Abstract
Obligate pathogenic and endosymbiotic bacteria typically experience gene loss due to functional redundancy, asexuality, and genetic drift. We hypothesize that reduced genomes increase their functional complexity through protein multitasking, in which many genes adopt new roles to counteract gene loss. Comparisons of interaction networks among six bacteria that have varied genome sizes (Mycoplasma pneumoniae, Treponema pallidum, Helicobacter pylori, Campylobacter jejuni, Synechocystis sp., and Mycobacterium tuberculosis) reveal that proteins in small genomes interact with proteins from a wider range of functions than do their orthologs in larger genomes. This suggests that surviving proteins form increasingly complex functional relationships to compensate for genes that are lost.
Keywords: protein–protein interactions, bacterial evolution, genome complexity, protein moonlighting
BACTERIAL genomes display a substantial diversity in size, with sequenced genomes ranging from only 138 kb in Tremblaya precipes to >13,000 kb in Sorangium cellulosum (Schneiker et al. 2007; McCutcheon and Von Dohlen 2011). Based on phylogenetic analyses, lineages harboring the smallest genomes are derived from large-genomed ancestors. Numerous bacterial phyla contain species with highly reduced genomes, indicating that small genomes have arisen multiple times throughout the evolution and diversification of bacteria. The bacterial taxa exhibiting the most extensive gene loss are usually those that form close, often obligate, associations with eukaryotic hosts (McCutcheon and Moran 2012).
The genes eliminated from the genomes of host-associated bacteria are of two general categories: the first includes those genes whose functions are rendered redundant in the nutrient-rich host environment. These superfluous genes, many of which serve in biosynthetic pathways, incur mutations and are eventually eliminated from the genome (McCutcheon and Moran 2012). The second includes genes that are useful, but not essential, and that have become debilitated or inactivated as a result of genetic drift (Wernegreen and Moran 1999; Moran 2002; McCutcheon and Moran 2012). Genetic drift is prevalent in host-associated bacteria because the population bottlenecks that occur during transmission between hosts drastically reduce effective population sizes (Ne) and hence the efficacy of selection acting on these bacteria. As a result, even beneficial genes will accumulate deleterious mutations. Because all bacterial genomes examined to date display a mutational bias toward deletions, all but the most essential genes are removed from contracting genomes.
The combination of relaxed selection and deletional bias together precipitate genome erosion, in which deleterious mutations accumulate and numerous potentially useful genes are inactivated and removed. As a result, the smallest genomes frequently lose genes that are not complemented by their hosts, and many of the proteins that persist evolve at elevated rates and are prone to misfolding (Wernegreen and Moran 1999; Moran 2002; McCutcheon and Moran 2012). The process of gene loss is evident in the genomes of several pathogens, which often lack individual enzymes from otherwise complete metabolic pathways; for example, Mycobacterium tuberculosis lacks a functional α-ketoglutarate dehydrogenase (kdh) despite having a functional tricarboxylic acid cycle (TCA cycle) (Tian et al. 2005; Wagner et al. 2011).
Genome reduction can alter the functional constraints on the genes remaining in the genome. The loss of a gene might disable a pathway, thereby removing constraints on (and expediting the loss of) other genes in the inactivated pathway (Dagan et al. 2006). Alternatively, the function of the lost genes might be complemented by others that can serve as compensatory alternatives (Pollack et al. 2002; Catrein and Herrmann 2011; Wagner et al. 2011). That the TCA cycle remains functional in M. tuberculosis despite the absence of KDH enzyme implies that another protein, which in this case is the multifunctional α-ketoglutarate decarboxylase (KGD), has taken on this compensatory role (Tian et al. 2005; Wagner et al. 2011).
The ability of proteins to “multitask” or “moonlight” to perform new or alternate functions is possibly a widespread phenomenon in bacterial pathogens (Henderson and Martin 2011). Many moonlighting proteins in pathogens are metabolic enzymes that can carry out additional catalytic side activities, or alternatively, completely novel functions, such as binding to unique host proteins (Henderson and Martin 2011). We hypothesize that in pathogens and other host-associated bacteria with contracting genomes, the selective pressures on those remaining genes shift and induce the evolution of multitasking. Several proteins in bacterial pathogens have been shown to take on new activities, apparently to compensate for gene loss (Catrein and Herrmann 2011), and we suspect that as genome sizes decrease, there will be a broad trend to increase the functional repertoire of genes retained in these genomes. By examining the genome-wide protein–protein interaction (PPI) data for six bacterial genomes, we find that the surviving proteins in smaller genomes assume new functions, as reflected in the number and diversity of their interaction partners, thereby increasing their overall functional complexity during the process of genome reduction.
Materials and Methods
We obtained information for all unique, binary PPIs in the genomes of M. tuberculosis, Synechocystis sp., Helicobacter pylori, Campylobacter jejuni, Treponema pallidum, Mycoplasma pneumonia, and Escherichia coli (Rain et al. 2001; Parrish et al. 2007; Sato et al. 2007; Titz et al. 2008; Kuhner et al. 2009; Peregrin-Alvarez et al. 2009; Wang et al. 2010).
For unbiased comparisons of the PPI complexity per protein, we focused on the functional relationships of proteins that are orthologous in all or most genomes. We identified 168 groups of orthologous proteins represented in at least six of the seven genomes [based on (i) inferences by OrthoMCL (Li et al. 2003), using default parameters, and (ii) the identity of the proteins’ biological names], for which PPI information is available in at least one genome (supporting information, Table S1).
We first examined the functional complexity of these broadly conserved proteins based only on their interactions with other broadly conserved proteins. The functional complexity of a particular protein’s interactions with other conserved proteins was assessed as the total number of Gene Ontology (GO) terms for “molecular function” (obtained from Dimmer et al. 2012) assigned to its complete set of conserved interacting partners. For comparison across genomes, we calculated the average functional complexity of interactions for each genome. In addition, average functional complexity of interactions was similarly measured using GO terms for “biological process” of interaction partners. For regressions, we applied the phylogenetic independent contrasts method of Felsenstein (1985), using Mesquite (Maddison and Maddison 2011), to eliminate the nonindependence of data points due to common ancestry. The topology and branch lengths of the phylogenetic model used in the regression analysis were derived from Ciccarelli et al. (2006).
We next examined the net functional complexity of the broadly conserved proteins within each genome. Net functional complexity is measured as the total number of “molecular function” GO terms that are assigned to the complete set of interaction partners, including the nonconserved proteins. A similar measurement of net functional complexity was obtained using the “biological process” GO terms of all proteins that interact with the conserved proteins.
It should be noted that regressions and statistical tests do not include complexity values for E. coli, which deviated greatly from the overall trend (12.48 using molecular function information, and 8.06 using biological process information for interactions between conserved proteins in E. coli). Unlike the other species, PPIs in E. coli have been determined using multiple methodologies, which together offer much higher PPI detection sensitivities. Consequently the E. coli PPI dataset is substantially more comprehensive, and as such, not directly comparable to the other PPI datasets. For example, the average number of conserved interaction partners per conserved protein in E. coli is 6.83 compared to 1.26–3.75 in other genomes (see Figure 1).
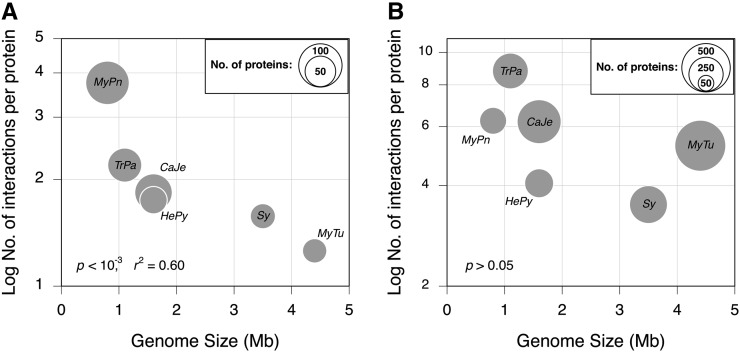
Figure 1 .
Relationship between genome size and the average number of interaction partners for the set of proteins that are broadly conserved across six bacterial genomes. Note that for Y-axis, the number of protein–protein interactions (PPIs) is limited to the set of 168 broadly conserved proteins. A is based on the PPIs that occur only among members of the set of broadly conserved proteins within each genome, whereas B is based on all PPIs recorded for the proteins in this set. In A, circle sizes represent the proportion of the 168 conserved proteins present in the corresponding genome, whereas in B, circle sizes are scaled to represent the total number of proteins in a genome that interact with the protein in the broadly conserved set (Table 1). Mpn, Mycoplasma pneumoniae; Tpa, Treponema pallidum; Hpy, Helicobacter pylori; Cje, Campylobacter jejuni; Sy, Synechocystis; Mtu, Mycobacterium tuberculosis.
The comparisons of the PPIs in these genomes could be affected by the different methodologies that are used to infer PPIs (Table 1) and by the fact that such genome-wide assays often fail to yield the complete set of PPIs (Von Mering et al. 2002; Yu et al. 2008). However, of the 168 conserved orthologs examined in this analysis, the number of proteins whose interactions have been assayed is similar in each genome (Table 1), suggesting that detection biases for this set of genes are negligible.
Table 1 . Features of the bacterial species analyzed.
| Genome | Genome size (Mb) | ORFs in genome | Lifestyle | Assay methoda | Conserved proteins with PPIsb | All proteins interacting with conserved proteins | Reference |
|---|---|---|---|---|---|---|---|
| Mycoplasma pneumoniae | 0.82 | 689 | Obligate intracellular pathogen | TAP-MS | 117 (99) | 153 | Kuhner et al. (2009) |
| Treponema pallidum | 1.13 | 1036 | Obligate intracellular pathogen | Y2H | 119 (61) | 271 | Titz et al. (2008) |
| Helicobacter pylori | 1.67 | 1627 | Obligate pathogen | Y2H | 103 (39) | 177 | Rain et al. (2001) |
| Campylobacter jejuni | 1.64 | 1623 | Obligate pathogen | Y2H | 140 (74) | 410 | Parrish et al. (2007) |
| Synochocystis sp. PCC6803 | 3.57 | 3179 | Freshwater photo and heterotroph | Y2H | 134 (31) | 304 | Sato et al. (2007) |
| Mycobacterium tuberculosis | 4.41 | 4003 | Facultative intracellular pathogen | B2H | 144 (21) | 552 | Wang et al. (2010) |
Y2H, yeast two-hybrid assay; TAP-MS, tandem affinity purification mass spectrometry; B2H, bacterial two-hybrid assay.
Shown are numbers of proteins from the conserved set of 168 proteins for which PPIs have been recognized experimentally. In parentheses are numbers of proteins from this set that have PPIs with others from the conserved set.
Results
Because most proteins function through interactions with multiple other proteins, we examined differences in functional repertoires and complexity of proteins as indicated by the diversity of their interaction partners.
Complexity of interactions between conserved proteins
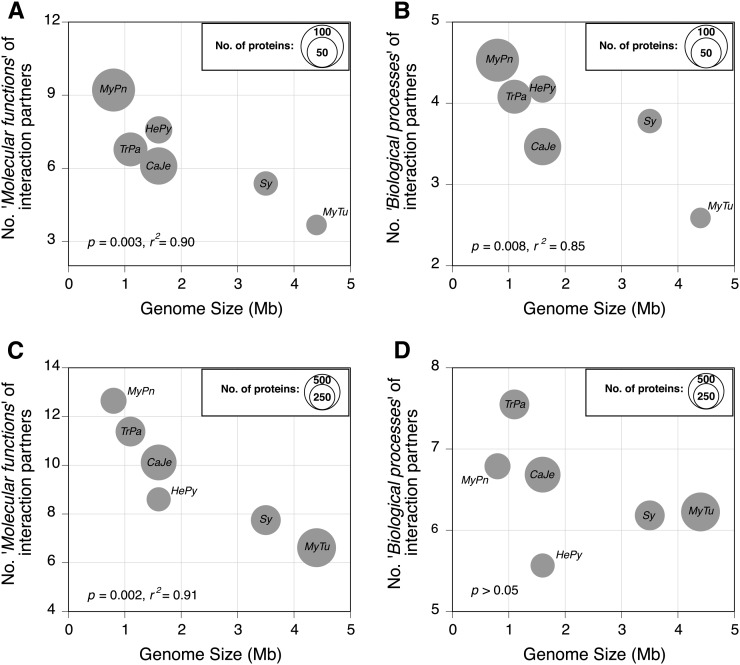
The average number of conserved interaction partners per protein is inversely proportional to the genome size, a trend dominated by the high degree of connectivity of M. pneumoniae proteins (P < 10−3, r2 = 0.60; Figure 1A). When considering the functional complexity of these same proteins as measured by their molecular function information, there is more consistent negative correlation between genome size and average (P = 0.003, r2 = 0.90; Figure 2A). Similarly, there is a negative relationship between genome size and functional complexity, as measured in terms of the number of biological processes in which interaction partners are involved (P = 0.008, r2 = 0.85; Figure 2B).
Figure 2 .
Relationship between genome size and the functional complexity of conserved proteins, as measured by the diversity of “molecular function” (A and C) and “biological process” (B and D) GO terms. A and B are based on the PPIs that occur only among members of the set of broadly conserved proteins within each genome, whereas C and D are based on all PPIs recorded for the proteins in this set. In A and B, circle sizes represent the proportion of the 168 conserved proteins present in the corresponding genome, whereas in C and D, circle sizes are scaled to represent the total number of proteins in a genome that interact with the protein in the broadly conserved set (Table 1). Mpn, Mycoplasma pneumoniae; Tpa, Treponema pallidum; Hpy, Helicobacter pylori; Cje, Campylobacter jejuni; Sy, Synechocystis; Mtu, Mycobacterium tuberculosis.
Although there is commensurately less information about the PPIs within this conserved set of orthologous proteins in the two larger genomes (M. tuberculosis and Synechocystis), the inverse relationships between protein functional complexity and genome size persist, but no longer reaches significance, after removing these two genomes from the analyses.
Net complexity of conserved proteins
The net complexity of conserved proteins was determined from the functional diversity of the complete set of interaction partners. Because smaller genomes encode fewer nonconserved proteins, and will often lose some biological pathways entirely (McCutcheon and Moran 2012), net functional complexity is expected to decrease in proportion to genome size unless the conserved proteins become more functionally diversified.
Genome size is significantly associated with the average number of interaction partners per conserved protein (Figure 1B), and there is no signification correlation between genome size and net functional complexity, as measured using the biological processes of interaction partners (Figure 2D). However, the net functional complexity of conserved proteins, as measured using the molecular function information of their interaction partners, is inversely related to the genome size (P = 0.002, r2 = 0.91; Figure 2C). These trends suggest that conserved proteins in the smaller genomes have diversified functionally in response to gene loss.
Taken together, these trends suggest that proteins in smaller genomes, when compared to their orthologs in larger genomes, are less restrained by their canonical roles and exhibit signals of functional innovation. This increase in functional complexity is accomplished through both the recruitment of new proteins and the reorganization of established interactions.
How might the different experimental procedures used to determine PPI networks (Table 1) affect these results? Such methodological differences might be expected to override or obscure any underlying biological trend. Despite this, we found that for a large set of broadly conserved genes, measures of functional complexity change according to genome size and not according to the experimental method, indicating that methodological biases, if any, do not impact the association.
Discussion
The finding that orthologs in smaller genomes are more functionally complex suggests that as beneficial genes are lost during the course of genome reduction, there is selection on the remaining genes to diversify. Despite the many sporadic cases of protein moonlighting and multitasking that have been reported (Henderson and Martin 2011), we detect broad genome-wide trends toward increasing protein functional diversity in smaller genomes. Moreover, the need for moonlighting activities may be exacerbated in pathogens and symbionts, which are predominantly asexual and unable to acquire genetic material horizontally. Although the question of how genome reductions lead to the changes in protein interactions has been explored previously (Ochman et al. 2007; Tamames et al. 2007; Lercher and Pal 2008; Mendonca et al. 2011), this study addresses whether the proteins remaining in reduced genomes are functionally altered or diversifying to compensate for the gene loss.
Although the new selective pressures instigated by gene loss can promote the functional diversification of proteins in reduced genomes, the genetic drift experienced by these genomes will also influence the evolution of these proteins, as suggested by a recent hypothesis put forward for evolution of complexity in eukaryotes (Fernandez and Lynch 2011). Drift allows the fixation of amino acid replacements that cause slightly deleterious misfolding of the proteins, leading to promiscuous PPIs. Some of these novel interactions may be functionally beneficial, leading to rapid increases in the functional complexity of the proteins (Fernandez and Lynch 2011). For proteins in reduced genomes, this hypothesis predicts a general disposition to misfold, and to be multifunctional, as is true for Mycoplasmas (Wong and Houry 2004; Catrein and Herrmann 2011). In addition to drift, multitasking can also arise from biotic interactions among organisms, as is clear from the host-invasive and immune-system–evasive moonlighting activities of some core metabolic proteins in pathogens (Henderson and Martin 2011).
Nearly all genes in the vast majority of genomes have been assigned function using a comparative method, in which a similarity in gene sequence is equated to a similarity in gene function. This approach relies on the assumption that gene function remains largely unmodified over evolutionary timescales. However, our findings indicate that even broadly conserved proteins in reduced genomes have expanded their functional repertoires in response to gene loss, implying that alignment-based approaches to assigning gene function will vastly underestimate the functional capabilities of genes and genomes. The increase in complexity of genes in reduced genomes is also reflected in other aspects of gene function and regulation in that small-genomed bacteria display levels of structural and transcriptional complexity that have not been observed in larger bacterial genomes (Yu et al. 2008; Guell et al. 2009; Ochman and Raghavan 2009).
At present, the large evolutionary distances between the genomes for which experimental information is available makes it challenging to trace the evolutionary changes occurring in individual proteins in their paths toward functional diversification. However, determination of PPI networks in closely related bacteria of different genome sizes and lifestyles can elucidate the route toward functional diversification. Moreover, resolution of the three-dimensional structures of multiple moonlighting variants of proteins, such as the multitasking forms of glyceraldehyde 3-phosphate dehydrogenase (Henderson and Martin 2011), will divulge the key structural changes that increase functional complexity.
The determination of lineage-specific innovations in protein function is necessary for understanding the evolution and physiology of pathogens. Protein moonlighting can allow pathogens to circumvent some of the current targets for antibiotics; but alternatively, knowledge of pathogen-specific modifications of biochemical processes may help with the development of highly targeted drugs.
Supplementary Material
Acknowledgments
We thank Jeff Lawrence for helpful discussions and comments on the manuscript. This work was supported in part by National Institutes of Health grant GM74738 (to H.O.) and by a grant from the John Templeton Foundation. This work was also supported in part by the facilities and staff of the Yale University Faculty of Arts and Sciences High Performance Computing Center.
Footnotes
Communicating editor: J. Lawrence
Literature Cited
- Catrein I., Herrmann R., 2011. The proteome of Mycoplasma pneumoniae, a supposedly “simple” cell. Proteomics 11: 3614–3632 [DOI] [PubMed] [Google Scholar]
- Ciccarelli F. D., Doerks T., Von Mering C., Creevey C. J., Snel B., et al. , 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287 [DOI] [PubMed] [Google Scholar]
- Dagan T., Blekhman R., Graur D., 2006. The “domino theory” of gene death: gradual and mass gene extinction events in three lineages of obligate symbiotic bacterial pathogens. Mol. Biol. Evol. 23: 310–316 [DOI] [PubMed] [Google Scholar]
- Dimmer E. C., Huntley R. P., Alam-Faruque Y., Sawford T., O’donovan C., et al. , 2012. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 40: D565–D570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., 1985. Phylogenies and the comparative method. Am. Nat. 1: 1–15 [Google Scholar]
- Fernandez A., Lynch M., 2011. Non-adaptive origins of interactome complexity. Nature 474: 502–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guell M., Van Noort V., Yus E., Chen W. H., Leigh-Bell J., et al. , 2009. Transcriptome complexity in a genome-reduced bacterium. Science 326: 1268–1271 [DOI] [PubMed] [Google Scholar]
- Henderson B., Martin A., 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79: 3476–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner S., Van Noort V., Betts M. J., Leo-Macias A., Batisse C., et al. , 2009. Proteome organization in a genome-reduced bacterium. Science 326: 1235–1240 [DOI] [PubMed] [Google Scholar]
- Lercher M. J., Pal C., 2008. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol. Biol. Evol. 25: 559–567 [DOI] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J., Jr, Roos D. S., 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W. P., Maddison D. R., 2011. Mesquite: a modular system for evolutionary analysis. Version 2.75. http://mesquiteproject.org
- McCutcheon J. P., Von Dohlen C. D., 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21: 1366–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon J. P., Moran N. A., 2012. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10: 13–26 [DOI] [PubMed] [Google Scholar]
- Mendonca A. G., Alves R. J., Pereira-Leal J. B., 2011. Loss of genetic redundancy in reductive genome evolution. PLOS Comput. Biol. 7: e1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108: 583–586 [DOI] [PubMed] [Google Scholar]
- Ochman H., Raghavan R., 2009. Systems biology. Excavating the functional landscape of bacterial cells. Science 326: 1200–1201 [DOI] [PubMed] [Google Scholar]
- Ochman H., Liu R., Rocha E. P., 2007. Erosion of interaction networks in reduced and degraded genomes. J. Exp. Zoolog. B Mol. Dev. Evol. 308: 97–103 [DOI] [PubMed] [Google Scholar]
- Parrish J. R., Yu J., Liu G., Hines J. A., Chan J. E., et al. , 2007. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 8: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peregrin-Alvarez J. M., Xiong X., Su C., Parkinson J., 2009. The modular organization of protein interactions in Escherichia coli. PLOS Comput. Biol. 5: e1000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack J. D., Myers M. A., Dandekar T., Herrmann R., 2002. Suspected utility of enzymes with multiple activities in the small genome Mycoplasma species: the replacement of the missing “household” nucleoside diphosphate kinase gene and activity by glycolytic kinases. OMICS 6: 247–258 [DOI] [PubMed] [Google Scholar]
- Rain J. C., Selig L., De Reuse H., Battaglia V., Reverdy C., et al. , 2001. The protein-protein interaction map of Helicobacter pylori. Nature 409: 211–215 [DOI] [PubMed] [Google Scholar]
- Sato S., Shimoda Y., Muraki A., Kohara M., Nakamura Y., et al. , 2007. A large-scale protein protein interaction analysis in Synechocystis sp. PCC6803. DNA Res. 14: 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiker S., Perlova O., Kaiser O., Gerth K., Alici A., et al. , 2007. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 25: 1281–1289 [DOI] [PubMed] [Google Scholar]
- Tamames J., Moya A., Valencia A., 2007. Modular organization in the reductive evolution of protein-protein interaction networks. Genome Biol. 8: R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J., Bryk R., Shi S., Erdjument-Bromage H., Tempst P., et al. , 2005. Mycobacterium tuberculosis appears to lack alpha-ketoglutarate dehydrogenase and encodes pyruvate dehydrogenase in widely separated genes. Mol. Microbiol. 57: 859–868 [DOI] [PubMed] [Google Scholar]
- Titz B., Rajagopala S. V., Goll J., Hauser R., Mckevitt M. T., et al. , 2008. The binary protein interactome of Treponema pallidum–the syphilis spirochete. PLoS ONE 3: e2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Mering C., Krause R., Snel B., Cornell M., Oliver S. G., et al. , 2002. Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417: 399–403 [DOI] [PubMed] [Google Scholar]
- Wagner T., Bellinzoni M., Wehenkel A., O’hare H. M., Alzari P. M., 2011. Functional plasticity and allosteric regulation of alpha-ketoglutarate decarboxylase in central mycobacterial metabolism. Chem. Biol. 18: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Wang Y., Cui T., Zhang C., Yang M., Huang Y., et al. , 2010. Global protein-protein interaction network in the human pathogen Mycobacterium tuberculosis H37Rv. J. Proteome Res. 9: 6665–6677 [DOI] [PubMed] [Google Scholar]
- Wernegreen J. J., Moran N. A., 1999. Evidence for genetic drift in endosymbionts (Buchnera): analyses of protein-coding genes. Mol. Biol. Evol. 16: 83–97 [DOI] [PubMed] [Google Scholar]
- Wong P., Houry W. A., 2004. Chaperone networks in bacteria: analysis of protein homeostasis in minimal cells. J. Struct. Biol. 146: 79–89 [DOI] [PubMed] [Google Scholar]
- Yu H., Braun P., Yildirim M. A., Lemmens I., Venkatesan K., et al. , 2008. High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.