Abstract
Peroxisomes are organelles that sequester certain metabolic pathways; many of these pathways generate H2O2, which can damage proteins. However, little is known about how damaged or obsolete peroxisomal proteins are degraded. We exploit developmentally timed peroxisomal content remodeling in Arabidopsis thaliana to elucidate peroxisome-associated protein degradation. Isocitrate lyase (ICL) is a peroxisomal glyoxylate cycle enzyme necessary for early seedling development. A few days after germination, photosynthesis begins and ICL is degraded. We previously found that ICL is stabilized when a peroxisome-associated ubiquitin-conjugating enzyme and its membrane anchor are both mutated, suggesting that matrix proteins might exit the peroxisome for ubiquitin-dependent cytosolic degradation. To identify additional components needed for peroxisome-associated matrix protein degradation, we mutagenized a line expressing GFP–ICL, which is degraded similarly to endogenous ICL, and identified persistent GFP-ICL fluorescence (pfl) mutants. We found three pfl mutants that were defective in PEROXIN14 (PEX14/At5g62810), which encodes a peroxisomal membrane protein that assists in importing proteins into the peroxisome matrix, indicating that proteins must enter the peroxisome for efficient degradation. One pfl mutant was missing the peroxisomal 3-ketoacyl-CoA thiolase encoded by the PEROXISOME DEFECTIVE1 (PED1/At2g33150) gene, suggesting that peroxisomal metabolism influences the rate of matrix protein degradation. Finally, one pfl mutant that displayed normal matrix protein import carried a novel lesion in PEROXIN6 (PEX6/At1g03000), which encodes a peroxisome-tethered ATPase that is involved in recycling matrix protein receptors back to the cytosol. The isolation of pex6-2 as a pfl mutant supports the hypothesis that matrix proteins can exit the peroxisome for cytosolic degradation.
Keywords: organelle quality control, peroxin, peroxisome, PEX6 AAA ATPase, protein degradation
PEROXISOMES are single-membrane-bound organelles that house essential metabolic pathways in plants and other eukaryotes. For example, peroxisome biogenesis defects underlie the Zellweger spectrum of human congenital disorders, which often are fatal in infancy (reviewed in Wanders and Waterham 2005). Similarly, peroxisomes are essential for plant embryogenesis and development following germination (reviewed in Hu et al. 2012). The essential role of plant peroxisomes likely reflects the importance of peroxisomal enzymes, which catalyze key steps in photorespiration, fatty acid β-oxidation, jasmonate production, and conversion of the protoauxin indole-3-butyric acid (IBA) to the active auxin indole-3-acetic acid (IAA) (reviewed in Hu et al. 2012).
Peroxisomes import matrix proteins from the cytosol with the assistance of peroxin (PEX) proteins. Most matrix proteins are directed to the peroxisome by a C-terminal peroxisome-targeting signal 1 (PTS1) that binds the cytosolic receptor PEX5 (Keller et al. 1987). PEX5-cargo complexes dock with the PEX13 and PEX14 membrane peroxins (reviewed in Azevedo and Schliebs 2006; Williams and Distel 2006) at the peroxisome membrane. Other matrix proteins use an N-terminal PTS2 to bind the cytosolic receptor PEX7 (Osumi et al. 1991; Swinkels et al. 1991). In plants and mammals, PEX7 depends on PEX5 (Matsumura et al. 2000; Hayashi et al. 2005; Woodward and Bartel 2005) for cargo delivery to the PEX13 and PEX14 docking peroxins (reviewed in Lazarow 2006). After matrix proteins are delivered, yeast PEX5 is ubiquitinated in the peroxisome membrane by the ubiquitin-conjugating enzyme PEX4 and the ubiquitin-protein ligase PEX12 (Platta et al. 2009). Ubiquitinated PEX5 is retrotranslocated to the cytosol with the assistance of the peroxisome-tethered ATPases PEX1 and PEX6 (reviewed in Fujiki et al. 2012; Grimm et al. 2012) to be reused in further rounds of import.
Many metabolic pathways sequestered in peroxisomes produce hydrogen peroxide (H2O2). For example, H2O2 is generated by the acyl-CoA oxidases acting in fatty acid β-oxidation (Eastmond et al. 2000b; Adham et al. 2005) and the glycolate oxidases acting in photorespiration (Fahnenstich et al. 2008). H2O2 can damage proteins (Van Den Bosch et al. 1992; Willekens et al. 1997), but little is known about how damaged or obsolete peroxisomal proteins are degraded. Three possible mechanisms for peroxisomal matrix protein degradation can be envisioned: degradation within the organelle by resident proteases, degradation of the entire organelle via autophagy, or retrotranslocation out of the organelle followed by cytosolic degradation.
Many organelles, including mitochondria and chloroplasts, contain proteases that degrade damaged or misfolded proteins (reviewed in Leidhold and Voos 2007). Several proteases are found in Arabidopsis peroxisomes (Reumann et al. 2004, 2007; Helm et al. 2007; Lingard and Bartel 2009). For example, DEG15 cleaves PTS2 proteins from their targeting signal after import (Helm et al. 2007; Schumann et al. 2008) and the LON2 ATP-dependent protease is needed for sustained matrix protein import (Lingard and Bartel 2009). Although a fungal LON isoform contributes to degradation of oxidatively damaged peroxisomal matrix proteins (Bartoszewska et al. 2012), no resident peroxisomal proteases have been implicated in matrix protein degradation in plants.
A second possibility for peroxisomal protein degradation is removal of the entire organelle by autophagy or pexophagy, a specialized form of autophagy. For example, yeast use pexophagy to degrade excess peroxisomes by encasing the peroxisome in a membrane for fusion with the vacuole (reviewed in Manjithaya et al. 2010). Although autophagy occurs in Arabidopsis (reviewed in Li and Vierstra 2012), pexophagy has not been reported in plants.
A third potential mechanism for peroxisome-associated protein degradation is modeled after ER-associated protein degradation (ERAD), the process by which misfolded proteins are ubiquitinated and retrotranslocated from the ER lumen to the cytosol for proteasomal degradation (reviewed in Hoseki et al. 2010). Peroxins needed for PEX5 ubiquitination and retrotranslocation resemble ERAD components (Gabaldon et al. 2006; Schluter et al. 2006), suggesting that damaged peroxisomal proteins may be retrotranslocated out of the peroxisome and degraded in the cytosol by the 26S proteasome (Zolman et al. 2005).
Some evidence in Arabidopsis is consistent with a retrotranslocation model for matrix protein degradation. Isocitrate lyase (ICL) and malate synthase (MLS) are peroxisomal glyoxylate cycle enzymes that enable carbon from acetyl-CoA to be utilized in gluconeogenesis, thus providing energy for germinating seedlings (reviewed in Graham 2008). In Arabidopsis, ICL and MLS are degraded a few days after germination (Zolman et al. 2005; Lingard et al. 2009). Mutation of the PEX4 ubiquitin-conjugating enzyme along with PEX22, which tethers PEX4 to the peroxisome (pex4-1 pex22-1; Zolman et al. 2005), partially stabilizes MLS, ICL, and a GFP–ICL translational fusion without markedly impairing matrix protein import (Zolman et al. 2005; Lingard et al. 2009). Stabilization of these glyoxylate cycle enzymes in pex4-1 pex22-1 suggests a role for PEX4-mediated ubiquitination in promoting matrix protein degradation.
To identify additional components necessary for the turnover of damaged or unnecessary peroxisomal proteins, we initiated a forward genetic screen for Arabidopsis mutants exhibiting delayed GFP–ICL degradation. We identified several mutants with prolonged GFP–ICL fluorescence that also stabilized endogenous ICL. Characterization of these mutants confirmed that matrix proteins must enter the peroxisome to be subject to efficient degradation and is consistent with the possibility that damaged or obsolete matrix proteins can exit the peroxisome for cytosolic degradation.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana accession Columbia (Col-0) or Col-0 transformed with ICLp:GFP–ICL, which drives a GFP–ICL fusion protein from the ICL 5′-regulatory region (Lingard et al. 2009), was used as wild type (as indicated in figure legends). pex14-1, pex14-2/SALK_007441, pex14-3/SALK_072373, pex14-4 (Monroe-Augustus et al. 2011), ped1-96 (Lingard and Bartel 2009), lon2-2/SALK_043857 (Lingard and Bartel 2009), pex6-1 (Zolman and Bartel 2004), and pex6-1 lines transformed with pBINPEX6, 35S:HsPEX6, and 35S:PEX5 (Zolman and Bartel 2004) were previously described. Prior to phenotypic analyses, pfl7/ped1-7, pfl20/lon2-6, pfl47/pex6-2, pfl49/pex14-6, and pfl175/pex14-5 were backcrossed to wild-type Col-0 at least once. Unidentified mutants (pfl29, pfl99, and pfl106) and the second pex14-5 isolate (pfl164) were characterized using nonbackcrossed progeny of original isolates. pex6-2 carrying pBINPEX6, 35S:HsPEX6, or 35S:PEX5 were generated by Agrobacterium tumefaciens-mediated transformation (Clough and Bent 1998) of backcrossed pex6-2 lacking the ICLp:GFP-ICL transgene. Homozygous lines were selected in the progeny of transformants by following resistance to kanamycin (pBINPEX6) or glufosinate ammonium (Basta) (35S:HsPEX6 and 35S:PEX5).
Surface-sterilized seeds were plated on plant nutrient (PN) medium (Haughn and Somerville 1986) supplemented with 0.5% (w/v) sucrose (PNS) and solidified with 0.6% (w/v) agar. Hormone stocks were dissolved in ethanol at 10 or 100 mM and media normalized to the same ethanol content were used as controls. For assays of light-grown seedlings, seeds were stratified for 1 day, plated on the indicated auxin concentrations, and grown for 8 days at 22° under continuous illumination through yellow long-pass filters, which slow indolic compound breakdown (Stasinopoulos and Hangarter 1990), unless otherwise indicated. For assays with dark-grown seedlings, seeds were stratified for 1 day, allowed to begin germination in the absence of hormones under yellow light for 1 day, plated on the indicated media, returned to yellow light for 1 day, and placed in darkness for 4 or 5 days, after which hypocotyl lengths were measured. For lateral root assays, seeds were stratified for 1 day, plated on PNS, and grown in yellow light for 4 days. Four-day-old seedlings were then transferred to a mock or IBA-containing PNS plate and grown under yellow light for 4 additional days, after which the primary root length was measured and the number of lateral roots that had emerged from the primary root were counted.
Mutant isolation and recombination mapping
Seeds from Col-0 lines transformed with ICLp:GFP-ICL (Lingard et al. 2009) were mutagenized with ethyl methanesulfonate (EMS; Normanly et al. 1997). M2 seeds were surface sterilized (Last and Fink 1988), stratified for 0–1 day, plated on PNS (∼1000 seeds per 100 × 100 × 15 mm square plate), and grown in white light. Mutants displaying GFP–ICL fluorescence at 7–9 days were selected using a Leica MZ FLIII fluorescence stereomicroscope equipped for GFP detection, transferred to a fresh PNS plate to recover, and moved to soil for seed production. For retesting, M3 progeny seeds were stratified for 3 days to promote uniform germination and assayed for prolonged fluorescence after 6–7 days of growth under white light. Lines displaying prolonged fluorescence were retained as persistent GFP-ICL fluorescence (pfl) mutants.
Mutants isolated from Col-0 carrying the ICLp:GFP-ICL construct were outcrossed to Landsberg erecta (Ler) for recombination mapping. F2 seedlings from pfl47 and pfl106 outcrosses were screened on PNS for prolonged GFP–ICL fluorescence compared to the unmutagenized parent lines and F3 progeny of mapping plants were confirmed to have prolonged fluorescence. F2 seedlings from pfl7 and pfl99 outcrosses were screened for sucrose dependence. F2 seedlings from pfl20 and pfl29 outcrosses were screened for IBA-resistant root elongation. DNA was isolated from individuals in the mapping populations showing the mutant phenotype and assayed using published and newly developed PCR-based polymorphic markers (Table 1).
Table 1 . Markers used in recombination mapping.
| Fragment size (bp) | ||||||
|---|---|---|---|---|---|---|
| Marker | Nearest gene | Enzyme | Col-0 | Ler | Primer sequences (or reference) | |
| nga63 | At1g09920 | — | 111 | 89 | Bell and Ecker (1994) | |
| MAR109 | At1g17290 | BsaBI | 597 | 353, 244 | TTTTTGGGGGTTCTCAGGTTATC | |
| GCACGCTCAATCGAATCAGAAC | ||||||
| LCS1114 | At1g28500 | DdeI | 231, 207 | 438 | AGAAAATGAGAAGCCCCTGGATAAG | |
| CGCGGCTCTGTTCTTGATGTTTC | ||||||
| SEC202 | At2g41230 | HpaI | 413 | 296, 117 | TGAATTATGACGCAGCTGGAAGAAAAGAGAC | |
| TAAAGAGGCGTAAATAGAATGAGG | ||||||
| SEC242 | At2g46250 | TaqI | 112 | 85, 27 | GGACTTCAGCCCATGTATTCACCT | |
| CGTCAACGGATCACCTCAACCTA | ||||||
| SEC321 | At3g07810 | RsaI | 137, 29 | 166 | AAAAACAATAAAGATGCAGAATGGCTACT | |
| TTTGATTATCCTCGTCTTCTTTCTGGAATG | ||||||
| F24K9 | At3g11590 | BglII | 200, 30 | 230 | AATTTAAAATTATATGCAAACTAATTAGAT | |
| GTAGCTAAAAAGTTGCTGCAAGCAAGGAAA | ||||||
| SO191 | At5g37780 | — | 180 | 200 | Copenhaver et al. (1998) | |
| ILL3 | At5g54140 | NdeI | 360 | 260, 100 | Davies et al. (1999) | |
Immunoblot analysis
To avoid complications in assessing the timing of ICL or MLS degradation that would arise if a mutant exhibited delayed germination, time-course immunoblots used seedlings that had germinated within 24 hr after plating. Protein was extracted from seedlings grown under continuous white light on PNS for the indicated number of days. To extract protein, frozen tissue was ground with a pestle and two volumes of 2× loading buffer (Invitrogen, Carlsbad, CA) was added. Samples were centrifuged and 20 μl of supernatant was transferred to a fresh tube with 2.1 μl of 0.5 M dithiothreitol and heated at 100° for 5 min. Samples were loaded onto NuPAGE 10% Bis-Tris gels (Invitrogen) next to prestained protein markers (P7708S, New England Biolabs, Beverly, MA) and Cruz Markers (Santa Cruz Biotechnology, Santa Cruz, CA). After electrophoresis, proteins were transferred for 30 min at 24 V to a Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech, Piscataway, NJ) using NuPAGE transfer buffer (Invitrogen). After transfer, membranes were rocked for 1 hr at 4° in blocking buffer (8% nonfat dry milk, 20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween-20) and incubated overnight at 4° with primary antibodies diluted in blocking buffer: rabbit α-GFP (1:300 dilution, BD Biosciences 8372-2), rabbit α-ICL (1:2,000 dilution, Maeshima et al. 1988), rabbit α-HPR (1:1000 dilution, Agrisera AS11 1797 or Kleczkowski and Randall 1988), rabbit α-MLS (1:25,000 dilution; Olsen et al. 1993), rabbit α-PEX5 (1:100 dilution; Zolman and Bartel 2004), rabbit α-PEX6 (1:1,000 dilution; Ratzel et al. 2011), rabbit α-PEX7 (1:800 dilution; Ramón and Bartel 2010), rabbit α-PEX14 (1:2,500 dilution, Agrisera AS08 372), rabbit α-PMDH2 (1:2,000 dilution; Pracharoenwattana et al. 2007), rabbit α-thiolase (PED1 isoform, 1:10,000 dilution; Lingard et al. 2009), or mouse α-HSC70 (1:20,000–1:30,000 dilution, StressGen Bioreagents SPA-817), followed by a 4-hr incubation with horseradish peroxidase-linked goat α-rabbit or α-mouse IgG secondary antibody (1:5000 dilution in blocking buffer, Santa Cruz Biotechnology, SC2030 or SC-2031). Horseradish peroxidase was visualized by incubation with LumiGlo reagent (Cell Signaling Technology, Danvers, MA) or WesternBright ECL reagent (Advansta, Menlo Park, CA).
RNA analysis
RNA was isolated from 8-day-old light-grown Col-0 and pex14-6 seedlings using the TRI Reagent RNA Extraction method according to the instructions of the manufacturer (Sigma, St. Louis, MO) and dissolved in DEPC-treated water. cDNA was synthesized from RNA using a 3′ gene-specific primer (PED2-6; Table 2) and SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). The pex14-6 cDNA was PCR amplified across the exon 1–2 junction using PED2-1 and PED2-4 primers (Table 2) and sequenced using PED2-1.
Table 2 . Primers used for amplification and sequencing of candidate genes.
| Gene/accession no. | Primer names | Primer sequences |
|---|---|---|
| PEX14/At5g62810 | PED2-1 and | CATCCTCATCATCTCTCATCAT |
| PED2-2 | CTTAATGGCCTAACCATTTTATCCCAC | |
| PED2-3 and | GGGTTACTTGGCATAGTCCTCTAAAGACG | |
| PED2-4 | GGACGTGTGTCATAATCAACATTGCTG | |
| PED2-5 and | GGTAACATTAGGAACCTTGATATGTGATGC | |
| PED2-6 | GGCAAACCTCATAAAGTATCAATAACCCG | |
| PED1/At2g33150 | PED1-11 and | TTCCCGCACATTCTGATGATGACC |
| PED1-12 | AGAAATGGCTGCCACCCAAA | |
| PED1-13 and | GACATAGGCATTGATAGAGAAGACGAATCT | |
| PED1-8 | AAGTACCAGCAGTAGTGGTGCCAT | |
| PED1-14 and | TGTTGACCCGAAGACTGGTGATGAGAAAC | |
| PED1-15 | AGATATATCTCGGCTGTGGATTTCTTAAGG | |
| PED1-16 and | ATCTGGGCTTCAGGCTGTTGCTGAT | |
| PED1-17 | TGCCTTTCTGTGCGAGTCAACCTA | |
| LON2/At5g47040 | LON2-1s and | CTCATAAGTGTTGCCTTTCGCTAAATCCC |
| LON2-2s | CCACATTCACTTTCCTGCTGG | |
| LON2-3s and | CTTATCTTTGATGCCACCAACAGGCAGAAC | |
| LON2-4s | GTCTGTATTGACTGTTACCCTTAACGG | |
| LON2-5s and | CAGATGGCGCATAGCTATCTTAAG | |
| LON2-6s | GATGGTGTGTGACTGTGGACCAACTTG | |
| LON2-7s and | GGCTAAACCATAGTGATCACTGTCAAGACG | |
| LON2-8s | TAGTTTCGACTTAGAGCTTATTTGG | |
| LON2-9s and | CTGGATCTTGTTTTACTTGCTCCAACTc | |
| LON2-10s | GAAACAGTGGAGCTCCCGAGTAGGTTAGCG | |
| LON2-11s and | GATCGAAGCGTAAGAATGTTAGGAATTGAG | |
| LON2-12s | GTAATGTAATGGGCCTTAGTCTCATTGTTTC | |
| PEX6/At1g03000 | PEX6-7 and | ATCCTCTTCAGTCTTCATCGGTTCG |
| PEX6-8 | CGATGTACGAGGGATTTCAGGCAAGATA | |
| PEX6-9 and | ACTCTGGTTCTTTGGTATGTCCTTCTC | |
| PEX6-10 | CTAAATTCAACTACATGCAGCCCCAACCTC | |
| PEX6-11 and | CCAGGTACATTTGCTTCGGTTTC | |
| PEX6-12 | GCGATTAGCAGCACTTGATGTCC | |
| PEX6-13 and | GATTTTCATTTCTTTCCTTGGTTCTC | |
| PEX6-14 | AATGGCTTACTTACTTTCCCTGTTCC | |
| PEX6-15 and | AAATGTGAAATGGGATGATGTTGGTG | |
| PEX6-16 | AAACACAAACCTAATATAACAAACTGATGAT |
Confocal microscopy
Four-day-old seedlings carrying ICLp:GFP-ICL (Lingard et al. 2009) were mounted in water under a cover glass. Images were collected using a Carl Zeiss LSM 710 laser scanning confocal microscope equipped with a Meta detector. Samples were imaged through a 40× oil immersion objective after excitation with a 488-nm argon laser; GFP emission was collected between 494 and 560 nm. Each image is an average of 8 exposures using a 70-μm pinhole, corresponding to a 1.8-μm optical slice.
Sequencing and genotyping of mutant lesions
Candidate genes in mapping intervals were PCR amplified from mutant genomic DNA using primers listed in Table 2. Amplicons were purified using Zymo PCR purification kit (Zymo Research, Irvine, CA) and sequenced directly (Lone Star Labs, Houston, TX) with the primers used for amplification.
The PEX14 gene (At5g62810) was amplified from pfl49, pf164, and pf175 genomic DNA using three oligonucleotide pairs (Table 2). The resulting overlapping fragments covered the gene from 75 bp upstream of the translation start site to 29 bp downstream of the stop codon. The lesion identified in pfl49 changed PEX14 G73 (where 1 is the first nucleotide of the initiator codon) to an A, which altered a splice site. The lesion identified in pfl164 and pfl175 (which are likely siblings as they were isolated from the same pool of mutagenized seeds) changed PEX14 G904 (where 1 is the first nucleotide of the initiator codon) to an A, which changed Trp152 to a stop codon.
The PED1 gene (At2g33150) was amplified from pfl7 genomic DNA using four oligonucleotide pairs (Table 2). The resulting overlapping fragments covered the gene from 72 bp upstream of the translation start site to 274 bp downstream of the stop codon. The lesion identified in pfl7 changed G2624 (where 1 is the first nucleotide of the initiator codon) of PED1 to an A, which altered a splice site.
The LON2 gene (At5g47040) was amplified from pfl20 genomic DNA using six oligonucleotide pairs (Table 2). The resulting overlapping fragments covered the gene from 1307 bp upstream of the translation start site to 625 bp downstream of the stop codon. The lesion identified in pfl20 changed G2809 (where 1 is the first nucleotide of the initiator codon) of LON2 to an A, which created an amino acid change (Gly445Arg) and destroyed an MnlI restriction site.
The PEX6 gene (At1g0300) was amplified from pfl47 genomic DNA using five oligonucleotide pairs (Table 2). The resulting overlapping fragments covered the gene from 151 bp upstream of the translation start site to 346 bp downstream of the stop codon. The lesion identified in pfl47 changed C1156 (where 1 is the first nucleotide of the initiator codon) of PEX6 to a T, which created an amino acid change (Leu328Phe).
Identified mutations were followed in the progeny of crosses using PCR amplification with the primers listed in Table 3 followed by digestion of the resultant amplicons with the restriction enzymes indicated in Table 3. The pex6-1 mutation was followed as previously described (Zolman and Bartel 2004).
Table 3 . PCR-based markers for determining genotypes of identified mutations.
| Product size (bp) | |||||
|---|---|---|---|---|---|
| Mutant | Primer names | Primer sequences | Enzyme | Wt | Mutant |
| pfl7/ped1-7 | PED1-7 | GAAATTCCAGCCAAGTAAGTGATG | DpnII | 125 | 100, 25 |
| PED1–DpnIIa | CGTAGCTTTGTAAGTAATTATTACCGA | ||||
| pfl20/lon2-6 | LON2-14 | AATTTGTTCGCTTATCTTTGGGTGGTGT | MlnI | 57, 42 | 99 |
| LON2-15 | CTCCCCAAGTTCCTCATCAGCATAAGC | ||||
| pfl47/pex6-2 | PEX6-19 | AGGAACCTTTGATCTATACACCAGT | AvaII | 62, 29 | 91 |
| PEX6–AvaIIa | AGTGAATCACTCCCAAACCGCCCTGGTC | ||||
| pfl49/pex14-6 | PED2-1 | CATCCTCATCATCTCTCATCAT | AvaII | 150, 30 | 180 |
| PED2–AvaIIa | GAAATAATGATTAGAAGGTGTAAATTGGAC | ||||
| pfl164/175/ pex14-5 | PED2-2 | CATCCTCATCATCTCTCATCAT | RsaI | 156, 25 | 181 |
| PED2–RsaI | GACTGGGAGGTAATTTTGTATG | ||||
This is a dCAPS oligonucleotide (Michaels and Amasino 1998; Neff et al. 1998); the underlined nucleotide differs from wild-type sequence to create a restriction site in either the mutant or wild-type PCR amplicon.
Results
Screening for mutants with stabilized GFP–ICL
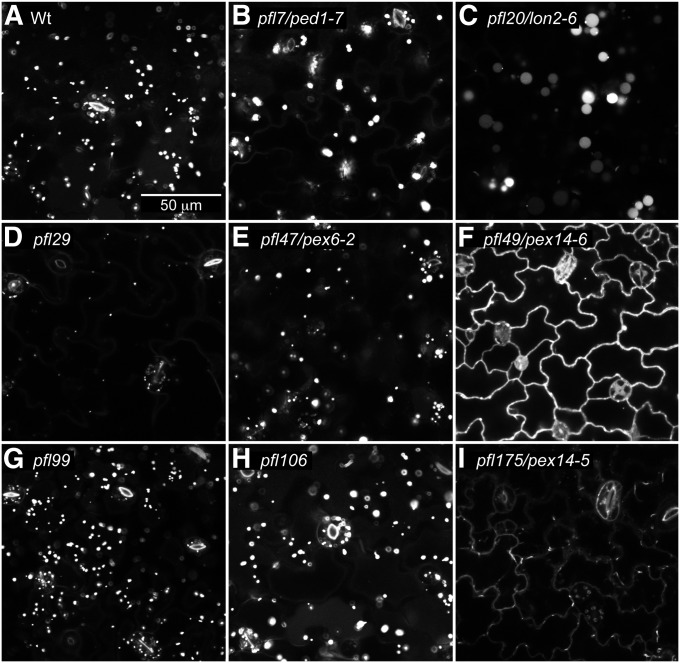
ICL is a peroxisomal matrix protein that is degraded a few days after Arabidopsis seedling germination (Zolman et al. 2005; Lingard et al. 2009). GFP–ICL driven from the endogenous ICL promoter is degraded with similar kinetics as unmodified ICL; GFP–ICL fluorescence, like ICL protein, is no longer apparent 5–6 days after plating (Lingard et al. 2009). To isolate mutants with defects in peroxisome-associated protein degradation, we screened for mutants that exhibited prolonged GFP–ICL fluorescence. We mutagenized lines carrying the ICLp:GFP–ICL construct with EMS and screened ∼44,500 of the resulting M2 seedlings for GFP–ICL fluorescence that remained visible 7–9 days after plating. We selected 175 putative mutants exhibiting prolonged fluorescence. Of these, 105 died or were infertile, 49 did not display prolonged fluorescence in the M3 generation, and 21 appeared to prolong GFP–ICL fluorescence in the M3 generation. We used confocal microscopy to examine GFP–ICL localization in several of these pfl mutants. As summarized in Table 4, we found three with extensive cytosolic GFP–ICL fluorescence (Figure 1, F and I), three with punctate GFP–ICL fluorescence similar to the unmutagenized parent (Figure 1, C, E, and G), and three with partially punctate and partially cytosolic GFP–ICL fluorescence (Figure 1, B, D, and H). In addition to these matrix protein localization defects, we observed some aberrations in peroxisome appearance in the mutants. For example, peroxisomes appeared clustered in pfl7 and pfl106 (Figure 1, B and H), larger in pfl20 (Figure 1C), and smaller in pfl29 (Figure 1D) compared to wild type (Figure 1A).
Table 4 . Classification of persistent fluorescence mutants.
| Class | Isolate | GFP–ICL fluorescencea | ICL stabilization | Sucrose dependence | IBA resistance | PTS2 processing defectb | |||
|---|---|---|---|---|---|---|---|---|---|
| Light | Dark | Light | Dark | Lateral roots | |||||
| — | Wt | P | No | No | No | No | No | No | No |
| 3 | pex4-1 pex22-1 (control) | P | Yes | Yes | Yes | Yes | Yes | Yes | Slight |
| 2 | pfl7/ped1-7 | C and P | Yes | Yes | Yes | Yes | Yes | Yes | Slight |
| 3 | pfl20/lon2-6 | P | No | Slight | Slight | Slight | Slight | Yes | Yes |
| 2 | pfl29 | C and P | Yes | slight | No | Slight | Yes | NT | No |
| 3 | pfl47/pex6-2 | P | Yes | slight | No | Slight | Slight | Yes | No |
| 1 | pfl49/pex14-6 | C | Yes | sight | No | Yes | Yes | Yes | Yes |
| 3 | pfl99 | P | Yes | Yes | Slight | No | No | NT | No |
| 2 | pfl106 | C and P | Yes | Slight | No | No | No | NT | No |
| 1 | pfl164/pfl175/pex14-5 | C | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
NT, not tested.
In 4-day-old seedlings; C, cytosolic; P, punctate.
In 8-day-old seedlings.
Figure 1 .
Localization of GFP–ICL using confocal microscopy separates pfl mutants into three categories: (1) cytosolic, (2) both cytosolic and punctate, and (3) punctate patterns. Cotyledon epidermal cells of 4-day-old light-grown Wt (Col-0 transformed with ICLp:GFP-ICL) (A) or pfl mutant (B–I) seedlings were imaged for GFP using confocal microscopy. pfl20/lon2-6 (C), pfl47/pex6-2 (E), and pfl99 (G) display punctate GFP–ICL fluorescence characteristic of peroxisomal localization. Cytosolic GFP–ICL is visible at the cell margins in pfl49/pex14-6 (F) and pfl175/pex14-5 (I). pfl7/ped1-7 (B), pfl29 (D), and pfl106 (H) display both punctate and cytosolic localization. This experiment was repeated twice with similar results. Scale bar, 50 µm.
Peroxisome function in persistent GFP–ICL fluorescence (pfl) mutants
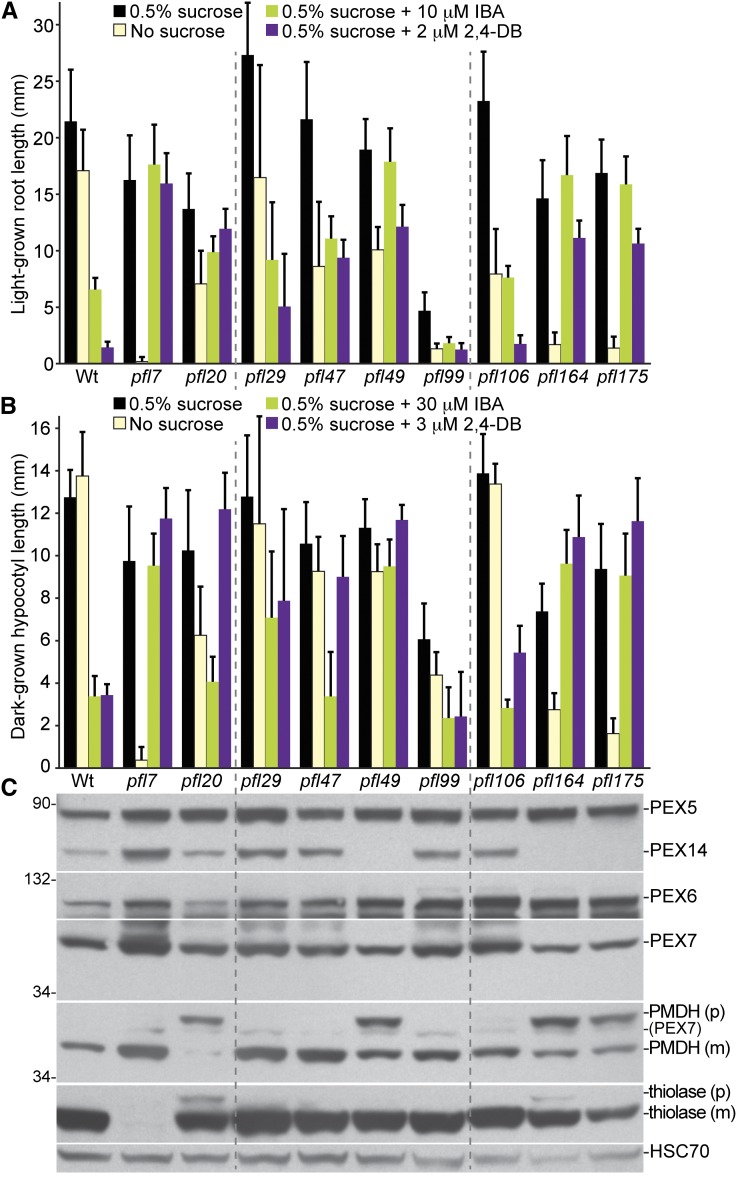
Because defects in peroxisomal matrix protein import often are accompanied by defects in peroxisomal metabolism, we tested peroxisome function in the pfl mutants using sucrose dependence and IBA resistance assays, which indirectly assess the efficiency of peroxisomal β-oxidation. Peroxisomal fatty acid β-oxidation provides energy for early seedling development prior to the onset of photosynthesis. Certain peroxisome-defective mutants, such as pxa1-1 (Zolman et al. 2001), arrest or develop slowly following germination because fatty acids are inefficiently metabolized. These defects can be partially bypassed by providing a fixed carbon source, such as sucrose, in the growth medium (Hayashi et al. 1998; Zolman et al. 2000). Four mutants (pfl7, pfl99, pfl164, and pfl175) displayed clear sucrose-dependent root elongation in the light (Figure 2A) and/or hypocotyl elongation in the dark (Figure 2B), suggesting inefficient β-oxidation of stored fatty acids, which could result from defects in peroxisome biogenesis or β-oxidation enzymes.
Figure 2 .
Most pfl mutants display physiological and/or molecular defects suggestive of peroxisomal defects. (A) Root lengths of 8-day-old pfl or Wt (Col-0) seedlings grown in yellow light in the presence or absence of sucrose or on sucrose-supplemented medium containing inhibitory concentrations of IBA or 2,4-DB are shown. Error bars show standard deviations of the means (n ≥ 12). (B) Hypocotyl lengths of 6-day-old pfl or Wt (Col-0) seedlings grown in the dark in the presence or absence of sucrose or on sucrose-supplemented medium containing inhibitory concentrations of IBA or 2,4-DB are shown. Error bars show standard deviations of the means (n ≥ 12). (C) Protein extracts from the 8-day-old seedlings grown in the light on 0.5% sucrose (in A) were processed for immunoblotting. The membrane was serially probed with antibodies to the indicated proteins. The positions of molecular mass markers (in kilodaltons) are indicated at the left. PMDH and thiolase (PED1) are synthesized as precursors (p) containing the PTS2 signal that is processed into the mature (m) protein in peroxisome. Residual PEX7 (PEX7) from a previous probing remains visible in the PMDH panel. HSC70 is a loading control. Experiments in A through C were repeated twice with similar results.
We also compared peroxisome function in the pfl mutants using IBA resistance assays. Because the protoauxin IBA is imported into peroxisomes and converted into the active auxin, IAA (reviewed in Strader and Bartel 2011), IBA application reduces primary root elongation (Zolman et al. 2000), inhibits hypocotyl elongation in dark-grown seedlings (Strader et al. 2008), and promotes proliferation of lateral roots in light-grown seedlings (Zolman et al. 2000, 2001, 2007, 2008). When IBA-to-IAA conversion is impaired, the auxin effects of IBA are diminished (Strader et al. 2010). Similarly, peroxisomal β-oxidation of the IBA analog 2,4-dichlorophenoxybutyric acid (2,4-DB) to the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D) generates auxin phenotypes (Hayashi et al. 1998). Typical peroxin mutants, such as pex6-1 (Zolman and Bartel 2004), display resistance to the inhibitory effects of IBA (and 2,4-DB) on root elongation in the light and hypocotyl elongation in the dark (Zolman and Bartel 2004; Strader et al. 2011). We found that seven pfl mutants (pfl7, pfl20, pfl29, pfl47, pfl49, pfl164, and pfl175) were IBA (and 2,4-DB) resistant in root (Figure 2A) and/or hypocotyl elongation (Figure 2B), suggesting inefficient β-oxidation of IBA to IAA (and 2,4-DB to 2,4-D) in these mutants.
Mutations in the gene encoding the PEX14 receptor-docking peroxin stabilize GFP–ICL
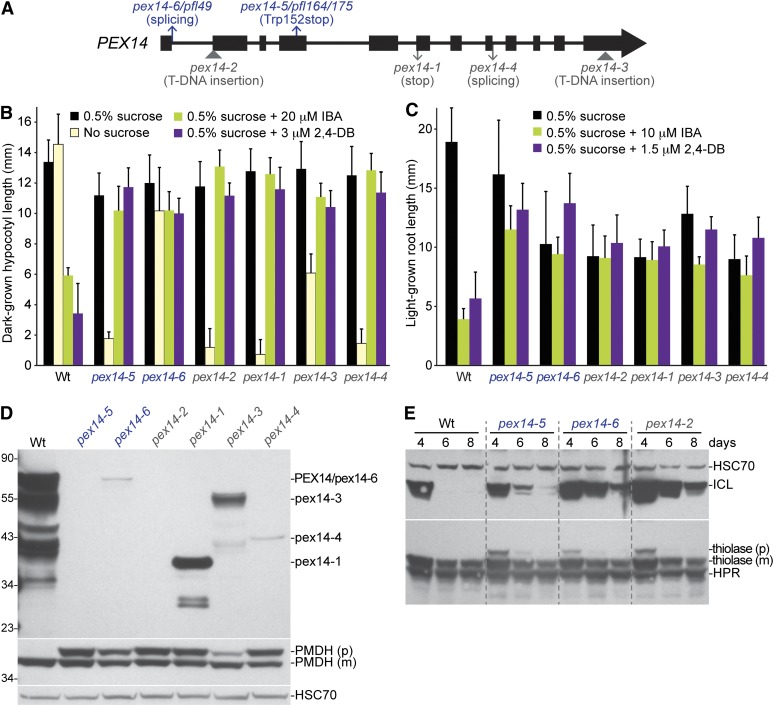
The three persistent GFP–ICL fluorescence mutants with predominantly cytosolic GFP–ICL (pfl49, pfl164, and pfl175; Figure 1 and Table 4) also displayed IBA and 2,4-DB resistance (Figure 2, A and B), consistent with inefficient matrix protein import. Because peroxins facilitate peroxisomal matrix protein import, we examined levels of several peroxins in the pfl mutants using immunoblot analysis. We found normal levels of PEX5, PEX6, and PEX7, but reduced levels of full-length PEX14 protein (Figure 2C) in all three mutants with predominantly cytosolic GFP–ICL (Figure 1, F and I). Upon sequencing the PEX14 gene (At5g62810) in these mutants, we identified two novel point mutations, which we renamed pex14-5 (pfl164 and pfl175) and pex14-6 (pfl49). pex14-5 changes a Trp to a stop codon in the fourth exon (Figure 3A). Overexposure of an anti-PEX14 immunoblot did not reveal PEX14 protein in pex14-5 seedlings (Figure 3D), suggesting that pex14-5 is a null allele. pex14-6 harbors a mutation in the last nucleotide of exon 1, which would change Glu25 to Lys and disrupt a slice site (Figure 3A), and accumulates a small amount of nearly full-length PEX14 protein (Figure 3D). We isolated RNA from the pex14-6 mutant and determined that the major pex14-6 splice product uses a cryptic 5′-donor site in the 5′-UTR, thereby skipping the first exon of PEX14.
Figure 3 .
pex14 mutants display physiological and molecular peroxisomal defects and stabilize ICL. (A) Positions of newly identified pfl alleles are shown above and characterized alleles are shown below a PEX14 gene model in which exons are shown as boxes and introns as lines. (B) Hypocotyl lengths of 7-day-old pfl or Wt (Col-0 transformed with ICLp:GFP-ICL) seedlings grown in the dark in the presence or absence of sucrose or on sucrose-supplemented medium containing inhibitory concentrations of IBA or 2,4-DB are shown. Error bars show standard deviations of the means (n ≥ 10). (C) Root lengths of 8-day-old seedlings pfl or Wt (Col-0 transformed with ICLp:GFP-ICL) grown under yellow-filtered light on sucrose-supplemented medium containing inhibitory concentrations of IBA or 2,4-DB are shown. Error bars show standard deviations of the means (n ≥ 10). (D) Protein extracts from the 8-day-old seedlings grown in the light on 0.5% sucrose in C were processed for immunoblotting. The membrane was serially probed with antibodies to the indicated proteins. The positions of molecular mass markers (in kilodaltons) are indicated at the left. An overexposed anti-PEX14 immunoblot revealed PEX14 protein in all pex14 alleles except pex14-5 and pex14-2. PMDH is synthesized as a precursor (p) with a cleavable PTS2 signal that is processed into mature (m) PMDH in the peroxisome; this cleavage is impaired in pex14 mutants. HSC70 is a loading control. (E) ICL is stabilized in pex14 mutants. Protein extracts from 4-, 6-, and 8-day-old light-grown Wt (Col-0) and pex14 seedlings were processed for immunoblotting. The membrane was serially probed with antibodies to the indicated proteins. Thiolase is synthesized as a precursor (p) with a cleavable PTS2 signal that is processed into mature (m) thiolase in the peroxisome. HSC70 is a loading control. Experiments in B through E were repeated twice with similar results.
We compared these two new pex14 alleles to several previously characterized pex14 alleles (Monroe-Augustus et al. 2011) and found that the new alleles conferred similar IBA and 2,4-DB resistance in both dark- and light-grown seedlings (Figure 3, B and C) along with transient defects in removal of the PTS2-containing presequence from the matrix protein thiolase (Figure 3E). Unlike other pex14 alleles, the pex14-6 allele was not dependent on sucrose in the dark (Figure 3B), suggesting that the low level of pex14-6 protein detected in this mutant (Figure 3D) retained partial PEX14 function.
To assess whether disruption of PEX14 stabilizes endogenous peroxisomal matrix proteins, we compared ICL stability in wild type, pex14-5, pex14-6, and pex14-2, a previously described pex14 null allele (Monroe-Augustus et al. 2011). We found that ICL protein was stabilized in all three pex14 alleles compared to wild type (Figure 3E). Our recovery of pex14 alleles as persistent GFP–ICL fluorescence mutants suggests that impaired peroxisome matrix protein import prevents access of GFP–ICL and ICL to the peroxisome-associated proteolysis machinery or the factors or conditions needed to target substrates to this machinery.
A mutation in a gene encoding the LON2 peroxisomal protease
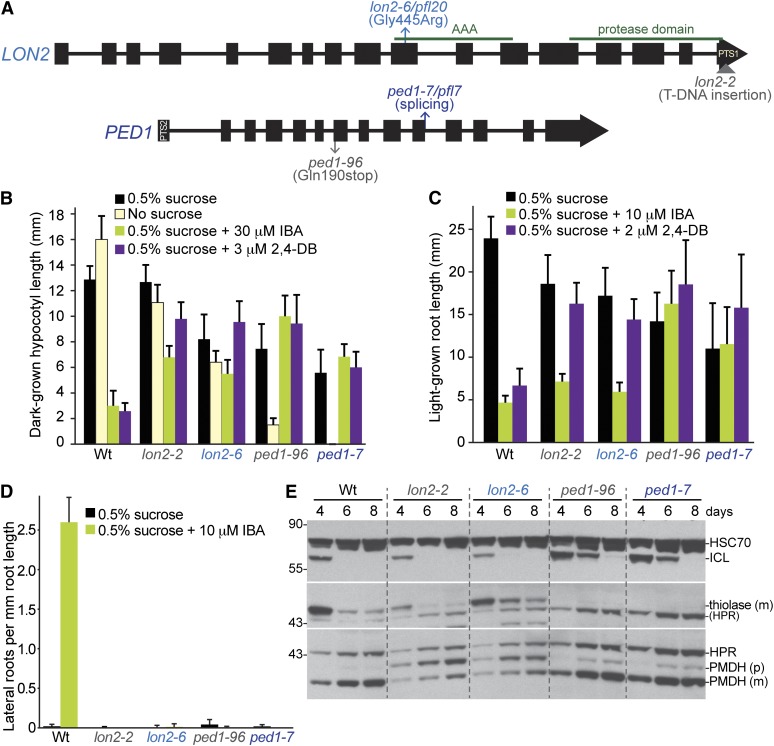
The pfl20 mutant displayed punctate GFP–ICL fluorescence in 4-day-old seedlings (Figure 1C). We used the associated phenotype of IBA-resistant primary root elongation (Figure 2A) to map the pfl20 lesion to an interval on the bottom of chromosome 5 (Figure 4) that included the gene encoding the LON2 (At5g47040) peroxisomal ATP-dependent protease (Lingard and Bartel 2009). We sequenced LON2 from pfl20 genomic DNA and found a point mutation in exon 10 (Figure 5A) that changed Gly445 to an Arg residue. The mutated Gly residue is in the AAA–ATPase domain (Figure 5A) between the Walker A and B domains and is invariant in LON isoforms from plants, fungi, bacteria, and mammals. Like other lon2 alleles (Lingard and Bartel 2009), pfl20 displayed moderate resistance to the inhibition of root and hypocotyl elongation by IBA (Figure 5, B and C), severe resistance to the promotive effects of IBA on lateral rooting (Figure 5D), and PMDH PTS2 processing defects (Figure 5E). We renamed this mutant lon2-6.
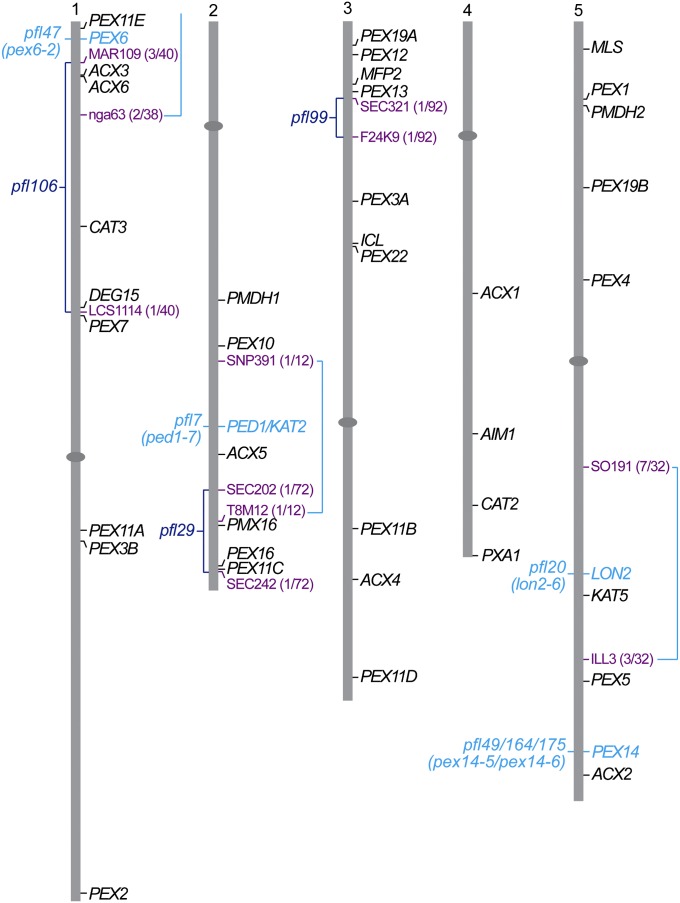
Figure 4 .
Map positions of pfl mutants were determined using recombination mapping. Map positions of genes encoding peroxins and selected additional peroxisomal proteins (in black) and bordering mapping markers used (in lavender) are shown to the right of the five Arabidopsis thaliana chromosome. Identified pfl mutants (in turquoise) and unidentified pfl mutants (in dark blue) are shown to the left of the chromosomes with mapping intervals bracketed and the number of recombinants/number of chromosomes scored shown in parentheses (in lavender).
Figure 5 .
Both lon2 and ped1 mutants display physiological and molecular peroxisomal defects, but only ped1 mutants stabilize ICL. (A) Positions of newly identified pfl alleles are shown above and characterized alleles are shown below gene models of LON2 and PED1. Green lines above the LON2 gene model delineate regions encoding the central AAA domain and the C-terminal protease domain. LON2 and PED1 encode proteins that are targeted to peroxisomes via a C-terminal PTS1 signal or an N-terminal PTS2 signal, respectively. (B) Hypocotyl lengths of 6-day-old pfl or Wt (Col-0) seedlings grown in the dark in the presence or absence of sucrose or on sucrose-supplemented medium containing inhibitory concentrations of IBA or 2,4-DB are shown. Error bars show standard deviations of the means (n ≥ 12). (C) Root lengths of 8-day-old pfl or Wt (Col-0) seedlings grown under yellow-filtered light on sucrose-supplemented medium containing inhibitory concentrations of IBA or 2,4-DB are shown. Error bars show standard deviations of the means (n ≥ 15). (D) Lateral roots per millimeter of root length of 8-day-old pfl or Wt (Col-0) seedlings 4 days after transfer to sucrose-containing medium with or without 10 µM IBA are shown. Error bars show standard deviations of the means (n ≥ 8). (E) ICL is stabilized in ped1 mutants but is degraded similarly to wild type in lon2 mutants. Protein extracts from 4-, 6-, and 8-day-old light-grown Wt (Col-0) and mutant seedlings were processed for immunoblotting. The membrane was serially probed with antibodies to the indicated proteins. The positions of molecular mass markers (in kilodaltons) are indicated at the left. PMDH and thiolase are synthesized as precursors (p) with a cleavable PTS2 signal that are processed into mature (m) versions in the peroxisome. Residual HPR (HPR) from a previous probing remains visible in the thiolase panel. HSC70 is a loading control. Experiments in B, C, and E were repeated twice with similar results.
Because LON2 is a peroxisomal protease (Ostersetzer et al. 2007), it was a candidate for participation in peroxisome-associated protein degradation. However, previously characterized lon2 T-DNA insertion alleles do not dramatically stabilize ICL or MLS (Lingard and Bartel 2009). To examine whether peroxisomal matrix proteins were stabilized in the lon2-6 mutant, we compared ICL stability in wild type, lon2-6, and lon2-2, a previously characterized (Lingard and Bartel 2009) lon2 allele disrupted by a T-DNA insertion near the 3′ end of the gene (Figure 5A). Indeed, we found that ICL was not stabilized in either lon2 mutant compared to wild type (Figure 5E). Some lon2 alleles display uneven germination (Lingard and Bartel 2009), suggesting that a germination delay of the M2 seedling may explain our original isolation of the pfl20/lon2-6 mutant in the persistent GFP–ICL fluorescence screen.
Mutations in the PED1 gene encoding a peroxisomal thiolase stabilize GFP–ICL
pfl7 displayed a combination of peroxisomal and cytosolic GFP–ICL fluorescence (Figure 1B) and the classical peroxisome-defective phenotypes of IBA- and 2,4-DB-resistant root and hypocotyl elongation and sucrose-dependent seedling development (Figure 2, A and B). We used the sucrose-dependence phenotype to map the pfl7 lesion to a region of chromosome 2 that included PED1 (At2g33150; Figure 4), which encodes a peroxisomal 3-ketoacyl-CoA thiolase, also known as KAT2, that is implicated in fatty acid, IBA, and 2,4-DB β-oxidation (Hayashi et al. 1998; Germain et al. 2001). In addition, we did not detect thiolase protein on immunoblots of pfl7 seedling extracts (Figure 2C). We sequenced PED1 from pfl7 DNA and found a point mutation in the first nucleotide of intron 10 that is predicted to disrupt PED1 splicing (Figure 5A). The nature of the lesion was consistent with the lack of full-length thiolase (PED1) protein detected in immunoblots of pfl7 seedling extracts (Figures 2C and 5E), and we renamed pfl7 as ped1-7.
We found that ped1-7 displayed β-oxidation defects similar in severity to ped1-96 (Figure 5, B-D), a previously isolated ped1 null allele (Lingard and Bartel 2009). However, matrix protein import defects have not been reported for ped1-96 (Lingard and Bartel 2009), the original ped1 allele (Hayashi et al. 1998), or the kat2-1 T-DNA insertion allele of PED1 (Germain et al. 2001). Because ped1-7 partially mislocalized GFP–ICL to the cytosol (Figure 1B), we examined PTS2 processing in ped1 mutants. We found that both ped1-7 and ped1-96 displayed a slight defect in PTS2 processing of PMDH (Figure 5E), consistent with the slight defect in matrix protein import revealed by the partial mislocalization of GFP–ICL to the cytosol in ped1-7 (Figure 1B).
To examine whether disruption of the PED1 thiolase stabilizes endogenous peroxisomal matrix proteins, we compared ICL stability in wild type, ped1-7, and ped1-96. We found that ICL was similarly stabilized in both ped1 alleles (Figure 5E). To examine whether this stabilization might reflect a developmental delay caused by the reduced fatty acid β-oxidation that would result from reduced thiolase activity, we monitored the timing of appearance of the photorespiration enzyme hydroxypyruvate reductase (HPR) in the ped1 mutants. In wild-type seedlings, HPR appears during the transition to photosynthetic growth as ICL is degraded (Lingard et al. 2009). We found that HPR appeared in ped1 mutants with similar timing as in wild type (Figure 5E), suggesting that ped1 developmental delays did not account for the observed ICL stabilization (Figure 5E).
The PEX6 AAA–ATPase is required for efficient peroxisome-associated degradation
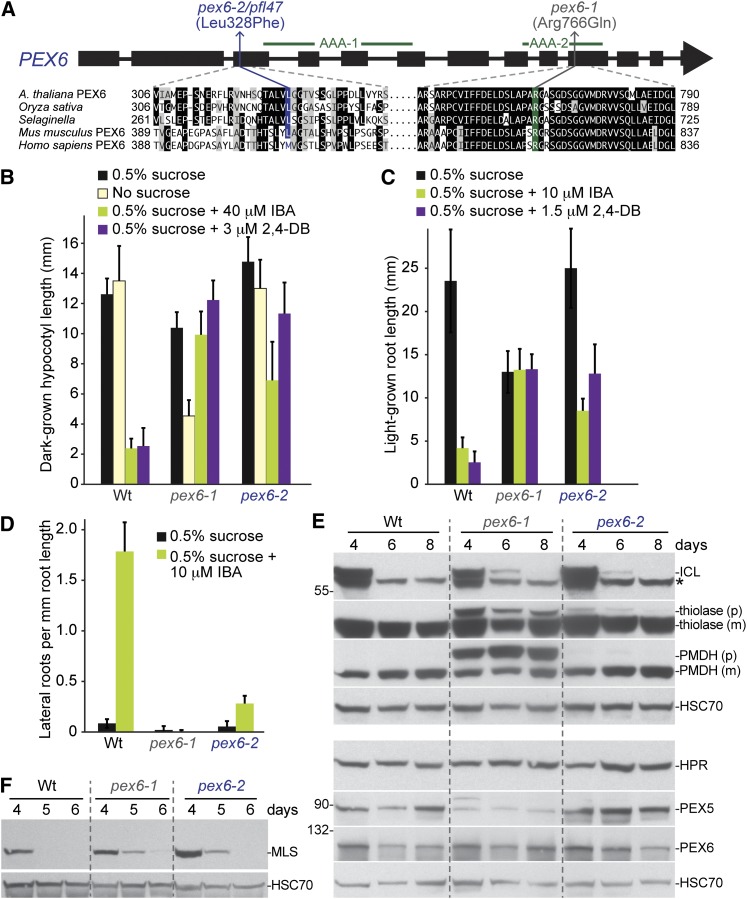
The pfl47 mutant displayed normal peroxisomal localization of GFP–ICL (Figure 1E) and normal levels of assayed peroxins (Figure 2C). We used the persistent GFP–ICL fluorescence phenotype to map pfl47 to a region at the top of chromosome 1 that included the PEX6 gene (At1g03000; Figure 4). Upon sequencing PEX6 from pfl47 DNA, we found a point mutation in exon 3 that changed Leu328 to a Phe residue (Figure 6A). We renamed pfl47 as pex6-2. We compared the phenotypes of pex6-2 to those of pex6-1, a different missense allele isolated in a screen for mutants displaying IBA-resistant root elongation that also is sucrose dependent and displays a marked PTS2 processing defect (Zolman and Bartel 2004). Unlike pex6-1, pex6-2 developed normally in the absence of sucrose in the dark (Figure 6B), was only moderately resistant to the inhibitory effects of IBA on hypocotyl (Figure 6B) or root (Figure 6C) elongation, processed the PTS2 proteins thiolase and PMDH nearly normally (Figures 2C and 6E), and displayed a wild-type root length on sucrose-supplemented medium (Figure 6C). Both pex6 alleles displayed clear resistance to the inhibitory effects of 2,4-DB on hypocotyl elongation in the dark (Figure 6B) and to the promotive effects of IBA on lateral root formation in the light (Figure 6D). Moreover, both alleles similarly stabilized ICL and MLS (Figure 6, E and F). Because the pex6-2 phenotypes were not identical to those of pex6-1, we introduced a wild-type genomic copy of PEX6 (Zolman and Bartel 2004) into pex6-2 using Agrobacterium-mediated transformation to ensure that the identified pex6-2 lesion was responsible for the phenotypes observed. We found that this PEX6p:PEX6 construct rescued the 2,4-DB (Figure 7A) and IBA resistance (Figure 7B) of pex6-2 and pex6-1, confirming that the identified pex6-2 lesion caused the peroxisome-defective phenotypes observed.
Figure 6 .
pex6-2 and pex6-1 display partially overlapping physiological and molecular peroxisomal defects and stabilize ICL and MLS. (A) The positions of the newly identified pfl47/pex6-2 allele and the characterized pex6-1 allele are shown above a gene model of PEX6. Green lines above the gene model delineate regions encoding the two PEX6 AAA domains. Arabidopsis PEX6 regions containing the pex6-2 and pex6-1 lesions are shown below the gene model aligned with orthologs from Oryza sativa (NP_001053886), Selaginella moellendorffii (XP_002979987), Mus musculus (NP_663463), and Homo sapiens (NP_000278). (B) Hypocotyl lengths of 6-day-old pfl or Wt (Col-0 transformed with ICLp:GFP-ICL) seedlings grown in the dark in the presence or absence of sucrose or on sucrose-supplemented medium containing inhibitory concentrations of IBA or 2,4-DB are shown. Error bars show standard deviations of the means (n ≥ 10). (C) Root lengths of 8-day-old pfl or Wt (Col-0 transformed with ICLp:GFP-ICL) seedlings grown under yellow-filtered light on sucrose-supplemented medium containing inhibitory concentrations of IBA or 2,4-DB are shown. Error bars show standard deviations of the means (n ≥ 8). (D) Lateral roots per millimeter root length of 8-day-old pfl or Wt (Col-0) seedlings 4 days after transfer to sucrose-containing medium with or without 10 µM IBA are shown. Error bars show standard deviations of the means (n ≥ 8). (E) Both pex6 alleles stabilize ICL, whereas only pex6-1 displays reduced PEX5 levels or severe PTS2 processing defects. Protein extracts from 4-, 6-, and 8-day-old light-grown Wt (Col-0 transformed with ICLp:GFP-ICL) or mutant seedlings were processed for immunoblotting. Membranes from duplicate gels were serially probed with antibodies to the indicated proteins to obtain the top four panels and the bottom four panels. The positions of molecular mass markers (in kilodaltons) are indicated at the left. PMDH and thiolase are synthesized as precursors (p) with a cleavable PTS2 signal that is processed into mature (m) versions in the peroxisome. An asterisk marks a cross-reacting band detected by the ICL antibody that is not present in an icl null mutant (Lingard et al. 2009). HSC70 is a loading control. (F) Both pex6 alleles stabilize MLS. Protein extracts from 4-, 5-, and 6-day-old Wt (Col-0 transformed with ICLp:GFP-ICL) or mutant light-grown seedlings were processed for immunoblotting with antibodies to MLS and HSC70, a loading control. Experiments in B through F were repeated at least twice with similar results.
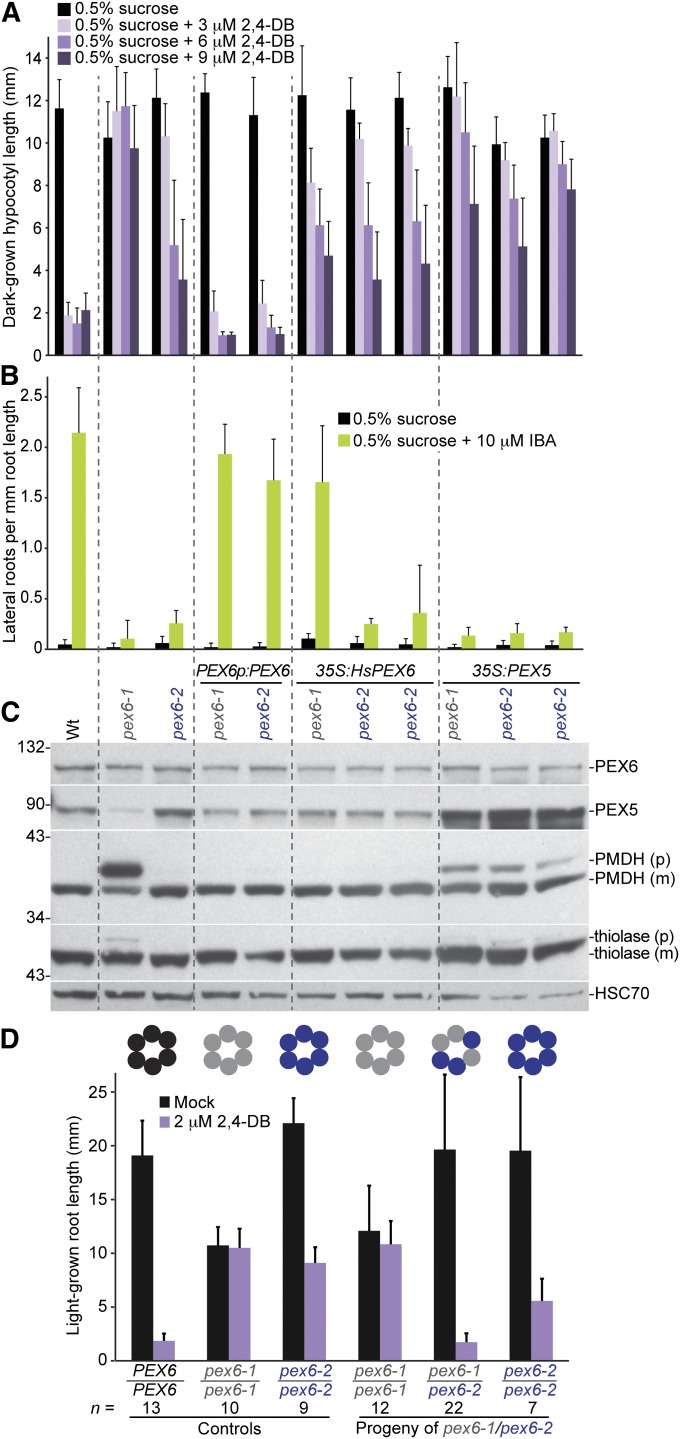
Figure 7 .
pex6 complementation analysis. (A) The 2,4-DB resistance of pex6-1 is fully rescued by the pBINPEX6 genomic Arabidopsis PEX6 construct (PEX6p:PEX6) and partially rescued by expression of a human PEX6 cDNA (35S:HsPEX6) or Arabidopsis PEX5 overexpression (35S:PEX5), whereas pex6-2 2,4-DB resistance is rescued by the genomic PEX6 construct, unaffected by expression of human PEX6 (two transformants shown), and enhanced by Arabidopsis PEX5 overexpression (two transformants shown). Hypocotyl lengths of 6-day-old Wt (Col-0) or mutant seedlings grown in the dark on sucrose-supplemented medium containing increasing concentrations of 2,4-DB are shown. Error bars show standard deviations of the means (n ≥ 15). (B) The IBA resistance of both pex6-1 and pex6-2 lateral root production is fully rescued by a genomic Arabidopsis PEX6 construct but not by Arabidopsis PEX5 overexpression. Human PEX6 expression restores IBA-responsive lateral rooting to pex6-1 but not to pex6-2 (two transformants shown). Lateral roots per millimeter root length of 8-day-old Wt (Col-0) or mutant seedlings 4 days after transfer to sucrose-containing medium with or without 10 µM IBA are shown. Error bars show standard deviations of the means (n ≥ 8). (C) The PTS2 processing defect and reduced PEX5 levels of pex6-1 are rescued by a genomic Arabidopsis PEX6 construct and by expression of human PEX6 and are partially rescued by Arabidopsis PEX5 overexpression; pex6-2 acquires PTS2 processing defects when Arabidopsis PEX5 is overexpressed. Protein extracts from the 8-day-old light-grown control seedlings from B were processed for immunoblotting. The membrane was serially probed with antibodies to the indicated proteins. The positions of molecular mass markers (in kilodaltons) are indicated at the left. PMDH and thiolase are synthesized as precursors (p) with a cleavable PTS2 signal that are processed into mature (m) proteins in the peroxisome. HSC70 is a loading control. (D) pex6-1 and pex6-2 exhibit intragenic complementation of 2,4-DB resistant root elongation. Control and F2 progeny were plated on media without and with 2,4-DB and root lengths of 8-day-old seedlings were measured. The genotype of each seedling was then determined. The number of seedlings (n) of each genotype is indicated. This intragenic complementation suggests that the pex6-1 and pex6-2 missense lesions affect different PEX6 functions and that mixed oligomers with both pex6-1 (gray circles) and pex6-2 (purple circles) can carry out PEX6 (black circles) functions. Experiments in A, C, and D were repeated at least twice with similar results.
To further define the extent of the differences between the pex6-1 and pex6-2 alleles, we compared the effects of overexpressing human PEX6 or Arabidopsis PEX5 in these mutants. The pex6-1 mutation alters an Arg residue in the second AAA domain (Figure 6A) that is conserved in human PEX6 (Zolman and Bartel 2004), whereas the Leu residue mutated in pex6-2 is a Met in the human protein and is in a less conserved region (Figure 6A). Expression of a human PEX6 cDNA from the cauliflower mosaic virus 35S promoter (35S:HsPEX6) rescues the IBA resistance and sucrose dependence of pex6-1 (Zolman and Bartel 2004). Similarly, we found that expressing this human PEX6 cDNA restored pex6-1 sensitivity to lateral root promotion by IBA (Figure 7B), rescued pex6-1 PTS2 processing defects (Figure 7C), and partially restored sensitivity of pex6-1 hypocotyls to 2,4-DB in the dark (Figure 7A). In marked contrast, 35S:HsPEX6 did not rescue the strong resistance of pex6-2 lateral roots to IBA (Figure 7B) or the partial resistance of pex6-2 hypocotyls to 2,4-DB (Figure 7A). Our observation that 35S:HsPEX6 failed to rescue the pex6-2 phenotypes assayed suggests that the function(s) disrupted by the pex6-2 mutation is not conserved in the human protein (Figure 6A), unlike the pex6-1 mutation (Zolman and Bartel 2004).
pex6-1 exhibits reduced PEX5 levels (Zolman and Bartel 2004), probably because PEX5 is polyubiquitinated and degraded when it is not efficiently removed from the peroxisome by PEX6. PEX5 overexpression from the 35S promoter (35S:PEX5) partially suppresses the sucrose dependence and growth defects of pex6-1 without restoring IBA sensitivity (Zolman and Bartel 2004). In addition, we found that PEX5 overexpression partially restored PTS2 processing in pex6-1 (Figure 7C). Unlike pex6-1, we found normal PEX5 levels in pex6-2 (Figures 2C, 6E, and 7C). In contrast to the beneficial effects of PEX5 overexpression in pex6-1 (Figure 7, A and C), PEX5 overexpression enhanced pex6-2 2,4-DB resistance (Figure 7A) and induced a PTS2 processing defect in pex6-2 (Figure 7C). These enhancements of pex6-2 defects by PEX5 overexpression suggest that unlike pex6-1, pex6-2 defects are not caused by lack of PEX5 available to escort proteins into the peroxisome.
Because the pex6-1 and pex6-2 alleles performed differently in a variety of assays (Figure 6, B–E, and Figure 7, A–C), we assessed the ability of each pex6 lesion to complement the defects of the other. F2 plants from a cross of pex6-1 and pex6-2 were assayed for 2,4-DB resistance in roots and individual plants were genotyped. Surprisingly, we found that pex6-1/pex6-2 seedlings were as sensitive to 2,4-DB as wild-type PEX6/PEX6 seedlings were (Figure 7D). This intragenic complementation is consistent with our observation that both pex6 alleles accumulated wild-type levels of pex6 protein (Figures 6E and 7C) and implied that the two missense mutations affect separable functions of PEX6.
Additional pfl mutants
We used recombination mapping to localize pfl29, pfl99, and pfl106 to distinct chromosome regions (Figure 4). We mapped the pfl29 lesion to the bottom of chromosome 2, using persistent GFP–ICL fluorescence phenotype, pfl99 to the top of chromosome 3 using the associated phenotype of sucrose dependence, and pfl106 to a region on chromosome 1 using the persistent GFP–ICL fluorescence phenotype (Figure 4). These mapping data indicate that additional loci can mutate to confer GFP–ICL stabilization. Map-based cloning of these additional loci is ongoing.
Discussion
Three classes of pfl mutants based on subcellular GFP–ICL localization
Although much is known about how matrix proteins enter peroxisomes (reviewed in Hu et al. 2012), little is known about how these matrix proteins are ultimately degraded. The developmentally controlled degradation of the glyoxylate cycle enzymes ICL and MLS provides model substrates with which to unravel peroxisome-associated degradation. We have begun isolating and characterizing mutants with impaired degradation of a GFP–ICL reporter, anticipating that analysis of the defective genes will elucidate the mechanism of peroxisomal matrix protein degradation. We selected mutants that retained GFP–ICL fluorescence longer than wild type, and subcellular GFP–ICL localization has allowed us to separate the mutants into different classes. The first class contains mutants with predominantly cytosolic GFP–ICL, mutants in the second class display both cytosolic and punctate GFP–ICL, and the third class includes mutants with predominantly punctate GFP–ICL (Table 4).
Import into the peroxisome is needed for efficient ICL degradation
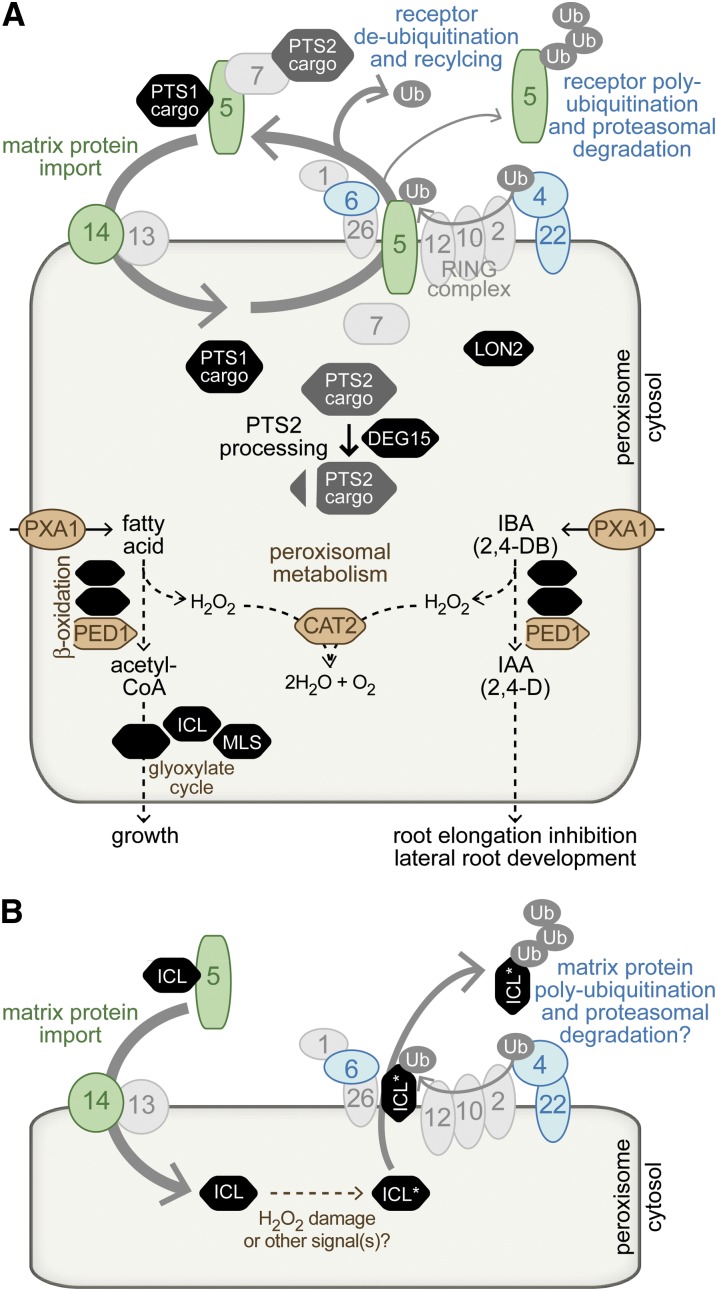
The pex14-5 and pex14-6 mutants are members of the first class of pfl mutants (Table 4). As illustrated in Figure 8A, PEX14 is a peroxisomal membrane protein (Hayashi et al. 2000) that acts with PEX13 as the PEX5–PEX7 docking complex (Schell-Steven et al. 2005) and may assist PEX5 in forming a transient matrix protein import pore (Meinecke et al. 2010). Whereas pex14-5 resembles previously described pex14 null alleles (Hayashi et al. 2000; Monroe-Augustus et al. 2011), pex14-6 is unique among described Arabidopsis pex14 mutants in displaying sucrose independence (Figure 3B), suggesting that residual pex14-6 protein (Figure 3D) retains some PEX14 function. The viability of the pex14-5 apparent null allele (Figure 3D) confirms a recent report that PEX14, unlike its docking partner PEX13 (Boisson-Dernier et al. 2008), is not required for Arabidopsis viability (Monroe-Augustus et al. 2011). All of the assayed pex14 alleles similarly stabilize ICL (Figure 3E). ICL and MLS also are stabilized in the pex5-10 mutant (Lingard et al. 2009), another peroxin mutant that displays severe matrix protein import defects (Khan and Zolman 2010). These demonstrations that ICL and MLS must enter the peroxisome to be efficiently degraded suggest that either the degradation machinery or the machinery needed to target ICL for destruction is peroxisome associated (Figure 8B).
Figure 8 .
Arabidopsis peroxisomal matrix protein degradation is influenced by proteins implicated in matrix protein import, receptor recycling, and peroxisomal metabolism. (A) Likely functions of Arabidopsis peroxins (numbered ovals) in peroxisome matrix protein import based on data from Arabidopsis and other systems (reviewed in Hu et al. 2012). Matrix proteins are targeted to the peroxisome via a C-terminal PTS1 or an N-terminal PTS2, which are recognized in the cytosol by the PEX5 and PEX7 receptors, respectively. Receptors dock with the membrane peroxins PEX13 and PEX14, deliver cargo, and are recycled. PEX5 recycling requires the ubiquitin-conjugating enzyme PEX4 and a RING-finger complex composed of PEX2, PEX10, and PEX12. The PEX6 and PEX1 AAA–ATPases promote retrotranslocation of ubiquitinated PEX5 out of the peroxisome; in the absence of efficient recycling, PEX5 can be multi-ubiquitinated and degraded in the proteasome. Once in the peroxisome, PTS2 proteins are processed by the peroxisomal protease DEG15 (Helm et al. 2007; Schumann et al. 2008). Both PTS2 and PTS1 proteins contribute to peroxisome metabolism, including fatty acid and IBA β-oxidation, exemplified by PED1 (Hayashi et al. 1998; Zolman et al. 2000), the glyoxylate cycle, exemplified by ICL (Eastmond et al. 2000a) and MLS (Cornah et al. 2004), and H2O2 decomposition by catalases including CAT2. PXA1 is a membrane protein that likely transports fatty acids and IBA into the peroxisome (Zolman et al. 2001). Mutants defective in proteins shown in color alter the degradation rate of glyoxylate cycle enzymes, including proteins involved in matrix protein import (green), receptor recycling components (blue), and proteins involved in peroxisomal metabolism (brown). (B) A model for peroxisomal matrix protein degradation. Efficient ICL degradation requires PEX5 (Lingard et al. 2009) and PEX14 (this work), implying that ICL import into the peroxisome precedes ICL degradation. Once in the peroxisome, peroxisome metabolism influences the ICL degradation rate, perhaps by modulating the extent of H2O2 damage. For example, ICL degradation is slowed in ped1 (this work) and pxa1 (Lingard et al. 2009) and is enhanced in a cat2 mutant (Lingard et al. 2009). The stabilization of ICL (and MLS) in the pex4-1 pex22-1 mutant (Lingard et al. 2009) and pex6 mutants (this work) is consistent with the possibility that ICL may exit the peroxisome for cytosolic degradation in the proteasome.
Peroxisomal metabolism can influence ICL degradation
We found that PED1 promotes efficient peroxisomal matrix protein degradation (Figure 5E). PED1 is a peroxisomal thiolase (Figure 8A) needed for β-oxidation of fatty acids to acetyl-CoA (Hayashi et al. 1998) and of IBA to IAA (Zolman et al. 2000). We were surprised to find that PED1 also was needed for efficient matrix protein import, as judged by both incomplete removal of the PTS2-containing sequence from PMDH (Figure 5E) and partial GFP–ICL mislocalization to the cytosol (Figure 1B) in ped1 mutants. ped1 mutants have larger peroxisomes than wild type (Hayashi et al. 1998); perhaps this altered geometry physically impairs matrix protein import. Alternatively, there may exist an undiscovered feedback mechanism linking matrix protein import with peroxisomal metabolism. In either case, ICL stabilization in ped1 mutants might result from inefficient import of ICL into the peroxisome matrix, as in the pex14 and pex5-10 mutants discussed above. Arguing against this possibility is our observation that lon2 mutants, which display more severe PTS2 processing defects than ped1 mutants display, fail to stabilize ICL (Figure 5E). An alternative possibility is that reduced β-oxidation in ped1 lowers peroxisomal H2O2, reducing oxidative damage and slowing degradation. Indeed, ICL and MLS are similarly stabilized in a pxa1 mutant (Lingard et al. 2009) that shows complete sucrose dependence, strong IBA resistance (Zolman et al. 2001), and reduced H2O2 levels (Eastmond 2007) due to a reduced ability to move β-oxidation substrates into the peroxisome (Figure 8A). Conversely, ICL and MLS degradation is hastened in the cat2 mutant (Lingard et al. 2009), which is missing one of the peroxisomal catalases that decompose H2O2. A third nonexclusive possibility is that ICL and MLS degradation may be linked to the depletion of seedling fatty acid stores, which also would explain our observations that ICL degradation is delayed in several mutants with impaired fatty acid β-oxidation. For example, ICL and MLS degradation might be inhibited by fatty acids or β-oxidation intermediates or might be promoted by sucrose or other downstream metabolites of β-oxidation.
The PEX6 ATPase is needed for efficient matrix protein degradation
As illustrated in Figure 8A, PEX4 is a ubiquitin-conjugating enzyme that in yeast and plants is tethered to the peroxisome by PEX22 (Koller et al. 1999; Zolman et al. 2005) and in yeast provides ubiquitin to RING finger peroxins that ubiquitinate the matrix protein receptor PEX5 (Thoms and Erdmann 2006; Platta et al. 2007, 2009). PEX6 and PEX1 are AAA–ATPases that in yeast and mammals assist in the retrotranslocation of ubiquitinated PEX5 from the peroxisome (Figure 8A), thus recycling PEX5 for further import rounds (reviewed in Fujiki et al. 2012; Grimm et al. 2012); PEX5 is poly-ubiquitinated and degraded in the proteasome when PEX6 is not functional (Platta et al. 2007). Arabidopsis PEX6 likely functions similarly to its yeast and mammalian orthologs, as the pex6-1 allele has decreased PEX5 levels and is partially rescued by PEX5 overexpression (Figure 7C and Zolman and Bartel 2004).
By screening for GFP–ICL stabilization, we identified a novel pex6 allele, pex6-2, that shares only a subset of pex6-1 defects, including IBA and 2,4-DB resistance (Figure 6, B-D). Unlike pex6-1, pex6-2 did not require sucrose for normal development in the dark (Figure 6B), processed PTS2 proteins nearly as efficiently as wild type (Figure 6E), and had normal PEX5 levels (Figures 2C, 6E, and 7C). Moreover, pex6-2 physiological and molecular defects were exacerbated rather than rescued by PEX5 overexpression (Figure 7, A and C). Interestingly, the pex6-1 and pex6-2 lesions were able to complement one another (Figure 7D). PEX6 is thought to function as a hexamer (reviewed in Fujiki et al. 2012; Grimm et al. 2012), and this intragenic complementation suggests that the pex6-1 and pex6-2 missense lesions affect different PEX6 functions, allowing mixed pex6-1 pex6-2 oligomers to carry out all PEX6 functions. Like pex4-1 pex22-1 and pex6-1 (Lingard et al. 2009), pex6-2 stabilized ICL and MLS (Figure 6, E and F). The stabilization of ICL and MLS without dramatic effects on other peroxisomal processes such as matrix protein import (Figure 1E) suggests that ICL and MLS stabilization in pex6-2 does not result from a failure to import ICL and MLS into the peroxisome, as in pex14 alleles. Rather, it seems feasible that peroxisomal matrix proteins require the PEX5-recycling machinery, including PEX6 and PEX4, to move from the peroxisome to the cytosol for proteasomal degradation (Figure 8B).
Multiple genes contribute to efficient peroxisomal matrix protein degradation
By screening for mutants exhibiting GFP–ICL stabilization, we have begun identifying genes needed for matrix protein degradation and deciphering the peroxisome-associated matrix protein degradation pathway (Figure 8B). We found that matrix proteins need to enter the peroxisome to be subject to efficient degradation and that the metabolic status of the peroxisome affects the degradation rate. Moreover, several peroxins involved in ubiquitinating and retrotranslocating PEX5 are needed for efficient degradation, consistent with the intriguing possibility that matrix proteins may leave the peroxisome for proteasomal degradation in the cytosol. The progress reported here also reveals several gaps in our understanding of peroxisome-associated matrix protein degradation that remain to be elucidated, including how matrix proteins are recognized for degradation and how metabolic status is linked to degradation rate. Several pfl mutants for which the defective genes have not been identified displayed neither IBA resistance nor sucrose dependence, but rather appeared to have wild-type β-oxidation phenotypes (Table 4). Identification of the genes defective in these mutants may provide additional insights into how peroxisomal proteins are degraded.
Acknowledgments
We thank John Harada (University of California, Davis) for the MLS antibody, Masayoshi Maeshima (Nagoya University, Japan) for the ICL antibody, Douglas Randall (University of Missouri, Columbia) for the HPR antibody, and Steven Smith and Itsara Pracharoenwattana (University of Western Australia) for the PMDH2 antibody. We are grateful to Wendell Fleming for assistance with confocal microscopy, Daniel Wagner for stereomicroscope use, Lucia Strader and Mauro Rinaldi for recombination mapping markers, and Emily Liljestrand for assistance in mapping lon2-6. We thank the Arabidopsis Biological Resource Center at Ohio State University for seeds from Salk Institute insertion lines and Lisa Farmer, Wendell Fleming, Kim Gonzalez, Yun-Ting Kao, and Mauro Rinaldi for critical comments on the manuscript. This research was supported by the National Institutes of Health (R01GM079177) and the Robert A. Welch Foundation (C-1309). Confocal microscopy was performed on equipment obtained through a Shared Instrumentation Grant from the National Institutes of Health (S10RR026399-01). M.J.L. was supported in part by a postdoctoral fellowship from the U.S. Department of Agriculture (2008-20659).
Footnotes
Communicating editor: S. Poethig
Literature Cited
- Adham A. R., Zolman B. K., Millius A., Bartel B., 2005. Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in β-oxidation. Plant J. 41: 859–874 [DOI] [PubMed] [Google Scholar]
- Azevedo J. E., Schliebs W., 2006. Pex14p, more than just a docking protein. Biochim. Biophys. Acta 1763: 1574–1584 [DOI] [PubMed] [Google Scholar]
- Bartoszewska M., Williams C., Kikhney A., Opalinski L., Van Roermund C. W., et al. , 2012. Peroxisomal proteostasis involves a Lon family protein that functions as protease and chaperone. J. Biol. Chem. 287: 27380–27395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C. J., Ecker J. R., 1994. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Frietsch S., Kim T.-H., Dizon M. B., Schroeder J. I., 2008. The peroxin loss-of-function mutation abstinence by mutual consent disrupts recognition between male and female gametophytes. Curr. Biol. 18: 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Copenhaver G. P., Browne W. E., Preuss D., 1998. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl. Acad. Sci. USA 95: 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornah J. E., Germain V., Ward J. L., Beale M. H., Smith S. M., 2004. Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J. Biol. Chem. 279: 42916–42923 [DOI] [PubMed] [Google Scholar]
- Davies R. T., Goetz D. H., Lasswell J., Anderson M. N., Bartel B., 1999. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P. J., 2007. MONODEHYROASCORBATE REDUCTASE4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 19: 1376–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P. J., Germain V., Lange P. R., Bryce J. H., Smith S. M., et al. , 2000a. Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl. Acad. Sci. USA 97: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P. J., Hooks M., Graham I. A., 2000b. The Arabidopsis acyl-CoA oxidase gene family. Biochem. Soc. Trans. 28: 95–99 [PubMed] [Google Scholar]
- Fahnenstich H., Scarpeci T. E., Valle E. M., Flugge U. I., Maurino V. G., 2008. Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol. 148: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y., Nashiro C., Miyata N., Tamura S., Okumoto K., 2012. New insights into dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p in shuttling of PTS1-receptor Pex5p during peroxisome biogenesis. Biochim. Biophys. Acta 1823: 145–149 [DOI] [PubMed] [Google Scholar]
- Gabaldon T., Snel B., Van Zimmeren F., Hemrika W., Tabak H., et al. , 2006. Origin and evolution of the peroxisomal proteome. Biol. Direct 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain V., Rylott E. L., Larson T. R., Sherson S. M., Bechntold N., et al. , 2001. Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid β-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J. 28: 1–12 [DOI] [PubMed] [Google Scholar]
- Graham I. A., 2008. Seed storage oil mobilization. Annu. Rev. Plant Biol. 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Grimm I., Saffian D., Platta H. W., Erdmann R., 2012. The AAA-type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1823: 150–158 [DOI] [PubMed] [Google Scholar]
- Haughn G. W., Somerville C., 1986. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204: 430–434 [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M., 1998. 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Nito K., Toriyama-Kato K., Kondo M., Yamaya T., et al. , 2000. AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19: 5701–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Yagi M., Nito K., Kamada T., Nishimura M., 2005. Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J. Biol. Chem. 280: 14829–14835 [DOI] [PubMed] [Google Scholar]
- Helm M., Lück C., Prestele J., Hierl G., Huesgen P. F., et al. , 2007. Dual specificities of the glyoxysomal/peroxisomal processing protease DEG15 in higher plants. Proc. Natl. Acad. Sci. USA 104: 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoseki J., Ushioda R., Nagata K., 2010. Mechanism and components of endoplasmic reticulum-associated degradation. J. Biochem. 147: 19–25 [DOI] [PubMed] [Google Scholar]
- Hu J., Baker A., Bartel B., Linka N., Mullen R. T., et al. , 2012. Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. A., Gould S., Deluca M., Subramani S., 1987. Firefly luciferase is targeted to peroxisomes in mammalian cells. Proc. Natl. Acad. Sci. USA 84: 3264–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan B. R., Zolman B. K., 2010. pex5 Mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis. Plant Physiol. 154: 1602–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski L. A., Randall D. D., 1988. Purification and characterization of a novel NADPH(NADH)-dependent hydroxypyruvate reductase from spinach leaves: comparison of immunological properties of leaf hydroxypyruvate reductases. Biochem. J. 250: 145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller A., Snyder W. B., Faber K. N., Wenzel T. J., Rangell L., et al. , 1999. Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J. Cell Biol. 146: 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last R. L., Fink G. R., 1988. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240: 305–310 [DOI] [PubMed] [Google Scholar]
- Lazarow P. B., 2006. The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta 1763: 1599–1604 [DOI] [PubMed] [Google Scholar]
- Leidhold C., Voos W., 2007. Chaperones and proteases–guardians of protein integrity in eukaryotic organelles. Ann. N. Y. Acad. Sci. 1113: 72–86 [DOI] [PubMed] [Google Scholar]
- Li F., Vierstra R. D., 2012. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Lingard M. J., Bartel B., 2009. Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 151: 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard M. J., Monroe-Augustus M., Bartel B., 2009. Peroxisome-associated matrix protein degradation in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 4561–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima M., Yokoi H., Asahi T., 1988. Evidence for no proteolytic processing during transport of isocitrate lyase into glyoxysomes in castor bean endosperm. Plant Cell Physiol. 29: 381–384 [Google Scholar]
- Manjithaya R., Nazarko T. Y., Farre J. C., Subramani S., 2010. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 584: 1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T., Otera H., Fujiki Y., 2000. Disruption of the interaction of the longer isoform of Pex5p, Pex5pL, with Pex7p abolishes peroxisome targeting signal type 2 protein import in mammals: study with a novel PEX5-impaired Chinese hamster ovary cell mutant. J. Biol. Chem. 275: 21715–21721 [DOI] [PubMed] [Google Scholar]
- Meinecke M., Cizmowski C., Schliebs W., Kruger V., Beck S., et al. , 2010. The peroxisomal importomer consitutes a large and highly dynamic pore. Nat. Cell Biol. 3: 273–277 [DOI] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M., 1998. A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 14: 381–385 [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus M., Ramón N. M., Ratzel S. E., Lingard M. J., Christensen S. E., et al. , 2011. Matrix proteins are inefficiently imported into Arabidopsis peroxisomes lacking the receptor-docking peroxin PEX14. Plant Mol. Biol. 77: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M. M., Neff J. D., Chory J., Pepper A. E., 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Normanly J., Grisafi P., Fink G. R., Bartel B., 1997. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9: 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L. J., Ettinger W. F., Damsz B., Matsudaira K., Webb M. A., et al. , 1993. Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell 5: 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer O., Kato Y., Adam Z., Sakamoto W., 2007. Multiple intracellular locations of Lon protease in Arabidopsis: evidence for the localization of AtLon4 to chloroplasts. Plant Cell Physiol. 48: 881–885 [DOI] [PubMed] [Google Scholar]
- Osumi T., Tsukamoto T., Hata S., Yokota S., Miura S., et al. , 1991. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem. Biophys. Res. Commun. 181: 947–954 [DOI] [PubMed] [Google Scholar]
- Platta H. W., El Magraoui F., Schlee D., Grunau S., Girzalsky W., et al. , 2007. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177: 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H. W., Magraoui F. E., Baumer B. E., Schlee D., Girzalsky W., et al. , 2009. Pex2 and Pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol. Cell. Biol. 29: 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I., Cornah J. E., Smith S. M., 2007. Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J. 50: 381–390 [DOI] [PubMed] [Google Scholar]
- Ramón N. M., Bartel B., 2010. Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol. Cell. Biol. 21: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzel S. E., Lingard M. J., Woodward A. W., Bartel B., 2011. Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic 12: 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Ma C., Lemke S., Babujee L., 2004. AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 136: 2587–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Babujee L., Ma C., Wienkoop S., Siemsen T., et al. , 2007. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19: 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell-Steven A., Stein K., Amoros M., Landgraf C., Volkmer-Engert R., et al. , 2005. Identification of a novel, intraperoxisomal PEX14-binding site in PEX13: association of PEX13 with the docking complex is essential for peroxisomal matrix protein import. Mol. Cell. Biol. 25: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter A., Fourcade S., Ripp R., Mandel J. L., Poch O., et al. , 2006. The evolutionary origin of peroxisomes: an ER-peroxisome connection. Mol. Biol. Evol. 23: 838–845 [DOI] [PubMed] [Google Scholar]
- Schumann H., Huesgen P. F., Gietl C., Adamska I., 2008. The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant J. 148: 1847–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos T. C., Hangarter R. P., 1990. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Bartel B., 2011. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 4: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Monroe-Augustus M., Bartel B., 2008. The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L., Culler Hendrickson A., Cohen J., Bartel B., 2010. Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 153: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Wheeler D. L., Christensen S. E., Berens J. C., Cohen J. D., et al. , 2011. Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23: 984–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels B. W., Gould S. J., Bodnar A. G., Rachubinski R. A., Subramani S., 1991. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 10: 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S., Erdmann R., 2006. Peroxisomal matrix protein receptor ubiquitination and recycling. Biochim. Biophys. Acta 1763: 1620–1628 [DOI] [PubMed] [Google Scholar]
- Van Den Bosch H., Schutgens R. B., Wanders R. J., Tager J. M., 1992. Biochemistry of peroxisomes. Annu. Rev. Biochem. 61: 157–197 [DOI] [PubMed] [Google Scholar]
- Wanders R. J. A., Waterham H. R., 2005. Peroxisomal disorders I: biochemistry and genetics of peroxisome biogenesis disorders. Clin. Genet. 67: 107–133 [DOI] [PubMed] [Google Scholar]
- Willekens H., Chamnongpol S., Davey M., Schraudner M., Langebartels C., et al. , 1997. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 16: 4806–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C., Distel B., 2006. Pex13p: Docking or cargo handling protein? Biochim. Biophys. Acta 1763: 1585–1591 [DOI] [PubMed] [Google Scholar]
- Woodward A. W., Bartel B., 2005. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Bartel B., 2004. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc. Natl. Acad. Sci. USA 101: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Yoder A., Bartel B., 2000. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Silva I. D., Bartel B., 2001. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Monroe-Augustus M., Silva I. D., Bartel B., 2005. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B. K., Nyberg M., Bartel B., 2007. IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 64: 59–72 [DOI] [PubMed] [Google Scholar]
- Zolman B. K., Martinez N., Millius A., Adham A. R., Bartel B., 2008. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180: 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]