Abstract
A key question in developmental biology addresses the mechanism of asymmetric cell division. Asymmetry is crucial for generating cellular diversity required for development in multicellular organisms. As one of the potential mechanisms, chromosomally borne epigenetic difference between sister cells that changes mating/cell type has been demonstrated only in the Schizosaccharomyces pombe fission yeast. For technical reasons, it is nearly impossible to determine the existence of such a mechanism operating during embryonic development of multicellular organisms. Our work addresses whether such an epigenetic mechanism causes asymmetric cell division in the recently sequenced fission yeast, S. japonicus (with 36% GC content), which is highly diverged from the well-studied S. pombe species (with 44% GC content). We find that the genomic location and DNA sequences of the mating-type loci of S. japonicus differ vastly from those of the S. pombe species. Remarkably however, similar to S. pombe, the S. japonicus cells switch cell/mating type after undergoing two consecutive cycles of asymmetric cell divisions: only one among four “granddaughter” cells switches. The DNA-strand–specific epigenetic imprint at the mating-type locus1 initiates the recombination event, which is required for cellular differentiation. Therefore the S. pombe and S. japonicus mating systems provide the first two examples in which the intrinsic chirality of double helical structure of DNA forms the primary determinant of asymmetric cell division. Our results show that this unique strand-specific imprinting/segregation epigenetic mechanism for asymmetric cell division is evolutionary conserved. Motivated by these findings, we speculate that DNA-strand–specific epigenetic mechanisms might have evolved to dictate asymmetric cell division in diploid, higher eukaryotes as well.
Keywords: Schizosaccharomyces japonicus, fission yeast, mating-type switching, asymmetric cell division, epigenetic differentiation mechanism
THE fission yeast genus Schizosaccharomyces has four known species: Schizosaccharomyces pombe, S. japonicus, S. octosporus, and S. cryophilus. Another fission yeast variant, S. kambucha, is grouped with S. pombe for having a <1% nucleotide sequence difference between them (Singh and Klar 2002; Rhind et al. 2011). The S. japonicus, composed of S. japonicus var. japonicus and S. japonicus var. versatilis varieties, is the most distant from the other three fission yeasts based on their physiology and phylogenic comparison (Helston et al. 2010; Rhind et al. 2011). The S. japonicus cells divide by medial fission, like the well-studied S. pombe cells. Unlike S. pombe, but like other molds, S. japonicus can also form invasive growth filaments in agar. The two types of growth modes result from different nutritional conditions and can be changed by inducing DNA damage (Furuya and Niki 2010). Stressing major differences in their cell biology, S. japonicus partially dissolves nuclear membrane during mitosis and its cell produces eight-spored ascus, while S. pombe undergoes closed mitosis and produces four-spored ascus.
Our knowledge of fission yeast mating-type switching comes from extensive studies conducted for >50 years with S. pombe (reviewed in Leupold 1950; Arcangioli and Thon 2004; Egel 2005; Klar 2007). Its cells exist in a haploid state as one of two cell types, called Plus (P) and Minus (M), with homothallic (called h90) stock cells switch to the opposite mating type efficiently (Leupold 1955; Egel and Eie 1987). Remarkably, of the four “granddaughter” cells derived from a single cell, only one cell switches in nearly 90% of pedigrees. This is called the “one-in-four” granddaughters switching “rule” (Miyata and Miyata 1981; Egel and Eie 1987; Klar 1990). This rule results from asymmetric cell division occurring in each of the two consecutive generations. Such a growth pattern is analogous to the mammalian stem cell asymmetric cell division, in which one daughter cell behaves like the parental cell, while the other daughter cell differentiates to a different cell type. Remarkably, the asymmetric cell division is dictated by the specific daughter cell inheriting template vs. first time synthesized, plus “Watson” vs. “Crick” DNA strand at the mat1 locus from the parental cell (Klar 1987a; Dalgaard and Klar 1999; Dalgaard and Klar 2000). The S. pombe has provided a powerful model system for investigating the chromosomally borne epigenetic mechanism of asymmetric cell division, which might be required for cellular differentiation in higher organisms as well.
The S. pombe mating-type switching process requires three loci, called “cassettes”: mating-type locus (mat1) contains either a P or an M allele, and that determines the cell type, and two “donor” loci. The donor loci region, which is located 15.0 kb away from mat1, contains mat2-P and mat3-M cassettes. The donor cassettes are separated by an 11.0-kb region and 4.3 kb of that consists of centromeric repeat sequences (Grewal and Klar 1997). The donors are used as a source of genetic information and a copy of one of the donor loci is transposed to substitute the mat1 allele resulting in a cell-type switch. The mat1-P allele encodes two mating-type genes, Pc and Pi, and the mat1-M cassette encodes Mc and Mi genes (Kelly et al. 1988). The mat1 locus transcription is induced by nutritional starvation, but both donor loci remain permanently silenced by heterochromatic assembly over the region (Klar 2007). The three cassettes share extensive sequence homology. On the centromeric-proximal side of all three mat cassettes resides a 135-bp homology box (H2), while the other side has a 59-bp homology box 1 (H1). Both mat2-P and mat3-M share an additional 57-bp H3 box next to the H2 box (Egel 2005; Klar 2007). The three cassettes reside in direct orientation and encompass a 30-kb region in the middle of chromosome 2, which also contains genes unrelated to the mating process.
Thus far the S. pombe mating is the only system where the double helical structure of DNA has been demonstrated to confer asymmetric cell division. For some time we have been searching for another biological system that is amenable experimentally to decipher whether or not a DNA-strand–based asymmetric cell division mechanism operates there. The Schizosaccharomyces genome project from the Broad Institute of Harvard University and Massachusetts Institute of Technology, had recently sequenced and annotated the four fission yeast genomes, including S. japonicus var. japonicus (Rhind et al. 2011). Mating-type genes and other loci are conserved among fission yeasts, but the S. japonicus mating-type locus’s structure is not well defined. After the DNA sequence of S. japonicus was known, our study was initiated to determine the pattern of cell-type switching in cell pedigrees, to further define the structure of mating-type loci, and to characterize the mechanism of mat1 switching of S. japonicus. We found that S. japonicus cells switch by following the one-in-four rule, the pattern first discovered in the S. pombe species. The S. japonicus mating-type loci structure showed that the donor loci region is embraced by 2.4-kb inverted repeat sequences. Our comparative analyses of the mating-type loci of the two fission yeasts showed that both use the same strategy of generating a DNA double-strand break (DSB) to initiate cell-type switching despite exhibiting vast variations in mat cassette DNA sequences and their genomic organization. Therefore, these two mating systems represent an incredible example of conservation of the DNA-strand–based epigenetic mechanism of asymmetric cell division in organisms that have diverged far apart during evolution.
Materials and Methods
Strains and culture conditions
The Sj1 S. japonicus var. versatilis culture was kindly provided by J. Hyams (University College, London). This culture contained a mixture of homothallic and other derivatives that failed to undergo meiosis and sporulation. The homothallic Sj4 strain, a prototrophic strain, was derived as an isolate from the Sj1 stock. Heterothallic mutants, Sj2 and Sj3, were isolated as spontaneously generated iodine-staining–negative derivatives of Sj4. We used the culture conditions for S. japonicus research that are employed for research with S. pombe cultures (Moreno et al. 1991). The S. pombe yeast extract adenine (YEA) and pombe minimum adenine (PMA) medium were used for S. japonicus growth and sporulation analyses, respectively (Moreno et al. 1991).
Cloning and sequencing
Partial sequences of the S. japonicus mating-type cassettes, including the Pc, Pi, Mc, and Mi genes and H1 and H2 boxes, were obtained from the Broad Institute database (http://www.broadinstitute.org/annotation/genome/schizosaccharomyces_group/MultiHome.html). Based on the sequence available in the database, our PCR and sequence analysis showed that the structure between mat2-P and mat3-M in our Sj4 isolate was >99% identical to that of the database. The sequences flanking each cassette were cloned by genome walking with an inverse PCR strategy (Ochman et al. 1988) and confirmed by PCR and/or by Southern blot analysis.
Genome DNA preparation and Southern blot analyses
Genome DNA preparation and Southern blot experiments were carried out as described previously (Sambrook et al. 1989; Moreno et al. 1991). The mat1M probe was prepared by amplifying sequences from genomic DNA with mat1 flanking primers AACGCCTCAATATGTCTAAACCAAGTGT and TTAGGGCAGTGTGGCTCTCGGCAT and the H3 probe by using primers TGTTGGAAAACACGGGTGGGATTA and CAATGATCCCTTGTGAGCGTTGC.
Results
S. japonicus var. versatilis follows the one-in-four switching rule
As stated in the Introduction, the S. pombe sister cells exhibit different developmental potential, based strictly on inheritance of specific mat1 DNA strands from the parental cell. Chromosome replication generates differentiated sister chromatids such that one specific member of the two is imprinted. The cell inheriting the template (arbitrarily named) Watson strand-containing chromatid is switching competent, and its sister cell, inheriting the template Crick strand-containing chromatid, is always incompetent (Klar 1987a, 1990, 2007). The difference in strands is the result of the DNA sequence- and strand-specific imprint moiety installed at mat1 (Klar 1987a, 1990). In the next cell division, replication of the imprinted chromosome generates the DSB in the specific chromatid, which initiates recombination required for mat1 switching. Most interestingly, it takes two consecutive cell divisions to complete the entire developmental program of this single-celled organism. Specifically, the one-in-four cell-type switching pattern of S. pombe results from two consecutive asymmetric cell divisions occurring in cell pedigrees. As a result, mating between sister or cousin cells of opposite mating type (due to switching) produces only a single zygote among the four progeny of a single cell while they are growing undisturbed on the surface of solid medium conducive for mating. This observation of producing a single zygote indicates that only one among four granddaughter progeny of a single cell ever switches (Miyata and Miyata 1981; Egel and Eie 1987; Klar 1990). The zygote formation is recorded by light microscopic observations (Miyata and Miyata 1981).
We determined the efficiency and the switching pattern of S. japonicus var. versatilis strain Sj4 cells by quantifying zygote formation among four granddaughter progeny of a single cell. Luckily, unlike S. pombe, S. japonicus cell pairs of opposite mating type readily mate without requiring further growth and starvation on PMA medium. We exploited this property of S. japonicus to determine the efficiency and the pattern of switching, as indicated by mating and zygote formation among progeny of single cells when they multiply on the surface of solid PMA medium. Individual cells were planted on the surface of solid medium by micromanipulation, and the efficiency of zygote formation in the four grandchildren of a cell was recorded. Notably, S. japonicus switched by conforming to the one-in-four rule (Figure 1, A and B) first observed in the switching pattern of S. pombe: 63% of pedigrees produced one zygote and never two zygotes (Figure 1C). Therefore, the S. japonicus’s switching program is similar to that of S. pombe, although the rate of switching is lower (63%) as compared with S. pombe (nearly 90%) (Miyata and Miyata 1981; Egel and Eie 1987; Klar 1990). Because of the vast divergence of DNA sequence, it is not surprising these organisms differ in switching rates. For example, the probability of zygote formation of compatible cells might be reduced in S. japonicus as compared to that of S. pombe under very different growth conditions employed for determining their switching rates. In addition, the efficiency of nonrandom choice of donor loci for mat1 switching, called directionality (Thon and Klar 1993), might vary between these organisms. This switching pattern of S. japonicus is consistent with the notion that two consecutive asymmetric cell divisions cause only one in four granddaughters to switch. However, to rigorously demonstrate that only the cell inheriting the Watson strand is competent for switching, integration of an extra inverted cassette with subsequent analysis of broken molecules and their switching mode is required, just as what was performed decades ago in experiments with S. pombe (Klar 1987b, 1990).
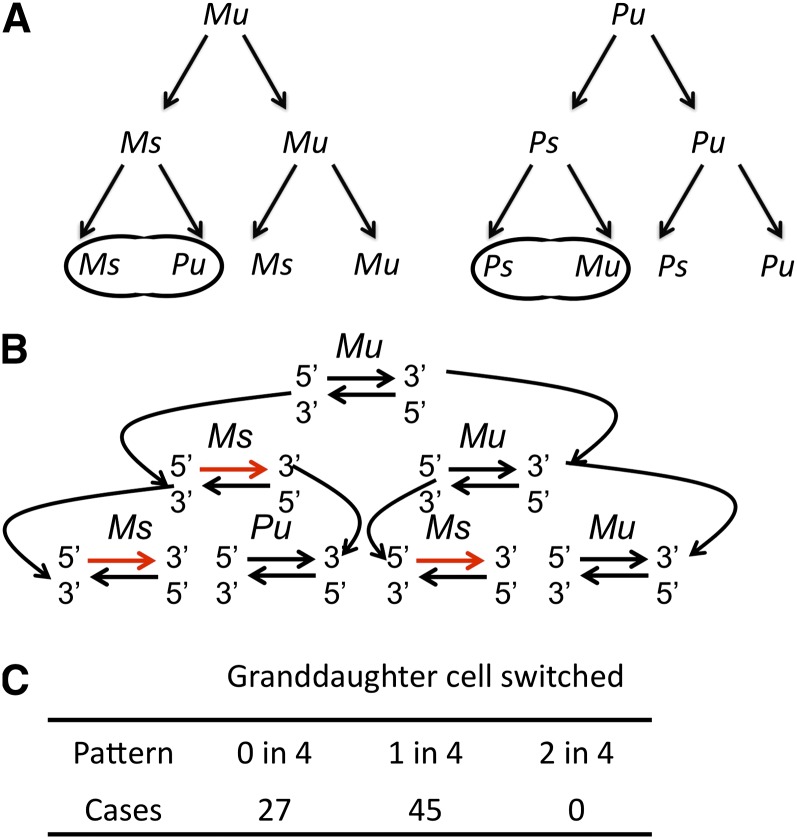
Figure 1 .
The mating-type switching pattern of the S. japonicus Sj4 strain. (A) The drawings depict the one-in-four switching pattern of mat1-M (M) and mat1-P (P) cells. Mu/Pu, unswitchable cell, and of Ms/Ps, switchable cell, were as defined previously for the S. pombe switching system (Klar 1987a). Zygotes formed by mating between the progeny of a single cell at the four-cell stage reflected a single switched cell in the pedigree. (B) The strand-specific imprint and its inheritance by the specific daughter cell diagrammed to depict the pattern of switching in cell pedigrees described in A. Only the M cell switching and strand inheritance pattern is diagrammed, and an equivalent pattern is observed in P cells (not drawn). The imprinted mat1 strand is shown as a red line. The figure was modified from (Dalgaard and Klar 2000). (C) The table lists the number of pedigrees observed with an indicated switching pattern.
Identification and characterization of heterothallic derivatives of S. japonicus
The S. pombe ascospores synthesize starch, while the mitotically growing cells do not. Therefore, homothallic (i.e., switching proficient) colonies, capable of switching, mating, and sporulation, stain black in color when exposed to iodine vapors, and the heterothallic (i.e., non-switching) colonies stain yellow because they do not produce (starch-containing) ascospores (Bresch et al. 1968). The iodine-vapor staining procedure is a powerful simple tool used to evaluate switching ability of cells contained in individual colonies. We found that Sj4 colonies growing from ascospores in PMA (growth and sporulating medium) also stained black (Figure 2A). This result showed that the colony-staining phenotype is stable both in mitosis and in meiosis in our stock of S. japonicus var. versatilis.
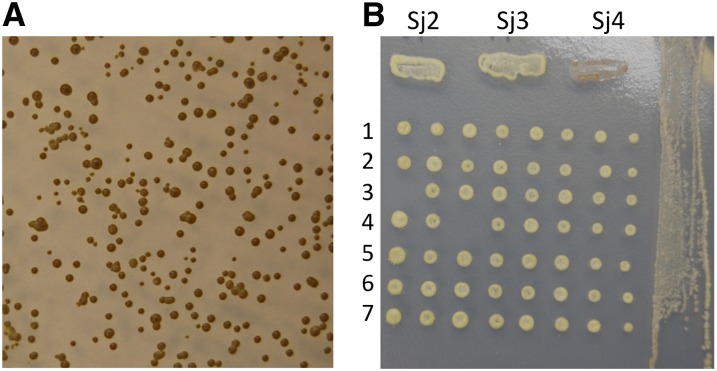
Figure 2 .
The iodine-staining phenotype of S. japonicus var. versatilis. (A) Picture of iodine-staining phenotype of colonies originating from random meiotic spores of the Sj4 strain. (B) Picture of iodine-staining phenotype of segregants of the Sj2 × Sj3 cross. The eight segregants from individual octads were arranged in a horizontal row on YEA growth medium by micromanipulation. The colonies of segregants dissected from seven asci are shown. After growth for 3 days, they were replicated onto plates containing PMA medium. After 3 days of growth and sporulation there, iodine-staining phenotype of colonies and culture patches was photographed. At the top, patches of heterothallic Sj2, Sj3, and homothallic Sj4 stocks are shown for comparing their staining phenotype.
To further characterize the mating-type switching phenomenon of this organism, we isolated nonswitching derivatives of it. Two spontaneous mutants from Sj4, named Sj2 and Sj3 (presumably nonswitching, called heterothallic), stained yellow when exposed to iodine vapors (Figure 2B). While observed by light microscopy, neither derivative produced asci under appropriate growth medium conditions. The reason for their switching defect was investigated. The Sj2 and Sj3 cells could mate with each other, indicating that these strains possess opposite mating types. Each ascus produced by the cross contained eight ascospores. The ascus dissection showed that all segregants were of heterothallic type because they stained yellow when exposed to iodine vapors. Segregants of two octads were tested for mating capability with the Sj2 and Sj3 parental stocks. Both octads showed the Mendelian segregation pattern for mating type: four mated with Sj2, and the other four mated with Sj3. Thus, the Sj2 and Sj3 stocks were judged to be stable heterothallic strains with opposite mating types. A previous report indicated that some homothallic S. japonicus var. japonicus colonies show light iodine staining, but their staining was not correlated with their mating-type switching property (Furuya and Niki 2009). We presume that S. japonicus var. versatilis we used for our studies differs from S. japonicus var. japonicus in the iodine-staining phenotype. The iodine-staining property has been an extremely useful tool for research with S. pombe cellular differentiation and it should make the S. japonicus var. versatilis a very good alternative model system for further investigating the evolution of the mechanism of cellular differentiation in general. To exploit the iodine-staining feature of our stock for future studies, we thought it necessary to first characterize the structure of mating-type cassettes of S. japonicus var. versatilis and compare it to those from the recently described S. japonicus var. japonicus strain (Furuya and Niki 2009; Rhind et al. 2011).
Structure of the S. japonicus mating-type genes
Overall, mating-type genes have been conserved across the fission yeast group, but only partial mat2-P and mat3-M sequences for S. japonicus were available from the Broad Institute database (Rhind et al. 2011). Thus far, the mat1 locus sequence has not been defined altogether, but Southern blot analysis suggests that it likely exists in the genome (Rhind et al. 2011). To characterize the molecular structure of all the cassettes, we cloned three mat cassettes using PCR-based methods and assembled the donor loci region from cloned fragments of the S. japonicus var. versatilis strain (Figure 3A; accession nos. JQ735907 and JQ735908). Next, we performed Southern blot analysis by digesting genomic DNA with multiple restriction enzymes to further validate the structure we produced from our cloned sequences and from the information derived from the fission yeast genomic database of S. japonicus var. japonicus strain. The results verified our assembled contigs (Figure 3, B and C). By following the definitions of mating-type cassettes structure of S. pombe (Beach 1983; Beach and Klar 1984; Kelly et al. 1988), we designate the 81 bp as the homology box H1, and the 418-bp sequence that flanks each of the three mating-type cassettes as H2. Another 2014-bp homology sequence, extending from the H2 boxes of both mat2-P and mat3-M donor loci cassettes, was named H3. Overall, the cell-type switching involves substitution events between the 1.2-kb mat1 P-specific and the 1.1-kb mat1 M-specific sequences.
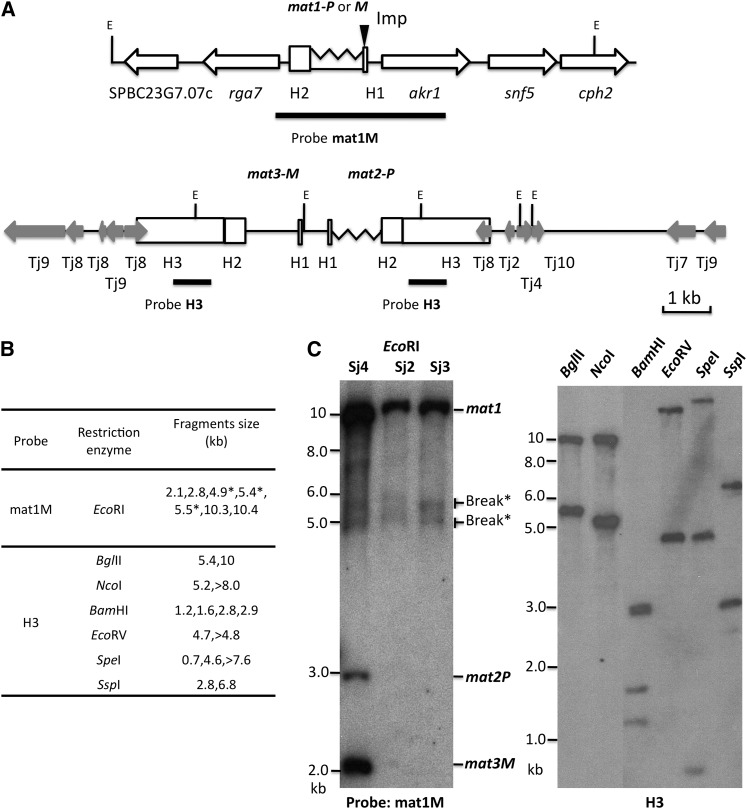
Figure 3 .
The structure of S. japonicus mating-type loci. (A) Diagrams of the mat1 and donor loci: Sequence accession nos. JQ735907 and JQ735908. The proposed Imprint site (Imp) located at the end of the H1 element is indicated by a solid triangle. The probes used for Southern blot experiments (mat1M and H3) are shown in boldface lines. The physical distance and orientation of mat1 with respect to the donor loci are unknown. The EcoRI restriction enzyme sites are indicated with the letter E. The S. pombe genes situated around mat1 are indicated with open arrows. The S. japonicus retrotransposon homologs, with nucleotides identity >70%, that flank the donor loci, are shown with shaded arrows. (B) Sizes of the predicted Southern blot bands are listed. The bands resulting from the DSB due to mat1 imprint in a fraction of DNA molecules are marked with *. (C) Southern blot analysis of the mating-type loci. The genomic DNA was digested with EcoRI (left) and hybridized with the mat1M probe. The mat cassette fragment sizes are indicated. Right shows hybridization with the H3 probe. The size markers are indicated on the left side of both Southern blot panels.
The gene order of chromosomes is largely conserved within the fission yeast groups (Rhind et al. 2011). We determined the S. japonicus gene order of our cloned mat1-region sequences. The S. japonicus Rho-GTPase–activating protein 7 encoding gene (rga7) is located on the H2-element side of mat1, similar to other fission yeasts. However, akr1, snf5, and cph2 genes, located on the mat1 H1 side, are not linked to mat1 in the other three yeasts; they are located on chromosome 1 of S. pombe.
In an arrangement that is fundamentally different from that of S. pombe (Kelly et al. 1988; Grewal and Klar 1997), the mat2-P and mat3-M genes of S. japonicus exist in the opposite orientation, separated by only a short 0.7-kb region, a configuration similar to that of S. octosporus and S. cryophilus (Rhind et al. 2011). Compared to the 57-bp length of S. pombe H3 homology element, the S. japonicus H3 is 2.4-kb long and consists of an unrelated DNA sequence. The S. pombe donor loci region is flanked by the 2.1-kb inverted repeats (called IL-R and IL-L) that function as barriers for spreading heterochromatin emanating from the mat2–mat3 region (Noma et al. 2001; Singh and Klar 2002; Thon et al. 2002). The S. japonicus did not have other long, inverted repeat elements within our cloned donor loci sequences. Perhaps the 2.4-kb inverted repeat sequence within the H3 box in S. japonicus helps maintain both mat2 and mat3 genes in the silenced state. Furthermore, the sequences flanking the donor cassettes are rich in repeat sequences, which may also impose silencing on this region. Most of the repeats flanking the donor loci (Figure 3B) belong to the group of gypsy-type Tj1–Tj10 retrotransposon elements (Rhind et al. 2011). These elements are confined to the centromeric and telomeric regions in S. japonicus. The S. japonicus mat1 locus is genetically linked to the centromere and to donor loci (Furuya and Niki 2009; Rhind et al. 2011). The donor loci may also be located within the centromeric regions of other fission yeasts. Thus, the overall mating-type cassette structure, DNA sequence, and the genomic location of these newly sequenced fission yeasts are very much different from those of the S. pombe species. The location of the mat2 and mat3 donor loci within the centromere of S. japonicus is intriguing. Perhaps this location promotes both donor loci gene silencing and facilitates their recombinational interaction for mat1 gene conversion, which is required for mat1 switching. Our study describes the structure of all three cassettes of S. japonicus, while the previous study only described mat2-P and mat3-M loci (Rhind et al. 2011). In addition, comparing our results presented in Figure 3A, the previous study (Rhind et al. 2011) misassembled mat1 and donor loci together perhaps due to repetitive H2 sequences existing in all cassettes.
Sj2 and Sj3 heterothallic derivatives lack donor loci
Our mating-type loci structure described above was defined from the homothallic Sj4 strain. Using PCR analysis, we found that our heterothallic Sj2 strain contains mat1-P, and Sj3 contains mat1-M information, and both strains lack the mat2 and mat3 loci. Southern analysis with the mat1M probe and additionally with the H3 sequence confirmed that the donor loci information was missing in both heterothallic strains (Figure 3C). Spontaneous recombination events in repeated sequences flanking the mat2–mat3 region in Sj4 stock had likely generated our heterothallic derivatives. The donor loci have been deleted in our heterothallic strains, a result similar to the independently derived and previously described heterothallic derivatives of S. japonicus var. japonicus strain (Furuya and Niki 2009). Since our S. japonicus var. versatilis Sj4 colonies stain black with the iodine-vapor staining procedure (Figure 2A), it was very easy for us to isolate our Sj2 and Sj3 heterothallic derivatives by screening for nonstaining colonies from the stock. In contrast, isolation of heterothallic derivatives from iodine-vapors nonstaining S. japonicus var. japonicus strain required microscopic observations with each of about 20,000 colonies (Furuya and Niki 2009).
S. japonicus cells generate imprint at the H1 box
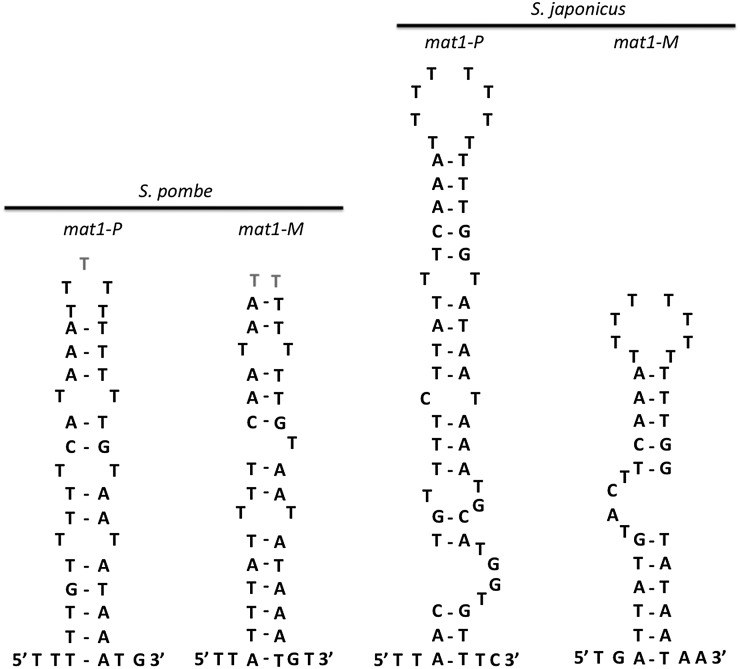
Located in the T-rich sequence at the mat1:H1 junction, the imprint site in S. pombe is sensitive to alkaline treatment and generates the DSB during genomic DNA extraction from yeast cells (Beach and Klar 1984; Kelly et al. 1988; Nielsen and Egel 1989; Kaykov and Arcangioli 2004; Vengrova and Dalgaard 2006). The same T-rich region is also present at the equivalent position in S. japonicus and is conserved across the fission yeast group (Figure 4). On the Southern blot, the sizes of weak bands (identified by * in Figure 3, B and C) matched a predicted DNA break at the T-rich region. Previous work with S. pombe hypothesized that the palindrome-like sequence around the imprint site (Figure 5) is potentially important for imprint formation (Nielsen and Egel 1989; Vengrova and Dalgaard 2004). Although recent mutagenesis analysis showed that the palindrome structure is not essential for imprint formation (Sayrac et al. 2011), we also found a similar palindrome-like structure around the presumed imprint site in S. japonicus (Figure 5). The significance of this structure for mat1 switching would require further analysis.
Figure 4 .
Alignment of 300-bp sequence flanking the H1 region of four fission yeast species. SP, S. pombe; SJ, S. japonicus var. japonicus; SC, S. cryophilus; and SO, S. octosporus. The highly conserved nucleotides are identified by *. The S. pombe H1, SAS1α, and SAS1β sites are depicted in boldface type and shaded letters. The S. pombe SAS2 region is indicated by the italicized bases. The three motifs (mut3, mut5, and mut7) required for efficient mat1 switching (Kaykov et al. 2004) are underlined.
Figure 5 .
The palindromic structures near the imprint sites of S. pombe and S. japonicus. Mfold (Zuker 2003) software was used for the structure analysis in which the G–T pairing is permitted. The S. pombe structure was adapted from Nielsen and Egel (1989).
The Sap1 (switch-activating protein 1) binding site, called SAS site, located near the S. pombe H1 region, is required for efficient mat1 switching (Arcangioli and Klar 1991). A highly diverged potential SAS site is also found near the mat1 in S. octosporus and S. cryophilus (Rhind et al. 2011). Additionally, a few motifs located within the H1 region were identified by mutagenesis and proposed to control replication pause at the mat1 pause site 1 (MPS) and/or to protect the imprint site from DNA repair (Kaykov et al. 2004). An ∼300-nt long sequence from the S. japonicus, S. pombe, S. octosporus, and S. cryophilus species, including the H1 and mat1 flanking sequence, was aligned for sequence comparison (Figure 4). Most parts of the H1 region and the SAS1β motif exhibited high conservation between them. Three previously identified mutation motifs (mut3, mut5, and mut7) (Kaykov et al. 2004) partially overlap with the conserved sequence motifs (Figure 4). Some very short, three to four conserved nucleotides were found within the mapped SAS2 region (Arcangioli and Klar 1991). We did not find significant sequence conservation of S. japonicus H2 and H3 regions with the three other yeasts. The observation of conserved H1 and other nearby located motifs suggests that they play an important role for the imprint synthesis. More likely, the four fission yeasts use the same replication-coupled mechanism to form the DSB that initiates mat1 switching, despite the vastly different structure of mating-type cassettes and physiology (e.g., some produce four-spored ascus, while others produce eight-spored ascus) of these organisms. Furthermore, conservation of the switching mechanism in these organisms is all the more telling (see Discussion) given their low level of DNA sequence conservation. For example, between the S. pombe and S. japonicus cassettes, Furuya and Niki (2009) noted 27% identity with mat-Pi, 53% identity with mat-Pc, and 29% identity with mat-Mc genes.
The imprint likely induces meiotic mat1 gene conversion in S. japonicus
The imprint in S. pombe was shown to initiate efficient mat1 gene conversion in crosses of donor-deleted strains (Klar and Miglio 1986): ∼20% of tetrads produced 3:1 type of mat1 locus conversions through recombination between homologs. Thus, the imprint, which is used usually for mat1 switching in somatic cells, efficiently initiates meiotic mat1 gene conversion. Such meiotic analysis was crucial to genetically identify the mat1 linked imprint moiety, and the genes that perform the imprint, through conventional meiotic analysis (Klar and Bonaduce 1993). These genetic results initially led to the suggestion of the DNA strand-segregation mechanism (Figure 1B) as a mechanism for explaining the one-in-four switching pattern observed in cell pedigrees (Klar 1987a, 1990).
Considering the point of vast evolutionary divergence between the two yeasts, does the imprint installed in mitosis induce meiotic mat1 gene conversion in S. japonicus as well? For that, we crossed our Sj2 and Sj3 heterothallic donor-deleted strains and subjected it to octad dissection analysis. In our cross only the mat1 marker was segregating because the stock is not known to have auxotrophic markers. Should we observe “aberrant” meiotic mat1 segregation, it would have been difficult to assess whether they resulted from bona fide gene conversions or from erroneously mixing of spores of different octads during their dissection. To avoid this complication, we first picked intact zygotic asci and dissected them after 4 hr once they had released their ascospores due to spore germination. If aberrant segregation events were observed, they should be ascribed to gene conversion recombination process. With this strategy, we analyzed 166 octads: 158 were of the 4P:4M type, three of 6P:2M, and five of the 2P:6M type. Thus, 4.8% of octads gene converted mat1 allele in both directions. In our limited analysis we did not find any case of the 5:3 type of postmeiotic segregation, perhaps because >1.1-kb heteroduplex between mat1 sequence does not escape correction in meiosis, alternatively, such a heteroduplex intermediate is not formed during switching. We concluded that the imprint likely induces meiotic mat1 conversion in S. japonicus, but at a reduced rate from that found in S. pombe (20%).
The mat cassettes are tightly linked genetically
In our cross of donor-deleted strains, mat1 conversion can occur only through mat1 to mat1 interaction between homologous chromosomes. Results of a genetic cross where only one of the parents was deleted for both donor loci, mat1-P (donor-deleted) X mat2-P− donor locus mutant, was reported previously (Furuya and Niki 2009): among 78 octads analyzed, two had changed mat1-P to mat1-M in donor-deleted segregants. The authors attributed these two cases to mat1 gene conversion events occurring by interaction of mat1-P in one chromosome with mat1-M and/or mat3-M information residing in the homolog. However, crossover events occurring in the mat1 and donor loci interval can also explain their origin. By following the latter possibility, we calculate that mat1 and donor loci are genetically linked tightly, separated only by ≤1.3 (2/(2 × 78)) cM meiotic distance.
Discussion
Multiple mechanisms of asymmetric cell division are best described in model organisms, such as worms, flies, and yeasts (Gonczy 2008). Our more extensive analyses of the structure of mating-type cassettes of S. japonicus var. versatilis species confirm findings and extend those of the recently published studies with S. japonicus var. japonicus (Furuya and Niki 2009; Rhind et al. 2011). In addition, we describe here the pattern of switching in cell pedigrees and define the epigenetic mechanism of asymmetric cell division of this organism and compare it to that of the well-studied S. pombe system. Overall, our study provides important insight into the evolution of epigenetic mechanisms underlying asymmetric cell division. Because of the experimentally very useful iodine-staining feature of S. japonicus var. versatilis we describe and exploit here in our study (Figure 2A), this organism should become a highly experimentally tractable model for exploring the evolution of molecular bases of asymmetric cell division and gene silencing. Such species-specific comparative evolutionary analysis is crucial for fully understanding strategies evolved to accomplish asymmetric cell division underpinning the regulation of form in multicellular organisms. Knowledge gained from comparison of mechanisms of two haploid yeasts motivates us to advance implications of our work concerning developmental biology of higher organisms.
Asymmetric cell division is crucial to produce diversity during development and to produce self-renewing stem cells. Prior to this study, the S. pombe mat1 switching had been the only example in which DNA strand–specific imprinting was established to produce differentiated sister chromatids, constituting a unique mechanism of asymmetric cell division (Klar 1987a, 1990). Our work, presented here, suggests that a parallel DNA-strand–based mechanism likely operates in the highly diverged S. japonicus species. Indeed, the genome of S. pombe contain 36% GC while that of S. japonicus contain 44% GC. The two fission yeast show 55% amino acid identity between protein orthologs, a divergence similar to that between humans and the cephalochordate amphioxus (Rhind et al. 2011). Such a conservation of the asymmetric cell division mechanism is remarkable given that these organisms have diverged so far apart in evolution. In contrast, the mother–daughter cell differentiation bias in an evolutionarily distinct budding yeast, Saccahromyces cerevisiae, is not due to inheritance of specific strands of DNA at the HO gene (Klar 1987b). Hence, two among these three systems specifically examined for the existence of DNA-strand–based mechanism produced asymmetric cell division through DNA structure and the replication history of DNA strands at the specific locus. Notably, cellular differentiation is coordinated with the progression of cell cycle in the parental cell in all these yeast-mating systems, a feature likely to be very important in biology at large. This analysis raises the obvious question: How general is such a simple but elegant mechanism of cellular differentiation simply based on the double-helical structure of DNA? We note that determining the existence of such a mechanism, especially in diploid multicellular organisms, is impossible because it requires identification of inherited strands of specific chromosome(s) by specific daughter cells during asymmetric cell division of individual cells during embryogenesis. Therefore, it is unlikely that such a mechanism would have been discovered thus far in investigations of higher organisms. As a result, controversial models invoking distribution of morphogen gradients are instead proposed to control cellular differentiation and organogenesis in higher organisms, but lately cell intrinsic models have been favored (Vandenberg and Levin 2010).
We emphasize that generating asymmetric cell division in fission yeast, which is a haploid and single-celled eukaryotic organism, does not require selective segregation of sister chromatids of a specific chromosome to a specific daughter cell. Here, the daughter cell simply acquires the cell fate epigenetic determinant borne by the chromosome it inherits from the parental cell, and acts accordingly. However, for such a mechanism to function in a diploid organism, selective segregation of differentiated sister chromatids of both homologs of a specific chromosome (or a specific set of chromosomes perhaps varying by the cell type) to specific daughter cells at a specific cell division is necessarily required. Genetic evidence in support of such a strand-specific imprinting and selective chromatid segregation (the SSIS) model as a general mechanism for asymmetric cell division, and specifically to explain vertebrates’ visceral organ and human brain hemispheric laterality and psychoses development, has been presented (Klar 1994, 2004, 2008; Armakolas and Klar 2007; Furusawa 2011). In studies of model systems, such as flies and worms, it is well documented that mechanisms controlling cellular polarity and spindle positioning at the time of cell division are coupled with asymmetric cell division (Gonczy 2008). Nearly all studies have invoked unequal carryover or stability of proteins, RNA, or unequal microenvironmental exposure of sister cells as mechanisms of asymmetric cell division. However, such hypotheses only move the question a step back aspiring to search for the primary determinant of asymmetric cell division. To confer a particular epigenetic state of gene expression on sister cells, all such mechanisms might be required ultimately to facilitate nonrandom chromatid partitioning in mitoses. Indeed, a model identical to the SSIS model has been recently proposed for the development of neuronal laterality in the Caenorhibitis elegans worm (Nakano et al. 2011). Discovery of the DNA-strand–based mechanism is most satisfying because it highlights the identity of the primary basis of developmental asymmetry, of being physical in nature, not based on differential regulation of regulators of differentiation. Moreover, by hypothesis, the differentiation program of daughter cells is a carryover of decisions taken by the mother cell. In sum, our study provides a second example of an organism that exploits the inherent chirality of the double-helical structure of DNA to achieve asymmetric cell division. Future studies should be initiated to investigate whether an analogous epigenetic mechanism of cellular differentiation ever operates in muticellular organisms. Moreover, the work described here should form the basis of future research for defining the mechanism of asymmetric cell division and gene silencing in this model organism highly amenable to classical genetics and molecular biology tools. New episomal vectors and efficient transformation procedure for S. japonicus have been developed (Aoki et al. 2010). More recently, a collection of mutant strains tagged with selection markers in the entire S. japonicus genome and a cosmid library were generated (Furuya et al. 2012). The developed tools would greatly help future investigations to identify mating-type switching genes and to elucidate the mechanism of mating-type differentiation in S. japonicus.
Acknowledgments
We thank Jeremy Hyams for providing the Sj1 stock to us. The Intramural Research Program of the National Institutes of Health, Frederick National Laboratory for Cancer Research supports this research.
Footnotes
Communicating editor: A. Houben
Literature Cited
- Aoki K., Nakajima R., Furuya K., Niki H., 2010. Novel episomal vectors and a highly efficient transformation procedure for the fission yeast Schizosaccharomyces japonicus. Yeast 27: 1049–1060 [DOI] [PubMed] [Google Scholar]
- Arcangioli B., Klar A. J., 1991. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J. 10: 3025–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B., Thon G., 2004. Mating-types cassettes: structure, switching and silencing, pp. 129–147 in Molecular Biology of Schizosaccharomyces pombe, edited by R. Egel. Springer Verlag, Berlin.
- Armakolas A., Klar A. J., 2007. Left-right dynein motor implicated in selective chromatid segregation in mouse cells. Science 315: 100–101 [DOI] [PubMed] [Google Scholar]
- Beach D. H., 1983. Cell type switching by DNA transposition in fission yeast. Nature 305: 682–688 [Google Scholar]
- Beach D. H., Klar A. J., 1984. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J. 3: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresch C., Muller G., Egel R., 1968. Genes involved in meiosis and sporulation of fission yeast. Mol. Gen. Genet. 102: 301–306 [DOI] [PubMed] [Google Scholar]
- Dalgaard J. Z., Klar A. J., 1999. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400: 181–184 [DOI] [PubMed] [Google Scholar]
- Dalgaard J. Z., Klar A. J., 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102: 745–751 [DOI] [PubMed] [Google Scholar]
- Egel R., 2005. Fission yeast mating-type switching: programmed damage and repair. DNA Repair (Amst.) 4: 525–536 [DOI] [PubMed] [Google Scholar]
- Egel R., Eie B., 1987. Cell lineage asymmetry in Schizosaccharomyces pombe: unilateral transmission of a high-frequency state for mating-type switching in diploid pedigrees. Curr. Genet. 12: 429–433 [Google Scholar]
- Furusawa M., 2011. Implications of double-stranded DNA structure for development, cancer and evolution. Open J. Genet. 1: 78–87 [Google Scholar]
- Furuya K., Niki H., 2009. Isolation of heterothallic haploid and auxotrophic mutants of Schizosaccharomyces japonicus. Yeast 26: 221–233 [DOI] [PubMed] [Google Scholar]
- Furuya K., Niki H., 2010. The DNA damage checkpoint regulates a transition between yeast and hyphal growth in Schizosaccharomyces japonicus. Mol. Cell. Biol. 30: 2909–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K., Aoki K., Niki H., 2012. Construction of an insertion marker collection of Sz. japonicus (IMACS) for genetic mapping and a fosmid library covering its genome. Yeast 29: 241–249 [DOI] [PubMed] [Google Scholar]
- Gonczy P., 2008. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9: 355–366 [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Klar A. J., 1997. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146: 1221–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helston R. M., Box J. A., Tang W., Baumann P., 2010. Schizosaccharomyces cryophilus sp. nov., a new species of fission yeast. FEMS Yeast Res. 10: 779–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaykov A., Arcangioli B., 2004. A programmed strand-specific and modified nick in S. pombe constitutes a novel type of chromosomal imprint. Curr. Biol. 14: 1924–1928 [DOI] [PubMed] [Google Scholar]
- Kaykov A., Holmes A. M., Arcangioli B., 2004. Formation, maintenance and consequences of the imprint at the mating-type locus in fission yeast. EMBO J. 23: 930–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M., Burke J., Smith M., Klar A., Beach D., 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 7: 1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., 1987a Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature 326: 466–470 [DOI] [PubMed] [Google Scholar]
- Klar A. J., 1987b The mother-daughter mating type switching asymmetry of budding yeast is not conferred by the segregation of parental HO gene DNA strands. Genes Dev. 1: 1059–1064 [DOI] [PubMed] [Google Scholar]
- Klar A. J., 1990. The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands. EMBO J. 9: 1407–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., 1994. A model for specification of the left-right axis in vertebrates. Trends Genet. 10: 392–396 [DOI] [PubMed] [Google Scholar]
- Klar A. J., 2004. An epigenetic hypothesis for human brain laterality, handedness, and psychosis development. Cold Spring Harb. Symp. Quant. Biol. 69: 499–506 [DOI] [PubMed] [Google Scholar]
- Klar A. J., 2007. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu. Rev. Genet. 41: 213–236 [DOI] [PubMed] [Google Scholar]
- Klar A. J., 2008. Support for the selective chromatid segregation hypothesis advanced for the mechanism of left-right body axis development in mice. Breast Dis. 29: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Bonaduce M. J., 1993. The mechanism of fission yeast mating-type interconversion: evidence for two types of epigenetically inherited chromosomal imprinted events. Cold Spring Harb. Symp. Quant. Biol. 58: 457–465 [DOI] [PubMed] [Google Scholar]
- Klar A. J., Miglio L. M., 1986. Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell 46: 725–731 [DOI] [PubMed] [Google Scholar]
- Leupold U., 1950. The inheritance of homothally and heterothally in Schizosaccharomyces pombe. C. R. Trav. Lab. Carlsberg Ser. Physiol. 24: 381–480 [Google Scholar]
- Leupold U., 1955. Methods concerning genetics of Schizosaccharomyces pombe. Schweiz. Z. Allg. Pathol. Bakteriol. 18: 1141–1146 [PubMed] [Google Scholar]
- Miyata H., Miyata M., 1981. Mode of conjugation in homothallic cells of Schizosaccharomyces pombe. J. Gen. Appl. Microbiol. 27: 365–371 [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nakano S., Stillman B., Horvitz H. R., 2011. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell 147: 1525–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen O., Egel R., 1989. Mapping the double-strand breaks at the mating-type locus in fission yeast by genomic sequencing. EMBO J. 8: 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K., Allis C. D., Grewal S. I., 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293: 1150–1155 [DOI] [PubMed] [Google Scholar]
- Ochman H., Gerber A. S., Hartl D. L., 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N., Chen Z., Yassour M., Thompson D. A., Haas B. J., et al. , 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Maniatis T., Fritsch E. F., 1989. Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- Sayrac S., Vengrova S., Godfrey E. L., Dalgaard J. Z., 2011. Identification of a novel type of spacer element required for imprinting in fission yeast. PLoS Genet. 7: e1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G., Klar A. J., 2002. The 2.1-kb inverted repeat DNA sequences flank the mat2,3 silent region in two species of Schizosaccharomyces and are involved in epigenetic silencing in Schizosaccharomyces pombe. Genetics 162: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G., Klar A. J., 1993. Directionality of fission yeast mating-type interconversion is controlled by the location of the donor loci. Genetics 134: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G., Bjerling P., Bunner C. M., Verhein-Hansen J., 2002. Expression-state boundaries in the mating-type region of fission yeast. Genetics 161: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg L. N., Levin M., 2010. Far from solved: a perspective on what we know about early mechanisms of left-right asymmetry. Dev. Dyn. 239: 3131–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova S., Dalgaard J. Z., 2004. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 18: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova S., Dalgaard J. Z., 2006. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 7: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]