Abstract
Background
The p21 codon 31 single nucleotide polymorphism (SNP), rs1801270, has been linked to cervical cancer but with controversial results. The aims of this study were to investigate the role of p21 SNP-rs1801270 and other untested p21 SNPs in the risk of cervical cancer in a Chinese population.
Methods
We genotyped five p21 SNPs (rs762623, rs2395655, rs1801270, rs3176352, and rs1059234) using peripheral blood DNA from 393 cervical cancer patients and 434 controls.
Results
The frequency of the rs1801270 A allele in patients (0.421) was significantly lower than that in controls (0.494, p = 0.003). The frequency of the rs3176352 C allele in cases (0.319) was significantly lower than that in controls (0.417, p < 0.001).The allele frequency of other three p21 SNPs showed not statistically significantly different between patients and controls. The rs1801270 AA genotype was associated with a decreased risk for the development of cervical cancer (OR = 0.583, 95%CI: 0.399 - 0.853, P = 0.005). We observed that the three p21 SNPs (rs1801270, rs3176352, and rs1059234) was in linkage disequilibrium (LD) and thus haplotype analysis was performed. The AGT haplotype (which includes the rs1801270A allele) was the most frequent haplotype among all subjects, and both homozygosity and heterozygosity for the AGT haplotype provided a protective effect from development of cervical cancer.
Conclusions
We show an association between the p21 SNP rs1801270A allele and a decreased risk for cervical cancer in a population of Chinese women. The AGT haplotype formed by three p21 SNPs in LD (rs1801270, rs3176352 and rs1059234) also provided a protective effect in development of cervical cancer in this population.
Keywords: P21, Single nucleotide polymorphism, Cervical cancer, Haplotype
Background
Cervical cancer is the second most common cancer in women worldwide [1,2]. The human papilloma virus (HPV) appears to be a necessary factor in the development of almost all cases (> 90%) of cervical cancer [3]. The HPV E6 and E7 proteins are viral genes expressed in virtually all HPV-positive cervical carcinomas, and many experiments have shown that these are cooperative viral oncoproteins [4] that inactivate p53 and retinoblastoma (pRb) tumor suppressors, promoting carcinogenesis [4,5].
HPV infection is relatively common while only a small fraction of those infected develop cancer, suggesting that additional environmental, genetic, or immunological factors contribute to the progression to cervical cancer [6,7]. Cell cycle progression is regulated by cyclin-dependent kinases, crucial for normal growth and differentiation. Disruption of cell cycle control is common in cancer cells and is believed to play an essential role in cancer initiation and development. The cyclin-dependent kinase inhibitor p21 (also known as WAF1or CIP1) is encoded by the CDKN1A gene located on chromosome 6p21.2 [8,9]. The p21 protein binds to and inhibits the activity of cyclin-CDK2 or -CDK4 complexes, and disrupts cell cycle progression at G1 phase [10,11]. The expression of p21 is induced by the binding of tumor suppressor protein p53 to the p21 promoter [12-14]. The p21 protein can also interact with proliferating cell nuclear antigen (PCNA), a DNA polymerase accessory factor, and plays a regulatory role in S phase DNA replication and DNA damage repair [15]. Therefore, alteration in the p21 functional and/or promoter regions may adversely affect the regulation of cellular proliferation and increase susceptibility to cancer.
Identification of several genetic variants in p21 have been associated with cervical cancer [16,17]. The p21 single nucleotide polymorphism (SNP) rs1801270C/A, which occurs in codon 31, results in an amino acid substitution of arginine for serine. This polymorphism is located in a highly conserved region of p21 and is expected to affect its molecular function [18]. Prior studies have linked the p21 codon 31 SNP (rs1801270) to cervical cancer, with conflicting results [19,20]. In this study, we genotyped five p21 SNPs (rs1801270 at codon 31, rs762623 and rs2395655 in the promoter region, rs3176352 in an intron, and rs1059234 in the 3’ non-coding region) in 393 cervical cancer patients and 434 cancer-free controls to look for any associations between SNP alleles or genotypes and cervical cancer in a Chinese population.
Methods
Before beginning this study, the study protocol was approved by the Ethics Committee of our hospital(Shengjing Hospital of China Medical University, Shenyang, China).
Subjects
We ascertained 393 patients with cervical cancer between October, 2008 and September, 2011 at the Shengjing Hospital of China Medical University. Diagnosis of cervical cancer was confirmed by routine histopathological examination. A total of 434 control samples were randomly selected from the physical examination center from the same hospital and during the same period. The selection criteria for the controls included no individual history of cancer, no history of cervical intraepithelial neoplasia, normal cervical cytology and HPV examination, or normal pathological examination of the cervix within the past two years. HPV examination was performed as described previously [21]. At recruitment, written informed consent was obtained from all study subjects.
Age, smoking status, HPV status, and pathological stage of all 393 cervical cancer cases and 434 controls are presented in Table 1. The two groups seemed to be adequately matched in terms of age and smoking. Among the patients, 89.3% were HPV positive. The patients’ disease severity was assessed using the FIGO (1988) cervical cancer staging system (Table 1).
Table 1.
Demographic and clinical data of the study group
| |
Patients (n=393) |
Controls (n=434) |
Pa |
|---|---|---|---|
| No. (%) | No. (%) | ||
| Age |
|
|
|
| ≤35 |
82 (20.9) |
105 (24.4) |
0.377 |
| 36-50 |
219 (55.7) |
241 (55.5) |
|
| >50 |
92 (23.4) |
88 (20.3) |
|
| Smoking |
|
|
|
| Never |
371 (94.4) |
419 (96.5) |
0.177 |
| Ever |
22 (5.6) |
15 (3.5) |
|
| HPV status |
|
|
|
| HPV+ |
351 (89.3) |
0 (0) |
|
| HPV- |
42 (10.7) |
434 (100) |
|
| Clinical stageb |
|
|
|
| Stage 0 |
213 (54.2) |
|
|
| Stage I |
138 (35.1) |
|
|
| Stage II |
34 (8.7) |
|
|
| Stage III |
7 (1.8) |
|
|
| Stage IV | 1 (0.3) |
Genotype analysis
Genomic DNA was isolated from peripheral blood using the Tiangen Blood Genome Kit. Detailed primer and reaction information for the determination of the p21 SNP genotypes is given in Additional file 1: Table S1. Briefly, genotyping of rs762623 and rs2395655 was determined by DNA sequencing (Additional file 2: Figure S1). Genotyping of rs1801270, rs3176352, and rs1059234 was determined using a PCR-RFLP method. Each PCR reaction consisted of 0.1 mg DNA, 1 U Taq polymerase, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.0 mM MgCl2, 20 mM of each dNTP, and 0.2 μM of each primer. PCR products were digested with the indicated restriction enzymes and visualized on agarose gels (Additional file 3: Figure S2, Additional file 4: Figure S3, Additional file 5: Figure S4).
Statistical analysis
Genotype and allele frequencies were used to assess conformity to Hardy–Weinberg equilibrium. Differences in age and smoking status between patients and controls were evaluated using the Chi-square test. The statistical effect of this study was calculated by Gpower 14 software (Faul & Erdfelder, Bonn, Germany). Post-hoc analysis (affect scale set to 0.2 and α set to 0.05) determined the statistical power of this study to be 0.86. Linkage disequilibrium (LD) between five loci of p21 was estimated by the likelihood-ratio test. A Bayesian statistical method was used to estimate the haplotypes for each subject based on their known genotypes. These analyses were implemented in Haploview software (version 4.2; Massachusetts Institute of Technology, Cambridge, MA). Haplotype construction and haplotype frequency analysis were performed by PHASE (version 2.1) software [22]. All other statistical analyses were performed using SPSS 13.0 software (SPSS Inc., Chicago, USA). After Bonferroni correction, differences were deemed significant at p < 0.01.
Results
Association of p21 SNP alleles and genotypes with cervical cancer risk
The frequencies of each allele of the five p21 SNPs (rs1801270, rs762623, rs2395655, rs3176352, and rs1059234) for patients and controls are given in Table 2. After performing Bonferroni correction to eliminate bias of multiple comparison, we found that the frequency of the rs1801270 A allele in cases (0.421) was statistically significantly lower than that found in controls (0.494; p = 0.003). Frequency of the rs3176352 C allele in cases (0.319) was also statistically significantly lower than that in controls (0.417; p < 0.001). The frequency of the rs1059234 alleles between cases and controls was close to statistical significance (p = 0.055). The frequencies of the other two polymorphisms were not statistically significant between the two groups (Table 2).
Table 2.
Allele frequencies for all five p21 SNPs in cervical cancer patients and controls
| |
Patient alleles |
control alleles |
|
|
|---|---|---|---|---|
| |
(n = 786) |
(n = 868) |
|
|
| SNP | No. (%) | No. (%) | OR (95% CI) | P* |
| rs762623 |
|
|
|
|
| A |
151 (19.2) |
167 (19.2) |
1.001 (0.782-1.281) |
0.993 |
| G |
635 (80.8) |
701 (80.8) |
|
|
| rs2395655 |
|
|
|
|
| A |
399 (50.8) |
406(46.8) |
1.139 (0.938-1.383) |
0.115 |
| G |
387 (49.2) |
462 (53.2) |
|
|
| rs1801270 |
|
|
|
|
| A |
331 (42.1) |
429 (49.4) |
0.744 (0.613-0.904) |
0.003 |
| C |
455 (57.8) |
439 (50.6) |
|
|
| rs3176352 |
|
|
|
|
| C |
251(31.9) |
317 (41.7) |
0.656 (0.533-0.807) |
0.000 |
| G |
535 (68.1) |
551 (58.3) |
|
|
| rs1059234 |
|
|
|
|
| C |
422 (53.6) |
425 (48.9) |
1.208 (0.996-1.466) |
0.055 |
| T | 364 (46.3) | 443 (51.0) |
* P value was adjusted for age and smoking by multiple logistic regressions. After Bonferroni correction, significant P value was < 0.01.
The genotype distributions among both patients and controls (Table 3) were all in accordance with Hardy–Weinberg equilibrium. We used an unconditional logistic regression model adjusted for age and smoking to find any association between genotypes and the risk of cervical cancer (Table 3). We found that the rs1801270 AA genotype was associated with a decreased risk for the development of cervical cancer (OR = 0.583, 95% CI: 0.399 - 0.853, P = 0.005). This observation is consistent with the increased frequency of the rs1801270 A allele among controls. However, we did not observe a similar protective effect of the rs3176352 CC genotype even though the rs3176352 C allele frequency was lower in controls. No other genotypes of p21 SNPs were found to be associated with the risk of cervical cancer (Table 3).
Table 3.
Genotype frequencies for five p21 SNPs in cervical cancer patients and controls
| |
Patients |
controls |
|
|
|---|---|---|---|---|
| |
(n=393) |
(n=434) |
|
|
| SNP | No. (%) | No. (%) | OR (95% CI) | P* |
| rs762623 |
|
|
|
|
| AA |
9 (2.3) |
16 (3.7) |
0.627 (0.272-1.447) |
0.274 |
| AG |
133 (33.8) |
135 (31.1) |
1.096(0.816-1.472) |
0.542 |
| GG |
251 (63.9) |
283 (65.2) |
reference |
|
| rs2395655 |
|
|
|
|
| AA |
96 (24.4) |
92 (21.2) |
1.393 (0.936-2.074) |
0.102 |
| AG |
207 (52.7) |
222 (51.2) |
1.245 (0.892-1.738) |
0.197 |
| GG |
90 (22.9) |
120 (27.6) |
reference |
|
| rs1801270 |
|
|
|
|
| AA |
76 (19.3) |
117 (27.0) |
0.583(0.399-0.853) |
0.005 |
| AC |
179 (45.5) |
195 (44.9) |
0.811 (0.590-1.114) |
0.196 |
| CC |
138 (35.1) |
110 (28.1) |
reference |
|
| rs3176352 |
|
|
|
|
| CC |
37 (9.4) |
56 (12.9) |
0.637 (0.339-1.016) |
0.058 |
| CG |
177 (45.0) |
205 (47.2) |
0.883 (0.623-1.114) |
0.218 |
| GG |
179 (45.5) |
173 (39.9) |
reference |
|
| rs1059234 |
|
|
|
|
| CC |
131 (33.3) |
102 (23.5) |
1.392 (0.957-2.023) |
0.083 |
| CT |
160 (40.7) |
221 (50.9) |
0.786 (0.560-1.101) |
0.161 |
| TT | 102 (26.0) | 111 (25.6) | reference |
* P value was adjusted for age and smoking by multiple logistic regressions. After Bonferroni correction, significant P value was <0.01.
Haplotype analysis of p21 SNPs
Linkage disequilibrium (LD) analyses gave D’ values and r2 values (Additional file 6: Table S2), which suggested that three of the p21 SNP loci (rs1801270, rs3176352, and rs1059234) were in LD (Figure 1). The most common haplotypes were AGT, CCC, CGC, and CGT, accounting for 94.1% of the haplotypes observed in all individuals studied (Table 4). The frequency of AGT haplotype in cases (37.88%) was slightly lower than that in controls (44.42%), while the frequency of the CGC and CGT haplotypes were slightly higher in cases (20.92% and 7.07%, respectively) than in controls (11.63% and 3.91%, respectively; Table 4).
Figure 1.
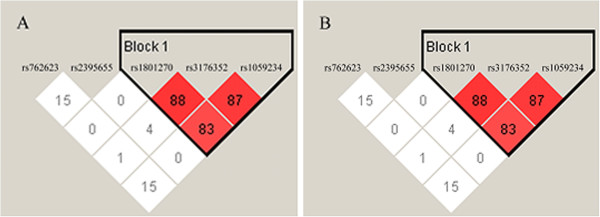
Linkage disequilibrium (LD) plot and block structure of five p21 SNPs in 393 cervical cancer patients (A) and 434 cancer-free controls (B) showing LD of three SNPs. In both groups, the haplotype block is based on confidence intervals D’. Each diamond represents the pairwise magnitude of LD, with red indicating strong LD (D’ > 0.8) and logarithm of odds score (LOD) ≥ 2.0.
Table 4.
Haplotypes of the three p21 SNPs (rs1801270, rs3176352, and rs1059234) in patients and controls
|
Haplotype |
Case (n=393) |
Control (n=434) |
|
|---|---|---|---|
| ht | Haplotype | Percentage (%)(95% CI) | Percentage (%)(95% CI) |
| 1 |
AGT |
37.88 (37.68-38.07) |
44.42 (44.18-44.65) |
| 2 |
CCC |
29.30 (29.03-29.57) |
33.11 (32.85-33.36) |
| 3 |
CGC |
20.92 (20.69-21.15) |
11.63 (11.41-11.86) |
| 4 |
CGT |
7.07 (6.88-7.26) |
3.91 (3.71-4.11) |
| 5 | AGC | 2.20 (2.05-2.36) | 3.52 (3.40-3.63) |
Haplotypes with overall estimated frequency < 3% are not shown.
To determine any effect of a particular haplotype on the risk of cervical cancer, we looked at the frequency distribution of the four common p21 haplotypes among cases and controls. To further confirm the association between the p21 haplotype and the risk of cervical cancer, we selected the haplotypes of which the sum frequency in case and control is more than 5% and analyzed the association between the numbers of variants in each haplotype (ht1-AGT, ht2-CCC, ht3-CGC and ht4-CGT) and the risk of cervical cancer (Table 3). We found that homozygosity or heterozygostiy for the AGT haplotype provides a protective effect (Table 5). The OR value of the homozygous AGT haplotype is 0.643 (95% CI: 0.434 - 0.954, P = 0.028), while the OR value of the heterozygous AGT haplotype is 0.636 (95% CI: 0.467 - 0.865, P = 0.004). Homozygosity of the CGC haplotype significantly increased the risk of cervical cancer (OR=3.047; 95% CI: 1.788-5.192, P=0.000), heterozygostiy for the CGC haplotype had no effect on risk of cervical cancer (P = 0.321). Heterozygosity for the CGT haplotype was more common among patients, suggesting an increased risk of cervical cancer among CGT heterozygotes (OR = 1.846; 95% CI: 1.156 - 2.945, P = 0.010). Homozygosity for the CGT haplotype was not statistically significantly different among groups, possibly because of the small sample size (P=0.338). The frequency of the CCC haplotype was not statistically significantly different among the two groups and was therefore not associated with risk of cervical cancer in this population.
Table 5.
Haplotypes of the three p21 SNPs in LD (rs1801270, rs3176352, and rs1059234) in patients and controls
| |
Patients |
controls |
|
|
|---|---|---|---|---|
| |
(n=393) |
(n=434) |
|
|
| haplotype | No. (%) | No. (%) | OR (95% CI) | P*a |
| ht1-AGT |
|
|
|
|
|
ht1/ht1 |
67 (17.0) |
87 (20.0) |
0.643 (0.434-0.954) |
0.028 |
|
ht1/- |
166 (42.2) |
215(49.5) |
0.636 (0.467-0.865) |
0.004 |
| −/− |
160(40.7) |
132(30.4) |
reference |
|
| ht2- CCC |
|
|
|
|
| ht2/ht2 |
26 (6.6) |
41 (9.4) |
0.628 (0.368-1.072) |
0.088 |
| ht2/- |
183 (46.6) |
210 (48.4) |
0.865 (0.650-1.151) |
0.321 |
| −/− |
184 (46.8) |
183 (42.2) |
reference |
|
| ht3-CGC |
|
|
|
|
|
ht3/ht3 |
51 (13.0) |
21 (4.8) |
3.047 (1.788-5.192) |
0.000 |
| ht3/- |
59 (15.0) |
56 (12.9) |
1.322 (0.888-1.970) |
0.169 |
| −/− |
283 (72.0) |
357 (82.3) |
reference |
|
| ht4- CGT |
|
|
|
|
| ht4/ht4 |
3(0.8) |
1(0.2) |
3.054 (0.311-29.936) |
0.338 |
|
ht4/- |
50 (12.7) |
32 (7.4) |
1.846 (1.156-2.945) |
0.010 |
| −/− | 340 (86.5) | 401 (92.4) | reference |
* P value was adjusted for age and smoking by multiple logistic regressions.
Discussion
This study looked for an association between five p21 SNPs (rs1801270, rs762623, rs2395655, rs3176352, rs1059234) and the risk of cervical cancer in a Chinese population. Studies have suggested an association between the p21 codon 31 polymorphism (rs1801270) and susceptibility to cancer, but with conflicting results [17,20,23,24]. Roh [17], Lee [23] and Jiang [24] found A allele seem to be a risk factor of cervical cancer, while Bhattacharya [20] and Tian [25] demonstrated C allele seem to be the risk factor. We found that the rs1801270 AA genotype and A allele occur more frequently in cancer-free controls, and seem to have a significant protective effect in Chinese women. Our results with the rs1801270 SNP are consistant with some previous reports [20,25], but differ from others [17,19,23,26], which found a deleterious effect of the rs1801270 A allele in cervical cancer. This discrepancy could be due to ethnic differences or sample size. Further, large-scale studies will be needed to validate the role of the p21 codon 31 SNP in cervical cancer.
Of the five p21 SNPs studied, three (rs1801270, rs3176352, and rs1059234) were found to be in linkage disequilibrium (LD). Haplotype analysis of these three p21 SNPs showed that haplotype AGT (which includes the rs1801270 A allele) occurs less frequently in cervical cancer patients than in controls. Our results suggest that both homozygosity and heterozygosity of the haplotype AGT may reduce the risk of cervical cancer in Chinese women. However, the heterozygosity of the haplotype CGT increases the risk of cervical cancer. In the current study, we found the association of the other two loci (rs3176352 and rs1059234) and cervical cancer tended to statistical significance. However, the role of the two p21 polymorphisms needs to be studied in a larger cervical cancer patient cohort in the future.
The molecular mechanism underlying the protective effect of the rs1801270 A allele in cervical cancer is unclear. This C/A SNP within the p21 codon 31 results in a substitution of arginine for serine in a conserved region of the protein and may affect the DNA binding affinity and thus physiological function of p21 [27]. The p21 protein plays a role in cell cycle regulation. It binds to cyclin-CDK complexes to arrest cell cycle progression at G1 phase. The transcription of p21 is partially regulated by p53, which binds to the p21 promoter and induces expression of p21. Therefore, functional domains as well as the promoter region of p21 could be affected by SNPs and lead to altered p21 functionality [26]. The rs2395655 SNP is located in the transcription factor E2F binding region. The rs1059234 SNP occurs in the 3’-non coding region and may affect p21 mRNA stability. Although our results do not find an association between these SNPs and cervical cancer, the molecular mechanism(s) through which p21 SNPs affect risk of cervical cancer is a field that should be pursued further.
Studies of p21 expression in cervical cancer have reported higher levels of expression of p21 in cancer tissue compared to normal tissue [28,29], while others showed there is no association between the expression of p21 and the pathological features of cervical cancer [30]. After infection with HPV, the E6 protein of HPV promotes p53 degradation and thus inhibits p21 expression and function. Loss of p21 results in decreased ability of the cell to stop cell cycle progression, possibly leading to carcinogenesis. However, the molecular mechanisms that mediate carcinogenesis are very complex, and this is not the only plausible explanation of the relationship between p21, p53, HPV, and cancer.
Conclusion
In conclusion, our study showed a significant association between the A allele of the p21 rs1801270 SNP and decreased risk of cervical cancer in a Chinese population. Furthermore, a haplotype containing the rs1801270 A allele was also associated with decreased risk of cervical cancer in this population. The molecular mechanisms underlying this association remain to be determined.
Abbreviations
HPV: Human Papillomavirus; SNP: Single nucleotide polymorphism; HWE: Hardy–Weinberg Equilibrium; MAF: Minor allele frequency; PCR-RFLP: Polymerase chain reaction-restriction fragment length polymorphism; LD: Linkage disequilibrium; FIGO: International Federation of Gynecology and Obstetrics.
Competing interests
The authors declare no conflict of interest.
Authors’ contributions
NW participated in the design of the study and sequencing. SW contributed to PCR-RFLP. QZ carried out sample collection and management. YL participated in statistical analysis. HW carried out DNA extraction. WL participated in sequencing. SZ participated in the design of the study. YD participated in statistical analysis. OY contributed to PCR-RFLP. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table S1. Methods used for p21 SNP genotyping.
Figure S1. Sample sequence tracing of rs762623 and rs2395655 SNPs showing homozygosity for each allele (top and bottom) or heterozygosity (middle).
Figure S2. PCR-RFLP analysis of p21 SNP rs1801270A/C. The 310 bp PCR products were digested by BglI, and presence of the C allele resulted in two bands of sizes 239 and 71 bp. Electrophoresis was performed in 3% agarose gel.
Figure 3. PCR-RFLP analysis of p21 SNP rs3176352A/C. The 448 bp PCR products were digested by ApaLI, and presence of the C allele resulted in two bands of sizes 289 and 159 bp. Electrophoresis was performed in 2.5% agarose gel.
Figure S4. PCR-RFLP analysis of p21 SNP rs1059234T/C. The 480 bp PCR products were digested by PstI, and presence of the C allele resulted in two bands of sizes 291 and 189 bp. Electrophoresis was performed in 2.5% agarose gel. (TIFF 571 kb)
Table S2. Pairwise linkage disequilibrium analysis for all five p21 SNPs for all subjects studied.
Contributor Information
Ning Wang, Email: nonaware@sina.com.
Shizhuo Wang, Email: zhuo1014@hotmail.com.
Qiao Zhang, Email: zhangqiao1983@sina.com.
Yanming Lu, Email: luym@sj-hospital.org.
Heng Wei, Email: weiheng-1986@163.com.
Wei Li, Email: lw0118461@126.com.
Shulan Zhang, Email: zsl0909@sina.com.
Duo Yin, Email: yind@sj-hospital.org.
Yangling Ou, Email: ouyang1964@163.com.
Acknowledgments
This study was supported by the National Nature Science Foundation of China (No. 30973191 and 81202047), Science and Technology Program of Liaoning Province (No.2008225004), Peak Medical Construction Special Project of Liaoning Province (No.2010696), Science and Technology Program of Shenyang City (No.F11-262-9-15) and Free Researcher Project of Shengjing Hospital (No.200806).
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008. GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kumme JA, Shah KV. et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M. et al. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudenon S, Huibregtse JM. HPV E6, E6AP and cervical cancer. BMC Biochem. 2008;9(1):S4. doi: 10.1186/1471-2091-9-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson PK, Sparen P, Gyllensten UB. Genetic link to cervical tumours. Nature. 1999;400:29–30. doi: 10.1038/21801. [DOI] [PubMed] [Google Scholar]
- Apple RJ, Erlich HA, Klitz W, Manos MM, Becker TM, Wheeler CM. HLA DR-DQ associations with cervical carcinoma show papillomavirus-type specificity. Nat Genet. 1994;6:157–162. doi: 10.1038/ng0294-157. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM. et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Serfas MS, Tyner AL. p21–negative regulator of the cell cycle. Proc Soc Exp Biol Med. 1996;213:138–149. doi: 10.3181/00379727-213-44046. [DOI] [PubMed] [Google Scholar]
- Harada K, Ogden GR. An overview of the cell cycle arrest protein, p21(WAF1) Oral Oncol. 2000;36:3–7. doi: 10.1016/s1368-8375(99)00049-4. [DOI] [PubMed] [Google Scholar]
- Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal. 2010;22:1003–1012. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Raber ML, Lawson BR, Dummer W, Barnhouse M, Koundouris S, Wilson CB. et al. Role of cyclin kinase inhibitor p21 in systemic autoimmunity. J Immunol. 2001;167:4067–4074. doi: 10.4049/jimmunol.167.7.4067. [DOI] [PubMed] [Google Scholar]
- Prives C, Gottifredi V. The p21 and PCNA partnership: a new twist for an old plot. Cell Cycle. 2008;7:3840–3846. doi: 10.4161/cc.7.24.7243. [DOI] [PubMed] [Google Scholar]
- Santos AM, Sousa H, Pinto D, Portela C, Pereira D, Catarino R. et al. Linking TP53 codon 72 and P21 nt590 genotypes to the development of cervical and ovarian cancer. Eur J Cancer. 2006;42:958–963. doi: 10.1016/j.ejca.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Roh J, Kim M, Kim J, Park N, Song Y, Kang S. et al. Polymorphisms in codon 31 of p21 and cervical cancer susceptibility in Korean women. Cancer Lett. 2001;165:59–62. doi: 10.1016/s0304-3835(01)00401-3. [DOI] [PubMed] [Google Scholar]
- Chedid M, Michieli P, Lengel C, Huppi K, Givol D. A single nucleotide substitution at codon 31 (Ser/Arg) defines a polymorphism in a highly conserved region of the p53-inducible gene WAF1/CIP1. Oncogene. 1994;9:3021–3024. [PubMed] [Google Scholar]
- Harima Y, Sawada S, Nagata K, Sougawa M, Ostapenko V, Ohnishi T. Polymorphism of the WAF1 gene is related to susceptibility to cervical cancer in Japanese women. Int J Mol Med. 2001;7:261–264. doi: 10.3892/ijmm.7.3.261. [DOI] [PubMed] [Google Scholar]
- Bhattacharya P, Sengupta S. Lack of evidence that proline homozygosity at codon 72 of p53 and rare arginine allele at codon 31 of p21, jointly mediate cervical cancer susceptibility among Indian women. Gynecol Oncol. 205;99:176–182. doi: 10.1016/j.ygyno.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang S, Ruan Q, Ji Y, Ma L, Zhang Y. Prevalence and type distribution of human papillomavirus in women with cervical lesions in Liaoning Province, China. Int J Gynecol Cancer. 2010;20:147–153. doi: 10.1111/IGC.0b013e3181c20860. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Lee SJ, Namkoong SE, Um SJ, Sull JW, Jee SH. et al. Gene-gene and gene-environmental interactions of p53, p21, and IRF-1 polymorphisms in Korean women with cervix cancer. Int J Gynecol Cancer. 2004;14:118–125. doi: 10.1111/j.1048-891x.2004.014040.x. [DOI] [PubMed] [Google Scholar]
- Jiang P, Liu J, Li W, Zeng X, Tang J. Role of p53 and p21 polymorphisms in the risk of cervical cancer among Chinese women. Acta Biochim Biophys Sin (Shanghai) 2010;42:671–676. doi: 10.1093/abbs/gmq069. [DOI] [PubMed] [Google Scholar]
- Tian Q, Lu W, Chen H, Ye F, Xie X. The nonsynonymous single-nucleotide polymorphisms in codon 31 of p21 gene and the susceptibility to cervical cancer in Chinese women. Int J Gynecol Cancer. 2009;19:1011–1014. doi: 10.1111/IGC.0b013e3181a8b950. [DOI] [PubMed] [Google Scholar]
- Roh JW, Kim BK, Lee CH, Kim J, Chung HH, Kim JW. et al. P53 codon 72 and p21 codon 31 polymorphisms and susceptibility to cervical adenocarcinoma in Korean women. Oncol Res. 2010;18:453–459. doi: 10.3727/096504010x12671222663719. [DOI] [PubMed] [Google Scholar]
- Facher EA, Becich MJ, Deka A, Law JC. Association between human cancer and two polymorphisms occurring together in the p21Waf1/Cip1 cyclin-dependent kinase inhibitor gene. Cancer. 1997;79:2424–2429. doi: 10.1002/(sici)1097-0142(19970615)79:12<2424::aid-cncr19>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Werness BA, Wang HQ, Chance J, Goldstein DJ. p53-independent expression of p21waf1/cip1 in preinvasive and invasive squamous neoplasms of the uterine cervix. Mod Pathol. 1997;10:578–584. [PubMed] [Google Scholar]
- Milde-Langosch K, Riethdorf S, Kraus-Poppinghaus A, Riethdorf L, Loning T. Expression of cyclin-dependent kinase inhibitors p16MTS1, p21WAF1, and p27KIP1 in HPV-positive and HPV-negative cervical adenocarcinomas. Virchows Arch. 2001;439:55–61. doi: 10.1007/s004280100439. [DOI] [PubMed] [Google Scholar]
- Graflund M, Sorbe B, Karlsson M. Immunohistochemical expression of p53, bcl-2, and p21(WAF1/CIP1) in early cervical carcinoma: correlation with clinical outcome. Int J Gynecol Cancer. 2002;12:290–298. doi: 10.1046/j.1525-1438.2002.01113.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Methods used for p21 SNP genotyping.
Figure S1. Sample sequence tracing of rs762623 and rs2395655 SNPs showing homozygosity for each allele (top and bottom) or heterozygosity (middle).
Figure S2. PCR-RFLP analysis of p21 SNP rs1801270A/C. The 310 bp PCR products were digested by BglI, and presence of the C allele resulted in two bands of sizes 239 and 71 bp. Electrophoresis was performed in 3% agarose gel.
Figure 3. PCR-RFLP analysis of p21 SNP rs3176352A/C. The 448 bp PCR products were digested by ApaLI, and presence of the C allele resulted in two bands of sizes 289 and 159 bp. Electrophoresis was performed in 2.5% agarose gel.
Figure S4. PCR-RFLP analysis of p21 SNP rs1059234T/C. The 480 bp PCR products were digested by PstI, and presence of the C allele resulted in two bands of sizes 291 and 189 bp. Electrophoresis was performed in 2.5% agarose gel. (TIFF 571 kb)
Table S2. Pairwise linkage disequilibrium analysis for all five p21 SNPs for all subjects studied.


