Abstract
Traditionally microorganisms were considered to be autonomous organisms that could be studied in isolation. However, over the last decades cell-to-cell communication has been found to be ubiquitous. By secreting molecular signals in the extracellular environment microorganisms can indirectly assess the cell density and respond in accordance. In one of the best-studied microorganisms, Bacillus subtilis, the differentiation processes into a number of distinct cell types have been shown to depend on cell-to-cell communication. One of these cell types is the spore. Spores are metabolically inactive cells that are highly resistant against environmental stress. The onset of sporulation is dependent on cell-to-cell communication, as well as on a number of other environmental cues. By using individual-based simulations we examine when cell-to-cell communication that is involved in the onset of sporulation can evolve. We show that it evolves when three basic premises are satisfied. First, the population of cells has to affect the nutrient conditions. Second, there should be a time-lag between the moment that a cell decides to sporulate and the moment that it turns into a mature spore. Third, there has to be environmental variation. Cell-to-cell communication is a strategy to cope with environmental variation, by allowing cells to predict future environmental conditions. As a consequence, cells can anticipate environmental stress by initiating sporulation. Furthermore, signal production could be considered a cooperative trait and therefore evolves when it is not too costly to produce signal and when there are recurrent colony bottlenecks, which facilitate assortment. Finally, we also show that cell-to-cell communication can drive ecological diversification. Different ecotypes can evolve and be maintained due to frequency-dependent selection.
Author Summary
Biological systems are characterized by communication; humans talk, insects produce pheromones and birds sing. Over the last decades it has been shown that even the simplest organisms on earth, the bacteria, communicate. Despite the prevalence of communication, it is often hard to explain how communicative systems evolve. In bacteria, communication results from the secretion of molecular signals that accumulate in the environment. Cells can assess the concentration of these signals, which indicate cell density, and respond in accordance. This form of cell-to-cell communication is responsible for the regulation of numerous bacterial behaviors, such as sporulation. Spores are metabolically inactive cells that are highly resistant against environmental stress. It is adaptive for a cell to sporulate when it struggles to survive. We show, via individual-based simulations, that cell-to-cell communication evolves because it allows cells to predict future environmental conditions. As a consequence, cells are capable of anticipating environmental stress by initiating sporulation before conditions are actually harmful. Furthermore, our model shows that cell-to-cell communication can even drive ecological diversification, since it facilitates the evolution of individuals that specialize on distinct ecological conditions.
Introduction
Complex systems in biology often come about through the communication of their parts, such as pheromone communication in insect societies and language in humans. Communication has been found to be ubiquitous in microorganisms as well [1]–[4]. Due to self-produced molecular signals that are secreted in the environment, cells can monitor the population density, which can quantitatively affect a cell's gene expression or trigger a differentiation process. In 1994, Fuqua and colleagues were the first to characterize this form of cell-to-cell communication as quorum-sensing signaling [5]. Quorum-sensing signaling has been shown to regulate a multitude of bacterial processes, such as extracellular enzyme production, antibiotic production and biofilm formation [6]–[11]. In one of the best-studied microorganisms, Bacillus subtilis, the differentiation of a number of cell types has been shown to depend on cell-to-cell communication [12]–[14]. These cell types emerge during the developmental process of biofilm formation and are presumably needed to survive the harsh environmental conditions that are present in the soil [10], [15], [16]. The most remarkable survival strategy among these cell types is that of the spore [17], [18].
A spore is a metabolically inactive cell that compartmentalized its DNA together with some essential proteins to survive starvation or other environmental stressors [18], [19]. Spore formation is an energy-expensive process that can take 6 to 8 hours and involves the expression of hundreds of genes [19], [20]. The initiation of sporulation is primarily dependent on the activation of a single transcription factor called Spo0A [14], [21]–[24]. When the level of activated Spo0A is sufficiently high, the sporulation process will be initiated [25]–[28]. The level of activated Spo0A is indirectly affected by a number of environmental and physiological cues, of which some are self-produced quorum-sensing signals [13], [22], [29]. These signals are assumed to accumulate in the environment and thereby give an indication of the cell density. As a consequence, the fraction of cells that initiate sporulation is higher for higher cell densities [29]–[33]. Even though these quorum-sensing signals affect the proportion of cells that initiate sporulation, they themselves are not sufficient for initiating sporulation since starvation is absolutely required [34]–[36]. Bischofs and colleagues (2009) mathematically modeled the regulatory mechanisms that integrate the quorum-sensing signals with other environmental cues, including those that are indicative of starvation [36]. They showed that the quorum-sensing signals allow for a density-dependent normalization of certain environmental cues. For example, when a cell can sense the amount of nutrients that are left in the environment, quorum-sensing signaling makes it possible to estimate the amount of nutrients that are left per cell. They concluded that these density-dependent normalizations might be adaptive for cellular decision-making, such as determining when to initiate sporulation (see also [34]).
However, despite the detailed knowledge of the regulatory mechanisms that underlie the sporulation process, little is known about their evolutionary origin. Why does cell-to-cell communication evolve and under which ecological and developmental conditions is it selected for? Here we examine, by using individual-based simulations, how three conditions, which inevitably relate to sporulation [15], [19], [20], [34], [37], [38], affect the evolution of cell-to-cell communication: environmental variation in nutrient conditions, costs of sporulation and time expenditure of sporulation. Even though our model is inspired by sporulation in B. subtilis, it is aimed to be conceptual and therefore does not include mechanistic details. The model is made such that it allows for the evolution of various developmental strategies, in which a cell's sensitivity and response to environmental cues can evolve.
Throughout the paper we discuss different versions of our model, which gradually increase in complexity. First we study the evolution of cell-to-cell communication under clonally-growing colonies. Next we allow for within colony-variation by initiating colonies with multiple individuals. Under these conditions multiple ecotypes evolve that transiently coexist over time due to negative frequency-dependent selection. Finally, we examine the evolution of cell-to-cell communication when signal production is costly. Under these conditions cooperative dilemmas emerge naturally and we find that different ecotypes evolve, which use different communicative strategies to time the onset of sporulation. The evolutionary significance of these strategies can only be understood by considering their ecological context.
Model
We assume that cells are scattered throughout the soil. Only in a few locations these cells can grow and form colonies, because only in these areas there are nutrients available to do so. During colony growth cells consume nutrients in order to perform cell division and cell differentiation. A cell can differentiate into two cell types—a signal-producing cell or a spore—or it could remain undifferentiated. Eventually, all the nutrients will be depleted and a colony enters a starvation period. This period can only be survived by the spores. It is therefore crucial for a cell to initiate sporulation on time (i.e. when the nutrients that are needed to complete the sporulation process are still available). To decide when to initiate sporulation a cell could make use of two environmental cues: the nutrient concentration and the amount of quorum-sensing signal. The spores that eventually survive the starvation period migrate and germinate in new nutrient rich areas, where they form new colonies. Over evolutionary time, a cell's responsiveness to the environmental cues can evolve and thereby the timing of sporulation can evolve as well. We examine under which ecological and developmental conditions there is selection for cells that use quorum-sensing signaling to time the onset of sporulation. The system is studied by using individual-based simulations, which we describe in the following paragraphs.
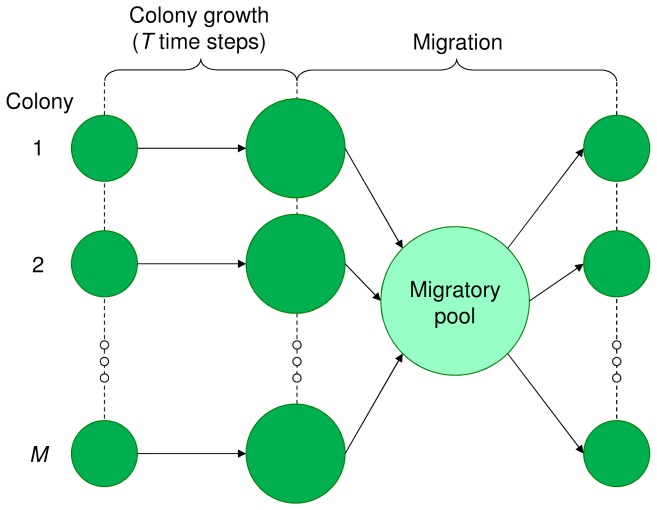
We assume that the population of cells is divided into 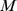 subpopulations, each representing a colony (i.e. biofilm or pellicle). Each colony is established by
subpopulations, each representing a colony (i.e. biofilm or pellicle). Each colony is established by 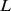 individuals. A colony is said to grow clonally when it is established by only one individual (
individuals. A colony is said to grow clonally when it is established by only one individual (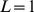 ). At the onset of colony growth there is a single nutrient input, which for each colony is taken from a normal distribution that is given by
). At the onset of colony growth there is a single nutrient input, which for each colony is taken from a normal distribution that is given by 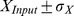 . Thus, the nutrient could be different for each colony. After receiving the nutrient input colonies are allowed to grow for a fixed number of time steps (
. Thus, the nutrient could be different for each colony. After receiving the nutrient input colonies are allowed to grow for a fixed number of time steps (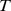 ); during this period cells consume nutrients in order to perform cell division and differentiation. At the end of a nutritional cycle all individuals (cells and spores) enter migration. The nutritional cycles of all colonies are synchronized such that the individuals from all colonies enter migration at the same time, forming a single migratory pool (see figure 1). Since migration occurs passively, we assume that all individuals have the same chance to establish a new colony. Thus,
); during this period cells consume nutrients in order to perform cell division and differentiation. At the end of a nutritional cycle all individuals (cells and spores) enter migration. The nutritional cycles of all colonies are synchronized such that the individuals from all colonies enter migration at the same time, forming a single migratory pool (see figure 1). Since migration occurs passively, we assume that all individuals have the same chance to establish a new colony. Thus, 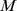 new colonies are established by choosing, for each colony separately,
new colonies are established by choosing, for each colony separately, 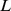 random individuals from the migratory pool. After this, the new colonies simultaneously start the next nutritional cycle.
random individuals from the migratory pool. After this, the new colonies simultaneously start the next nutritional cycle.
Figure 1. Nutritional cycle and population structure.
The colonies (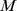 in total) are first allowed to grow for a fixed number of time steps (
in total) are first allowed to grow for a fixed number of time steps (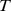 ). Then, all individuals (spores and cells) enter migration, forming a single migratory pool. From this pool,
). Then, all individuals (spores and cells) enter migration, forming a single migratory pool. From this pool, 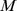 new colonies are established by taking, for each colony separately,
new colonies are established by taking, for each colony separately, 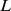 random individuals. The complete cycle from the establishment of the colonies to the eventual migration of the individuals is called the nutritional cycle and is repeated over time. Notice that for clonally growing colonies
random individuals. The complete cycle from the establishment of the colonies to the eventual migration of the individuals is called the nutritional cycle and is repeated over time. Notice that for clonally growing colonies 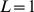 , hence only a single genotype establishes a new colony.
, hence only a single genotype establishes a new colony.
Within a nutritional cycle three different cellular processes can occur at any time step (for each cell in the colony). First, a cell gets the opportunity to differentiate. A cell can differentiate into two different cell types—a signal-producing cell or a spore—or it could remain undifferentiated. A signal-producing cell secretes a fixed amount of signal in the environment. The more cells that produce signal, the higher the amount of extracellular signal. At the same time, the signal is degraded with a fixed rate 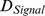 . Thus, the amount of signal changes over time depending on the number of cells that are producing it. A cell could also initiate sporulation. Sporulation is an irreversible process that takes a fixed number of time steps (
. Thus, the amount of signal changes over time depending on the number of cells that are producing it. A cell could also initiate sporulation. Sporulation is an irreversible process that takes a fixed number of time steps (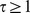 ) and during which a fixed amount of nutrients is consumed (
) and during which a fixed amount of nutrients is consumed (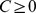 ), which is needed for making the spore. Thus, a sporulating cell consumes
), which is needed for making the spore. Thus, a sporulating cell consumes  nutrients per time step. When there is an insufficient amount of nutrients in the environment, the sporulation process cannot be completed; in this case a cell inevitably dies. After completing the sporulation process, a mature and resistant spore is formed. A spore cannot divide, but has a much lower death rate than a cell. A spore germinates at the onset of a new nutritional cycle. Since sporulation requires
nutrients per time step. When there is an insufficient amount of nutrients in the environment, the sporulation process cannot be completed; in this case a cell inevitably dies. After completing the sporulation process, a mature and resistant spore is formed. A spore cannot divide, but has a much lower death rate than a cell. A spore germinates at the onset of a new nutritional cycle. Since sporulation requires  time steps, a cell can be in one, out of
time steps, a cell can be in one, out of 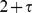 , phenotypic states. It can be an undifferentiated cell, a signal producing cell or a sporulating cell, of which the latter is subsequently composed of
, phenotypic states. It can be an undifferentiated cell, a signal producing cell or a sporulating cell, of which the latter is subsequently composed of  states that indicate the number of time steps a cell has been sporulating (
states that indicate the number of time steps a cell has been sporulating (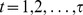 ). At the final time step of sporulation (
). At the final time step of sporulation (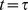 ) a cell turns into a spore. The cell's decision to differentiate into a signal-producing cell or spore depends in our model on two environmental cues—the amount of nutrients and signal—and on a cell's genotype (which we describe later).
) a cell turns into a spore. The cell's decision to differentiate into a signal-producing cell or spore depends in our model on two environmental cues—the amount of nutrients and signal—and on a cell's genotype (which we describe later).
The second cellular process that a cell can undergo, after having had the opportunity to differentiate, is division. All cells, excluding spores, have a certain chance of dividing. This chance is dependent on the amount of nutrients that are present in the environment (for details see equation S1). The more nutrients that are present in the environment, the greater the chance of cell division, with a maximum chance of 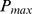 . During each cell division a fixed amount of nutrients (
. During each cell division a fixed amount of nutrients (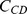 ) is consumed. At each cell division there is a certain probability that the dividing cell incurs a mutation (the mutation process is described later).
) is consumed. At each cell division there is a certain probability that the dividing cell incurs a mutation (the mutation process is described later).
The third and last cellular process that can occur at any particular time step is that of cell death. Both cells and spores have a fixed chance of dying, which is independent of the nutrient concentration. The death rate of a spore is much lower than that of a cell (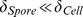 ). Hence, it is better to be a cell when nutrients are plentiful, because the chance of having cell division outweighs the chance of having cell death. On the contrary, when the nutrients are depleted, it is better to be a spore because spores have a smaller chance of dying than cells. The fitness of a genotype therefore depends on the timing of sporulation. When a genotype sporulates too early—at a nutrient concentration that is too high—it loses reproductive potential, since not all the nutrients are utilized. When a genotype sporulates too late—at a nutrient concentration that is too low—it has an increased risk of dying, especially when, due to nutrient scarcity, the sporulation process cannot be completed.
). Hence, it is better to be a cell when nutrients are plentiful, because the chance of having cell division outweighs the chance of having cell death. On the contrary, when the nutrients are depleted, it is better to be a spore because spores have a smaller chance of dying than cells. The fitness of a genotype therefore depends on the timing of sporulation. When a genotype sporulates too early—at a nutrient concentration that is too high—it loses reproductive potential, since not all the nutrients are utilized. When a genotype sporulates too late—at a nutrient concentration that is too low—it has an increased risk of dying, especially when, due to nutrient scarcity, the sporulation process cannot be completed.
A crucial part of the model is the cell differentiation process. We aim to model it such that various developmental strategies can evolve. This requires to have sufficient degrees of freedom. On the other hand, we want to restrict the number of evolvable variables, in order to keep the model simple and tractable. The combination of these requirements resulted in a cell differentiation process that could be described by two Boolean decision-making steps, which are affected by the amount of nutrients and signal. The cell should decide to initiate sporulation or not and when it does not sporulate, a cell should decide if it wants to produce signal or not. These two decisions can be expressed by the following two inequalities (see figure 2):
| (1a) |
| (1b) |
Inequality 1a shows when a cell initiates sporulation and inequality 1b shows when a cell initiates signal production. We assume that the decision to initiate sporulation is dominant over the decision to produce signal. Thus when both inequalities hold, only the sporulation process is initiated. The left hand side of each inequality contains the environmental cues: the amount of nutrients (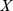 ) and the amount of extracellular signal (
) and the amount of extracellular signal (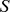 ). Since nutrients are consumed and signal can be produced and degraded over time, the values of these environmental cues change during colony growth. The effect of an environmental cue on the differentiation process depends on what we call the connection weight,
). Since nutrients are consumed and signal can be produced and degraded over time, the values of these environmental cues change during colony growth. The effect of an environmental cue on the differentiation process depends on what we call the connection weight, 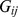 ; here
; here  is the environmental cue (1 is the amount of nutrients and 2 is the amount of signal in the environment) that is affecting differentiation process
is the environmental cue (1 is the amount of nutrients and 2 is the amount of signal in the environment) that is affecting differentiation process 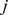 (1 is sporulation and 2 is signal production). For example,
(1 is sporulation and 2 is signal production). For example, 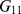 determines how the amount of nutrients affects the initiation of sporulation. When a connection weight is positive, its corresponding environmental cue stimulates the differentiation process. When the connection weight is negative, the environmental cue inhibits the differentiation process. The absolute value of a connection weight shows the impact that a certain environmental cue has on the differentiation process. The right hand side of both inequalities is the activation threshold,
determines how the amount of nutrients affects the initiation of sporulation. When a connection weight is positive, its corresponding environmental cue stimulates the differentiation process. When the connection weight is negative, the environmental cue inhibits the differentiation process. The absolute value of a connection weight shows the impact that a certain environmental cue has on the differentiation process. The right hand side of both inequalities is the activation threshold, 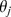 ; here
; here 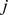 is the differentiation process to which the activation threshold belongs (1 is sporulation and 2 is signal production). The activation threshold shows how much stimulus from the environmental cues is required before the differentiation process is initiated. For example, when
is the differentiation process to which the activation threshold belongs (1 is sporulation and 2 is signal production). The activation threshold shows how much stimulus from the environmental cues is required before the differentiation process is initiated. For example, when 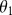 is positive a cell only sporulates when the stimulus from the nutrients (
is positive a cell only sporulates when the stimulus from the nutrients (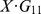 ) plus the stimulus from the signal (
) plus the stimulus from the signal (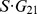 ) is bigger than the activation threshold (
) is bigger than the activation threshold (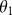 ). On the contrary, when
). On the contrary, when 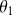 is negative a cell sporulates by default (when
is negative a cell sporulates by default (when 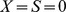 ) and sporulation can only be prevented if the environmental cues inhibit the sporulation process (i.e. negative connection weights). The activation thresholds could be viewed as a normalization of the connection weights. Namely, one could divide both sides of inequality 1a and 1b by the absolute values of, respectively,
) and sporulation can only be prevented if the environmental cues inhibit the sporulation process (i.e. negative connection weights). The activation thresholds could be viewed as a normalization of the connection weights. Namely, one could divide both sides of inequality 1a and 1b by the absolute values of, respectively, 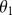 and
and 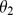 , without altering the behavior of a genotype. Therefore the model could be simplified by fixing the activation thresholds (i.e. preventing mutations to occur in the activation thresholds), as long as it does not affect the strategies that can evolve. In the first two sections of the results we applied this simplification to the model and only allowed the connection weights to mutate. To show that this simplification did not affect the evolutionary outcome of the model we performed all simulations under non-simplified conditions and show the results in the supplementary information (figure S3). In the last section we did not fix the activation thresholds, because when signal production is assumed to be costly, the evolutionary outcome would be constrained by fixing the activation thresholds. We call the collection of connection weights (
, without altering the behavior of a genotype. Therefore the model could be simplified by fixing the activation thresholds (i.e. preventing mutations to occur in the activation thresholds), as long as it does not affect the strategies that can evolve. In the first two sections of the results we applied this simplification to the model and only allowed the connection weights to mutate. To show that this simplification did not affect the evolutionary outcome of the model we performed all simulations under non-simplified conditions and show the results in the supplementary information (figure S3). In the last section we did not fix the activation thresholds, because when signal production is assumed to be costly, the evolutionary outcome would be constrained by fixing the activation thresholds. We call the collection of connection weights (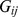 ) and activation thresholds (
) and activation thresholds (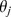 ) the genotype of an individual. In essence, the genotype describes how a cell responds to each combination of environmental cues.
) the genotype of an individual. In essence, the genotype describes how a cell responds to each combination of environmental cues.
Figure 2. Regulatory network that regulates cell differentiation.
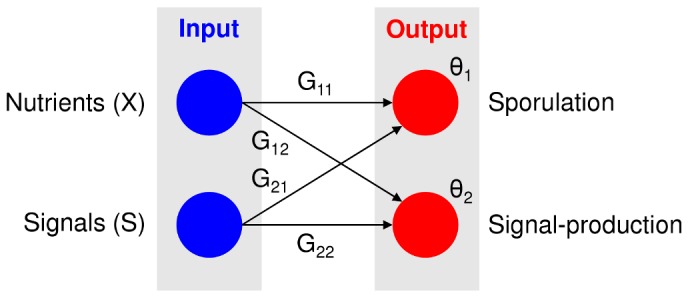
The left side shows the environmental cues that a cell can sense (the blue nodes): the nutrient concentration (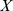 ) and the amount of signal (
) and the amount of signal (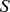 ). The right side shows the different cell types into which a cell can differentiate (the red nodes). A cell could differentiate into a sporulating or signal-producing cell. Each connection (
). The right side shows the different cell types into which a cell can differentiate (the red nodes). A cell could differentiate into a sporulating or signal-producing cell. Each connection (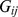 ) shows how the associated environmental cue affects the differentiation process (black arrows). Each activation threshold (
) shows how the associated environmental cue affects the differentiation process (black arrows). Each activation threshold (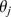 ) shows how much stimulus is required before the associated cell differentiation process occurs. The regulatory network corresponds to inequality 1a and 1b, which are described in the main text.
) shows how much stimulus is required before the associated cell differentiation process occurs. The regulatory network corresponds to inequality 1a and 1b, which are described in the main text.
When a cell division occurs each of the genotypic variables (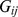 and
and 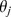 ) has a certain chance to mutate (
) has a certain chance to mutate (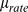 ). When a mutation occurs, a small value taken from the normal distribution
). When a mutation occurs, a small value taken from the normal distribution 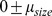 is added to the genotypic variable. Every mutation is taken independently from the same normal distribution, irrespective of the genotypic variable that mutates. All evolutionary simulations are initiated with the same monomorphic population of cells that do not produce signal and are not sensitive to it (
is added to the genotypic variable. Every mutation is taken independently from the same normal distribution, irrespective of the genotypic variable that mutates. All evolutionary simulations are initiated with the same monomorphic population of cells that do not produce signal and are not sensitive to it (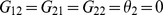 ). In addition, the initial cells are assumed to sporulate, to prevent the population from going extinct. The initial cells sporulate at a nutrient concentration of 500 (
). In addition, the initial cells are assumed to sporulate, to prevent the population from going extinct. The initial cells sporulate at a nutrient concentration of 500 (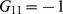 and
and 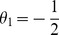 ; all input variables that are perceived by the cells are divided by 1000 as normalization, which is done consistently throughout the paper). Similar results would however be obtained if sporulation would occur at another nutrient concentration, as long as the initial population does not go extinct in the first growth cycle. By assuming that both
; all input variables that are perceived by the cells are divided by 1000 as normalization, which is done consistently throughout the paper). Similar results would however be obtained if sporulation would occur at another nutrient concentration, as long as the initial population does not go extinct in the first growth cycle. By assuming that both 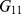 and
and 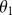 are negative, we assume that nutrients inhibit the sporulation process and that when this inhibition is too weak (e.g. when
are negative, we assume that nutrients inhibit the sporulation process and that when this inhibition is too weak (e.g. when 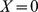 ) a cell initiates sporulation. Thus, we are not examining the evolution of sporulation, but the evolution of cell-to-cell communication as a mechanism to time the onset of sporulation.
) a cell initiates sporulation. Thus, we are not examining the evolution of sporulation, but the evolution of cell-to-cell communication as a mechanism to time the onset of sporulation.
Results
A cell should turn into a spore when the growth rate of a spore exceeds that of a cell. The effective growth rate is given by the birth rate (i.e. chance of cell division; equation S1) minus the death rate (i.e. chance of cell death; 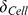 and
and 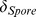 for respectively cells and spores). Since a spore cannot divide, its effective growth rate is
for respectively cells and spores). Since a spore cannot divide, its effective growth rate is 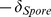 , which is approximately equal to 0 (assuming that
, which is approximately equal to 0 (assuming that 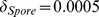 ). A cell should therefore turn into a spore when the chance of having cell death exceeds the chance of having cell division. The chance of cell division is subsequently dependent on the nutrient concentration (see equation S1). Thus, there is a critical nutrient concentration at which a cell should turn into a spore (see equation S2). However, sporulation costs time and during sporulation nutrients are consumed [19], [20]. In other words, the decision to sporulate has to be made in advance, before the critical nutrient concentration is reached. We examine why and when a cell uses quorum-sensing signals for its decision to sporulate. Moreover, we examine under which conditions cell-to-cell communication evolves. This is done for different variants of the model with increasing complexity. First, we examine if cell-to-cell communication evolves under the assumption that colonies grow clonally. Second, we examine how within-colony variation affects the evolution of cell-to-cell communication. Third and last, we examine if cell-to-cell communication evolves when signal production is costly.
). A cell should therefore turn into a spore when the chance of having cell death exceeds the chance of having cell division. The chance of cell division is subsequently dependent on the nutrient concentration (see equation S1). Thus, there is a critical nutrient concentration at which a cell should turn into a spore (see equation S2). However, sporulation costs time and during sporulation nutrients are consumed [19], [20]. In other words, the decision to sporulate has to be made in advance, before the critical nutrient concentration is reached. We examine why and when a cell uses quorum-sensing signals for its decision to sporulate. Moreover, we examine under which conditions cell-to-cell communication evolves. This is done for different variants of the model with increasing complexity. First, we examine if cell-to-cell communication evolves under the assumption that colonies grow clonally. Second, we examine how within-colony variation affects the evolution of cell-to-cell communication. Third and last, we examine if cell-to-cell communication evolves when signal production is costly.
Clonally growing colonies
In this section we examine the evolution of cell-to-cell communication under the assumption that colonies grow clonally, meaning that colonies are initiated by a single individual (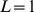 ). Genetic variation can only arise in these colonies via mutations. Moreover, for simplicity as explained before, we also assume that only the connection weights (
). Genetic variation can only arise in these colonies via mutations. Moreover, for simplicity as explained before, we also assume that only the connection weights (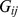 ) can mutate (similar results are however obtained when the activation thresholds are allowed to mutate as well; see figure S3). Under these conditions, the timing of sporulation depends on
) can mutate (similar results are however obtained when the activation thresholds are allowed to mutate as well; see figure S3). Under these conditions, the timing of sporulation depends on 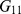 and
and 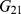 and the differentiation into a signal-producing cell solely depends on
and the differentiation into a signal-producing cell solely depends on 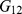 and
and 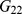 (the activation thresholds,
(the activation thresholds, 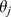 , are fixed over evolutionary time). To evolve cell-to-cell communication a cell should acquire two properties over evolutionary time. First, a cell should produce signal. Thus, before initiating sporulation a cell has to differentiate into a signal-producing cell. Second, a cell should be sensitive to the signal (
, are fixed over evolutionary time). To evolve cell-to-cell communication a cell should acquire two properties over evolutionary time. First, a cell should produce signal. Thus, before initiating sporulation a cell has to differentiate into a signal-producing cell. Second, a cell should be sensitive to the signal (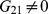 ), meaning that the nutrient concentration at which a cell initiates sporulation has to depend on the amount of signal. Irrespectively of the order in which these properties evolve, when both are present there is cell-to-cell communication. To examine if both properties can evolve in our model, we ran individual-based simulations that were initiated with a monomorphic population of cells that did not produce signal and were not sensitive to the signal (
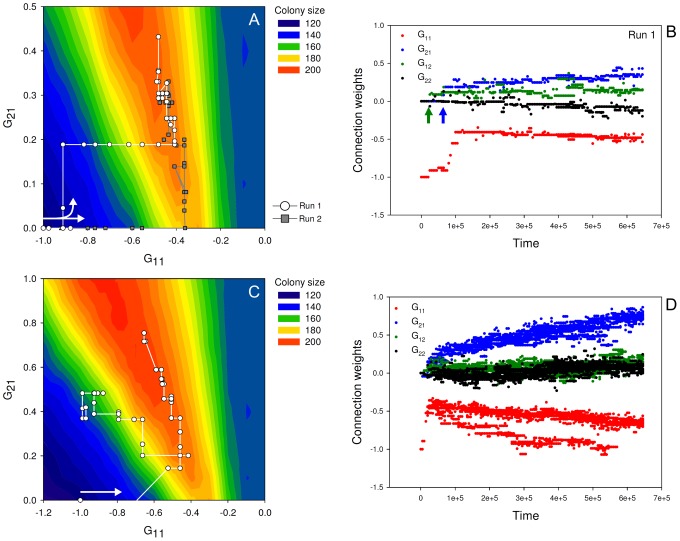
), meaning that the nutrient concentration at which a cell initiates sporulation has to depend on the amount of signal. Irrespectively of the order in which these properties evolve, when both are present there is cell-to-cell communication. To examine if both properties can evolve in our model, we ran individual-based simulations that were initiated with a monomorphic population of cells that did not produce signal and were not sensitive to the signal (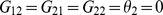 ). Figure 3A shows two independent evolutionary trajectories projected on an adaptive landscape (for more replicates see figure S1).
). Figure 3A shows two independent evolutionary trajectories projected on an adaptive landscape (for more replicates see figure S1).
Figure 3. The evolution of cell-to-cell communication under clonal and non-clonal growth conditions.
The upper panel (plot A and B) shows the clonal growth conditions (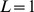 ). The lower panel (plot C and D) shows the non-clonal growth conditions (
). The lower panel (plot C and D) shows the non-clonal growth conditions (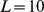 ). The left plot in each panel (plot A & C) shows the evolutionary trajectory (645.000 time steps with 5.000 time step intervals) plotted on an adaptive landscape. Thereby illustrating the evolution of cell-to-cell communication. The right plot (plot B & D) in each panel shows the connection weights of the most-abundant genotypes (present in the population in more than 100 copies). These figures thereby show both the evolution of signal sensitivity and signal production. In addition, they show how sporulation depends on the nutrient concentration (
). The left plot in each panel (plot A & C) shows the evolutionary trajectory (645.000 time steps with 5.000 time step intervals) plotted on an adaptive landscape. Thereby illustrating the evolution of cell-to-cell communication. The right plot (plot B & D) in each panel shows the connection weights of the most-abundant genotypes (present in the population in more than 100 copies). These figures thereby show both the evolution of signal sensitivity and signal production. In addition, they show how sporulation depends on the nutrient concentration (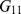 ). The adaptive landscapes (background coloration of plot A and C) are generated by growing each genotype—meaning each combination of
). The adaptive landscapes (background coloration of plot A and C) are generated by growing each genotype—meaning each combination of 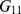 and
and 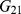 —clonally and taking the average colony size at the end of a nutritional cycle as fitness measurement (assuming that
—clonally and taking the average colony size at the end of a nutritional cycle as fitness measurement (assuming that 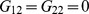 ,
, 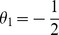 and
and 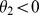 ). The white arrows within plot A and C show the onset of the evolutionary trajectory, as well as the direction of evolution. The arrows in plot B indicate when signal production (green arrow) and signal sensitivity (blue arrow) evolved. The parameter settings are the following:
). The white arrows within plot A and C show the onset of the evolutionary trajectory, as well as the direction of evolution. The arrows in plot B indicate when signal production (green arrow) and signal sensitivity (blue arrow) evolved. The parameter settings are the following: 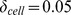 ,
, 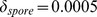 ,
, 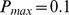 ,
, 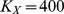 ,
, 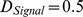 ,
, 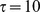 ,
, 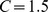 ,
, 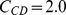 ,
, 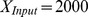 ,
, 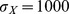 ,
, 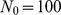 ,
, 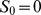 ,
, 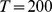 ,
, 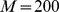 ,
, 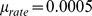 and
and 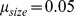 .
.
The adaptive landscape is constructed by showing for each possible genotype—meaning each combination of 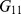 and
and 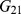 —the average colony size that is obtained at the end of a nutritional cycle. When solely examining the adaptive landscape, one expects that cell-to-cell communication would evolve, because the best-performing genotypes that are signal-sensitive (
—the average colony size that is obtained at the end of a nutritional cycle. When solely examining the adaptive landscape, one expects that cell-to-cell communication would evolve, because the best-performing genotypes that are signal-sensitive (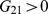 ) have a higher fitness than those that are signal-insensitive (
) have a higher fitness than those that are signal-insensitive (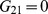 ). The two evolutionary trajectories that are plotted on the adaptive landscape are called run 1 and run 2 (both runs were performed under the same parameter settings). In both runs cell-to-cell communication evolved, which means that both signal-production and signal-sensitivity evolved. The evolutionary trajectories of figure 3A and S1 closely match the adaptive landscape and hence the adaptive landscape can be used to predict the outcome of evolution. The adaptive landscape only shows the selective advantage of cell-to-cell communication for
). The two evolutionary trajectories that are plotted on the adaptive landscape are called run 1 and run 2 (both runs were performed under the same parameter settings). In both runs cell-to-cell communication evolved, which means that both signal-production and signal-sensitivity evolved. The evolutionary trajectories of figure 3A and S1 closely match the adaptive landscape and hence the adaptive landscape can be used to predict the outcome of evolution. The adaptive landscape only shows the selective advantage of cell-to-cell communication for 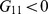 and
and 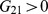 since nothing interesting happens outside this quadrant. In other words, nutrients are expected to inhibit sporulation (i.e. a cell only sporulates when there is nutrient scarcity), while signal is expected to stimulate sporulation (i.e. a cell sporulates earlier when it occurs in a bigger population). A limitation of the adaptive landscape of figure 3A is that it does not show the other two connection weights,
since nothing interesting happens outside this quadrant. In other words, nutrients are expected to inhibit sporulation (i.e. a cell only sporulates when there is nutrient scarcity), while signal is expected to stimulate sporulation (i.e. a cell sporulates earlier when it occurs in a bigger population). A limitation of the adaptive landscape of figure 3A is that it does not show the other two connection weights, 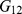 and
and 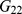 .
. 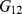 and
and 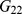 determine when a cell differentiates into a signal-producing cell (see figure 2). Signal production is, next to signal-sensitivity, essential for the evolution of cell-to-cell communication. To examine how signal production evolved we plotted the values of all connection weights (corresponding to the most-abundant genotypes), of run 1, along a time-axis (see figure 3B).
determine when a cell differentiates into a signal-producing cell (see figure 2). Signal production is, next to signal-sensitivity, essential for the evolution of cell-to-cell communication. To examine how signal production evolved we plotted the values of all connection weights (corresponding to the most-abundant genotypes), of run 1, along a time-axis (see figure 3B).
Figure 3B shows that signal production evolves after about 20.000 time steps (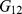 becomes positive; as indicated by the green arrow). About 40.000 time steps later signal-sensitivity evolves as well (
becomes positive; as indicated by the green arrow). About 40.000 time steps later signal-sensitivity evolves as well (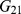 becomes positive; as indicated by the blue arrow). In other words, signal production emerges before the occurrence of signal-sensitivity. Hence there was no selective advantage for signal production at the moment it evolved. Signal production evolved because a neutral mutation in
becomes positive; as indicated by the blue arrow). In other words, signal production emerges before the occurrence of signal-sensitivity. Hence there was no selective advantage for signal production at the moment it evolved. Signal production evolved because a neutral mutation in 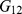 hitchhiked along with a beneficial mutation in
hitchhiked along with a beneficial mutation in 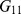 . Genetic hitchhiking is relatively prevalent, because there is no genetic recombination. In addition, there are no costs for signal production in this version of the model. Thus, cell-to-cell communication evolves by the sequential evolution of signal production and signal-sensitivity.
. Genetic hitchhiking is relatively prevalent, because there is no genetic recombination. In addition, there are no costs for signal production in this version of the model. Thus, cell-to-cell communication evolves by the sequential evolution of signal production and signal-sensitivity.
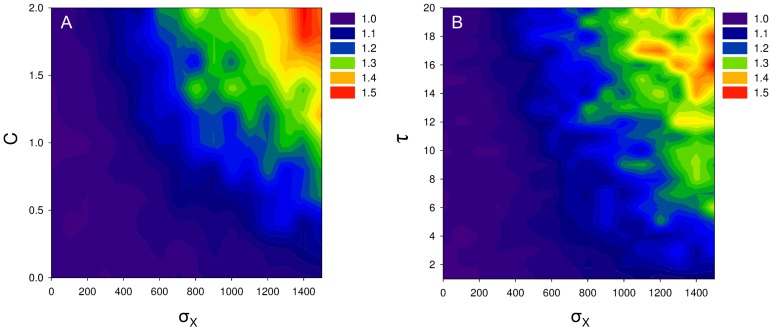
The question we are interested in though, is why cell-to-cell communication evolved at all. By sensing signal a cell can assess the colony size at the onset of sporulation. This estimate gives an indication of the amount of nutrients that will be consumed by the colony during sporulation. As explained before, a cell should turn into a spore when the chance of having cell death exceeds that of cell division, which is associated with a critical nutrient concentration (for details see equation S2). Since sporulation requires time, a cell has to anticipate or predict if the nutrient concentration at the end of sporulation matches this critical nutrient concentration. To make this prediction it is necessary to assess the amount of nutrients that will be consumed during sporulation. Since the total amount of nutrient consumption depends on the number of cells within a colony, it is advantageous for a cell to sense quorum-sensing signals. When the colony is big, a high amount of nutrients will be consumed during sporulation due to which a cell should initiate sporulation relatively early (i.e. at a high nutrient concentration). On the contrary, when the colony is small, a small amount of nutrients will be consumed and therefore a cell should initiate sporulation relatively late (i.e. at a low nutrient concentration). Thus, cell-to-cell communication allows a cell to predict the total amount of nutrient consumption during sporulation and, thereby, a cell can anticipate future environmental changes. There are three requirements that should be satisfied for cell-to-cell communication to evolve (corresponding to the parameter values in our model; see figure 4): (i) the colony size should affect the nutrient concentration during sporulation by, for example, nutrient consumption (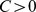 ); (ii) there should be a time-lag between the moment that a cell decides to sporulate and the moment that it turns into a mature spore (
); (ii) there should be a time-lag between the moment that a cell decides to sporulate and the moment that it turns into a mature spore (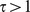 ); and (iii) there should be environmental variation (
); and (iii) there should be environmental variation (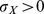 ). High values of
). High values of  ,
,  and
and 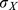 (e.g.
(e.g. 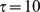 ,
, 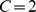 and
and 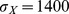 ) can result in a
) can result in a 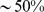 fitness advantage for cells that sense quorum-sensing signals over those that do not (figure 4).
fitness advantage for cells that sense quorum-sensing signals over those that do not (figure 4).
Figure 4. Selective advantage of cell-to-cell communication.
Plot A shows the relative fitness benefit of cell-to-cell communication under different parameter conditions of 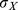 and
and  (meaning the amount of environmental variation in the nutrient input and the amount of nutrients required for completing a single sporulation process). Plot B shows the relative fitness benefit of cell-to-cell communication under different parameter conditions of
(meaning the amount of environmental variation in the nutrient input and the amount of nutrients required for completing a single sporulation process). Plot B shows the relative fitness benefit of cell-to-cell communication under different parameter conditions of 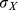 and
and  (meaning the amount of environmental variation in the nutrient input and the time-lag between the decision to sporulate and actually being a spore). The relative fitness is defined as the relative colony size of colonies that contain communicative cells over those that do not. Thus, when the relative fitness is bigger than one there is selection for cell-to-cell communication. For plot A we assume that
(meaning the amount of environmental variation in the nutrient input and the time-lag between the decision to sporulate and actually being a spore). The relative fitness is defined as the relative colony size of colonies that contain communicative cells over those that do not. Thus, when the relative fitness is bigger than one there is selection for cell-to-cell communication. For plot A we assume that 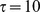 and for plot B we assume that
and for plot B we assume that 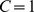 , hence the horizontal lines at which
, hence the horizontal lines at which 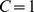 in plot A and
in plot A and 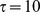 in plot B are replicates of the same parameter conditions. The other parameter settings are the following:
in plot B are replicates of the same parameter conditions. The other parameter settings are the following: 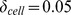 ,
, 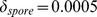 ,
, 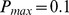 ,
, 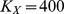 ,
, 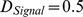 ,
, 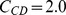 ,
, 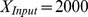 ,
, 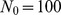 ,
, 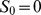 and
and 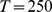 .
.
The first requirement for the evolution of cell-to-cell communication is that the colony size should affect the nutrient concentration (figure 4A). For example, when each cell consumes a fixed amount of nutrients during sporulation ( ), the total nutrient consumption depends on the colony size. When there is no nutrient consumption during sporulation (
), the total nutrient consumption depends on the colony size. When there is no nutrient consumption during sporulation (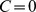 ) the optimal time at which to initiate sporulation does not depend on the colony size and hence cell-to-cell communication does not evolve. Second, cell-to-cell communication only evolves when there is a time-lag between the moment that a cell decides to sporulate and the moment that it turns into a spore (figure 4B). In other words, sporulation should require time. When sporulation does not require time, there is no need to assess the nutrient consumption since a cell could turn into a spore instantaneously. Thus, cell-to-cell communication only evolves when
) the optimal time at which to initiate sporulation does not depend on the colony size and hence cell-to-cell communication does not evolve. Second, cell-to-cell communication only evolves when there is a time-lag between the moment that a cell decides to sporulate and the moment that it turns into a spore (figure 4B). In other words, sporulation should require time. When sporulation does not require time, there is no need to assess the nutrient consumption since a cell could turn into a spore instantaneously. Thus, cell-to-cell communication only evolves when 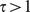 . The third and last requirement for the evolution of cell-to-cell communication is the presence of environmental variation (figure 4). When there is no variation (
. The third and last requirement for the evolution of cell-to-cell communication is the presence of environmental variation (figure 4). When there is no variation (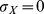 ), the amount of nutrients at the onset of a nutrient cycle is always the same. As a consequence, the changes in the nutrient concentration over time correlate with those of the colony size, since all colonies are initiated with the same number of cells, which reproduce at the same rate. Under these conditions, the nutrient concentration could be used as an accurate indication of the colony size, which makes the use of quorum-sensing signals superfluous, since these give an indication of the colony size as well. Only when the correlation between the nutrient concentration and colony size is relatively weak, the amount of signal could be used as a unique indication of the colony size. For this reason, there is stronger selection for cell-to-cell communication for higher levels of
), the amount of nutrients at the onset of a nutrient cycle is always the same. As a consequence, the changes in the nutrient concentration over time correlate with those of the colony size, since all colonies are initiated with the same number of cells, which reproduce at the same rate. Under these conditions, the nutrient concentration could be used as an accurate indication of the colony size, which makes the use of quorum-sensing signals superfluous, since these give an indication of the colony size as well. Only when the correlation between the nutrient concentration and colony size is relatively weak, the amount of signal could be used as a unique indication of the colony size. For this reason, there is stronger selection for cell-to-cell communication for higher levels of 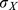 . Alternative conditions that weaken the correlation between the colony size and nutrient concentration can have a similar effect. For example, one could vary the initial colony sizes; colonies would still be clonal but different colonies would be initiated by different numbers of cells (see figure S8).
. Alternative conditions that weaken the correlation between the colony size and nutrient concentration can have a similar effect. For example, one could vary the initial colony sizes; colonies would still be clonal but different colonies would be initiated by different numbers of cells (see figure S8).
Within-colony variation
In most laboratory experiments sporulation is studied in isogenic populations. However, it is plausible that multiple genotypes can co-occur in a single colony [39]. In this section we examine how the developmental mechanisms that determine the onset of sporulation evolve when multiple genotypes can initiate a single colony (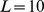 ). This is done for the same conditions as those described in the previous section (i.e. only the connection weights,
). This is done for the same conditions as those described in the previous section (i.e. only the connection weights, 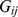 , are allowed to mutate; see figure S3 for simulations in which also the activation thresholds could mutate).
, are allowed to mutate; see figure S3 for simulations in which also the activation thresholds could mutate).
In figure 3C the evolutionary trajectory of a single run is shown on the adaptive landscape. Figure 3D shows, for the same evolutionary run, the connection weights of the most-abundant genotypes along a time-axis (for more replicates see figure S2). In contrast to the previous section, there is a bifurcation event during the evolutionary process that results in two coexisting ecotypes (an ecotype is a cluster of genotypes that is adapted to specific ecological condition). One of these ecotypes eventually goes extinct (see figure 3D and S2). Both ecotypes produce quorum-sensing signal and are sensitive to it. The ecotypes only differ in their responsiveness towards the nutritional conditions in the environment (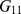 ). In one ecotype the value of
). In one ecotype the value of 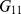 is lower than in the other, meaning that the nutrients more strongly inhibit the sporulation process (see figure 3D and S2). This ecotype is therefore called the late sporulating ecotype (i.e. sporulation is initiated at a low nutrient concentration), while the other one is called the early sporulating ecotype (i.e. sporulation is initiated at a high nutrient concentration).
is lower than in the other, meaning that the nutrients more strongly inhibit the sporulation process (see figure 3D and S2). This ecotype is therefore called the late sporulating ecotype (i.e. sporulation is initiated at a low nutrient concentration), while the other one is called the early sporulating ecotype (i.e. sporulation is initiated at a high nutrient concentration).
How can the late and early sporulating ecotypes stably coexist? In the absence of cell-to-cell communication, a genotype can only efficiently make use of the available nutrients for a limited range of nutrient inputs (i.e. nutrient concentration at the onset of a nutritional cycle; see figure S4 and S5B). When the nutrient input is higher than this particular range, a genotype would sporulate too late and when it is lower than this range a genotype would sporulate too early (see figure S4). When a genotype sporulates too early, not all the nutrients will be consumed. The leftovers can be used by other genotypes that sporulate slightly later and co-occur in the same colony. The late sporulating genotypes, in turn, cannot efficiently make use of the nutrients at high nutrient inputs, because they initiate sporulation too late. As a consequence, there is frequency-dependent selection in which the late sporulating ecotype has a selective advantage when the early sporulating ecotype is abundant and vice versa (see figure S6). Figure 3D shows that the early sporulating ecotype evolves first and later is accompanied by the late sporulating ecotype.
Over evolutionary time both the early and late sporulating ecotypes become more sensitive to the quorum-sensing signal (increase in 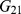 ) and thereby evolve cell-to-cell communication (figure 3D). In other words, both ecotypes evolve the ability to adjust the timing of sporulation to the nutrient input. This increases the range of nutrient inputs at which an ecotype could efficiently make use of the nutrients (see figure S5C). As a consequence, there is an increasing overlap in the range of nutrient inputs at which both ecotypes grow efficiently, hence strengthening the competition between them. Ultimately, only a single ecotype survives (see figure 3D and S2). This ecotype is a generalist, since it grows efficiently at most nutrient inputs due to the evolved cell-to-cell communication. Thus, over evolutionary time, the evolved specialists—the early and late sporulating ecotypes—are replaced by a generalist—a signaling ecotype—that can grow efficiently at most nutrient inputs.
) and thereby evolve cell-to-cell communication (figure 3D). In other words, both ecotypes evolve the ability to adjust the timing of sporulation to the nutrient input. This increases the range of nutrient inputs at which an ecotype could efficiently make use of the nutrients (see figure S5C). As a consequence, there is an increasing overlap in the range of nutrient inputs at which both ecotypes grow efficiently, hence strengthening the competition between them. Ultimately, only a single ecotype survives (see figure 3D and S2). This ecotype is a generalist, since it grows efficiently at most nutrient inputs due to the evolved cell-to-cell communication. Thus, over evolutionary time, the evolved specialists—the early and late sporulating ecotypes—are replaced by a generalist—a signaling ecotype—that can grow efficiently at most nutrient inputs.
Not surprisingly, when there is no environmental variation (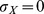 ), a bifurcation event cannot occur. In that case only a single ecotype evolves that outcompetes all others (see figure S7). Branching is most likely to occur for high levels of
), a bifurcation event cannot occur. In that case only a single ecotype evolves that outcompetes all others (see figure S7). Branching is most likely to occur for high levels of 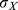 (see figure S7); the same conditions that select for cell-to-cell communication (see figure 3 and 4). Another condition under which a bifurcation event cannot occur is clonal growth, since it hampers the presence of within-colony variation. Within-colony variation allows for competition at the cellular-level and hence for the coexistence of multiple ecotypes. However, allowing for within-colony variation can also result in a conflict between the genotypes that are selected for at the colony-level and those that are selected for at the cellular-level. In particular, when signal production is costly conflicts are expected, since cells that do not produce the costly signal have a fitness advantage at the cellular-level but undermine the performance of the colony. In the next section we examine whether cell-to-cell communication evolves when signal production is costly.
(see figure S7); the same conditions that select for cell-to-cell communication (see figure 3 and 4). Another condition under which a bifurcation event cannot occur is clonal growth, since it hampers the presence of within-colony variation. Within-colony variation allows for competition at the cellular-level and hence for the coexistence of multiple ecotypes. However, allowing for within-colony variation can also result in a conflict between the genotypes that are selected for at the colony-level and those that are selected for at the cellular-level. In particular, when signal production is costly conflicts are expected, since cells that do not produce the costly signal have a fitness advantage at the cellular-level but undermine the performance of the colony. In the next section we examine whether cell-to-cell communication evolves when signal production is costly.
Costs for signal production
In this section we examine whether cell-to-cell communication can still evolve when signal production is costly. We assume that a signal-producing cell has a reduced chance of dividing by subtracting a fixed value (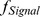 ) from the chance of having cell division (see equation S3). In contrast to the previous sections, all genotypic variables can mutate, to allow for a wider variety of communicative strategies. In this section we focus on a single representative evolutionary run (for more replicates see figure S9).
) from the chance of having cell division (see equation S3). In contrast to the previous sections, all genotypic variables can mutate, to allow for a wider variety of communicative strategies. In this section we focus on a single representative evolutionary run (for more replicates see figure S9).
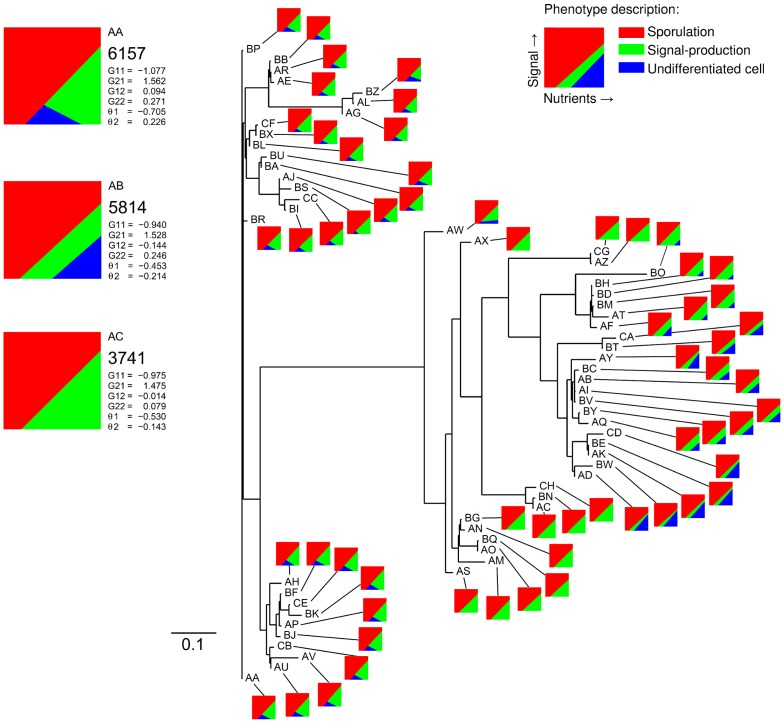
Figure 5 shows the outcome of this evolutionary run, by using a phenogram. The phenogram shows the dissimilarity between genotypes in a population that evolved for 550.000 time steps. The genotypes are named by letter-codes, which are ranked in alphabetic order and represent abundance, with genotype ‘AA’ being the most abundant and genotype ‘CH’ the least. Besides the letter-code, every genotype is connected to a small graph, which shows its phenotype for a range of environmental conditions. The population consists of multiple communicative strategies that cluster together. The three most-abundant genotypes partly reflect these clusters and are shown on the left side of the phenogram. Since, the phenogram does not show evolutionary descendance, the evolutionary lineages of the three most-abundant genotypes were used to construct an evolutionary tree. This tree is shown in figure 6. Hereafter, the phenotypes of the three most-abundant genotypes are called phenotype 1, 2 and 3; corresponding to the order in which they appear in figure 6.
Figure 5. Unrooted phenogram based on the most-abundant genotypes at time step 550.000.
This diagram shows the phenotypic population structure at time step 550.000 based on genotypic relatedness. The horizontal lines represent the distances between genotypes. The distance between two genotypes is given by the sum of absolute differences between the connection weights and activation thresholds of both genotypes. Thus, closely related genotypes cluster together in the tree diagram. Only horizontal distances are informative, thus the upper and lower clusters are closer related to each other than either of them are to the biggest cluster of genotypes in the middle. From the tree one cannot infer evolutionary descendance, because it is unrooted. The tree is constructed from the distance matrix of the 60 most-abundant genotypes using the Fitch-Margoliash method (from the PHYLIP v3.69 package). The letter-code that is given to each genotype represents abundance, with ‘AA’ being the most-abundant genotype and ‘CH’ the least-abundant genotype. The three most-abundant genotypes and their associated phenotypes are shown in the upper left corner (AA, AB & AC). For each of these genotypes, we show the abundance, connection weights, activation thresholds and phenotype description. The phenotypes are described by a small diagram that shows the behavior of a cell for different environmental conditions: red area is sporulation; green area is signal production; and blue area is no differentiation. The parameter settings that are used for this simulation are the following: 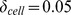 ,
, 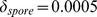 ,
, 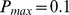 ,
, 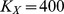 ,
, 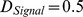 ,
, 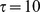 ,
, 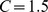 ,
, 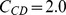 ,
, 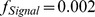 ,
, 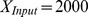 ,
, 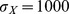 ,
, 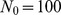 ,
, 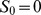 ,
, 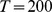 ,
, 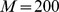 ,
, 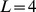 ,
, 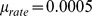 and
and 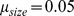 .
.
Figure 6. The evolution of different phenotypes shown by an evolutionary tree.
The phenotypes that are associated with the three most-abundant genotypes that were present at the end of the simulation (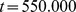 ) are called phenotype 1, 2 and 3, each belonging to a distinct ecotype. The phenotypes that are projected on the evolutionary tree correspond to the ancestral and evolved genotypes at respectively time step 0, 100.000, 300.000, 400.000, 500.000 and 550.000 (from the left to the right). Each phenotype is shown by a small graph that shows the behavior of a cell for different environmental conditions: red area is sporulation; green area is signal production; and blue area is no differentiation. For the parameter settings see figure 5.
) are called phenotype 1, 2 and 3, each belonging to a distinct ecotype. The phenotypes that are projected on the evolutionary tree correspond to the ancestral and evolved genotypes at respectively time step 0, 100.000, 300.000, 400.000, 500.000 and 550.000 (from the left to the right). Each phenotype is shown by a small graph that shows the behavior of a cell for different environmental conditions: red area is sporulation; green area is signal production; and blue area is no differentiation. For the parameter settings see figure 5.
All three phenotypes produce quorum-sensing signal for a range of parameter conditions (shown by the green areas in figure 6). Phenotype 2 produces quorum-sensing signal for all environmental conditions, except for those at which it sporulates. Since signal production is costly this phenotype is exploited by phenotype 1 and 3, which lack signal production for respectively high and low nutrient concentrations. As a consequence, phenotype 2 is always selected against at the cellular-level, irrespective of the population composition at the onset of a nutritional cycle. However, phenotype 2 is maintained in the population due to selection at the colony-level, in which the colonies that contain phenotype 2 often have a selective advantage over those that do not contain phenotype 2 (for details see table S1). This selective advantage results from the improved timing of sporulation. Thus, the selection pressures at the colony-level outweigh those at the individual-level. Since the other two phenotypes exploit phenotype 2 for different environmental conditions, they occupy different niches.
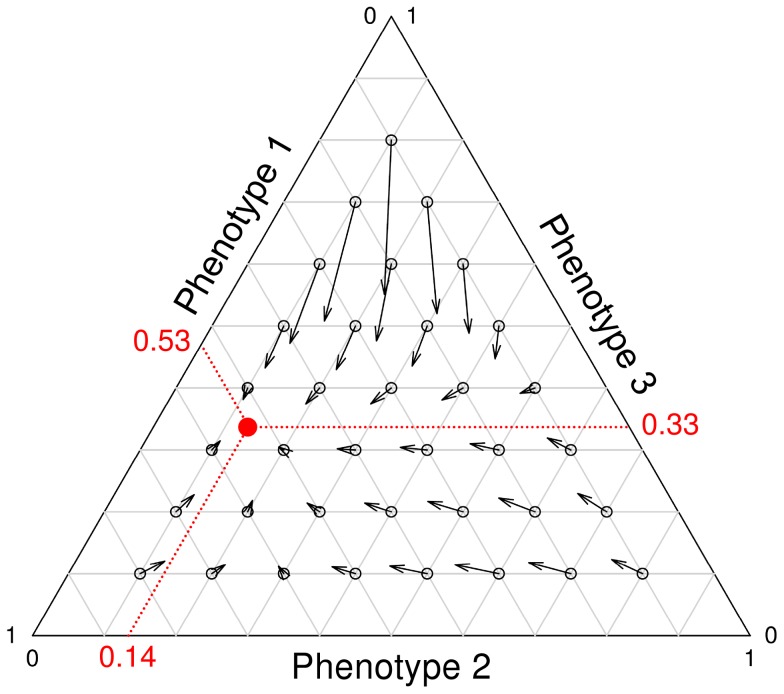
Figure 7 shows the selection pressures that act on each phenotype, given the frequency at which each phenotype occurs in the population (frequency over all colonies). The fitness measurements include the selection processes at the cellular- and colony-level. All phenotypes have a selective advantage when they are present in a low overall frequency. Thus, negative frequency-dependent selection is responsible for the stable coexistence of the three phenotypes. Since the three phenotypes are subject to a continuing process of evolution, it is unlikely that these specific phenotypes would coexist forever. Frequency-dependent selection does however assure the coexistence of multiple ecotypes, as shown by figure 5 and S9.
Figure 7. Selection pressures that act on the three most abundant phenotypes.
The direction of an arrow shows how the phenotype frequencies change over time. The length of an arrow indicates the speed of this change and hence the strength of selection. The red dot shows to the phenotype frequencies at equilibrium (i.e. the population state in which all phenotypes have exactly the same fitness). The frequency changes are determined from the onset of the current nutritional cycle to that of the next nutritional cycle. The calculations therefore include both cellular-level and colony-level selection. For the parameter settings see figure 5.
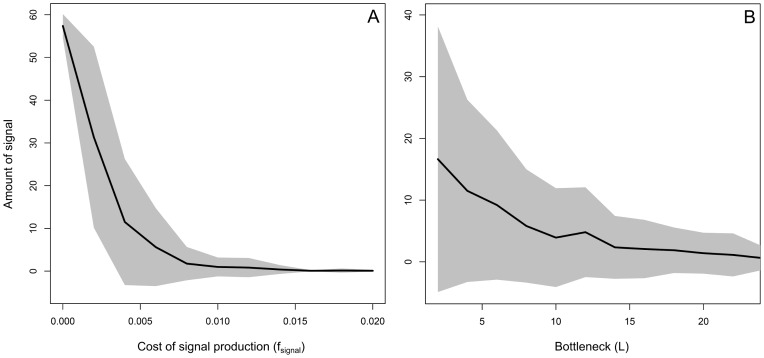
It is important to notice that the evolutionary simulation shown by figures 5, 6 and 7 assumes relatively low costs for signal production and a small bottleneck size. The costs of signal production are 2% of the maximal growth rate (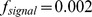 ), which means that a signal-producing cell has a 2% smaller chance to divide than an undifferentiated cell under the optimal growth conditions. The bottleneck size is given by the number of individuals that initiate a single colony (
), which means that a signal-producing cell has a 2% smaller chance to divide than an undifferentiated cell under the optimal growth conditions. The bottleneck size is given by the number of individuals that initiate a single colony (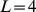 ). Smaller bottleneck sizes facilitate assortment, because signal-producing cells are more likely to end up in a colony that only contains signal-producers. As a consequence, signal-producing cells are less likely to be exploited by cells that lack signal production. Figure 8 shows how the evolution of cell-to-cell communication depends on
). Smaller bottleneck sizes facilitate assortment, because signal-producing cells are more likely to end up in a colony that only contains signal-producers. As a consequence, signal-producing cells are less likely to be exploited by cells that lack signal production. Figure 8 shows how the evolution of cell-to-cell communication depends on 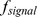 and
and 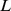 , by showing the average amount of signal that is present in a population that evolved for 550.000 time steps. As expected, cell-to-cell communication is more likely to evolve for smaller signal costs and stronger population bottlenecks.
, by showing the average amount of signal that is present in a population that evolved for 550.000 time steps. As expected, cell-to-cell communication is more likely to evolve for smaller signal costs and stronger population bottlenecks.
Figure 8. Evolution of signal production under various levels of signal costs and colony bottleneck sizes.
The plots show the amount of signal that is present in a population of cells that evolved for 550.000 time steps for different values of 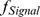 (plot A) and
(plot A) and 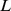 (plot B). The grey area shows the standard deviation. For every parameter setting, 50 independent runs were studied. ‘Signal’ gives the average amount of signal that is present in the environment per time step and colony.
(plot B). The grey area shows the standard deviation. For every parameter setting, 50 independent runs were studied. ‘Signal’ gives the average amount of signal that is present in the environment per time step and colony. 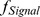 is the reduced chance of having cell division. Thus,
is the reduced chance of having cell division. Thus, 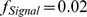 is equal to a 2% lower chance of having cell division. Notice that the maximum chance of having cell division is 10% (
is equal to a 2% lower chance of having cell division. Notice that the maximum chance of having cell division is 10% (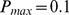 ).
). 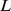 is the number of individuals that initiate a colony and hence the bottleneck size. For plot A we assumed that
is the number of individuals that initiate a colony and hence the bottleneck size. For plot A we assumed that 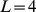 and for plot B we assume that
and for plot B we assume that 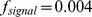 . Thus, the runs of plot B at
. Thus, the runs of plot B at 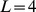 are performed under the same parameter settings as those of plot A at
are performed under the same parameter settings as those of plot A at 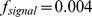 . The relatively large standard deviation in plot B can be explained by the co-existence of multiple communicative strategies, of which some produce signal, while others do not. Since the abundances of these strategies change over time, the amount of signal that is being present differs strongly between the runs. Furthermore, in some runs cell-to-cell communication does not evolve (e.g. at high values of
. The relatively large standard deviation in plot B can be explained by the co-existence of multiple communicative strategies, of which some produce signal, while others do not. Since the abundances of these strategies change over time, the amount of signal that is being present differs strongly between the runs. Furthermore, in some runs cell-to-cell communication does not evolve (e.g. at high values of 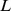 ). The other parameter settings are the following:
). The other parameter settings are the following: 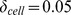 ,
, 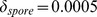 ,
, 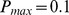 ,
, 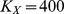 ,
, 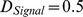 ,
, 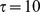 ,
, 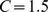 ,
, 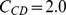 ,
, 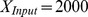 ,
, 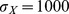 ,
, 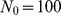 ,
, 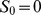 ,
, 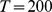 ,
, 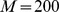 ,
, 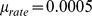 and
and 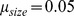 .
.
In conclusion, when signal production is costly, cell-to-cell communication can still evolve. However, signal-producing cells can be exploited by cells that lack signal production. This ultimately results in the evolution of ecological diversity, in which multiple ecotypes can coexist. Even though it is to be expected that signal production costs result in cheating (i.e. cells that do not produce signal), it is less intuitive that three ecotypes would evolve, including one that cheats for high nutrient inputs and another that cheats for low nutrient inputs. This coexistence is facilitated by negative frequency-dependent selection, which results from the selection processes at the cellular- and colony-level. Cell-to-cell communication only emerges in our simulations for relatively low costs of signal production and in the presence of population bottlenecks.
Discussion
We demonstrated that cell-to-cell communication can evolve to regulate the timing of sporulation. The evolution of cell-to-cell communication requires both the evolution of signal production and signal-sensitivity. By sensing quorum-sensing signals a cell can predict future environmental conditions and thereby anticipate a starvation period by initiating sporulation. To predict the environmental conditions a cell has to assess the rate of nutrient consumption, which depends on the colony size. Our model shows that three conditions, which inevitably relate to sporulation, are sufficient to explain the evolution of cell-to-cell communication: (i) the population size has to affect the nutrient concentration (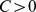 ); (ii) a cell has to predict future environmental conditions (
); (ii) a cell has to predict future environmental conditions (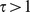 ; see also [40]–[42]); and (iii) there has to be environmental variation (
; see also [40]–[42]); and (iii) there has to be environmental variation (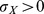 ). Irrespectively of how these conditions come about, when all three are satisfied and signal production is not too costly, cell-to-cell communication evolves. It is not our claim that these conditions are strictly necessary, but rather that they are sufficient for the evolution of cell-to-cell communication. In nature, the requirements for the evolution of cell-to-cell communication in sporulating bacteria might be less stringent, since additional advantages, besides the timing of cell differentiation, can facilitate the evolution of cell-to-cell communication (e.g. colony-level properties; [2]).
). Irrespectively of how these conditions come about, when all three are satisfied and signal production is not too costly, cell-to-cell communication evolves. It is not our claim that these conditions are strictly necessary, but rather that they are sufficient for the evolution of cell-to-cell communication. In nature, the requirements for the evolution of cell-to-cell communication in sporulating bacteria might be less stringent, since additional advantages, besides the timing of cell differentiation, can facilitate the evolution of cell-to-cell communication (e.g. colony-level properties; [2]).
In contrast to previous models on the evolution of cell-to-cell communication [43]–[46], our model shows that cell-to-cell communication can evolve as a mechanism to evaluate other environmental cues [34], [36]: neither the absolute signal concentration nor the absolute nutrient concentration determine the onset of sporulation. To understand when cell-to-cell communication evolves one has to understand how the information that results from quorum-sensing signaling is integrated with that of other environmental cues [47]–[49]. Moreover, we have demonstrated that cell-to-cell communication can even evolve when there is genetic variation within the colony and, in addition, when signal production is costly. Models on sporulation (or other persistence phenotypes) often exclude cell-to-cell communication as a mechanism to regulate sporulation [27], [50], [51]. This is because sporulation is mostly studied as a bet-hedging strategy: only a small fraction of genetically-identical cells sporulates under the same environmental conditions [28], [40]. Bet-hedging is a risk-spreading strategy that ensures the survival of a colony when there are severe and sudden environment changes [52], [53]. In our model a bet-hedging strategy cannot evolve, because cells always perceive accurate environmental information and lack developmental noise. Furthermore, bet-hedging is only beneficial when environmental changes are unpredictable [50], [54]. In our model, environmental changes might only become unpredictable when a cell is surrounded by different ecotypes, which differ in the amount of signal production and the timing of sporulation. It might therefore be interesting to extend the model, in order to examine how the evolution of bet-hedging affects that of cell-to-cell communication.
In our model, cell-to-cell communication represents a form of phenotypic plasticity, because it allows a cell to adjust the timing of sporulation in response to environmental changes [55]. Without cell-to-cell communication a cell can only grow efficiently for a limited range of nutrient inputs (figure S5). In that case, multiple ecotypes evolve that specialize on distinct ecological niches (e.g. the late and early sporulating ecotypes that evolved at the onset of our simulations, see figure 3C–D). However, by evolving cell-to-cell communication the range of nutrient inputs at which a cell grows efficiently increases. This ultimately results in competitive exclusion: the specialized ecotypes (i.e. narrow niche width)—such as the late and early sporulating ecotypes—are replaced by a single generalist (i.e. broad niche width) that can grow efficiently under most environment conditions due to cell-to-cell communication [56]–[58]. In our model phenotypic plasticity is a colony-level property, instead of a cellular property, since cells cannot respond to changes in environmental conditions without cooperation [59]: the amount of signal only gives an accurate indication of the colony size when all cells (or a constant fraction) produce quorum-sensing signals. The evolution of cell-to-cell communication therefore entails a cooperative dilemma (given that signal production is costly; [4], [60]–[62]). Cells that do not produce signal (i.e. public good) have an advantage over those that do, but at the same time they undermine the colony performance (see also [4], [63]–[66]). The cells that do not produce signal could therefore be called ‘cheaters’, while signal-producing cells are ‘cooperators’.
In our model, cheaters and cooperators evolved and stably coexisted due to frequency-dependent selection [43], [46], [67]–[71]. They have different communicative strategies [72] and therefore occupy distinct complementary niches (see figure 5 and 6). That is, the cheaters lack signal production for different subsets of environmental conditions. This emphasizes the importance of studying cell-to-cell communication under a wide range of environmental conditions, since a cooperator under one condition might be a cheater under another. The population structure (see figure 1), which results in two levels of selection, was essential for the maintenance of the different ecotypes [66], [73], [74]. Previous studies have shown that population structure can facilitate the evolution and maintenance of cooperation [69], [75]–[83]. The population structure makes individuals interact assortatively [84]: cooperators are therefore more likely to interact with other cooperators than cheaters. As a consequence, the benefits of cooperation mostly end up with cooperators, due to which there is a net selective advantage for cooperation. In our model the degree of assortment depends on the number of individuals that initialize a single colony (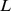 ) or, in other words, on the strength of the recurrent population bottlenecks [85], . We assumed that the colonies themselves are well-mixed, although within-colony structure—via the emergence of assortment—might have facilitated cooperation even more [87], [88]. When signal production is too costly, cell-to-cell communication does not evolve, because the selective advantage of cheaters at the cellular-level cannot be compensated by the selective advantage of cooperators at the colony-level. It is important to notice that our model only included signal production costs, even though plausible arguments could be made that the maintenance costs of a communicative system should be considered as well [89]. However, we do not expect that including maintenance costs would affect our results, since both cheaters and cooperators need to have a communicative system—and hence carry the associated costs—to sense the quorum-sensing signal.
) or, in other words, on the strength of the recurrent population bottlenecks [85], . We assumed that the colonies themselves are well-mixed, although within-colony structure—via the emergence of assortment—might have facilitated cooperation even more [87], [88]. When signal production is too costly, cell-to-cell communication does not evolve, because the selective advantage of cheaters at the cellular-level cannot be compensated by the selective advantage of cooperators at the colony-level. It is important to notice that our model only included signal production costs, even though plausible arguments could be made that the maintenance costs of a communicative system should be considered as well [89]. However, we do not expect that including maintenance costs would affect our results, since both cheaters and cooperators need to have a communicative system—and hence carry the associated costs—to sense the quorum-sensing signal.
Although our model is limited to sporulation, it could be extended to examine the role of cell-to-cell communication in the timing of other differentiation events as well, for example: motility, bioluminescence, conjugation, competence, matrix-production, biofilm formation, biofilm detachment, etc. (e.g. [11]–[14], [47], [90]–[94]). Every time there is a trade-off between the growth rate of two cell types (e.g. cells and spores) over two or more environmental niches that alternate over time (e.g. nutrient availability and nutrient scarcity), a cell has a selective advantage when it accurately times the developmental transitions between both cell types (see also [45]). When the population size affects the optimal time at which a cell should differentiate (e.g. when a cell must predict future nutrient conditions), cell-to-cell communication is expected to evolve in order to enhance a cell's developmental timing. The challenge for future studies is to unravel the developmental trade-off and ecological niches that underlie each of these differentiation events. Furthermore, our study emphasizes the importance of examining the integration of different environmental cues in cellular decision-making [49], [95]–[97]. The quorum-sensing threshold—and hence the critical population density—at which a differentiation event occurs can and mostly will strongly depend on other environmental conditions, such as nutrient availability [34], [48], [98], [99].
Supporting Information
The evolution of cell-to-cell communication under clonal growth conditions. These plots depict the evolutionary trajectories of 100 runs performed under the same conditions than those shown in figure 3A. The left plot shows for every 20.000 time steps the average evolved genotype, which is given by the mean value of 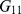 and
and 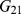 over 100 runs. The error bars show the standard deviations. In total 600.000 time steps of evolution are shown; starting from the dark-blue dot till the red dot. The right plot shows a subset of runs that evolved cell-to-cell communication, using the same color coding. For parameter settings see figure 3 of the main text.
over 100 runs. The error bars show the standard deviations. In total 600.000 time steps of evolution are shown; starting from the dark-blue dot till the red dot. The right plot shows a subset of runs that evolved cell-to-cell communication, using the same color coding. For parameter settings see figure 3 of the main text.
(TIF)
The evolution of cell-to-cell communication under non-clonal growth conditions. These plots show the evolved values of 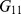 for 18 independent evolutionary runs. The conditions of these runs are the same as those shown in figure 3D, in which a early and late sporulating ecotype evolved. The simulations ran for 600.000 time steps, of which
for 18 independent evolutionary runs. The conditions of these runs are the same as those shown in figure 3D, in which a early and late sporulating ecotype evolved. The simulations ran for 600.000 time steps, of which 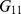 is shown for the most-abundant genotypes (present in the population in more than 100 copies) at 5.000 time steps intervals. For parameter settings see figure 3 of the main text.
is shown for the most-abundant genotypes (present in the population in more than 100 copies) at 5.000 time steps intervals. For parameter settings see figure 3 of the main text.
(TIF)
The evolution of cell-to-cell communication when the activation thresholds can evolve. The plots show the evolution of cell-to-cell communication under the same conditions as those shown in figure S1, S2 and 3. However, in contrast to these previous figures, the simulations in this figure allowed for the evolution of the activation thresholds (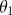 and
and 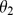 ). To facilitate a comparison between the plots in this figure and figure S1, S2 and 3, all shown connection weights are corrected for the evolved activation threshold. This is done by dividing the connection weights by two times the absolute value of the associated activation threshold (notice that for the previous figures we assumed that
). To facilitate a comparison between the plots in this figure and figure S1, S2 and 3, all shown connection weights are corrected for the evolved activation threshold. This is done by dividing the connection weights by two times the absolute value of the associated activation threshold (notice that for the previous figures we assumed that 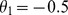 ). Plot A and B show the evolution of cell-to-cell communication under clonal growth conditions (see figure S1). Plot A shows for every 20.000 time steps the average evolved genotype, which is given by the mean value of
). Plot A and B show the evolution of cell-to-cell communication under clonal growth conditions (see figure S1). Plot A shows for every 20.000 time steps the average evolved genotype, which is given by the mean value of 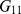 and
and 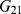 over 100 runs. The error bars show the standard deviations. In total 600.000 time steps of evolution are shown; starting from the dark-blue dot till the red dot. Plot B shows a subset of runs that evolved cell-to-cell communication, using the same color coding. Plot C shows the evolution of cell-to-cell communication under non-clonal growth conditions. The subplots show the evolved values of
over 100 runs. The error bars show the standard deviations. In total 600.000 time steps of evolution are shown; starting from the dark-blue dot till the red dot. Plot B shows a subset of runs that evolved cell-to-cell communication, using the same color coding. Plot C shows the evolution of cell-to-cell communication under non-clonal growth conditions. The subplots show the evolved values of 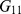 for the most-abundant genotypes (present in the population in more than 100 copies) at 5.000 time steps intervals. Only
for the most-abundant genotypes (present in the population in more than 100 copies) at 5.000 time steps intervals. Only 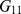 is shown, since this illustrates the evolution of the early and late sporulating ecotype, as shown in figure 3 and S2. For parameter settings see figure 3 of the main text.
is shown, since this illustrates the evolution of the early and late sporulating ecotype, as shown in figure 3 and S2. For parameter settings see figure 3 of the main text.
(TIF)
Growth of mixed colonies, consisting of early and late sporulating genotypes, at high and low nutrient inputs. The left plots show colony growth at a low nutrient input (i.e. 1000) and the right plots show the same for a high nutrient input (i.e. 2500). The early and late sporulating genotypes shown here do not have cell-to-cell communication and only differ with respect to the nutrient concentration at which sporulation is initiated: the early sporulating genotype sporulates at a nutrient concentration of 1000 (blue dashed line in the upper plot), while the late sporulating genotype sporulates at a nutrient concentration of 500 (red dashed line in the upper plot). The upper plots show the nutrient concentration (green line), the middle plots show the number of cells and the lower plots show the number of spores of the early (blue) and late (red) sporulating genotypes. Each line is the average of 1000 replicate runs and the shaded area shows the associated standard deviation. The parameter settings that are used for these simulations are the following: 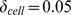 ,
, 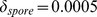 ,
, 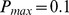 ,
, 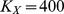 ,
, 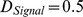 ,
, 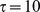 ,
, 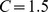 ,
, 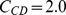 ,
, 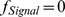 ,
, 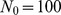 ,
, 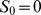 and
and 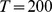 .
.
(TIF)
Overview of the colony performance at different nutrient inputs (i.e. nutrient concentration at onset of colony growth). Plot A: the distribution of nutrient inputs from which the nutrient input of a colony is taken in the evolutionary simulations (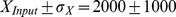 ). Plot B: The average colony size at the end of colony growth for two different genotypes that do not use cell-to-cell communication. The red line shows the average colony size for a late sporulating genotype (this genotype initiates sporulation when
). Plot B: The average colony size at the end of colony growth for two different genotypes that do not use cell-to-cell communication. The red line shows the average colony size for a late sporulating genotype (this genotype initiates sporulation when 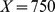 ) and the blue line shows the average colony size for an early sporulating genotype (this genotype initiates sporulation when
) and the blue line shows the average colony size for an early sporulating genotype (this genotype initiates sporulation when 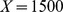 ). The dotted lines show the standard deviation from the average at each nutrient input. Plot C: The average colony size at the end of colony growth for the evolved genotype of figure 3. The evolved genotype is the dominant genotype that is present at the end of the simulation. This genotype evolved cell-to-cell communication (figure 3D) and has the following genotype:
). The dotted lines show the standard deviation from the average at each nutrient input. Plot C: The average colony size at the end of colony growth for the evolved genotype of figure 3. The evolved genotype is the dominant genotype that is present at the end of the simulation. This genotype evolved cell-to-cell communication (figure 3D) and has the following genotype: 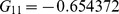 ;
; 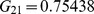 ;
; 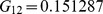 ;
; 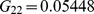 ;
; 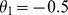 and
and 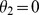 . Even though the optimal genotype is not reached yet (see figure 3C), this genotype performs considerably well under all possible nutrient inputs.
. Even though the optimal genotype is not reached yet (see figure 3C), this genotype performs considerably well under all possible nutrient inputs.
(TIF)
Invasion analysis of early and late sporulating genotype. The relative fitness of the early and late sporulating genotypes for different starting conditions: (A) 10% of early sporulating genotypes or (B) 10% of late sporulating genotypes. When a genotype's fitness is higher than one it is favored by selection and when it is lower than one it is selected against. There is frequency-dependent selection, since each genotype has a fitness advantage when it is rare: the early sporulating genotype has a fitness advantage when it is rare (A) and the late sporulating genotype has a fitness advantage when it is rare (B). The genotypes only differ in their sensitivity towards the nutrient concentration: 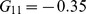 for the early sporulating genotype and
for the early sporulating genotype and 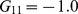 for the late sporulating genotype. Each bar shows the average fitness over 10 replicates and the error bars show the standard deviation. Each replicate consists of 200 colonies which are grown under the following conditions:
for the late sporulating genotype. Each bar shows the average fitness over 10 replicates and the error bars show the standard deviation. Each replicate consists of 200 colonies which are grown under the following conditions: 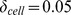 ,
, 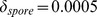 ,
, 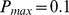 ,
, 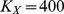 ,
, 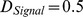 ,
, 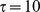 ,
, 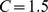 ,
, 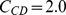 ,
, 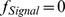 ,
, 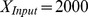 ,
, 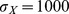 ,
, 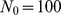 ,
, 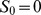 ,
, 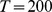 and
and 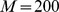 .
.
(TIF)
Pairwise invasibility plots for different levels of environmental variation. Each plot shows the invasibility of a mutant, given the presence a certain resident population. The genotypes differ in the nutrient concentration at which sporulation is initiated, which is shown for the resident genotype on the x-axis and for the mutant genotype on the y-axis. For each combination of mutant and resident, the invasibility of the mutant is tested by growing 800 colonies that are initiated with 10% of mutants. The mutant is said to invade when its average fitness is higher then that of the resident (red area), while it goes extinct when its fitness is lower (blue area). The black diagonal line shows when the resident and mutant have the same genotype and hence fitness. The different plots show the invasibility for various levels of environmental variation: 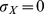 ,
, 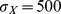 or
or 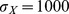 . For
. For 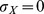 there is an evolutionary stable strategy (ESS) that cannot be invaded by mutants. This is illustrated by the white dot on the black diagonal line (the ESS is a resident genotype that sporulates at a nutrient concentration around 1100). The vertical white line, which is associated with the ESS, occurs exclusively in the blue region of the plot. This shows that none of the mutants can invade the ESS resident population. For
there is an evolutionary stable strategy (ESS) that cannot be invaded by mutants. This is illustrated by the white dot on the black diagonal line (the ESS is a resident genotype that sporulates at a nutrient concentration around 1100). The vertical white line, which is associated with the ESS, occurs exclusively in the blue region of the plot. This shows that none of the mutants can invade the ESS resident population. For 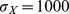 there is no ESS (as illustrated by the black dot), since one cannot draw a vertical white line that exclusively occurs in the blue region of the plot.
there is no ESS (as illustrated by the black dot), since one cannot draw a vertical white line that exclusively occurs in the blue region of the plot. 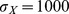 is also the condition for which we observe branching in the evolutionary simulations (see figure 3D and S2). The parameter settings that are used for these simulations are the following:
is also the condition for which we observe branching in the evolutionary simulations (see figure 3D and S2). The parameter settings that are used for these simulations are the following: 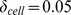 ,
, 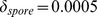 ,
, 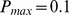 ,
, 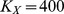 ,
, 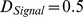 ,
, 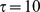 ,
, 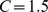 ,
, 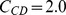 ,
, 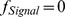 ,
, 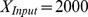 ,
, 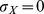 , 500 or 1000,
, 500 or 1000, 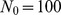 ,
, 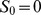 ,
, 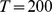 and
and 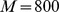 .
.
(TIF)
Selective advantage of cell-to-cell communication when varying the initial colony size. The relative fitness benefit of quorum-sensing signaling under different levels of variation in the initial colony size. In all other simulations we assumed that the initial colony size is constant. However, here we vary the initial colony size, which is taken from a normal distribution 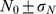 . The normal distribution of the colony size is truncated at 10 individuals, so that no colony was initiated with less than 10 individuals. The standard deviation is shown on the x-axis of this plot. Furthermore, we assume that there is no variation in the initial nutrient concentration (
. The normal distribution of the colony size is truncated at 10 individuals, so that no colony was initiated with less than 10 individuals. The standard deviation is shown on the x-axis of this plot. Furthermore, we assume that there is no variation in the initial nutrient concentration (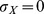 ). The bars show the average fitness of a quorum-sensing cell, relative to that of a cell that does not communicate. The error bars show the standard deviation over 100 replicate runs, each containing 200 colonies. When the relative fitness of a quorum-sensing cell is equal to 1 (the horizontal black line), there is no selective advantage for cell-to-cell communication. When it is higher than 1 there is a selective advantage for cell-to-cell communication. The parameter settings are the following:
). The bars show the average fitness of a quorum-sensing cell, relative to that of a cell that does not communicate. The error bars show the standard deviation over 100 replicate runs, each containing 200 colonies. When the relative fitness of a quorum-sensing cell is equal to 1 (the horizontal black line), there is no selective advantage for cell-to-cell communication. When it is higher than 1 there is a selective advantage for cell-to-cell communication. The parameter settings are the following: 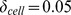 ,
, 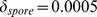 ,
, 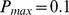 ,
, 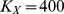 ,
, 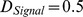 ,
, 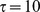 ,
, 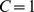 ,
, 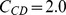 ,
, 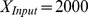 ,
, 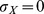 ,
, 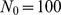 ,
, 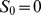 and
and 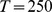 .
.
(TIF)
Unrooted phenograms. Four replicate studies for the evolution of cell-to-cell communication under the assumption of costly signal production. The simulations are performed under the same conditions as those shown in figure 5 of the main text and also the phenograms are constructed in accordance to figure 5. As shown in the main text, different genotypic clusters evolved and coexist, which consist of ‘cooperative’ and ‘cheating’ phenotypes. For each genotypic cluster a single representative phenotype is shown, which is produced by the most-abundant genotype within this cluster. For details see figure 5 of the main text.
(TIF)
The number of cells that are present at the end of colony growth for each phenotype, given the initial colony composition. On the left side of the table the colony composition is shown, because 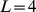 each phenotype could occur in 0%, 25%, 50%, 75% or 100% of the initial colony composition. The right side of the table shows the average number of cells that are present at the end of colony growth. The standard deviation is taken over 4 replicates, each replicate contains 200 colonies that are initiated with a nutrient input that is taken from the normal distribution,
each phenotype could occur in 0%, 25%, 50%, 75% or 100% of the initial colony composition. The right side of the table shows the average number of cells that are present at the end of colony growth. The standard deviation is taken over 4 replicates, each replicate contains 200 colonies that are initiated with a nutrient input that is taken from the normal distribution, 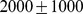 (for the 4 replicates the same nutrient inputs are used).
(for the 4 replicates the same nutrient inputs are used).
(PDF)
Supplementary material.
(PDF)
Acknowledgments
We thank Roberto Kolter, Hera Vlamakis, Bjorn Traag, Jan-Willem Veening and Franjo Weissing for their extensive advice, comments and support. Furthermore, we thank three anonymous reviewers for their helpful suggestions.
Funding Statement
The authors acknowledge support from the Groningen University Fund (Correspondence: 2010/719), the Marco Polo Fund (University of Groningen), the John Templeton Foundation, the National Science Foundation/National Institutes of Health joint program in mathematical biology (National Institutes of Health Grant R01GM078986), Bill and Melinda Gates Foundation (Grand Challenges Grant 37874) and the Milton Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Miller MB, Bassler BL (2001) Quorum sensing in bacteria. Annual Review of Microbiology 55: 165–199. [DOI] [PubMed] [Google Scholar]
- 2. Waters CM, Bassler BL (2005) Quorum sensing: Cell-to-cell communication in bacteria. Annual Review of Cell and Developmental Biology 21: 319–346. [DOI] [PubMed] [Google Scholar]
- 3. Camilli A, Bassler BL (2006) Bacterial small-molecule signaling pathways. Science 311: 1113–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Keller L, Surette MG (2006) Communication in bacteria: an ecological and evolutionary perspective. Nature Reviews Microbiology 4: 249–258. [DOI] [PubMed] [Google Scholar]
- 5. Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. Journal of Bacteriology 176: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parsek MR, Greenberg EP (1999) Quorum sensing signals in development of Pseudomonas aerug-inosa biofilms. Methods in Enzymology 310: 43–55. [DOI] [PubMed] [Google Scholar]
- 7. Kleerebezem M, Quadri LE (2001) Peptide pheromone-dependent regulation of antimicrobial peptide production in gram-positive bacteria: a case of multicellular behavior. Peptides 22: 1579–1596. [DOI] [PubMed] [Google Scholar]
- 8. Whitehead NA, Barnard AML, Slater H, Simpson NJL, Salmond GPC (2001) Quorum-sensing in gram-negative bacteria. FEMS Microbiology Reviews 25: 365–404. [DOI] [PubMed] [Google Scholar]
- 9. Parsek MR, Greenberg EP (2005) Sociomicrobiology: the connections between quorum sensing and biofilms. Trends in Microbiology 13: 27–33. [DOI] [PubMed] [Google Scholar]
- 10. Lopez D, Vlamakis H, Kolter R (2010) Biofilms. Cold Spring Harbor Perspectives in Biology 2: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sturme M, Kleerebezem M, Nakayama J, Akkermans A, Vaughan E, et al. (2002) Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek 81: 233–243. [DOI] [PubMed] [Google Scholar]
- 12. Lopez D, Vlamakis H, Kolter R (2009) Generation of multiple cell types in Bacillus subtilis . FEMS Microbiology Reviews 33: 152–163. [DOI] [PubMed] [Google Scholar]
- 13. Lopez D, Vlamakis H, Losick R, Kolter R (2009) Paracrine signaling in a bacterium. Genes & Development 23: 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lopez D, Kolter R (2010) Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis . FEMS Microbiology Reviews 34: 134–149. [DOI] [PubMed] [Google Scholar]
- 15. Earl AM, Losick R, Kolter R (2008) Ecology and genomics of Bacillus subtilis . Trends in Microbiology 16: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vlamakis H, Aguilar C, Losick R, Kolter R (2008) Control of cell fate by the formation of an architecturally complex bacterial community. Genes & Development 22: 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nicholson W, Munakata N, Horneck G, Melosh H, Setlow P (2000) Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiology and Molecular Biology Reviews 64: 548–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Setlow P (2007) I will survive: DNA protection in bacterial spores. Trends in Microbiology 15: 172–180. [DOI] [PubMed] [Google Scholar]
- 19. Piggot PJ, Hilbert DW (2004) Sporulation of Bacillus subtilis . Current Opinion in Microbiology 7: 579–586. [DOI] [PubMed] [Google Scholar]
- 20. Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, et al. (2004) The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis . PLoS Biology 2: e328 doi:10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grossman AD (1995) Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis . Annual Review of Genetics 29: 477–508. [DOI] [PubMed] [Google Scholar]
- 22. Sonenshein AL (2000) Control of sporulation initiation in Bacillus subtilis . Current Opinion in Microbiology 3: 561–566. [DOI] [PubMed] [Google Scholar]
- 23. Molle V, Fujita M, Jensen S, Eichenberger P, González-Pastor J, et al. (2003) The Spo0A regulon of Bacillus subtilis . Molecular microbiology 50: 1683–1701. [DOI] [PubMed] [Google Scholar]
- 24. Higgins D, Dworkin J (2012) Recent progress in Bacillus subtilis sporulation. FEMS Microbiology Reviews 36: 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fujita M, Gonzalez-Pastor JE, Losick R (2005) High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis . Journal of Bacteriology 187: 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fujita M, Losick R (2005) Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes & Development 19: 2236–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chastanet A, Vitkup D, Yuan GC, Norman TM, Liu JS, et al. (2010) Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis . Proceedings of the National Academy of Sciences of the United States of America 107: 8486–8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Jong IG, Veening JW, Kuipers OP (2010) Heterochronic phosphorelay gene expression as a source of heterogeneity in Bacillus subtilis spore formation. Journal of Bacteriology 192: 2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pottathil M, Lazazzera BA (2003) The extracellular Phr peptide-rap phosphatase signaling circuit of Bacillus subtilis . Frontiers in Bioscience 8: D32–D45. [DOI] [PubMed] [Google Scholar]
- 30. Grossman AD, Losick R (1988) Extracellular control of spore formation in Bacillus subtilis . Proceedings of the National Academy of Sciences of the United States of America 85: 4369–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lazazzera BA, Solomon JM, Grossman AD (1997) An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis . Cell 89: 917–925. [DOI] [PubMed] [Google Scholar]
- 32. Lazazzera BA (2001) The intracellular function of extracellular signaling peptides. Peptides 22: 1519–1527. [DOI] [PubMed] [Google Scholar]
- 33. Perego M, Brannigan JA (2001) Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides 22: 1541–1547. [DOI] [PubMed] [Google Scholar]
- 34. Lazazzera BA (2000) Quorum sensing and starvation: signals for entry into stationary phase. Current Opinion in Microbiology 3: 177–182. [DOI] [PubMed] [Google Scholar]
- 35. Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL (2001) Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes & Development 15: 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bischofs IB, Hug JA, Liu AW, Wolf DM, Arkin AP (2009) Complexity in bacterial cell-cell communication: Quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay. Proceedings of the National Academy of Sciences of the United States of America 106: 6459–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siala A, Hill IR, Gray TRG (1974) Populations of spore-forming bacteria in an acid forest soil, with special reference to bacillus subtilis. Journal of General Microbiology 81: 183–190. [Google Scholar]
- 38. Vilain S, Luo Y, Hildreth MB, Brozel VS (2006) Analysis of the life cycle of the soil saprophyte bacillus cereus in liquid soil extract and in soil. Applied and Environmental Microbiology 72: 4970–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davey ME, O'Toole GA (2000) Microbial biofilms: from ecology to molecular genetics. Microbiology and Molecular Biology Reviews 64: 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, et al. (2008) Bet-hedging and epigenetic inheritance in bacterial cell development. Proceedings of the National Academy of Sciences of the United States of America 105: 4393–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gross L (2012) Built-in timer delays differentiation. PLoS Biology 10: e1001254 doi:10.1371/journal.pbio.1001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Levine J, Fontes M, Dworkin J, Elowitz M (2012) Pulsed feedback defers cellular differentiation. PLoS biology 10: e1001252 doi:10.1371/journal.pbio.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brookfield JEY (1998) Quorum sensing and group selection. Evolution 52: 1263–1269. [DOI] [PubMed] [Google Scholar]
- 44. Brown SP, Johnstone RA (2001) Cooperation in the dark: signalling and collective action in quorum-sensing bacteria. Proceedings of the Royal Society of London Series B-Biological Sciences 268: 961–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nadell CD, Xavier JB, Levin SA, Foster KR (2008) The evolution of quorum sensing in bacterial biofilms. PLoS Biology 6: e14 doi:10.1371/journal.pbio.0060014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Czaran T, Hoekstra RF (2009) Microbial communication, cooperation and cheating: Quorum sensing drives the evolution of cooperation in bacteria. PLoS One 4: e6655 doi:10.1371/journal.pone.0006655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schuster M, Lostroh CP, Ogi T, Greenberg EP (2003) Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. Journal of Bacteriology 185: 2066–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meibom K, Blokesch M, Dolganov N, Wu C (2005) Schoolnik G (2005) Chitin induces natural competence in Vibrio cholerae . Science 310: 1824–1827. [DOI] [PubMed] [Google Scholar]
- 49. Ng W, Bassler B (2009) Bacterial quorum-sensing network architectures. Annual review of genetics 43: 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kussell E, Kishony R, Balaban NQ, Leibler S (2005) Bacterial persistence: A model of survival in changing environments. Genetics 169: 1807–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gardner A, West SA, Griffin AS (2007) Is bacterial persistence a social trait? PLoS One 2: e752 doi:10.1371/journal.pone.0000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Davidson CJ, Surette MG (2008) Individuality in bacteria. Annual Review of Genetics 42: 253–268. [DOI] [PubMed] [Google Scholar]
- 53. de Jong I, Haccou P, Kuipers O (2011) Bet hedging or not? A guide to proper classification of microbial survival strategies. Bioessays 33: 215–223. [DOI] [PubMed] [Google Scholar]
- 54. Donaldson-Matasci M, Lachmann M, Bergstrom C (2008) Phenotypic diversity as an adaptation to environmental uncertainty. Evolutionary Ecology Research 10: 493–515. [Google Scholar]
- 55. Scheiner SM (1993) Genetics and evolution of phenotypic plasticity. Annual Review of Ecology and Systematics 24: 35–68. [Google Scholar]
- 56. Ackermann M, Doebeli M (2004) Evolution of niche width and adaptive diversification. Evolution 58: 2599–2612. [DOI] [PubMed] [Google Scholar]
- 57. Leimar O, Hammerstein P, Van Dooren TJM (2006) A new perspective on developmental plasticity and the principles of adaptive morph determination. American Naturalist 167: 367–376. [DOI] [PubMed] [Google Scholar]
- 58. Leimar O (2009) Environmental and genetic cues in the evolution of phenotypic polymorphism. Evolutionary Ecology 23: 125–135. [Google Scholar]
- 59. Damore J, Gore J (2012) Understanding microbial cooperation. Journal of Theoretical Biology 299: 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. West S, Griffin A, Gardner A, Diggle S (2006) Social evolution theory for microorganisms. Nature Reviews Microbiology 4: 597–607. [DOI] [PubMed] [Google Scholar]
- 61. Diggle S, Gardner A, West S, Griffin A (2007) Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Philosophical Transactions of the Royal Society B: Biological Sciences 362: 1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maynard Smith J, Harper D (2003) Animal signals. New York, NY: Oxford University Press.
- 63. Velicer GJ, Kroos L, Lenski RE (2000) Developmental cheating in the social bacterium Myxococcus xanthus . Nature 404: 598–601. [DOI] [PubMed] [Google Scholar]
- 64. Diggle SP, Griffin AS, Campbell GS, West SA (2007) Cooperation and conict in quorum-sensing bacterial populations. Nature 450: 411–414. [DOI] [PubMed] [Google Scholar]
- 65. Velicer GJ, Vos M (2009) Sociobiology of the Myxobacteria. Annual Review of Microbiology 63: 599–623. [DOI] [PubMed] [Google Scholar]
- 66. Nowak MA (2012) Evolving cooperation. Journal of Theoretical Biology 299: 1–8. [DOI] [PubMed] [Google Scholar]
- 67.Maynard Smith J (1982) Evolution and the Theory of Games. Cambridge, UK: Cambridge University Press.
- 68.Hofbauer J, Sigmund K (1998) Evolutionary Games and Population Dynamics. Cambridge, UK: Cambridge University Press.
- 69. Nowak MA (2006) Five rules for the evolution of cooperation. Science 314: 1560–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hauert C, Michor F, Nowak M, Doebeli M (2006) Synergy and discounting of cooperation in social dilemmas. Journal of Theoretical Biology 239: 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Doebeli M, Hauert C, Killingback T (2004) The evolutionary origin of cooperators and defectors. Science 306: 859–862. [DOI] [PubMed] [Google Scholar]
- 72. Botero C, Pen I, Komdeur J, Weissing F (2010) The evolution of individual variation in communication strategies. Evolution 64: 3123–3133. [DOI] [PubMed] [Google Scholar]
- 73. Nowak MA, Sigmund K (2004) Evolutionary dynamics of biological games. Science 303: 793–799. [DOI] [PubMed] [Google Scholar]
- 74. Traulsen A, Nowak MA (2006) Evolution of cooperation by multilevel selection. Proceedings of the National Academy of Sciences of the United States of America 103: 10952–10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nowak MA, May RM (1992) Evolutionary games and spatial chaos. Nature 359: 826–829. [Google Scholar]
- 76. Nowak MA, Bonhoeffer S, May RM (1994) Spatial games and the maintenance of cooperation. Proceedings of the National Academy of Sciences of the United States of America 91: 4877–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rousset F (2004) Genetic structure and selection in subdivided populations. Princeton, NJ: Princeton University Press.
- 78. Ohtsuki H, Hauert C, Lieberman E, Nowak MA (2006) A simple rule for the evolution of cooperation on graphs and social networks. Nature 441: 502–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Taylor PD, Day T, Wild G (2007) Evolution of cooperation in a finite homogeneous graph. Nature 447: 469–472. [DOI] [PubMed] [Google Scholar]
- 80. Tarnita CE, Antal T, Ohtsuki H, Nowak MA (2009) Evolutionary dynamics in set structured populations. Proceedings of the National Academy of Sciences of the United States of America 106: 8601–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tarnita CE, Ohtsuki H, Antal T, Fu F, Nowak MA (2009) Strategy selection in structured populations. Journal of Theoretical Biology 259: 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Taylor PD, Grafen A (2010) Relatedness with different interaction configurations. Journal of Theoretical Biology 262: 391–397. [DOI] [PubMed] [Google Scholar]
- 83. Nowak MA, Tarnita CE, Antal T (2010) Evolutionary dynamics in structured populations. Philosophical Transactions of the Royal Society B: Biological Sciences 365: 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fletcher J, Doebeli M (2009) A simple and general explanation for the evolution of altruism. Proceedings of the Royal Society B: Biological Sciences 276: 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hauert C, Holmes M, Doebeli M (2006) Evolutionary games and population dynamics: maintenance of cooperation in public goods games. Proceedings of the Royal Society B: Biological Sciences 273: 2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cremer J, Melbinger A, Frey E (2012) Growth dynamics and the evolution of cooperation in microbial populations. Scientific Reports 2: 281 doi:10.1038/srep00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hallatschek O, Hersen P, Ramanathan S, Nelson D (2007) Genetic drift at expanding frontiers promotes gene segregation. Proceedings of the National Academy of Sciences 104: 19926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nadell CD, Foster KR, Xavier JB (2010) Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Computational Biology 6: e1000716 doi:10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ernande B, Dieckmann U (2004) The evolution of phenotypic plasticity in spatially structured environments: implications of intraspecific competition, plasticity costs and environmental characteristics. Journal of Evolutionary Biology 17: 613–628. [DOI] [PubMed] [Google Scholar]
- 90. Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, et al. (1998) The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280: 295–298. [DOI] [PubMed] [Google Scholar]
- 91. Zhu J, Mekalanos JJ (2003) Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae . Developmental Cell 5: 647–656. [DOI] [PubMed] [Google Scholar]
- 92. Lupp C, Ruby EG (2005) Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. Journal of Bacteriology 187: 3620–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dunny G (2007) The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution. Philosophical Transactions of the Royal Society B-Biological Sciences 362: 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sakuragi Y, Kolter R (2007) Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa . Journal of Bacteriology 189: 5383–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Perkins TJ, Swain PS (2009) Strategies for cellular decision-making. Molecular Systems Biology 5: 326 doi:10.1038/msb.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. van Hijum S, Medema M, Kuipers O (2009) Mechanisms and evolution of control logic in prokaryotic transcriptional regulation. Microbiology and Molecular Biology Reviews 73: 481–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Brennan M, Cheong R, Levchenko A (2012) How information theory handles cell signaling and uncertainty. Science 338: 334–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Long T, Tu K, Wang Y, Mehta P, Ong N, et al. (2009) Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biology 7: e1000068 doi:10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ireton K, Rudner DZ, Siranosian KJ, Grossman AD (1993) Integration of multiple developmental signals in bacillus subtilis through the Spo0A transcription factor. Genes & Development 7: 283–294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The evolution of cell-to-cell communication under clonal growth conditions. These plots depict the evolutionary trajectories of 100 runs performed under the same conditions than those shown in figure 3A. The left plot shows for every 20.000 time steps the average evolved genotype, which is given by the mean value of 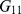 and
and 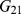 over 100 runs. The error bars show the standard deviations. In total 600.000 time steps of evolution are shown; starting from the dark-blue dot till the red dot. The right plot shows a subset of runs that evolved cell-to-cell communication, using the same color coding. For parameter settings see figure 3 of the main text.
over 100 runs. The error bars show the standard deviations. In total 600.000 time steps of evolution are shown; starting from the dark-blue dot till the red dot. The right plot shows a subset of runs that evolved cell-to-cell communication, using the same color coding. For parameter settings see figure 3 of the main text.
(TIF)
The evolution of cell-to-cell communication under non-clonal growth conditions. These plots show the evolved values of 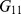 for 18 independent evolutionary runs. The conditions of these runs are the same as those shown in figure 3D, in which a early and late sporulating ecotype evolved. The simulations ran for 600.000 time steps, of which
for 18 independent evolutionary runs. The conditions of these runs are the same as those shown in figure 3D, in which a early and late sporulating ecotype evolved. The simulations ran for 600.000 time steps, of which 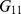 is shown for the most-abundant genotypes (present in the population in more than 100 copies) at 5.000 time steps intervals. For parameter settings see figure 3 of the main text.
is shown for the most-abundant genotypes (present in the population in more than 100 copies) at 5.000 time steps intervals. For parameter settings see figure 3 of the main text.
(TIF)
The evolution of cell-to-cell communication when the activation thresholds can evolve. The plots show the evolution of cell-to-cell communication under the same conditions as those shown in figure S1, S2 and 3. However, in contrast to these previous figures, the simulations in this figure allowed for the evolution of the activation thresholds (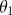 and
and 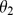 ). To facilitate a comparison between the plots in this figure and figure S1, S2 and 3, all shown connection weights are corrected for the evolved activation threshold. This is done by dividing the connection weights by two times the absolute value of the associated activation threshold (notice that for the previous figures we assumed that
). To facilitate a comparison between the plots in this figure and figure S1, S2 and 3, all shown connection weights are corrected for the evolved activation threshold. This is done by dividing the connection weights by two times the absolute value of the associated activation threshold (notice that for the previous figures we assumed that 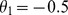 ). Plot A and B show the evolution of cell-to-cell communication under clonal growth conditions (see figure S1). Plot A shows for every 20.000 time steps the average evolved genotype, which is given by the mean value of
). Plot A and B show the evolution of cell-to-cell communication under clonal growth conditions (see figure S1). Plot A shows for every 20.000 time steps the average evolved genotype, which is given by the mean value of 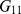 and
and 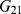 over 100 runs. The error bars show the standard deviations. In total 600.000 time steps of evolution are shown; starting from the dark-blue dot till the red dot. Plot B shows a subset of runs that evolved cell-to-cell communication, using the same color coding. Plot C shows the evolution of cell-to-cell communication under non-clonal growth conditions. The subplots show the evolved values of
over 100 runs. The error bars show the standard deviations. In total 600.000 time steps of evolution are shown; starting from the dark-blue dot till the red dot. Plot B shows a subset of runs that evolved cell-to-cell communication, using the same color coding. Plot C shows the evolution of cell-to-cell communication under non-clonal growth conditions. The subplots show the evolved values of 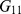 for the most-abundant genotypes (present in the population in more than 100 copies) at 5.000 time steps intervals. Only
for the most-abundant genotypes (present in the population in more than 100 copies) at 5.000 time steps intervals. Only 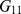 is shown, since this illustrates the evolution of the early and late sporulating ecotype, as shown in figure 3 and S2. For parameter settings see figure 3 of the main text.
is shown, since this illustrates the evolution of the early and late sporulating ecotype, as shown in figure 3 and S2. For parameter settings see figure 3 of the main text.
(TIF)
Growth of mixed colonies, consisting of early and late sporulating genotypes, at high and low nutrient inputs. The left plots show colony growth at a low nutrient input (i.e. 1000) and the right plots show the same for a high nutrient input (i.e. 2500). The early and late sporulating genotypes shown here do not have cell-to-cell communication and only differ with respect to the nutrient concentration at which sporulation is initiated: the early sporulating genotype sporulates at a nutrient concentration of 1000 (blue dashed line in the upper plot), while the late sporulating genotype sporulates at a nutrient concentration of 500 (red dashed line in the upper plot). The upper plots show the nutrient concentration (green line), the middle plots show the number of cells and the lower plots show the number of spores of the early (blue) and late (red) sporulating genotypes. Each line is the average of 1000 replicate runs and the shaded area shows the associated standard deviation. The parameter settings that are used for these simulations are the following: 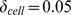 ,
, 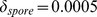 ,
, 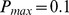 ,
, 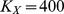 ,
, 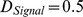 ,
, 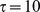 ,
, 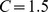 ,
, 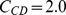 ,
, 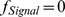 ,
, 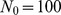 ,
, 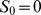 and
and 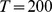 .
.
(TIF)
Overview of the colony performance at different nutrient inputs (i.e. nutrient concentration at onset of colony growth). Plot A: the distribution of nutrient inputs from which the nutrient input of a colony is taken in the evolutionary simulations (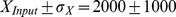 ). Plot B: The average colony size at the end of colony growth for two different genotypes that do not use cell-to-cell communication. The red line shows the average colony size for a late sporulating genotype (this genotype initiates sporulation when
). Plot B: The average colony size at the end of colony growth for two different genotypes that do not use cell-to-cell communication. The red line shows the average colony size for a late sporulating genotype (this genotype initiates sporulation when 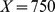 ) and the blue line shows the average colony size for an early sporulating genotype (this genotype initiates sporulation when
) and the blue line shows the average colony size for an early sporulating genotype (this genotype initiates sporulation when 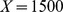 ). The dotted lines show the standard deviation from the average at each nutrient input. Plot C: The average colony size at the end of colony growth for the evolved genotype of figure 3. The evolved genotype is the dominant genotype that is present at the end of the simulation. This genotype evolved cell-to-cell communication (figure 3D) and has the following genotype:
). The dotted lines show the standard deviation from the average at each nutrient input. Plot C: The average colony size at the end of colony growth for the evolved genotype of figure 3. The evolved genotype is the dominant genotype that is present at the end of the simulation. This genotype evolved cell-to-cell communication (figure 3D) and has the following genotype: 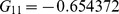 ;
; 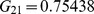 ;
; 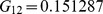 ;
; 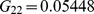 ;
; 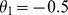 and
and 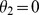 . Even though the optimal genotype is not reached yet (see figure 3C), this genotype performs considerably well under all possible nutrient inputs.
. Even though the optimal genotype is not reached yet (see figure 3C), this genotype performs considerably well under all possible nutrient inputs.
(TIF)
Invasion analysis of early and late sporulating genotype. The relative fitness of the early and late sporulating genotypes for different starting conditions: (A) 10% of early sporulating genotypes or (B) 10% of late sporulating genotypes. When a genotype's fitness is higher than one it is favored by selection and when it is lower than one it is selected against. There is frequency-dependent selection, since each genotype has a fitness advantage when it is rare: the early sporulating genotype has a fitness advantage when it is rare (A) and the late sporulating genotype has a fitness advantage when it is rare (B). The genotypes only differ in their sensitivity towards the nutrient concentration: 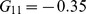 for the early sporulating genotype and
for the early sporulating genotype and 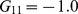 for the late sporulating genotype. Each bar shows the average fitness over 10 replicates and the error bars show the standard deviation. Each replicate consists of 200 colonies which are grown under the following conditions:
for the late sporulating genotype. Each bar shows the average fitness over 10 replicates and the error bars show the standard deviation. Each replicate consists of 200 colonies which are grown under the following conditions: 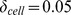 ,
, 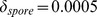 ,
, 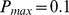 ,
, 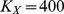 ,
, 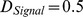 ,
, 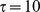 ,
, 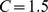 ,
, 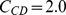 ,
,  ,
, 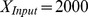 ,
, 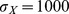 ,
, 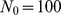 ,
, 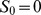 ,
, 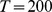 and
and 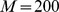 .
.
(TIF)
Pairwise invasibility plots for different levels of environmental variation. Each plot shows the invasibility of a mutant, given the presence a certain resident population. The genotypes differ in the nutrient concentration at which sporulation is initiated, which is shown for the resident genotype on the x-axis and for the mutant genotype on the y-axis. For each combination of mutant and resident, the invasibility of the mutant is tested by growing 800 colonies that are initiated with 10% of mutants. The mutant is said to invade when its average fitness is higher then that of the resident (red area), while it goes extinct when its fitness is lower (blue area). The black diagonal line shows when the resident and mutant have the same genotype and hence fitness. The different plots show the invasibility for various levels of environmental variation: 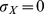 ,
, 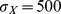 or
or 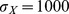 . For
. For 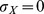 there is an evolutionary stable strategy (ESS) that cannot be invaded by mutants. This is illustrated by the white dot on the black diagonal line (the ESS is a resident genotype that sporulates at a nutrient concentration around 1100). The vertical white line, which is associated with the ESS, occurs exclusively in the blue region of the plot. This shows that none of the mutants can invade the ESS resident population. For
there is an evolutionary stable strategy (ESS) that cannot be invaded by mutants. This is illustrated by the white dot on the black diagonal line (the ESS is a resident genotype that sporulates at a nutrient concentration around 1100). The vertical white line, which is associated with the ESS, occurs exclusively in the blue region of the plot. This shows that none of the mutants can invade the ESS resident population. For 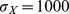 there is no ESS (as illustrated by the black dot), since one cannot draw a vertical white line that exclusively occurs in the blue region of the plot.
there is no ESS (as illustrated by the black dot), since one cannot draw a vertical white line that exclusively occurs in the blue region of the plot. 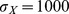 is also the condition for which we observe branching in the evolutionary simulations (see figure 3D and S2). The parameter settings that are used for these simulations are the following:
is also the condition for which we observe branching in the evolutionary simulations (see figure 3D and S2). The parameter settings that are used for these simulations are the following: 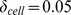 ,
, 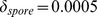 ,
, 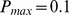 ,
, 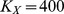 ,
, 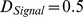 ,
, 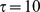 ,
, 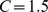 ,
, 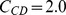 ,
,  ,
, 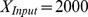 ,
, 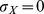 , 500 or 1000,
, 500 or 1000, 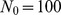 ,
, 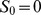 ,
, 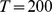 and
and 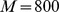 .
.
(TIF)
Selective advantage of cell-to-cell communication when varying the initial colony size. The relative fitness benefit of quorum-sensing signaling under different levels of variation in the initial colony size. In all other simulations we assumed that the initial colony size is constant. However, here we vary the initial colony size, which is taken from a normal distribution 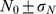 . The normal distribution of the colony size is truncated at 10 individuals, so that no colony was initiated with less than 10 individuals. The standard deviation is shown on the x-axis of this plot. Furthermore, we assume that there is no variation in the initial nutrient concentration (
. The normal distribution of the colony size is truncated at 10 individuals, so that no colony was initiated with less than 10 individuals. The standard deviation is shown on the x-axis of this plot. Furthermore, we assume that there is no variation in the initial nutrient concentration (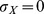 ). The bars show the average fitness of a quorum-sensing cell, relative to that of a cell that does not communicate. The error bars show the standard deviation over 100 replicate runs, each containing 200 colonies. When the relative fitness of a quorum-sensing cell is equal to 1 (the horizontal black line), there is no selective advantage for cell-to-cell communication. When it is higher than 1 there is a selective advantage for cell-to-cell communication. The parameter settings are the following:
). The bars show the average fitness of a quorum-sensing cell, relative to that of a cell that does not communicate. The error bars show the standard deviation over 100 replicate runs, each containing 200 colonies. When the relative fitness of a quorum-sensing cell is equal to 1 (the horizontal black line), there is no selective advantage for cell-to-cell communication. When it is higher than 1 there is a selective advantage for cell-to-cell communication. The parameter settings are the following: 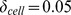 ,
, 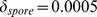 ,
, 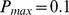 ,
, 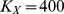 ,
, 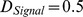 ,
, 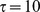 ,
, 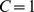 ,
, 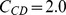 ,
, 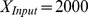 ,
, 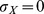 ,
, 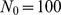 ,
, 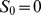 and
and 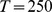 .
.
(TIF)
Unrooted phenograms. Four replicate studies for the evolution of cell-to-cell communication under the assumption of costly signal production. The simulations are performed under the same conditions as those shown in figure 5 of the main text and also the phenograms are constructed in accordance to figure 5. As shown in the main text, different genotypic clusters evolved and coexist, which consist of ‘cooperative’ and ‘cheating’ phenotypes. For each genotypic cluster a single representative phenotype is shown, which is produced by the most-abundant genotype within this cluster. For details see figure 5 of the main text.
(TIF)
The number of cells that are present at the end of colony growth for each phenotype, given the initial colony composition. On the left side of the table the colony composition is shown, because 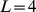 each phenotype could occur in 0%, 25%, 50%, 75% or 100% of the initial colony composition. The right side of the table shows the average number of cells that are present at the end of colony growth. The standard deviation is taken over 4 replicates, each replicate contains 200 colonies that are initiated with a nutrient input that is taken from the normal distribution,
each phenotype could occur in 0%, 25%, 50%, 75% or 100% of the initial colony composition. The right side of the table shows the average number of cells that are present at the end of colony growth. The standard deviation is taken over 4 replicates, each replicate contains 200 colonies that are initiated with a nutrient input that is taken from the normal distribution, 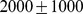 (for the 4 replicates the same nutrient inputs are used).
(for the 4 replicates the same nutrient inputs are used).
(PDF)
Supplementary material.
(PDF)