes. The slides were incubated for 48 hours at 37 °C and then washed in 4 M Urea/2× SSC at 45 °C for 5 minutes. Nuclei were counterstained with DAPI (200 ng/mL) (Vector Labs).
Statistics
Cytel Studio (StatXact-9) software was used for analysis. The exact Chi-square test was used to examine the association between age group and ATRX mutation. P-values less than 0.05 were considered statistically significant; all reported p-values were 2-sided. Proportions are reported with 95% Blyth-Still-Casella confidence intervals. No sample size calculation was performed as this was a retrospective exploratory study.
RESULTS
Whole Genome Sequencing
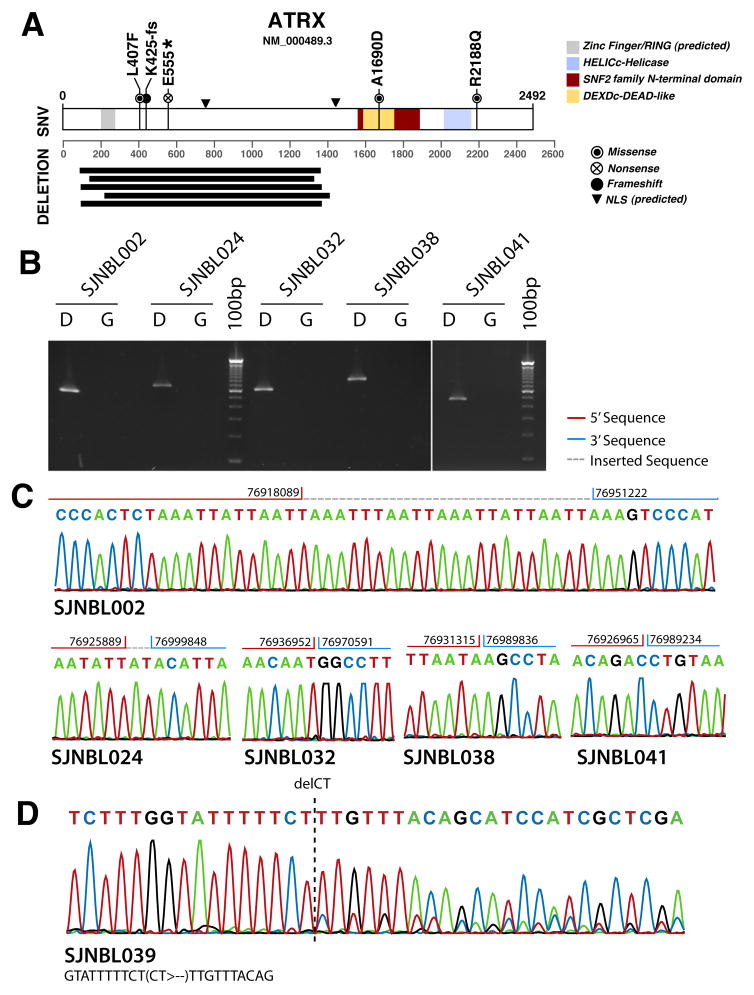
We performed WGS on a discovery cohort of 40 diagnostic neuroblastoma samples from infants (n=6), children (n=29), and adolescents and young adults (n=5) with stage-4 disease. Patient characteristics are presented in Table 1 and eTable1. Somatic mutations in ATRX including sequence mutations and structural variations were found in 25% (10/40) (95% CI, 13%–41%) of these patient’s tumors. Three of the mutations were missense mutations (L407F, A1690D and R2188Q), 1 of the mutations was a nonsense mutation (E555*), 1 was a frameshift mutation (K425-fs) and the remaining 5 mutations were structural variations resulting in an in-frame deletion in ATRX protein (Figure 1). The minimum overlapping region of the deletion involved exon 5 to exon 10, which encodes a predicted nuclear-localization signal (Fig. 1). All 10 somatic mutations were validated by PCR and Sanger sequencing. ATRX mutations were mutually exclusive of MYCN amplification in the discovery cohort (eTable 1).
Table 1.
Patient Characteristics
| All Patients (n=104) | Discovery Cohort (n=40) | Validation Cohort (n=64) | |
|---|---|---|---|
| N (%^) | N (%^) | N (%^) | |
| Sex | |||
| Male | 57 (55) | 26 (65) | 31 (48) |
| Female | 47 (45) | 14 (35) | 33 (52) |
| Ethnicity | |||
| White Non Hispanic | 72 (69) | 30 (75) | 42 (66) |
| White Hispanic | 2 (2) | 1 (3) | 1 (2) |
| Black Non Hispanic | 7 (7) | 6 (15) | 1 (2) |
| Black/African American | 8 (8) | 0 (0) | 8 (13) |
| Black Hispanic | 1 (1) | 1 (3) | 0 (0) |
| Asian/Far East/Indian Subcontinent | 3 (3) | 1 (3) | 2 (3) |
| Unknown | 10 (10) | 1 (3) | 9 (14) |
| Other | 1 (1) | 0 (0) | 1 (2) |
| Stage | |||
| 2B, then 4 | 1 (1) | 1 (3) | 0 (0) |
| 3, then 4 | 1 (1) | 0 (0) | 1 (2) |
| 4s, then 4 | 1 (1) | 1 (3) | 0 (0) |
| 3 | 1 (1) | 0 (0) | 1 (2) |
| 4 | 100 (96) | 38 (95) | 62 (97) |
| Age at Diagnosis | |||
| <18 months | 18 (17) | 6 (15) | 12 (19) |
| ≥ 18 months - <12 years | 54 (52) | 29 (73) | 25 (39) |
| ≥ 12 years | 32 (31) | 5 (13) | 27 (42) |
| MYCN amplification | |||
| Amplified | 24 (23) | 11 (28) | 13 (20) |
| Not amplified | 80 (77) | 29 (73) | 51 (80) |
| ATRX mutation/deletion | |||
| Yes | 23 (22) | 10 (25) | 13 (20) |
| No | 81 (78) | 30 (75) | 51 (80) |
| ALK mutation | |||
| Yes | 15 (14) | 6 (15) | 9 (14) |
| No | 89 (86) | 34 (85) | 55 (86) |
| †Telomere Length | |||
| Long | n/a | 20 (50) | n/a |
| Short | n/a | 20 (50) | n/a |
| 11q | |||
| Loss | n/a | 18 (45) | n/a |
| Gain | n/a | 3 (8) | n/a |
| No change | n/a | 19 (48) | n/a |
| 1p | |||
| Loss | n/a | 17 (43) | n/a |
| Gain | n/a | 4 (10) | n/a |
| No change | n/a | 17 (43) | n/a |
| * No change/weak loss | n/a | 1 (3) | n/a |
| * No change/weak gain | n/a | 1 (3) | n/a |
| 17q | |||
| Gain | n/a | 36 (90) | n/a |
| No change | n/a | 4 (10) | n/a |
| Relapse/Progression | |||
| Yes | 78 (75) | 28 (70) | 50 (78) |
| No | 26 (25) | 12 (30) | 14 (22) |
| Survival Status | |||
| Dead | 58 (56) | 21 (53) | 37 (58) |
| Alive | 46 (44) | 19 (48) | 27 (42) |
Percentages may not sum to 100% due to rounding
Telomere length is calculated from the genomic reads that correspond to the telomere sequence from the tumor and the matched germline as follows: (#telomere reads in tumor– #telomere reds in the germline sample)/#telomere reads in the germline sample. To be classified as Long this value must be > 0.05 and to be classified as short < −0.05
Weak gain: 0.25–0.75 copy gain; Weak loss: 0.25–0.75 copy loss
Figure 1. ATRX mutations in neuroblastoma.
(A) Diagram of the amino acid sequence of ATRX and the changes that result from the 5 single-nucleotide variations and 5 in-frame deletions found in the ATRX gene. (B) Photo of an ethidium bromide stained agarose gel with the PCR products for each of the 5 deletions shown in (A). D indicates the diagnostic tumor and G represents the germline DNA. (C) Sanger sequence traces of the PCR amplicons from (B) with junction breakpoints indicated. (D) Sanger sequence traces of the PCR amplicon spanning the frameshift mutation K425_E426fs.
ATRX is on the X chromosome and 7 of the 10 patients with ATRX mutations were males and would thus have only a mutant allele (eTable 1). One of the females with an ATRX mutation also sustained a loss of 1 copy of the X chromosome in the tumor, thereby eliminating the wild type allele of ATRX (eTable 1). The remaining 2 females with ATRX mutations were diagnosed as children (2.3 and 4.1 years of age) and had heterozygous in-frame deletions (Table 1 and eTable1).
Telomere Analysis
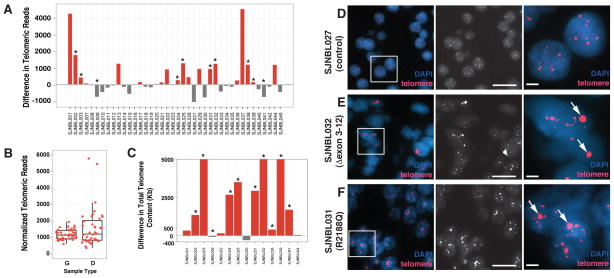
Among the patients whose tumors had ATRX mutations, 80% (8/10) (95% CI, 44%–96%) had evidence of longer telomeres based on WGS data (Tables 2 and Figure 2A,B). In contrast, only 40% (12/30) (95% CI, 24 – 59%) of the tumors with wild type ATRX had long telomeres (Table 1 and Table 3) although the difference was not statistically significant (P=0.065). Each of the 8 tumors that showed evidence of longer telomeres by WGS analysis were validated using the qPCR method and 1 of 2 samples with short telomeres by WGS had longer telomeres by qPCR (Fig. 2C and eTable 1). As expected, the 4 samples with wild type ATRX showed consistent results between the WGS and qPCR analysis (eTable 1).
Table 2.
ATRX & Telomere Results for the Discovery Cohort (n=40)
| ATRX Mutation | P-value | ||
|---|---|---|---|
| Telomere Length | Yes | No | |
| Long | 8 (80% [44%–96%]) | 12 (40% [24%–59%]) | 0.065 |
| Short | 2 (20% [4%–56%]) | 18 (60% [41%–76%]) | |
| Total | 10 | 30 | |
Proportions are presented with 95% confidence intervals in brackets
Figure 2. Telomere analysis in neuroblastoma.
(A) Histogram of the difference in telomeric reads for each of the 40 tumors in the discovery cohort. The samples with ATRX mutations are indicated by asterisks. The reference value for each tumor was the telomere length from matched normal DNA from the same patient. (B) Boxplot of normalized telomeric reads for the germline DNA (G) and the diagnostic tumor (D) from the WGS data for the 40 tumors and matched germline samples in the discovery cohort. The number of telomeric reads was normalized to the average genomic coverage for that particular sample. The upper and lower edge of the box represents the 75th and 25th percentile, respectively. The median is indicated as a horizontal line within the box and the vertical lines represent the lowest and highest values still within 1.5 of the interquartile range. (C) Histogram of the qPCR for telomeres in the 10 samples in the discovery cohort with ATRX mutations as well as 4 controls with wild type ATRX. Data are plotted as difference between tumor and germline for each patient. Red bars indicate those samples with increased telomere length and grey bars indicate those samples with shorter telomeres. (D–F) Images of control cells and 2 samples with mutant ATRX hybridized with the telomere FISH probe (red) and stained with DAPI to visualize the nucleus (blue). A low power overlay image is shown as well as a black and white image of the telomere FISH signal and a high magnification view highlighting the large ultrabright signal (arrows) in the ATRX mutant neuroblastoma cells. Scale bar: 5 μm for low magnification view and 1 μm for the high magnification view.
Table 3.
Genetic & Telomere Results by Age Group
| Discovery Cohort (n=40) | ||||
|---|---|---|---|---|
| Infants (<18 mos) (n=6) | Children (18 mos - <12 yrs) (n=29) | Adolescents & Young Adults (≥ 12 yrs) (n=5) | P-value | |
| ATRX Mutation | ||||
| Yes | 0 (0% [0–40%]) | 5 (17% [7–36%]) | 5 (100% [50–100%]) | <0.001 |
| No | 6 (100% [60–100%]) | 24 (83% [64–93%]) | 0 (0% [0–50%]) | |
| Telomere Length | ||||
| Long | 5 (83% [40–99%]) | 11 (38% [21–58%]) | 4 (80% [34–99%]) | 0.051 |
| Short | 1 (17% [1–60%]) | 18 (62% [42–79%]) | 1 (20% [1–66%]) | |
| Validation Cohort (n=64) | ||||
| Infants (<18 mos) (n=12) | Children (18 mos - <12 yrs) (n=25) | Adolescents & Young Adults (≥12 yrs) (n=27) | P-value | |
| ATRX Mutation | ||||
| Yes | 0 (0% [0–24%]) | 4 (16% [6–35%]) | 9 (33% [17–54%]) | 0.048 |
| No | 12 (100% [76–100%]) | 21 (84% [65–94%]) | 18 (67% [46–83%]) | |
| All Patients Combined (n=104) | ||||
| Infants (<18 mos) (n=18) | Children (18 mos - <12 yrs) (n=54) | Adolescents & Young Adults (≥ 12 yrs) (n=32) | P-value | |
| ATRX Mutation | ||||
| Yes | 0 (0% [0–17%]) | 9 (17% [9–29%]) | 14 (44% [28–62%]) | <0.001 |
| No | 18 (100% [83–100%]) | 45 (83% [71–91%]) | 18 (56% [38–72%]) | |
Telomere length data were available only for the discovery cohort
Proportions are presented with 95% confidence intervals in brackets
Telomere FISH analysis was performed on 28 patients in the discovery cohort that had available FFPE tumor specimens (eTable 1). All 8 of the ATRX mutant tumors contained a large ultrabright telomere FISH signal that is a hallmark of ALT 18,19 (Figure 2D–F and eTable 1). Only 1 of the 20 tumors with wild type ATRX had evidence of ALT (eTable 1).
ATRX Protein Localization
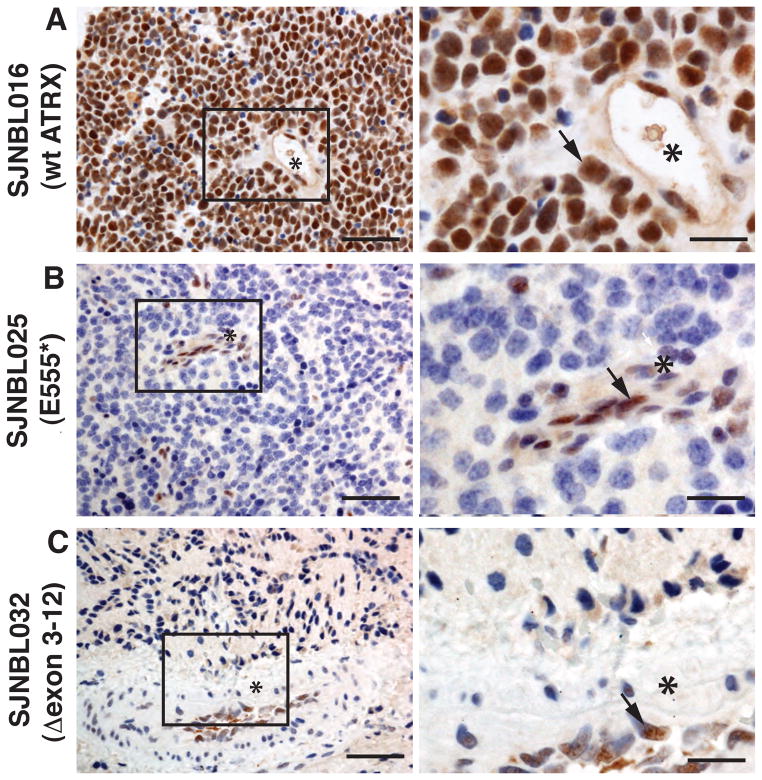
All 8 of the samples with ATRX mutations among the 28 with available tissue blocks had complete or mosaic loss of nuclear ATRX protein (eTable 1 and Figure 3). Only 1 of the 20 tumor samples with wild type ATRX had mosaic loss of ATRX protein in the nucleus (eTable 1). This sample did not have any evidence of ALT (eTable 1).
Figure 3. ATRX protein expression in neuroblastoma.
(A) Immunohistochemistry for ATRX protein (brown) in a neuroblastoma tumor with wild type ATRX. Hematoxylin was used as the counterstain, and it stains the unlabeled (ATRX negative) nuclei blue. Blood vessels are clearly visible (*) and representative nuclear staining of ATRX is highlighted (arrow). (B,C) Immunohistochemistry for ATRX protein on 2 neuroblastoma samples with ATRX mutations. The vascular endothelial cells lining the blood vessel (*) are immunopositive for ATRX (arrows) but the tumor cells are negative. Scale bar: 25 μm for low magnification and 5 μm for high magnification.
Age Association
All 5 samples (100%; 95% CI, 50%–100%) from adolescents and young adult patients in the discovery cohort had ATRX mutations, whereas no ATRX mutations were detected in samples obtained from infants (0/6) (0%; 95% CI, 0%–40%) (Table 3). Among children 18 months-12 years, ATRX mutations were identified in 17% (5/29) (95% CI, 7%–36%), with 4/5 patients living at least twice as long as their time to first relapse, similar to most indolent neuroblastoma seen in the adolescent and young adult group (eTable 1). A significant association (P<0.001) was observed among ATRX mutation and age group for the discovery cohort.
Validation Cohort
We analyzed the ATRX gene in tumors from an additional 64 patients with neuroblastoma (12 from infants, 25 from children, and 27 from adolescents and young adults; Table 3 and eTable 2). We identified 13 additional ATRX mutations in children (n=4) and adolescents and young adults (n=9) (eTable 2). No ATRX mutations were identified in infants from the validation cohort. The children with ATRX mutations were all over 5 years of age at diagnosis and 1 of the 3 who died had a protracted disease course (eTable 2). A significant association (P=0.048) was observed among ATRX mutation and age of disease diagnosis (Table 3). When the discovery set and validation set were combined, this age association was highly significant (P<0.001).
COMMENT
In this study ATRX mutations were found in 44% (95% CI, 28%–62%) of tumors from adolescent and young adult patients with neuroblastoma and none of the tumors (0%; 95% CI, 0%–17%) from infants with metastatic neuroblastoma. The children whose tumors had ATRX mutations were typically over 5 years of age or had a chronic or indolent course of disease. The ATRX mutations were characterized as missense, nonsense, frameshift, and in-frame deletion and they were mutually exclusive of MYCN amplification. ATRX mutations were associated with loss of nuclear ATRX protein, longer telomeres and ALT. Unlike PanNETs, a neuroendocrine tumor that is associated with a high frequency of ATRX and DAXX mutations, we did not detect any DAXX mutations in neuroblastomas. These results suggest that inactivation of the ATRX pathway correlates with older age at diagnosis and may provide a molecular marker and potential therapeutic target for neuroblastoma among adolescents and young adults. It may also delineate the subset of children with neuroblastoma who have a chronic but progressive clinical course. Specifically, patients with ATRX mutations, ultrabright telomere FISH signal characteristic of ALT, loss of nuclear ATRX protein and absence of MYCN gain may be more likely to have a chronic but progressive clinical course with standard therapeutic approaches and may require a different treatment strategy.
ATRX Function
ATRX is part of a multi-protein complex that includes DAXX and plays a role in regulating ATP-dependent chromatin remodeling, nucleosome assembly, and telomere maintenance. ATRX has been extensively studied in the α–thalassemia-mental retardation X-linked syndrome. 20 It is thought that mutations in patients with this syndrome retain partial activity of ATRX. In contrast, the identified ATRX mutations in PanNETs and neuroblastomas, as well as the DAXX mutations seen in PanNETs appear to be loss of function mutations. How these alterations leads to lengthened telomeres remains to be determined.
Beyond the proposed role in telomere maintenance, ATRX is also believed to play a role in epigenetic regulation of gene expression by controlling the deposition of histone H3.3 at transcriptionally silent regions of the genome. 21–24 This may lead to increased expression of oncogenes in tumors with ATRX mutations through epigenetic mechanisms.
ATRX, ALT and Long-Term Survival
In patients with neuroblastoma, the short-term survival for the adolescent and young adult group of patients is better than that among children, but the overall survival is worse. This reflects the chronic/indolent disease progression in this age group. Among patients with PanNET, 42.6% (29/68) had mutations in ATRX or DAXX and those mutations were associated with prolonged survival at 5 years after diagnosis. 25 However, all of the patients with ATRX/DAXX mutations died by 15 years after diagnosis reminiscent of the poor long-term outcome for adolescents and young adults with neuroblastoma. Although it is not yet known if the overall survival for patients with PanNET carrying the wild type ATRX/DAXX is better than those with ATRX/DAXX mutations by 15 years from diagnosis, it is a testable hypothesis that mutations in the ATRX/DAXX pathway may dictate a slower growth tumor in the short term, but in time they ultimately contribute to death.
ATRX and Neuroblastoma
Despite detailed clinical descriptions, 26,27 the definition of chronic/indolent neuroblastoma has been imprecise and sometimes inconsistent for optimal patient care. Since these tumors can respond differently to induction chemotherapy, stem cell transplant and antibody therapy, their inclusion in small pilot studies may be misleading, especially when stable disease is used as the treatment endpoint. While the age of 12 years at diagnosis is a convenient cut-off, it is now recognized that patients in the younger age group may also have a chronic/indolent clinical course. Unfortunately, their distinction from the rest of children with neuroblastoma is nearly always in retrospect. The absence of a thorough understanding of the molecular/genetic biology of this subset makes it difficult to identify and optimally treat these patients.
MYCN amplification and ALK mutations are among the most prevalent and biologically important genetic aberrations in neuroblastoma. 28,29 The oncogenic potential of MYCN is well known. 30 While MYCN amplification is generally found in high risk tumors, ALK mutation is equally represented among low stage and advanced stage patients. In fact, germ line ALK mutation are responsible for a subset of patients with hereditary neuroblastoma. 29 The identification of ATRX mutations now provides a new biomarker that may assist in identifying patients that develop a chronic but progressive clinical course, and thus may be candidates for altered risk-based therapies 6,31. Extending these results to additional neuroblastoma patients by cooperative groups world-wide will be required to assess the clinical utility of this new biomarker.
Strengths and Limitations
The major strength of our study is the relatively large sample size of patients with stage 4 neuroblastoma representing the three major age groups of this disease. Most of the other genetic or genomic studies on neuroblastoma have focused on the younger patients. Our analysis of infants, children, and adolescents and young adults from the same institution diagnosed during the same time period provided us with the unique opportunity to identify genetic lesions associated with age at diagnosis for neuroblastoma. The other strength of our study is the use of WGS in the discovery cohort. This provides us with the opportunity to identify both structural variations and sequence mutations in ATRX. These SVs resulting in focal deletions may be difficult to identify by SNP array analysis or exome capture approaches.
In this study, we used the Illumina paired-end sequencing technology to generate sequencing reads from short DNA fragments of the tumor and matched normal cells. This massively parallel sequencing technology provides multiple DNA sequencing reads across individual nucleotides in the genome. The average number of sequencing reads for each nucleotide (coverage) for the 40 tumors and 40 matched germline samples was 35.1× with a range from 26.7× to 46.5×. This coverage ensures that an average of 98.5% (96.1%~99.6%) of genomic regions and 93.3% (85.0%~98.8%) of exonic regions were covered by at least 10 high-quality sequence reads, which is sufficient to identify somatic mutations. However, one of the limitations of the current study is the possibility that a small proportion of somatic mutations were missed due to insufficient coverage.
Another limitation of our study is the different methods used to analyze ATRX mutations in the discovery and validation cohort. Our data on the discovery cohort is far more comprehensive than on the validation cohort because the discovery cohort utilized WGS while the validation cohort used PCR and Sanger sequencing. This may account for the lower rate of ATRX lesions in the validation cohort than the discovery cohort because the method used was not as sensitive in detecting deletions or low frequency SNVs. Also, we did not analyze the X-chromosome copy number in our validation cohort to determine if any of the females with ATRX mutations had lost all or part of the X chromosome. Similarly, we did not perform transcriptome sequencing for any of the samples in this study to determine if X-inactivation contributes to inactivation of the wild type allele of ATRX in female patients.
Future studies should focus on assembling larger international cohorts of patients to study the short-term and long-term outcomes for patients with neuroblastoma across all age groups with ATRX mutations in comparison to those without ATRX mutations. These data may be useful in defining a more relevant age cutoff for the adolescent and young adult group and help to identify more effectively those patients who will have an increased risk of developing chronic/indolent neuroblastoma. Moreover, future studies should focus on exploring novel therapeutics for treating patients with ATRX mutations and exploring the mechanistic connection between perturbations in this pathway, ALT, changes in gene expression that result from defects in histone H3.3 deposition and the unique form of disease found in these patients.
CONCLUSIONS
This analysis of the ATRX gene in 104 patients with advanced stage neuroblastoma provides age-group specific data on the frequency of ATRX pathway disruption and ALT. Additional studies with larger cohorts of patients will be required to determine if genetic analysis of ATRX mutation status in children will be useful to prospectively identify children likely to develop chronic or indolent neuroblastoma.
Supplementary Material
Acknowledgments
Michael A. Dyer, PhD had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Jinghui Zhang, PhD, Charles Lu, PhD, Alberto Pappo, MD, Li Ding, PhD, Bob Fulton, MS, Elaine R. Mardis, PhD, Richard K. Wilson, PhD, James R. Downing, MD and Michael A. Dyer, PhD are affiliated with the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project. Michael Rusch, BS (SJCRH) provided annotation of the focal deletions identified in ATRX. Robert Huether, PhD (SJCRH) provided interpretation of the functional impact of the ATRX mutations based on protein structure modeling. Clayton Naeve, PhD (SJCRH) provided computational infrastructure for data analysis. Elizabeth Chamberlain, BA (MSKCC) provided assistance with clinical data management. No compensation was provided to M.R., R.H., C.N. or E.C. The whole genome sequencing was supported as part of the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project. This work was supported, in part, by Cancer Center Support (CA21765) from the NCI; grants to M.A.D. from the NIH (EY014867 and EY018599); and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital. M.A.D. is a Howard Hughes Medical Institute Early Career Scientist. This work was also supported by the Catie Hoch Foundation, Band of Parents and the Robert Steel foundation (N.K.C.). None of the funding sources played any role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
References
- 1.Bowman LC, Hancock ML, Santana VM, et al. Impact of intensified therapy on clinical outcome in infants and children with neuroblastoma: the St Jude Children’s Research Hospital experience, 1962 to 1988. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1991 Sep;9(9):1599–1608. doi: 10.1200/JCO.1991.9.9.1599. [DOI] [PubMed] [Google Scholar]
- 2.Cotterill SJ, Pearson AD, Pritchard J, et al. Clinical prognostic factors in 1277 patients with neuroblastoma: results of The European Neuroblastoma Study Group ‘Survey’ 1982–1992. European journal of cancer. 2000 May;36(7):901–908. doi: 10.1016/s0959-8049(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 3.Ladenstein R, Lasset C, Hartmann O, et al. Impact of megatherapy on survival after relapse from stage 4 neuroblastoma in patients over 1 year of age at diagnosis: a report from the European Group for Bone Marrow Transplantation. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1993 Dec;11(12):2330–2341. doi: 10.1200/JCO.1993.11.12.2330. [DOI] [PubMed] [Google Scholar]
- 4.Kushner BH, Kramer K, Cheung NK. Chronic neuroblastoma. Cancer. 2002 Sep 15;95(6):1366–1375. doi: 10.1002/cncr.10800. [DOI] [PubMed] [Google Scholar]
- 5.Moroz V, Machin D, Faldum A, et al. Changes over three decades in outcome and the prognostic influence of age-at-diagnosis in young patients with neuroblastoma: a report from the International Neuroblastoma Risk Group Project. European journal of cancer. 2011 Mar;47(4):561–571. doi: 10.1016/j.ejca.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009 Jan 10;27(2):289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franks LM, Bollen A, Seeger RC, Stram DO, Matthay KK. Neuroblastoma in adults and adolescents: an indolent course with poor survival. Cancer. 1997 May 15;79(10):2028–2035. doi: 10.1002/(sici)1097-0142(19970515)79:10<2028::aid-cncr26>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 8.Kushner BH, Kramer K, LaQuaglia MP, Modak S, Cheung NK. Neuroblastoma in adolescents and adults: the Memorial Sloan-Kettering experience. Medical and pediatric oncology. 2003 Dec;41(6):508–515. doi: 10.1002/mpo.10273. [DOI] [PubMed] [Google Scholar]
- 9.Conte M, Parodi S, De Bernardi B, et al. Neuroblastoma in adolescents: the Italian experience. Cancer. 2006 Mar 15;106(6):1409–1417. doi: 10.1002/cncr.21751. [DOI] [PubMed] [Google Scholar]
- 10.Heaphy CM, de Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011 Jul 22;333(6041):425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Benavente CA, McEvoy J, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012 Jan 19;481(7381):329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012 Jan 12;481(7380):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Mullighan CG, Easton J, et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nature methods. 2011 Aug;8(8):652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson J, Gibson S, Williamson D, et al. Rapid and accurate determination of MYCN copy number and 1p deletion in neuroblastoma by quantitative PCR. Pediatric blood & cancer. 2006 Jun;46(7):820–824. doi: 10.1002/pbc.20311. [DOI] [PubMed] [Google Scholar]
- 15.Castle JC, Biery M, Bouzek H, et al. DNA copy number, including telomeres and mitochondria, assayed using next-generation sequencing. BMC genomics. 2010;11:244. doi: 10.1186/1471-2164-11-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic acids research. 2002 May 15;30(10):e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Callaghan N, Dhillon V, Thomas P, Fenech M. A quantitative real-time PCR method for absolute telomere length. BioTechniques. 2008 May;44(6):807–809. doi: 10.2144/000112761. [DOI] [PubMed] [Google Scholar]
- 18.Hampton T. Studies probe role of telomere length in predicting, modulating cancer risk. JAMA: the journal of the American Medical Association. 2011 Jun 8;305(22):2278–2279. doi: 10.1001/jama.2011.772. [DOI] [PubMed] [Google Scholar]
- 19.Pickett H, Reddel R. Alternative Lengthening of Telomeres in Human Cancer. In: Hiyama K, editor. Telomeres and Telomerase in Cancer. New York: Humana; 2009. [Google Scholar]
- 20.Gibbons R. Alpha thalassaemia-mental retardation, X linked. Orphanet journal of rare diseases. 2006;1:15. doi: 10.1186/1750-1172-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes & development. 2010 Jun 15;24(12):1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg AD, Banaszynski LA, Noh KM, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010 Mar 5;140(5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law MJ, Lower KM, Voon HP, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010 Oct 29;143(3):367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 24.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proceedings of the National Academy of Sciences of the United States of America. 2010 Aug 10;107(32):14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011 Mar 4;331(6021):1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollock BH, Krischer JP, Vietti TJ. Interval between symptom onset and diagnosis of pediatric solid tumors. The Journal of pediatrics. 1991 Nov;119(5):725–732. doi: 10.1016/s0022-3476(05)80287-2. [DOI] [PubMed] [Google Scholar]
- 27.Garaventa A, Parodi S, De Bernardi B, et al. Outcome of children with neuroblastoma after progression or relapse. A retrospective study of the Italian neuroblastoma registry. European journal of cancer. 2009 Nov;45(16):2835–2842. doi: 10.1016/j.ejca.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Seeger RC, Brodeur GM, Sather H, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. The New England journal of medicine. 1985 Oct 31;313(18):1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 29.Deyell RJ, Attiyeh EF. Advances in the understanding of constitutional and somatic genomic alterations in neuroblastoma. Cancer genetics. 2011 Mar;204(3):113–121. doi: 10.1016/j.cancergen.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. Embo J. 1997 Jun 2;16(11):2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.London WB, Castel V, Monclair T, et al. Clinical and biologic features predictive of survival after relapse of neuroblastoma: a report from the International Neuroblastoma Risk Group project. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011 Aug 20;29(24):3286–3292. doi: 10.1200/JCO.2010.34.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.