Abstract
Coral reefs are under considerable pressure from global stressors such as elevated sea surface temperature and ocean acidification, as well as local factors including eutrophication and poor water quality. Marine sponges are diverse, abundant and ecologically important components of coral reefs in both coastal and offshore environments. Due to their exceptionally high filtration rates, sponges also form a crucial coupling point between benthic and pelagic habitats. Sponges harbor extensive microbial communities, with many microbial phylotypes found exclusively in sponges and thought to contribute to the health and survival of their hosts. Manipulative experiments were undertaken to ascertain the impact of elevated nutrients and seawater temperature on health and microbial community dynamics in the Great Barrier Reef sponge Rhopaloeides odorabile. R. odorabile exposed to elevated nutrient levels including 10 µmol/L total nitrogen at 31°C appeared visually similar to those maintained under ambient seawater conditions after 7 days. The symbiotic microbial community, analyzed by 16S rRNA gene pyrotag sequencing, was highly conserved for the duration of the experiment at both phylum and operational taxonomic unit (OTU) (97% sequence similarity) levels with 19 bacterial phyla and 1743 OTUs identified across all samples. Additionally, elevated nutrients and temperatures did not alter the archaeal associations in R. odorabile, with sequencing of 16S rRNA gene libraries revealing similar Thaumarchaeota diversity and denaturing gradient gel electrophoresis (DGGE) revealing consistent amoA gene patterns, across all experimental treatments. A conserved eukaryotic community was also identified across all nutrient and temperature treatments by DGGE. The highly stable microbial associations indicate that R. odorabile symbionts are capable of withstanding short-term exposure to elevated nutrient concentrations and sub-lethal temperatures.
Introduction
The Great Barrier Reef (GBR) hosts high biodiversity and is the world’s largest coral reef ecosystem. At almost 2000 km long it was declared a World Heritage Area in 1981 [1]. Degradation of coastal marine ecosystems is occurring globally due to over-fishing, declining water quality and climate change [2], [3]. Despite the GBR’s protected status it is still exposed to anthropogenic and environmental pressures, making degradation due to terrestrial run-off the focus of intense management efforts [4]. Twenty-six major catchments, in which a wide range of industrial and agricultural activities take place, border the GBR [5]. Fertilisers used in cattle grazing, sugarcane production and horticulture can flow into the marine environment [6], with 80% of the total anthropogenic dissolved inorganic nitrogen (DIN) introduced into the GBR ecosystem thought to come from fertilisers (Table 1) [2], [7]. Moreover, nitrogen and phosphorus loads have increased by factors of approximately 6 and 9, respectively, since European settlement ca.1830 [8]. Catchment areas in the GBR are characterized by distinct wet/dry seasonal rainfall and are subject to intense cyclonic rainfall over periods of days to a few weeks [9]. River discharge of nutrients into the GBR therefore occurs almost entirely in large pulse events or flood plumes [6], [10] which generally affect reefs within 20 km of the coast (∼27% of all reefs). The resulting elevated nutrient levels can be 2–100 times higher than ambient [6], [10], [11] but are relatively short-lived, detectable for only 3–14 days after flood plume events [12]. Cyclonic events are increasing in frequency and intensity, with the most recent on the GBR (in January 2011) delivering extremely high levels of nutrients from agricultural and urban catchments to the reef environment [13]–[15]. Nutrient levels in ambient (non-flood) conditions are presented in Table 2.
Table 1. Total yearly inputs of anthropogenic nutrient loads into the GBR, 1 [2], 2 [121].
| Anthropogenic nutrient loads1,2 | Tonnes/year |
| Total nitrogen (TN) | 80000 |
| Dissolved inorganic nitrogen (DIN) | 11000 |
| Dissolved organic nitrogen (DON) | 6900 |
| Particulate nitrogen (PN) | 52000 |
| Total phosphorus (TP) | 16000 |
| Dissolved inorganic phosphorus (DIP) | 800 |
| Dissolved organic phosphorus (DOP) | 470 |
| Particulate phosphorus (PP) | 13000 |
Table 2. The level of nutrients in ambient (non flood) waters, reported from Pelorus Island from 2005–2011 1 [4].
| Nutrient | NH4 | NO3 | DON | PN | DOP | PP | DOC | POC |
| Ambient wet season1 | 0.13 | 0.06 | 5.64 | 1.08 | 0.14 | 0.1 | 70.66 | 9.44 |
| Ambient dry season1 | 0.04 | 0.03 | 5.49 | 0.74 | 0.17 | 0.06 | 59.25 | 6.93 |
Parameters are in µM for dissolved inorganic nutrients (NH4, NO3), dissolved organic nitrogen, phosphorus and carbon (DON, DOP, DOC) and particulate nitrogen, phosphorus and organic carbon (PN, PP, POC).
Marine sponges are important components of coastal and offshore coral reefs, exhibiting high diversity, high biomass and the ability to influence both benthic and pelagic processes [16]. Sponges also harbour extensive microbial communities which can comprise up to 35% of sponge tissue volume and include bacteria, archaea and eukarya [17], [18]. To date, 32 bacterial phyla and candidate phyla have been reported from sponges [19], [20], with some phylotypes appearing to occur exclusively in sponges and not in the surrounding environment i.e. so-called sponge-specific clusters (SCs) or sponge- and coral-specific clusters (SCCs) [17], [21]–[23]. In areas such as coral reefs, where dissolved nutrients and particulate organic matter are scarce, sponges may experience nitrogen limitation and symbiotic microorganisms are thought to contribute to nitrogen cycling within the host. Both autotrophic (such as Cyanobacteria) and heterotrophic symbionts may contribute to the nitrogen budget of sponges by fixing atmospheric nitrogen [24]–[26]. In low nutrient waters symbionts are thought to benefit by recycling nitrogenous waste excreted from the sponge host [18]. Ammonia-oxidising bacteria (AOB) of the genera Nitrosospira and Nitrosococcus [27] and the ammonia-oxidising archaea (AOA), such as “Candidatus Cenarchaeum symbiosum” [28], [29], which convert ammonia to nitrite, have all been identified in sponges. Nitrite-oxidising bacteria (NOB) such as Nitrospina and Nitrospira have also been detected in many sponge species [21], [30]–[33], as have denitrification and anaerobic ammonia oxidation (anammox) processes [31], [34]. Symbiosis between nitrogen-transforming microbes and sponges influences not only the ecology of the host but also the wider reef ecosystem (reviewed by [35]).
Elevated nutrient levels have been highlighted as a cause of coral reef decline, with some studies reporting an increase in the severity of coral diseases such as aspergillosis and yellow blotch [36]–[42]. Additionally, both resilience [43], [44] and sensitivity of coral-microbial associations due to elevated nutrient levels have been reported [45]–[47]. Despite the importance of sponge nitrogen cycling to coral reef ecosystems [35], very little research has addressed the sensitivity of sponge–microbial partnerships to nutrient enrichment, with the effects of eutrophication more widely reported for free-living microbial communities. In general, as the availability of nitrogen and phosphorus increases, phytoplankton and bacterial production increases which leads to a higher biological oxygen demand and increases the sedimentation rate of particulate matter [48]. In the early stages of nutrient loading within Chesapeake Bay [49], bacterioplankton communities remain dominated by SAR11, SAR86 and picocyanobacteria, however as anoxic conditions develop the bacterial community shifts to anaerobic members of the Firmicutes, Bacteroidetes and sulphur-oxidising Gammaproteobacteria. Another effect commonly observed in nutrient-rich environments is an increased abundance of prokaryotic cells; in natural seawater amendments (e.g. the addition of nutrient-rich deep waters to nutrient-depleted surface waters) an increase in the abundance of taxa such as SAR11 and marine Actinobacteria was reported [50]–[52]. Several studies have also addressed the effect of nutrient addition on bacterial community structure in the marine environment [53]–[57], however results have been variable due to different experimental methodologies and the high spatial and temporal variability of free-living marine communities [48]. Despite the variable microbial responses to experimental nutrient amendments, few microbial communities have been shown to be resistant to change after environmental disturbance.
Here we analyzed how the microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile responded to experimental nutrient exposures under ambient and elevated seawater temperature. While the interactive effects of multiple stressors have previously been explored in microbial biofilms [58], coral larvae [59], foraminifera [60], coral pathogens [41] and adult corals [61], the impact of combined anthropogenic stressors on marine sponges was unknown. Elevated seawater temperature has previously been shown to cause a shift in the dominant microbial community on marine sponges as well as a decline in sponge health [62]–[64], with some mass mortality events concomitant with anomalies in sea surface temperature [65], [66]. Previous experiments have demonstrated that adult R. odorabile exhibit necrosis and a loss of microbial symbionts within 72 h at 33°C [67], with subsequent experiments confirming a narrow thermal threshold for the host and symbiont community between 31–32°C [68], [69]. Here we investigated the combined effects of water quality and elevated seawater temperature by exposing sponges to a range of elevated nutrient levels under ambient (27°C) and sub-lethal (31°C) seawater temperatures.
Materials and Methods
Sample Collection and Experimental Design
18 R. odorabile individuals were collected by SCUBA from Pelorus Island on the Great Barrier Reef (GBR), Australia (18° 32.710′ S, 146° 29.273′ E) in July 2010. All necessary permits were obtained from the Great Barrier Reef Marine Park Authority for all of the described experimental studies. Donor sponges were cut into a total of 100 clones (each approximately 15 cm3) and randomly transferred to plastic racks which were secured with weights to the benthos [70]. The sponge clones were allowed to heal on the reef for 12 weeks before collection and transportation to an indoor aquarium at the Orpheus Island Research Station on the GBR (18°36.5′ S, 146°29.4′ E). Previous research has shown that there is little variability of the microbial communities between replicate clones that come from different donor individuals [71]. The experimental design incorporated three nutrient levels, ambient (unamended seawater), low and medium, and 2 temperatures (27 and 31°C) in 3 replicate 30 L flow-through (400 ml/min) aquaria per nutrient/temperature exposure, each holding 7 sponge clones. All tanks were illuminated using fluorescent tubes under a 12∶12 h diurnal cycle at 80 µmol quanta m_2 s_1 to reflect light intensity at 15 m on the reef. The experiment was conducted by randomly sampling one clone from each replicate tank, per treatment, at time points = 0, 1, 5, 7 and 12 days. Treatments were maintained for the first 7 days, then all nutrient dosing was stopped and temperatures were returned to 27°C for the final 5 days of the experiment, serving as a recovery period. Samples were snap-frozen in liquid nitrogen immediately after collection, and maintained at –80°C before DNA extraction and analysis.
Nutrient Concentrations and Calculations
This study used three nutrient levels: ambient (unamended seawater), low and medium. Stock solutions of Thrive® water-soluble plant fertiliser (NPK; 27∶5.5∶9 and trace elements) were continuously delivered by peristaltic pump (1 ml/min) into 30 L aquaria to final concentrations of (1) total inorganic nitrogen - ambient 1.3 µmol/L, low 1.9 µmol/L and medium 4.7 µmol/L, and (2) total nitrogen - ambient 6.5 µmol/L, low 7.1 µmol/L and medium 10 µmol/L (Table S1a). Nutrient levels were monitored throughout the experiment to ensure treatment levels were maintained (Table S1a). Seawater samples for analysis of dissolved nutrients (DIN, TDN and DOC) were hand-filtered through a 0.45 µm filter cartridge (Sartorius MiniSart) into acid-washed screw-cap plastic test tubes and stored frozen until later analysis. Samples for DOC analysis were filtered, acidified with 100 µL of HCl and stored frozen until analysis. Seawater samples for determination of particulate nutrients were collected by vacuum filtration on pre-combusted glass-fibre filters (Whatman GF/F). Filters were wrapped in pre-combusted aluminum foil envelopes and frozen until analysis. Dissolved and particulate nutrient levels (ammonium, nitrite, nitrate, phosphate, DOC, PN, POC) were analysed by the water quality laboratory at the Australian Institute of Marine Science (AIMS, Townsville) (Table S1a). Concentrations of dissolved inorganic nutrients and total dissolved nutrients were determined using a Bran and Luebbe AA3 segmented flow analyser using methods described by [72]. Concentrations of dissolved organic nutrients were determined by subtraction of the respective dissolved inorganic components (following UV irradiation of the samples to oxidise organic matter) from the levels of total dissolved nutrients.
DNA Extraction
All tissue samples were processed using an approach previously optimized for marine sponges [73]. Briefly, tissue samples were homogenised using lysing matrix E tubes (MPBio) in combination with a Mini-Beadbeater (Biospec Products, Bartleville, OK, USA). For DNA extraction, a Qiagen AllPrep DNA/RNA Mini kit (Cat. #80204) was used according to the manufacturer’s instructions. Purity and quantity of DNA were assessed using a NanoDrop 1000 spectrophotometer (Thermo Scientific) and gel electrophoresis of a 5 µL aliquot on a 1% agarose gel containing 0.5 µg ml−1 ethidium bromide. DNA was extracted from seawater filters by addition of 200 µL lysozyme (10 mg/ml), incubation at 37°C for 45 min, addition of 200 µL of proteinase K (0.2 mg/ml) in 1% SDS and incubation at 55°C for 1 h. Lysates were recovered into fresh Eppendorf tubes and nucleic acids extracted using the Qiagen AllPrep DNA/RNA Mini kit (Cat. #80204).
Denaturing Gradient Gel Electrophoresis (DGGE)
16S rRNA gene – Bacteria
The 16S rRNA genes from each sponge clone and seawater sample were amplified by PCR with primers 1055f: 5′-ATGGCTGTCGTCAGCT-3′ and 1392r: 5′-ACGGGCGGTGTGTAC-3′ [74]. The reverse primer was modified to incorporate a 40 bp GC clamp [75]. Cycling conditions were: 3 min at 95°C, followed by 30 cycles of 1 min at 94°C, 1 min at 54°C, 3 min at 72°C, with a final extension of 7 min at 72°C.
amoA gene from Ammonia-Oxidizing Archaea (AOA)
The amoA gene from ammonia-oxidizing archaea (AOA), which encodes for the catalytic subunit of ammonia monooxygenase [76], was targeted by amplifying a ∼635 bp fragment of the amoA gene using primers Arch-amoAF 5′-STAATGGTCTGGCTTAGACG-3′ and Arch-amoAR 5′-GCGGCCATCCATCTGTATGT-3′ [77]. The reverse primer was modified to incorporate a 40 bp GC clamp [75]. Cycling conditions were: 5 min at 95°C, followed by 30 cycles of 45 s at 95°C, 55 s at 55°C, 1 min at 72°C followed by a final extension of 5 min at 72°C. Amplification of ammonia-oxidizing bacteria (AOB) produced fragments of many different sizes so these data were excluded from further analysis.
18S rRNA gene – Eukarya
Changes in the eukaryotic microbial community in response to nutrient and temperature treatment were assessed using a eukaryote-specific primer set for the 18S rRNA gene. A subset of samples; Day 0 (control), Day 7 (ambient, low and medium nutrient exposures at both 27 and 31°C), with three replicates from each treatment, was screened by DGGE. The 18S rRNA gene was amplified by PCR with eukaryote-specific primers NS1f: 5′-GTA GTC ATA TGC TTG TCT C-3′ and NS2r: 5′- GGC TGC TGG CAC CAG ACT TGC-3′, [78]. The reverse primer was modified to incorporate a 40 bp GC clamp [75]. Cycling conditions were: 3 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 55°C, 1 min at 72°C followed by a final extension of 7 min at 72°C.
Products from all PCR reactions were applied to 8% w/v polyacrylamide (37.5∶1) gels containing denaturing gradients made from formamide and urea. 16S rRNA-bacterial and amoA-gene gels contained a gradient of 50–70%, while 18S rRNA gels had a 30–70% gradient. Gels were electrophoresed at 60°C for 17 h in 1×TAE (Tris-acetic acid EDTA) buffer at 65 V using the Ingeny D-Code system. Gels were stained with 1×Sybr Gold for 30 min, visualized under UV illumination and photographed (Quantity One; Bio-Rad, Gladesville, New South Wales, Australia).
Multidimensional Scaling (MDS)
Banding patterns from DGGE were transformed into presence/absence matrices and imported into PRIMER 6 (PRIMER-E, 2006, Ltd). MDS plots were created based on Bray-Curtis similarities, with 10000 iterations of bootstrapping. Hierarchical clustering of similarities was performed using the CLUSTER method and this information was superimposed onto the plots to create contours designating thresholds of similarity.
Amplification, Cloning, and Sequencing of 16S rRNA Genes from Archaea
Archaeal diversity in Day 7 ambient nutrient exposures was compared to Day 7 medium nutrient exposures, by clone library analysis. Two clone libraries were created (one for ambient and one for medium nutrient exposures) by combining all replicate sponges (A, B, C) and temperatures (27 and 31°C) within each of the ambient (n = 6) and medium (n = 6) nutrient treatments from Day 7. A portion of the archaeal 16S rRNA gene was amplified using the primers 21F 5′ TTCCGGTTGATCCYGCCGGA-3′ and 958R 5′TCCGGCGTTGAMTCCAATT-3′ [79]. Cycling conditions were 94°C for 1.5 min, 30 cycles of 94°C for 1 min, 54°C for 1.5 min, and 72°C for 2 min, and a final extension of 5 min at 72°C. Cloning was performed using the P-GemT Easy vector kit (Promega, Inc, Madison WI, USA) according to the manufacturer’s instructions. Clones containing the correct-sized insert were analyzed using amplified ribosomal DNA restriction analysis (ARDRA) [80], [81]. Restriction enzymes Hha1 and Hae III were used to analyze the diversity of archaeal 16S rRNA genes in each clone library. Digests were performed using 5 µL of PCR template with 1 µL of the respective enzyme and 3 µL of reaction buffer for 3 h at 37°C, followed by 20 min at 80°C to halt the reaction. Digested products were visualised on a 2% agarose gel to obtain ARDRA profiles. One to two clones representing each ARDRA banding pattern were selected for sequencing, which was performed on a capillary sequencer (Macrogen Inc, Seoul, South Korea).
Phylogenetic analysis of archaea
Archaeal 16S rRNA gene sequences were compared to available databases using the Basic Local Alignment Search Tool (BLAST) [82] to determine approximate phylogenetic affiliations. Chimeric sequences were identified using UCHIME [83] implemented in the program MOTHUR [84]. Sequences were aligned using the SINA Web Aligner [85] and then imported into the ARB programme package for manual editing using the SILVA database [22]. All subsequent phylogenetic analyses were performed in ARB. Maximum likelihood algorithms were used to calculate a phylogenetic tree, with maximum parsimony-based bootstraps (1000 resamplings) also calculated to assess the stability of observed branching patterns.
454 Pyrosequencing
A subset of samples (day 0 control and day 7 samples from all treatments) was screened by 454 pyrotag sequencing. The 16S rRNA Bacteria-specific sequences (targeting the V4–V5 region) were 454MID_533F (GTGCCAGCAGCYGCGGTMA) and 454_907R (CCGTCAATTMMYTTGAGTTT). Amplification primers were designed with FLX Titanium adapters. Forward primers contained the A adapter (CCA TCT CAT CCC TGC GTG TCT CCG AC) and the reverse primer contained the B (CCT ATC CCC TGT GTG CCT TGG CAG TC). A multiplex identifier (MID) was added to each of the forward 16S primer sequences (Roche Applied Sciences). Touchdown PCR conditions were as follows: 3 min at 94°C followed by 20 cycles of 30 s at 94°C, 30 s at 60°C (−0.5°C per cycle), 45 s at 72°C. This was followed by a further 10 cycles of 30 s at 94°C, 30 s at 50°C, 45 s at 72°C, with a final extension of 10 min at 72°C. For each sample PCR products were pooled from multiple reactions (100 µL total) and purified using AMPure XPbeads (Agencourt, Beckman Coulter, USA). Amplicon quality was checked on an Agilent Bioanalyzer 2100 DNA 1000 chip (Agilent Technologies). The number of molecules for each sample was calculated using size (bp) and concentration (ng/mL) data from the Qubit Quant-iT™ DNA high-sensitivity assay kit and a Qubit® fluorometer (Invitrogen) according to the manufacturer’s instructions. Pyrosequencing was performed using a 454/Roche GS Junior instrument (Roche, NJ, USA) at the School of Biological Sciences, University of Auckland, under the auspices of New Zealand Genomics Limited.
Processing of Raw Sequence Data
Sequences were processed using a combination of Mothur and custom PERL scripts [84]. Pyrosequencing flowgrams were filtered and denoised using the Mothur implementation of AmpliconNoise [86]. Sequences were removed from the analysis if they were <200 bp, contained ambiguous characters, had homopolymers longer than 8 bp, more than one MID mismatch, or more than two mismatches to the reverse primer sequence. Unique sequences were identified with Mothur, aligned against a SILVA alignment (available at http://www.mothur.org/wiki/Silva_reference_alignment). Sequences were chimera checked using UCHIME [83], then grouped into 97% operational taxonomic units (OTUs) based on uncorrected pairwise distance matrices. A representative sequence (defined in Mothur as the sequence with the minimum distance to the other sequences in the OTU) of each OTU was used for the taxonomic assignment using custom PERL scripts, similar to a previously used approach [19], [87]. For each tag sequence, a BLAST search [82] was performed against a manually modified SILVA database [22]. A Smith-Waterman algorithm was used to create pairwise global alignments between the 10 best hits against a tag sequence. For assignment the most similar sequence to the tag sequence (or multiple sequences if within a range of 0.1% sequence divergence) was used. Sequence similarity thresholds of 75%, 80%, 85%, 90% and 95% were applied for assignment at phylum, class, order, family and genus level, respectively. In cases where the taxonomy of the most similar sequences was inconsistent, a majority rule was applied and the tag was only assigned if at least 60% of all reference sequences shared the same taxonomic annotation at the respective taxonomic level. All previously published, sponge-derived sequences in the SILVA reference database were labelled as such [22] and it was noted when a tag sequence was assigned to a sponge-specific (SC) and/or sponge coral-specific sequence cluster (SCC). For assignment to an SC and/or SCC cluster a 75% sequence similarity threshold was applied.
Determining the Magnitude of Changes in Bacterial Community Structure
The magnitude of change in bacterial community structure was calculated for the 50 OTUs with the largest fold-change (positive or negative) by first normalising the number of reads per OTU, per sample and the values from three replicates were averaged. Treatment Day 7 values were then divided by Day 0 control values. Fold changes were subsequently Log (base 2)-transformed, so that positive (increases in relative abundance) and negative values (decreases) were weighted equally. Log (base 2)-transformation also means that the mapping space is equal and that positive and negative fold changes are comparable. Values for all samples were then ranked and the top 50 OTUs with the largest fold changes (be that positive or negative) were chosen for further analysis. Data was visualized as heatmaps using JColorGrid [88].
Results
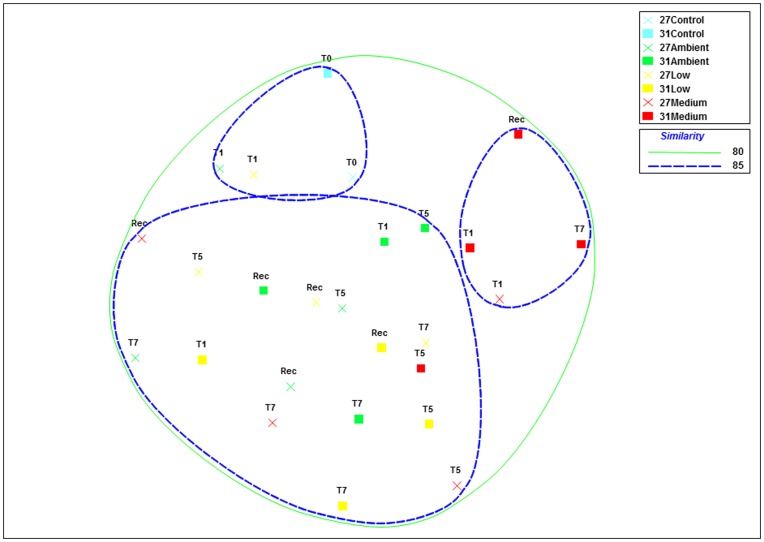
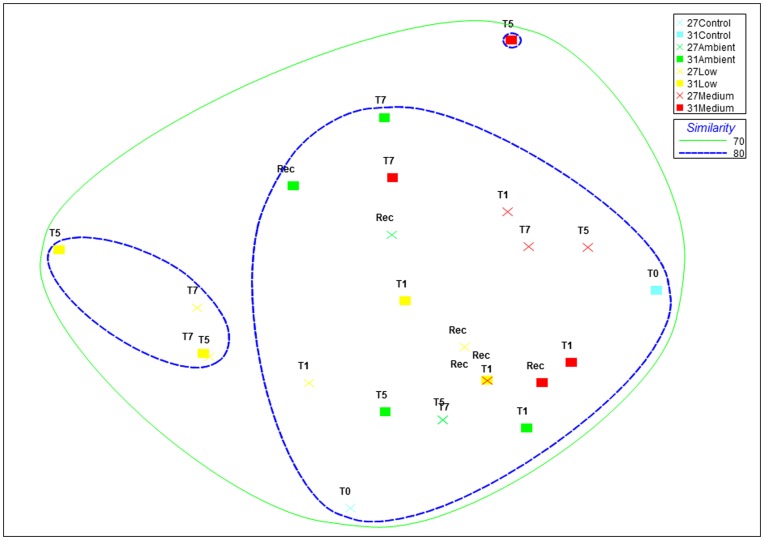
Nutrient exposures were selected using inshore data reported from the northern GBR and from previous research which demonstrated that our chosen exposure levels can cause stress in corals (Tables 1 and 2) [36], [45], [47], [89]. Our highest nutrient treatment levels resulted in 9-fold, 7.5-fold, 7-fold and 2.1-fold enrichments of ammonium, phosphate, nitrite and nitrate respectively (recorded from ambient exposures over the duration of the experiment (Table S1b)). R. odorabile clones exposed to all nutrient and temperature treatments remained visibly healthy throughout the duration of the experiment (Figure S1). 16S rRNA gene-targeting DGGE was used to screen all samples for changes in dominant members of the bacterial community. Most DGGE bands were highly conserved across all sponge clones, time periods, and nutrient/temperature exposures, revealing that microbial community composition is largely unaffected by the combined effects of elevated nutrients and sub-lethal temperature. A multidimensional scaling (MDS) plot combined with cluster analysis of DGGE profiles (Figure 1) revealed no clustering according to experimental treatment, with at least 80% similarity detected between all samples. As sponge health and the dominant microbial community did not appear to change at the highest nutrient/temperature treatment, only a subset of samples (day 0 control and day 7 samples from all treatments) were screened by 454 pyrotag sequencing to assess whether changes occur in the rare microbial biosphere.
Figure 1. MDS (multidimensional scaling) ordination of bacterial communities amongst samples as derived from 16S rRNA-DGGE profiles.
Banding patterns were transformed into a presence (scored as 1)/absence (scored as 0) matrix. MDS plots were generated using distance matrices to represent the relative distance between individual samples. Colours represent nutrient treatments: blue = control, green = ambient, yellow = low and red = medium nutrient level exposure. Crosses represent 27°C-exposed sponges and squares represent 31°C-exposed sponges. T = day of sampling and REC = recovery period (ambient nutrient levels and seawater temperatures). The final stress value of the plot was 0.21. Cluster analyses for similarity are indicated by colored contours at 80–85% similarity.
Nutrient and Sub-lethal Thermal Stress Effects on the Bacterial Biosphere
After noise reduction and quality filtering (see Material and Methods) a total of 35757 sequences was obtained from a total of 15 samples, with mean = 2384 (±346 (1SD)) sequences per sample (Table S2). For each sample, less than 0.05% (±0.07 (1SD)) of reads were taxonomically unassigned at phylum level. In total, 1743 OTUs (97% sequence similarity, Table S2) were identified, affiliated with 19 bacterial phyla. Rarefaction curves (Figure S2) indicated that, while further sequencing would have yielded a greater number of OTUs, diversity coverage was high with most curves approaching asymptotes. With the exception of sample 727LC, the observed number of OTUs was similar across all samples between 210–339 OTUs (Table S2).
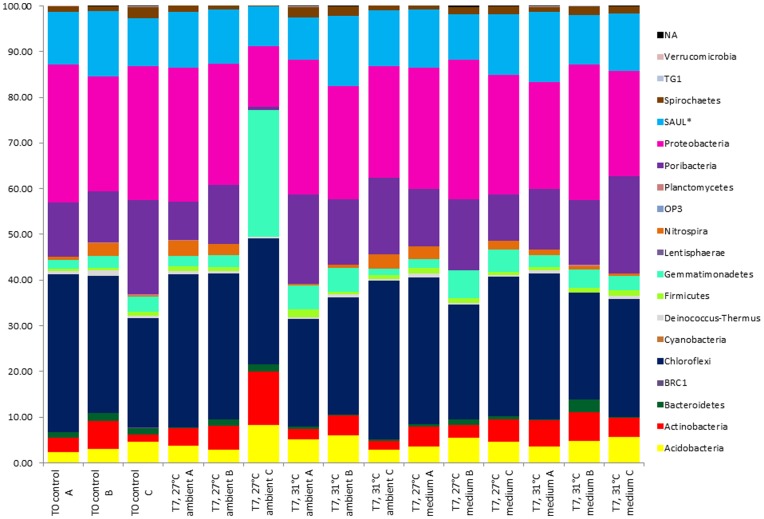
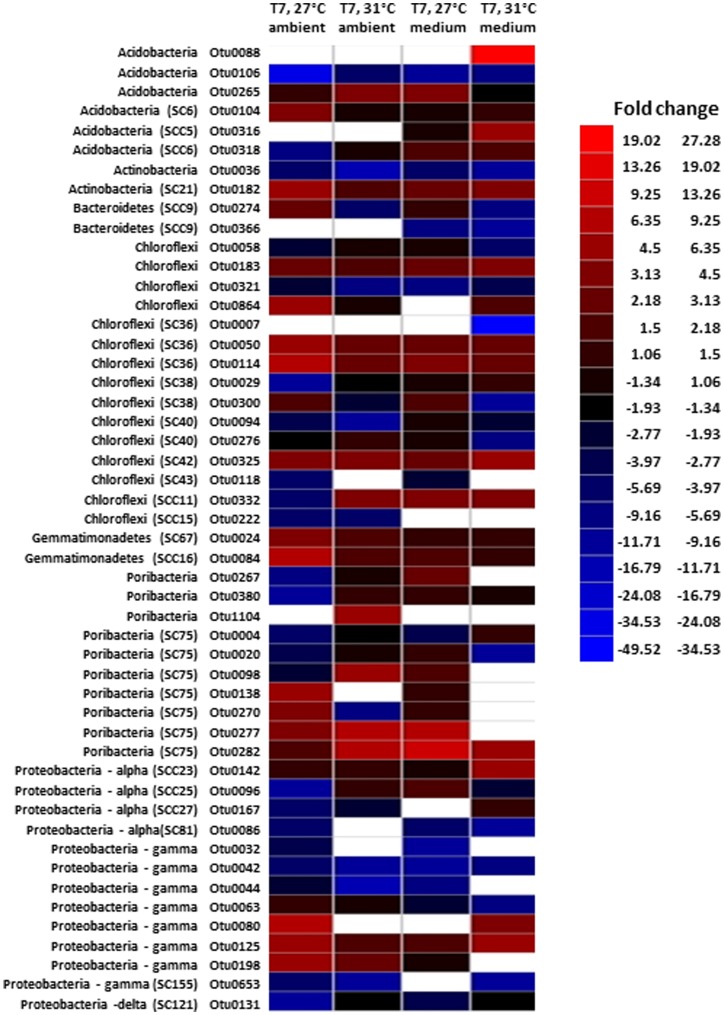
Phylum-level composition of bacterial communities was highly conserved across all samples (Figure 2). The dominant phyla were Chloroflexi, Proteobacteria, “Poribacteria” and Sponge-Associated Unidentified Lineage (SAUL), representing on average 29, 26, 14, and 12% of sequences, respectively, across all samples. Within the Proteobacteria most sequences were either Delta- or Gammaproteobacteria (13 and 10% respectively of total bacterial sequences). Phyla that were less abundant but found in all samples included the Acidobacteria (5%), Actinobacteria (5%) and Gemmatimonadetes (5%). Replicates from each treatment exhibited highly conserved bacterial community compositions, except for sample 727LC, which harbored the majority of phyla detected in the other samples but in markedly different proportions. Notably, the Gemmatimonadetes increased in proportion (27%) and there were very few “Poribacteria” (0.71%) in sample 727LC. Comparison of bacterial community composition of individual samples was tested, at the OTU level, using a non-metric multidimensional scaling (NMDS) plot, which showed all samples clustered tightly together with only sample 727LC separating on the ordination (Figure S3, stress = 0.09, R2 = 0.97). Unweighted Unifrac [90] analysis also revealed there were no significant differences between treatments at the OTU level. The 50 OTUs with the largest number of reads as determined by pyrosequencing were calculated (Figure S4) and together represented a range between 66–84% of total reads across all samples. These results support the phylum-level composition data, with most of the top 50 OTUs affiliated with the Chloroflexi, Proteobacteria and “Poribacteria”, and abundances of individual OTUs were highly similar across all treatments. The primary differences occurred in replicate 727LC, which had a very high abundance of reads (34% in total) assigned to OTU0014 (Gemmatimonadetes), OTU0024 (Gemmatimonadetes) and OTU0084 (Chloroflexi). As no substantial shifts in bacterial community composition at phylum or OTU level were observed in any other sample, values were calculated for the 50 OTUs with the largest (negative or positive) fold change, per nutrient/temperature exposure relative to Day 0 controls (Figure 3). Most of the top 50 OTU fold changes occurred in the Chloroflexi (30% of the top 50 fold-changes), Proteobacteria (26%) and “Poribacteria” (20%). Some trends were observed across all samples, for example OTUs belonging to the Gemmatimonadetes increasing in abundance and Deltaproteobacteria OTUs decreasing in abundance across all samples. Two OTUs (0088 and 0316) in the Actinobacteria only showed positive fold changes (abundance increases) in sponge clones exposed to the high nutrient treatments. BLAST searches revealed that these OTUs were closely affiliated to other sponge symbionts (data not shown). However, overall no major shifts in OTU fold change occurred due to elevated nutrient levels (with/without temperature treatment).
Figure 2. Distribution of 454 amplicon reads per phylum across nutrient/temperature treatments.
The number of reads per phylum is calculated as a percentage of the total reads in each sample. Samples are grouped in replicates (A, B, C), according to nutrient (ambient or medium)/temperature exposure (27 or 31°C) and ordered by sampling date (Days 0 and 7). *SAUL (sponge-associated unidentified lineage [87]). NA (not assigned).
Figure 3. Heatmap of the 50 OTUs (from Day 7) with the largest negative or positive fold-change from Day 0 controls.
Averages of three replicates were used for fold-change calculations. Treatment (ambient or medium nutrient exposure/27°C or 31°C) values were divided by the relevant control values (Day 0). Fold-changes were Log (base 2)-transformed, so that positive (increases in relative abundance) and negative values (decreases) were weighted equally. Key: Black squares representing no change from control value, red scale representing positive fold changes, blue scale representing negative fold changes and white squares indicating no data in control or treatment.
The proportion of reads assigned to one of the previously described SCs/SCCs [22] was highly similar across all nutrient and temperature exposures (Figure S5a) (63%–83%). Except for replicate sample 727LC, most SC clusters belonged to the Chloroflexi, Proteobacteria and “Poribacteria” (Figure S5b) and the majority of SCC were assigned to the Chloroflexi, Gemmatimonadetes and Alphaproteobacteria (Figure S5c). Of the top 50 OTUs with the largest (negative or positive) fold-change, 64% fell into SC/SCC clusters, mostly within these same phyla.
Nutrient and Sub-lethal Thermal Stress Effects on Archaeal Community Structure
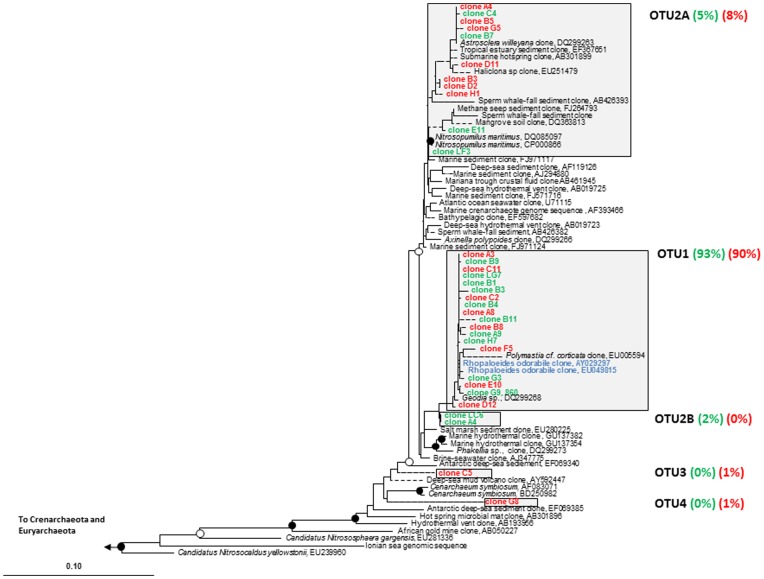
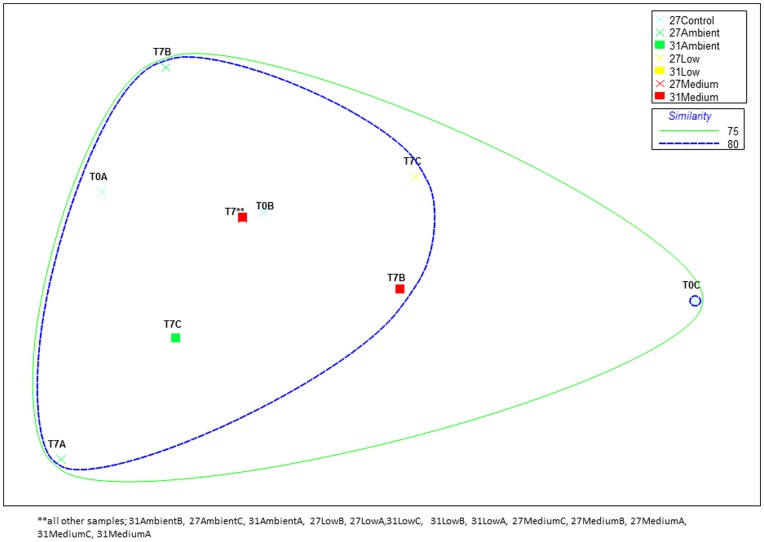
To determine the diversity of archaeal sequences, two 16S rRNA gene libraries were created, one from sponge clones kept in ambient nutrient conditions and a second from sponges exposed to the medium nutrient treatment. A total of 171 clones were screened by ARDRA analysis. ARDRA patterns revealed five unique OTUs (Figure 4), all associated with Thaumarchaeota (previously called Crenarchaeota Marine Group I). OTU1 and OTU2 were present in both ambient and medium nutrient treatment libraries, with OTU1 dominating both libraries (ambient = 93% and medium = 90%) and clustering with archaea previously reported from healthy R. odorabile. The ARDRA pattern that gave rise to OTU2 produced two distinct clusters (OTU2A and OTU2B), with OTU2B only present in the ambient nutrient library at 2%. The least common ARDRA patterns gave rise to OTUs 3 and 4, which were only present in the medium nutrient clone libraries at 1% each, with OTU4 related to Cenarchaeum symbiosum [28]. To determine whether nutrient enrichment altered potential functionality we also targeted the amoA gene in ammonia-oxidizing archaea (AOA) using DGGE. Variations in DGGE banding patterns were not correlated with any time points, nutrient or temperature treatments. MDS plots (Figure 5) clustered all samples together with at least 70% similarity, confirming that differences in AOA diversity are not related to nutrient or temperature stress.
Figure 4. Maximum likelihood phylogenetic tree from analysis of 16S rRNA gene sequences of archaeal clone libraries.
The tree is constructed based on long (≥1200 bp) sequences only; shorter sequences were added using the parsimony interactive tool in ARB and are indicated by dashed lines. Filled circles indicate bootstrap support (maximum parsimony, with 1000 resamplings) of ≥90%, and open circles represent ≥75% support. Bar, 10% sequence divergence. Sequences from medium nutrient clone library are in red and ambient library in green. Blue sequences represent archaea previously reported from healthy R. odorabile (Webster et al., 2001). The proportion of each OTU from treatment libraries (red or green) are in parentheses.
Figure 5. MDS (multidimensional scaling) ordination of R. odorabile -derived archaeal amoA genes as derived from DGGE profiles.
Banding patterns were transformed into a presence (scored as 1)/absence (scored as 0) matrix. MDS plots were generated using distance matrices to represent the relative distance between individual samples. Colours represent nutrient treatments: blue = control, green = ambient, yellow = low and red = medium nutrient level exposure. Crosses represent 27°C-exposed sponges and squares represent 31°C-exposed sponges. T = day of sampling and REC = recovery period (ambient nutrient levels and seawater temperatures). The final stress value of the plot was 0.18. Cluster analyses for similarity are indicated by colored contours at 70–80% similarity.
Nutrient and Sub-lethal Thermal Stress Effects on Community Structure of Microbial Eukaryotes
The majority of eukaryote-specific DGGE banding patterns were highly conserved regardless of nutrient/temperature treatment. MDS plots of DGGE banding pattern profiles confirmed eukaryotic community composition was highly similar across all treatments (Figure 6) and all samples clustered together with at least 75% similarity.
Figure 6. MDS (multidimensional scaling) ordination of the eukaryotic microbial communities of R. odorabile as derived from DGGE profiles.
Banding patterns were transformed into a presence (scored as 1)/absence (scored as 0) matrix. MDS plots were generated using distance matrices to represent the relative distance between individual samples. Colours represent nutrient treatments: blue = control, green = ambient, yellow = low and red = medium nutrient level exposure. Crosses represent 27°C-exposed sponges, squares represent 31°C-exposed sponges and T = day of sampling. The final stress value of the plot was 0.03. Cluster analyses for similarity are indicated by colored contours at 75–80% similarity.
Discussion
R. odorabile clones exposed to the combined effects of elevated nutrient levels and seawater temperature appeared visually similar to those maintained under ambient seawater conditions. The microbial communities of R. odorabile were not significantly affected by these environmental stressors, indicating that this sponge species is capable of withstanding short-term exposure to elevated nutrient concentrations and sub-lethal temperatures. R. odorabile is found throughout the GBR, occurring on inner-, mid- and outer-reef locations [91], [92]. The broad distribution of R. odorabile throughout the GBR exposes this species to a well-defined water quality gradient, with inner reef sponges experiencing higher nutrient loads, particularly during flood events, compared to mid and outer reef sponges [6], [93]. While reproductive output has been reported to decrease in female R. odorabile from inner reefs compared to outer reefs, these changes could not be directly linked to elevated nutrients or water turbidity [94]. Many cases have shown that microbial communities are sensitive to environmental perturbation [48], [95]. The evidence presented here, however, suggests that microbial communities within R. odorabile can resist these nutrient perturbations, even at temperatures of 31°C.
Eutrophication and poor water quality are major concerns for reef ecosystems globally. In addition to local factors, coral reefs are also faced with global stressors including elevated sea surface temperatures and ocean acidification [96], [97]. Despite this, the interacting effects of multiple environmental stressors on marine invertebrates are seldom investigated. Ambient levels of nitrogen and phosphorus recorded over the duration of the experiment were higher than those for nearby reefs, which is potentially due to input from the Orpheus Island Research Station. Sponges were exposed to nutrient concentrations 9-fold, 7.5-fold, 7-fold and 2.1-fold (ammonium, phosphate, nitrite and nitrate concentrations, respectively) above ambient yet showed no adverse health effects or changes in symbiosis. These results are further supported by a recent study that found nutrient enrichment does not affect the sponge Aplysina cauliformis or its symbiont community [98]. Healthy and Aplysina Red Band Syndrome (ARBS)-affected A. cauliformis were exposed to nutrient-enriched conditions (up to 4.8-fold and 2.1-fold increases of nitrate and phosphate respectively, from ambient levels over 7 days). A combination of terminal restriction fragment length polymorphism (T-RFLP), histology and chlorophyll fluorescence measurements [98] revealed no change in the bacterial communities of healthy sponges, nor an enhanced rate of disease progression in ARBS-affected sponges. However, nutrient enrichment levels similar to those in this experiment have been shown to exacerbate the onset and severity of coral diseases, including Black Band Disease [42], aspergillosis and Yellow Band Disease [36]. Although the mechanisms are unknown, this may be due to an enhancement of microbial growth rates [45] and/or increased pathogen virulence [36], [37].
The microbial community of R. odorabile analyzed by 454 pyrotag sequencing was highly conserved for the duration of the experiment at both phylum and OTU levels. Consistent with other sponge amplicon pyrosequencing studies [19], [20], [87], [99], communities were dominated by Chloroflexi, Proteobacteria, “Poribacteria” and SAUL at both the phylum and OTU level. Whilst the abundance data and fold change data for the majority of OTUs showed little correlation with particular nutrient/temperature treatments, replicate C at time 7 days, seawater temperature 27°C and ambient concentrations of nutrients (727LC) was an exception. In 727LC the differences were attributed to two Gemmatimonadetes OTUs and one OTU within the Chloroflexi. Given the high similarity of all other replicates and that this clone appeared visibly healthy, it is possible that this anomaly was a consequence of associated infauna being inadvertently sequenced with the sponge tissue. Sponge-specific clusters (SC) and sponge-coral-specific clusters (SCC) [17], [21], [22] are monophyletic clusters of 16S rRNA sequences found only in sponges (or sponges and corals) and not the surrounding environment such as seawater or sediments. We saw neither a significant increase nor decrease in the proportion of reads assigned to SCs or SCCs, with approximately 70% of all reads assigned to these clusters per sample across all treatments. Even though the roles of SCs/SCCs are still largely uncharacterized, it is predicted that their loss would be detrimental to the health and survival of the host sponge [100].
Nutrient enrichment and sub-lethal temperature stress did not alter the symbiotic archaeal associations within R. odorabile. Archaeal sequences were affiliated with Thaumarchaeota, which is consistent with archaeal sequences previously found in this species and in other sponge studies [101]–[103]. To address changes in community composition and potential functionality of sponge-associated archaea due to elevated nutrients and temperature, we also screened samples for changes in the amoA gene. In AOA and AOB the AmoA enzyme catalyses aerobic oxidation of ammonia to nitrite (the first step of nitrification). Analysis by qPCR in both the marine and terrestrial environment has suggested that AOA outnumber AOB [76], [104], [105] and this is also the case in at least some cold water marine sponges [29]. In this study the highest level of ammonium that sponges were exposed to was 9-fold higher than ambient levels. However, no shifts in the composition of AOA could be correlated with nutrient or temperature treatment, indicating that the diversity of AOA is stable under multiple environmental stressors.
Marine eukaryotic microbial communities mainly consist of algae, protozoa and marine fungi which play important roles in microbial food webs and in nutrient cycling [106], [107]. Within marine sponges, diatoms, dinoflagellates and fungi are known to live symbiotically although their functional roles within the sponge remain unclear [17], [108]. The effects of eutrophication on free-living eukaryotic microbial communities are widely reported: as the availability of nitrogen and phosphorus increases, primary production by algae is stimulated and the structures of phytoplankton communities change [109]–[112]. Ultimately these changes lead to increased turbidity in the water column coupled with oxygen depletion, which can greatly affect benthic communities [109]. Whilst the effect of nutrient amendment on host-associated microbial communities is less understood, nutrient enrichment studies with corals have shown increases in zooxanthellae abundance and indicate that these cells have preferential access to available CO2 which is then used for photosynthesis. Within the sponge Cymbastela concentrica, nutrient enrichment had no effect on symbiotic micro-algal growth as detected via chlorophyll concentration and sponge growth rate [113]. In the current study, the conserved eukaryotic community in R. odorabile across all nutrient and temperature treatments further highlights the stability of microbial associations within this sponge species. Based on eutrophication studies with free-living systems, one possible scenario resulting from nutrient elevation is an increase in the relative abundance of some species (particularly photosynthetic organisms). However, further exploration is required to investigate this.
Here we exposed sponges to seawater temperatures of 31°C which, based on Intergovernmental Panel on Climate Change (IPCC) scenarios, will occur before 2100 [114]. Previous studies assessing the response of R. odorabile to thermal stress have indicated sub-lethal effects at 31°C, including activation of the heat shock protein system [68] and a significant decrease in flow rate, filtration efficiency and choanocyte chamber density [115]. Whilst the bacterial community of R. odorabile is highly stable at 31°C, higher seawater temperatures cause a shift in the symbiont community which is concomitant with host tissue necrosis and mortality after four days at 32°C or three days at 33°C [67], [69]. Anthropogenic stressors such as water pollution have been shown to negatively interact with elevated seawater temperature, reducing coral larval metamorphosis [59] and increasing the persistence of the coral pathogen Serratia marcescens [41]. Research on coral bleaching thresholds also identified higher temperature sensitivity after exposure to increased DIN concentrations [116] with a recent study confirming that increased DIN, combined with limited phosphate concentrations, increases the susceptibility of corals to temperature- and light-induced bleaching. This is thought to occur due to an imbalanced DIN supply causing phosphate starvation of the symbiotic zooxanthellae [61]. In contrast, sub-lethal thermal stress in the current study did not appear to increase the susceptibility of R. odorabile to elevated nutrients.
Our study generated nutrient enrichment levels that are known to occur during major flood plume events and which have a destabilizing effect on the host-symbiont relationship in corals. The different response of sponges and corals to nutrient treatment may relate to the short timescale of exposure used in our study. However, the time scale of this study reflects real-time dispersal rates of elevated nutrients in the GBR environment. Many studies have reported the detrimental impacts of various water quality parameters on sponges and corals directly [36], [40], [42], [89], [94], [117]–[120]. In contrast, we report for the first time the effects of multiple environmental stressors on the important partnerships that reef invertebrates form with symbiotic microbes. We detected no changes in the bacterial, eukaryotic or archaeal community from any of the nutrient and/or temperature treatments, indicating that R. odorabile will be able to withstand nutrient pulses associated with flood plume events that are becoming more frequent in occurrence and severity [2], [13], [14], [93]. By assessing multiple stressors in combination, this study provides a first step for environmentally relevant sponge stress assessments which will enhance management strategies for GBR sponge populations.
Supporting Information
R. odorabile clones at the end of the recovery phase (12 days). Clones shown are (A) T = 12, ambient nutrient exposure and 31°C (B) T = 12, high nutrient exposure and 31°C. Large boxes show the entire sponge clone, smaller boxes show internal mesohyl tissue, from each respective sponge clone.
(TIF)
Bacterial diversity of all sponge samples amplified with 454 amplicon pyrosequencing. Rarefaction curves are based on OTUs at 97% sequence similarity. Calculations were performed in Mothur (Schloss et al., 2009)
(TIF)
Comparison of bacterial community composition of individual samples was tested, at the OTU level, using an nMDS plot. Lowest stress = 0.0904, R2 = 0.9711. Calculations were performed in Mothur (Schloss et al., 2009). Unweighted Unifrac analysis also revealed there were no significant differences between time points at the OTU level.
(TIF)
The relative abundance of the 50 most abundant OTUs (according to the sum of relative abundance across all samples). Samples are clustered according to phylogenetic affiliation. Scale is percentage of reads in each OTU (white = 0%). C = control with replicate A, B or C. A = ambient nutrient treatment with replicate A, B or C. M = medium nutrient treatment with replicate A, B or C
(TIF)
(a) The proportion of reads that were assigned to an SC or SCC per sample (b) The proportion of reads that were assigned to an SC per bacterial phylum and (c) The proportion of reads that were assigned to an SCC per bacterial phylum. The number of reads per phylum (b and c) is calculated as a percentage of the total reads that were assigned to a SC/SCC in each sample.
(TIF)
A. Average nutrient parameters measured in the experiment for ambient, low, medium and recovery nutrient exposure levels (n = 3 for each nutrient level). B. Average fold change in nutrient concentrations measured (for low, medium and recovery) during the course of the experiment from ambient values (n = 3 for each nutrient level).
(TIF)
Overview of the total number of sequence reads obtained per sample and the total numbers of OTUs obtained per sample are shown (after noise removal, quality filtering etc.) OTU information is reported at 97% similarity
(TIF)
Acknowledgments
We thank Florita Flores, Steve Whalan and Andrew Negri for their assistance with the aquarium facilities and for their technical advice and support.
Funding Statement
Research was supported by a University of Auckland Doctoral Scholarship to RLS and University of Auckland grant 3622989 to MWT. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brodie J, Devlin M, Haynes D, Waterhouse J (2011) Assessment of the eutrophication status of the Great Barrier Reef lagoon (Australia). Biogeochemistry 106: 281–302. [Google Scholar]
- 2. Brodie JE, Kroon FJ, Schaffelke B, Wolanski EC, Lewis SE, et al. (2012) Terrestrial pollutant runoff to the Great Barrier Reef: An update of issues, priorities and management responses. Mar Pollut Bull 65: 81–100. [DOI] [PubMed] [Google Scholar]
- 3. Brodie J, Waterhouse J (2012) A critical review of environmental management of the ‘not so Great’ Barrier Reef. Estuar Coast Shelf S 104–105: 1–22. [Google Scholar]
- 4. Schaffelke B, Carleton J, Skuza M, Zagorskis I, Furnas MJ (2012) Water quality in the inshore Great Barrier Reef lagoon: Implications for long-term monitoring and management. Mar Pollut Bull 65: 249–260. [DOI] [PubMed] [Google Scholar]
- 5. De’ath G, Fabricius K (2010) Water quality as a regional driver of coral biodiversity and macroalgae on the Great Barrier Reef. Ecol Appl 20: 840–850. [DOI] [PubMed] [Google Scholar]
- 6. Devlin MJ, Brodie J (2005) Terrestrial discharge into the Great Barrier Reef Lagoon: nutrient behavior in coastal waters. Mar Pollut Bull 51: 9–22. [DOI] [PubMed] [Google Scholar]
- 7. Webster AJ, Bartley R, Armour JD, Brodie JE, Thorburn PJ (2012) Reducing dissolved inorganic nitrogen in surface runoff water from sugarcane production systems. Mar Pollut Bull 65: 128–135. [DOI] [PubMed] [Google Scholar]
- 8. Kroon FJ (2012) Towards ecologically relevant targets for river pollutant loads to the Great Barrier Reef. Mar Pollut Bull 65: 261–266. [DOI] [PubMed] [Google Scholar]
- 9.Brodie J, Furnas MJ (1996) Cyclones, river flood plumes and natural water quality extremes in the Great Barrier Reef In; HM, Hunter, AG.Eyles and GE.Rayment; Downstream Effects of Land Use. Queensland Department of Natural Resources. 367–374.
- 10. Brodie J, Schroeder T, Rohde K, Faithful J, Masters B, et al. (2010) Dispersal of suspended sediments and nutrients in the Great Barrier Reef lagoon during river-discharge events: conclusions from satellite remote sensing and concurrent flood-plume sampling. Mar Freshw Res 61: 651–664. [Google Scholar]
- 11. Brodie J, Mitchell A (2005) Nutrients in Australian tropical rivers: changes with agricultural development and implications for receiving environments. Mar Freshw Res 56: 279–302. [Google Scholar]
- 12.Devlin MJ, Waterhouse J, Taylor J, Brodie JE (2001) Flood plumes in the Great Barrier Reef: spatial and temporal patterns in composition and distribution. GBRMPA Research Publication No 68.
- 13.Crow JM (2011) Queensland floods hit Great Barrier Reef. Nature News. Nature: doi:10.1038/news.2011.21.
- 14. Emanuel K (2005) Increasing destructiveness of tropical cyclones over the past 30 years. Nature 436: 686–688. [DOI] [PubMed] [Google Scholar]
- 15.Klotzbach PJ (2006) Trends in global tropical cyclone activity over the past twenty years (1986–2005). Geophys Res Lett 33.
- 16. Bell JJ (2008) The functional roles of marine sponges. Estuar Coast Shelf Sci 79: 341–353. [Google Scholar]
- 17. Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71: 295–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hentschel U, Piel J, Degnan SM, Taylor MW (2012) Genomic insights into the marine sponge microbiome. Nat Rev Microbiol 10: 641–654. [DOI] [PubMed] [Google Scholar]
- 19. Webster NS, Taylor MW, Behnam F, Lücker S, Rattei T, et al. (2010) Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol 12: 2070–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmitt S, Hentschel U, Taylor M (2012) Deep sequencing reveals diversity and community structure of complex microbiota in five Mediterranean sponges. Hydrobiologia 687: 341–351. [Google Scholar]
- 21. Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, et al. (2002) Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol 68: 4431–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simister RL, Deines P, Botté ES, Webster NS, Taylor MW (2012) Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ Microbiol 14: 517–524. [DOI] [PubMed] [Google Scholar]
- 23.Taylor MW, Tsai P, Simister R, Deines P, Botte E, et al.. (2012) “Sponge-specific” bacteria are widespread (but rare) in diverse marine environments. ISME J. doi: 10.1038/ismej.2012.111. [DOI] [PMC free article] [PubMed]
- 24. Wilkinson CR, Fay P (1979) Nitrogen fixation in coral reef sponges with symbiotic cyanobacteria. Nature 279: 527–529. [Google Scholar]
- 25. Weisz J, Hentschel U, Lindquist N, Martens C (2007) Linking abundance and diversity of sponge-associated microbial communities to metabolic differences in host sponges. Mar Biol 152: 475–483. [Google Scholar]
- 26. Mohamed NM, Colman AS, Tal Y, Hill RT (2008) Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ Microbiol 10: 2910–2921. [DOI] [PubMed] [Google Scholar]
- 27. Mohamed NM, Saito K, Tal Y, Hill RT (2010) Diversity of aerobic and anaerobic ammonia-oxidizing bacteria in marine sponges. ISME J 4: 38–48. [DOI] [PubMed] [Google Scholar]
- 28. Preston CM, Wu KY, Molinski TF, Delong EF (1996) A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci U S A 93: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Radax R, Hoffmann F, Rapp HT, Leininger S, Schleper C (2012) Ammonia-oxidizing archaea as main drivers of nitrification in cold-water sponges. Environ Microbiol 14: 909–923. [DOI] [PubMed] [Google Scholar]
- 30. Bayer K, Schmitt S, Hentschel U (2008) Physiology, phylogeny and in situ evidence for bacterial and archaeal nitrifiers in the marine sponge Aplysina aerophoba . Environ Microbiol 10: 2942–2955. [DOI] [PubMed] [Google Scholar]
- 31. Hoffmann F, Radax R, Woebken D, Holtappels M, Lavik G, et al. (2009) Complex nitrogen cycling in the sponge Geodia barretti . Environ Microbiol 11: 2228–2243. [DOI] [PubMed] [Google Scholar]
- 32. Diaz MC, Akob D, Cary CS (2004) Denaturing gradient gel electrophoresis of nitrifying microbes associated with tropical sponges. Boll Mus Ist Biol Univ Genova 68: 279–289. [Google Scholar]
- 33. Off S, Alawi M, Spieck E (2010) Enrichment and physiological characterization of a novel nitrospira-like bacterium obtained from a marine sponge. Appl Environ Microbiol 76: 4640–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schläppy M-L, Schöttner S, Lavik G, Kuypers M, de Beer D, et al. (2010) Evidence of nitrification and denitrification in high and low microbial abundance sponges. Mar Biol 157: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fiore CL, Jarett JK, Olson ND, Lesser MP (2010) Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol 18: 455–463. [DOI] [PubMed] [Google Scholar]
- 36. Bruno JF, Petes LE, Drew Harvell C, Hettinger A (2003) Nutrient enrichment can increase the severity of coral diseases. Ecol Lett 6: 1056–1061. [Google Scholar]
- 37.Kim K, Harvell CD (2002) Aspergillosis of sea fan corals: Disease dynamics in the Florida Keys. The Everglades, Florida Bay, and Coral Reefs of the Florida Keys: An Ecosystem Sourcebook: 813–824.
- 38. Fabricius KE (2005) Effects of terrestrial runoff on the ecology of corals and coral reefs: review and synthesis. Mar Pollut Bull 50: 125–146. [DOI] [PubMed] [Google Scholar]
- 39. Koop K, Booth D, Broadbent A, Brodie J, Bucher D, et al. (2001) ENCORE: The Effect of Nutrient Enrichment on Coral Reefs. Synthesis of Results and Conclusions. Mar Pollut Bull 42: 91–120. [DOI] [PubMed] [Google Scholar]
- 40. Kaczmarsky L, Richardson L (2011) Do elevated nutrients and organic carbon on Philippine reefs increase the prevalence of coral disease? Coral Reefs 30: 253–257. [Google Scholar]
- 41. Looney EE, Sutherland KP, Lipp EK (2010) Effects of temperature, nutrients, organic matter and coral mucus on the survival of the coral pathogen, Serratia marcescens PDL100. Environ Microbiol 12: 2479–2485. [DOI] [PubMed] [Google Scholar]
- 42. Voss J, Richardson L (2006) Nutrient enrichment enhances black band disease progression in corals. Coral Reefs 25: 569–576. [Google Scholar]
- 43. Garren M, Azam F (2012) Corals shed bacteria as a potential mechanism of resilience to organic matter enrichment. ISME J 6: 1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Garren M, Raymundo L, Guest J, Harvell CD, Azam F (2009) Resilience of coral-associated bacterial communities exposed to fish farm effluent. PLoS One 4: e7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kline DI, Kuntz NM, Breitbart M, Knowlton N, Rohwer F (2006) Role of elevated organic carbon levels and microbial activity in coral mortality. Mar Ecol Prog Ser 314: 119–125. [Google Scholar]
- 46. Smith JE, Shaw M, Edwards RA, Obura D, Pantos O, et al. (2006) Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol Lett 9: 835–845. [DOI] [PubMed] [Google Scholar]
- 47. Thurber RV, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, et al. (2009) Metagenomic analysis of stressed coral holobionts. Environ Microbiol 11: 2148–2163. [DOI] [PubMed] [Google Scholar]
- 48. Nogales B, Lanfranconi MP, Piña-Villalonga JM, Bosch R (2011) Anthropogenic perturbations in marine microbial communities. FEMS Microbiol Rev 35: 275–298. [DOI] [PubMed] [Google Scholar]
- 49. Crump BC, Peranteau C, Beckingham B, Cornwell JC (2007) Respiratory succession and community succession of bacterioplankton in seasonally anoxic estuarine waters. Appl Environ Microbiol 73: 6802–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morris RM, Vergin KL, Cho JC, Rappé MS, Carlson CA, et al. (2005) Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol Oceanogr 50: 1687–1696. [Google Scholar]
- 51. Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, et al. (2009) Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J 3: 283–295. [DOI] [PubMed] [Google Scholar]
- 52. Shi Y, McCarren J, DeLong EF (2012) Transcriptional responses of surface water marine microbial assemblages to deep-sea water amendment. Environ Microbiol 14: 191–206. [DOI] [PubMed] [Google Scholar]
- 53. Schäfer H, Bernard L, Courties C, Lebaron P, Servais P, et al. (2001) Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol Ecol 34: 243–253. [DOI] [PubMed] [Google Scholar]
- 54. Teira E, Martínez-García S, Calvo-Díaz A, Morán XAG (2010) Effects of inorganic and organic nutrient inputs on bacterioplankton community composition along a latitudinal transect in the Atlantic Ocean. Aquat Microb Ecol 60: 299–313. [Google Scholar]
- 55. Carlson CA, Giovannoni SJ, Hansell DA, Goldberg SJ, Parsons R, et al. (2002) Effect of nutrient amendments on bacterioplankton production, community structure, and DOC utilization in the northwestern Sargasso Sea. Aquat Microb Ecol 30: 19–36. [Google Scholar]
- 56. Sipura J, Haukka K, Helminen H, Lagus A, Suomela J, et al. (2005) Effect of nutrient enrichment on bacterioplankton biomass and community composition in mesocosms in the Archipelago Sea, northern Baltic. J Plankton Res 27: 1261–1272. [Google Scholar]
- 57. Øvreås L, Bourne D, Sandaa R-A, Casamayor EO, Benlloch S, et al. (2003) Response of bacterial and viral communities to nutrient manipulations in seawater mesocosms. Aquat Microb Ecol 31: 109–121. [Google Scholar]
- 58. Witt V, Wild C, Uthicke S (2012) Interactive climate change and runoff effects alter O2 fluxes and bacterial community composition of coastal biofilms from the Great Barrier Reef. Aquat Microb Ecol 66: 117–131. [Google Scholar]
- 59. Negri AP, Hoogenboom MO (2011) Water Contamination Reduces the Tolerance of Coral Larvae to Thermal Stress. PLoS One 6: e19703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Uthicke S, Vogel N, Doyle J, Schmidt C, Humphrey C (2012) Interactive effects of climate change and eutrophication on the dinoflagellate-bearing benthic foraminifer Marginopora vertebralis . Coral Reefs 31: 401–414. [Google Scholar]
- 61.Wiedenmann J, D’Angelo C, Smith EG, Hunt AN, Legiret F-E, et al.. (2012) Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nature Clim Change doi:10.1038/nclimate1661.
- 62.Lemoine N, Buell N, Hill A, Hill M (2007) Assessing the utility of sponge microbial symbiont communities as models to study global climate change: a case study with Halichondria bowerbanki. In: Custódio MR, Lôbo-Hajdu G, Hajdu E, Muricy G, editors. Porifera research: biodiversity, innovation and sustainability Série Livros 28, Museu Nacional, Rio de Janeiro.
- 63. López-Legentil S, Erwin PM, Pawlik JR, Song B (2010) Effects of sponge bleaching on ammonia-oxidizing archaea: distribution and relative expression of ammonia monooxygenase genes associated with the barrel sponge Xestospongia muta . Microb Ecol 60: 561–571. [DOI] [PubMed] [Google Scholar]
- 64. López-Legentil S, Song B, McMurray SE, Pawlik JR (2008) Bleaching and stress in coral reef ecosystems: hsp70 expression by the giant barrel sponge Xestospongia muta . Mol Ecol 17: 1840–1849. [DOI] [PubMed] [Google Scholar]
- 65. Cebrian E, Uriz MJ, Garrabou J, Ballesteros E (2011) Sponge Mass Mortalities in a Warming Mediterranean Sea: Are Cyanobacteria-Harboring Species Worse Off? PLoS One 6: e20211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Garrabou J, Coma R, Bensoussan N, Bally M, Chevaldonné P, et al. (2009) Mass mortality in Northwestern Mediterranean rocky benthic communities: effects of the 2003 heat wave. Glob Change Biol 15: 1090–1103. [Google Scholar]
- 67. Webster NS, Cobb RE, Negri AP (2008) Temperature thresholds for bacterial symbiosis with a sponge. ISME J 2: 830–842. [DOI] [PubMed] [Google Scholar]
- 68. Pantile R, Webster N (2011) Strict thermal threshold identified by quantitative PCR in the sponge Rhopaloeides odorabile . Mar Ecol Prog Ser 431: 97–105. [Google Scholar]
- 69.Simister R, Taylor MW, Tsai P, Fan L, Bruxner TJ, et al. (2012) Thermal stress responses in the bacterial biosphere of the Great Barrier Reef sponge, Rhopaloeides odorabile. Environ Microbiol: doi: 10.1111/1462–2920.12010. [DOI] [PubMed]
- 70. Louden D, Whalan S, Evans-Illidge E, Wolff C, de Nys R (2007) An assessment of the aquaculture potential of the tropical sponges Rhopaloeides odorabile and Coscinoderma sp. Aquaculture 270: 57–67. [Google Scholar]
- 71. Webster NS, Cobb RE, Soo R, Anthony SL, Battershill CN, et al. (2011) Bacterial community dynamics in the marine sponge Rhopaloeides odorabile under in situ and ex situ cultivation. Mar Biotechnol 13: 296–304. [DOI] [PubMed] [Google Scholar]
- 72.Ryle VD, Mueller HR, Gentien P (1981) Automated analysis of nutrients in tropical sea waters. AIMS Oceanography Series Tech Bulletin No3 AIMS OS 81 2: Townsville.
- 73. Simister RL, Schmitt S, Taylor MW (2011) Evaluating methods for the preservation and extraction of DNA and RNA for analysis of microbial communities in marine sponges. J Exp Mar Biol Ecol 397: 38–43. [Google Scholar]
- 74. Ferris MJ, Muyzer G, Ward DM (1996) Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol 62: 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Muyzer G, De Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, et al. (2006) Archaeal nitrification in the ocean. Proc Natl Acad Sci U S A 103: 12317–12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A 102: 14683–14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications: 315–322.
- 79. DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci U S A 89: 5685–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Martinez-Murcia AJ, Acinas SG, Rodriguez-Valera F (1995) Evaluation of prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol Ecol 17: 247–255. [Google Scholar]
- 81. Smit E, Leeflang P, Wernars K (1997) Detection of shifts in microbial community structure and diversity in soil caused by copper contamination using amplified ribosomal DNA restriction analysis. FEMS Microbiol Ecol 23: 249–261. [Google Scholar]
- 82. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 83. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 (16): 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ludwig W, Strunk O, Westram R, Richter L, Meier H, et al. (2004) ARB: A software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ (2011) Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12: 38 doi:10.1186/1471–2105–1112–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, et al. (2012) Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J 6: 564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Joachimiak MP, Weisman JL, May BCH (2006) JColorGrid: Software for the visualization of biological measurements. BMC Bioinformatics 7: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Humphrey C, Weber M, Lott C, Cooper T, Fabricius K (2008) Effects of suspended sediments, dissolved inorganic nutrients and salinity on fertilisation and embryo development in the coral Acropora millepora (Ehrenberg, 1834). Coral Reefs 27: 837–850. [Google Scholar]
- 90. Lozupone C, Knight R (2005) UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bannister R (2008) The ecology and feeding biology of the sponge Rhopaloeides odorabile. Thesis for the degree of Doctor of Philosophy in the School of Marine and Tropical Biology, Townsville, JCU.
- 92. Bannister RJ, Battershill CN, de Nys R (2010) Demographic variability and long-term change in a coral reef sponge along a cross-shelf gradient of the Great Barrier Reef. Mar Freshw Res 61: 389–396. [Google Scholar]
- 93. Bainbridge ZT, Wolanski E, Álvarez-Romero JG, Lewis SE, Brodie JE (2012) Fine sediment and nutrient dynamics related to particle size and floc formation in a Burdekin River flood plume, Australia. Mar Pollut Bull 65: 236–248. [DOI] [PubMed] [Google Scholar]
- 94. Whalan S, Battershill C, De Nys R (2007) Variability in reproductive output across a water quality gradient for a tropical marine sponge. Mar Biol 153: 163–169. [Google Scholar]
- 95. Allison SD, Martiny JBH (2008) Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci U S A 105: 11512–11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hoegh-Guldberg O, Mumby PJ, Hooten AJ, Steneck RS, Greenfield P, et al. (2007) Coral reefs under rapid climate change and ocean acidification. Science 318: 1737–1742. [DOI] [PubMed] [Google Scholar]
- 97. Pandolfi JM, Connolly SR, Marshall DJ, Cohen AL (2011) Projecting coral reef futures under global warming and ocean acidification. Science 333: 418–422. [DOI] [PubMed] [Google Scholar]
- 98. Gochfeld D, Easson C, Freeman C, Thacker R, Olson J (2012) Disease and nutrient enrichment as potential stressors on the Caribbean sponge Aplysina cauliformis and its bacterial symbionts. Mar Ecol Prog Ser 456: 101–111. [Google Scholar]
- 99. Lee OO, Wang Y, Yang J, Lafi FF, Al-Suwailem A, et al. (2011) Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J 5: 650–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Webster NS, Botté ES, Soo RM, Whalan S (2011) The larval sponge holobiont exhibits high thermal tolerance. Environ Microbiol Rep 3: 756–762. [DOI] [PubMed] [Google Scholar]
- 101. Meyer B, Kuever J (2008) Phylogenetic Diversity and Spatial Distribution of the Microbial Community Associated with the Caribbean Deep-water Sponge Polymastia cf. corticata by 16S rRNA, aprA, and amoA Gene Analysis. Microb Ecol 56: 306–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Webster NS, Watts JEM, Hill RT (2001) Detection and phylogenetic analysis of novel crenarchaeote and euryarchaeote 16s ribosomal RNA gene sequences from a Great Barrier Reef sponge. Mar Biotechnol 3: 600–608. [DOI] [PubMed] [Google Scholar]
- 103. Steger D, Ettinger-Epstein P, Whalan S, Hentschel U, De Nys R, et al. (2008) Diversity and mode of transmission of ammonia-oxidizing archaea in marine sponges. Environ Microbiol 10: 1087–1094. [DOI] [PubMed] [Google Scholar]
- 104. Leininger S, Urich T, Schloter M, Schwark L, Qi J, et al. (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442: 806–809. [DOI] [PubMed] [Google Scholar]
- 105. Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, et al. (2007) Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9: 1162–1175. [DOI] [PubMed] [Google Scholar]
- 106. Chen Y, Qin B, Teubner K, Dokulil MT (2003) Long-term dynamics of phytoplankton assemblages: Microcystis-domination in Lake Taihu, a large shallow lake in China. J Plankton Res 25: 445–453. [Google Scholar]
- 107. Hyde KD, Jones EBG, Leaño E, Pointing SB, Poonyth AD, et al. (1998) Role of fungi in marine ecosystems. Biodivers Conserv 7: 1147–1161. [Google Scholar]
- 108.Wilkinson CR (1992) Symbiotic interactions between marine sponges and algae. In: W. Reisser (ed.), Algae and symbioses: plants, animals, fungi, viruses, interactions explored. 111–151.
- 109. Meyer-Reil L-A, Köster M (2000) Eutrophication of Marine Waters: Effects on Benthic Microbial Communities. Mar Pollut Bull 41: 255–263. [Google Scholar]
- 110. Masó M, Garcés E (2006) Harmful microalgae blooms (HAB); problematic and conditions that induce them. Mar Pollut Bull 53: 620–630. [DOI] [PubMed] [Google Scholar]
- 111. Riegman R (1995) Nutrient-related selection mechanisms in marine phytoplankton communities and the impact of eutrophication on the planktonic food web. Water Sci Technol 32: 63–75. [Google Scholar]
- 112. Caron DA, Lim EL, Sanders RW, Dennett MR, Berninger U-G (2000) Responses of bacterioplankton and phytoplankton to organic carbon and inorganic nutrient additions in contrasting oceanic ecosystems. Aquat Microb Ecol 22: 175–184. [Google Scholar]
- 113. Roberts DE, Davis AR, Cummins SP (2006) Experimental manipulation of shade, silt, nutrients and salinity on the temperate reef sponge Cymbastela concentrica . Mar Ecol Prog Ser 307: 143–154. [Google Scholar]
- 114.IPCC (2007) Climate Change 2007: The physical basis. Contribution of working group I to the fourth assessment report of the Intergovernmental Panel on Climate Change: Geneva Cambridge, UK: Cambridge University Press.
- 115. Massaro AJ, Weisz JB, Hill MS, Webster NS (2012) Behavioral and morphological changes caused by thermal stress in the Great Barrier Reef sponge Rhopaloeides odorabile . J Exp Mar Biol Ecol 416–417: 55–60. [Google Scholar]
- 116. Wooldridge SA (2009) Water quality and coral bleaching thresholds: Formalising the linkage for the inshore reefs of the Great Barrier Reef, Australia. Mar Pollut Bull 58: 745–751. [DOI] [PubMed] [Google Scholar]
- 117. Ferrier-Pagès C, Gattuso JP, Dallot S, Jaubert J (2000) Effect of nutrient enrichment on growth and photosynthesis of the zooxanthellate coral Stylophora pistillata . Coral Reefs 19: 103–113. [Google Scholar]
- 118. Gerrodette T, Flechsig AO (1979) Sediment-induced reduction in the pumping rate of the tropical sponge Verongia lacunosa . Mar Biol 55: 103–110. [Google Scholar]
- 119. Bannister RJ, Battershill CN, de Nys R (2012) Suspended sediment grain size and mineralogy across the continental shelf of the Great Barrier Reef: Impacts on the physiology of a coral reef sponge. Cont Shelf Res 32: 86–95. [Google Scholar]
- 120. Cervino JM, Winiarski-Cervino K, Polson SW, Goreau T, Smith GW (2006) Identification of bacteria associated with a disease affecting the marine sponge Ianthella basta in New Britain, Papua New Guinea. Mar Ecol Prog Ser 324: 139–150. [Google Scholar]
- 121. Kroon FJ, Kuhnert PM, Henderson BL, Wilkinson SN, Kinsey-Henderson A, et al. (2012) River loads of suspended solids, nitrogen, phosphorus and herbicides delivered to the Great Barrier Reef lagoon. Mar Pollut Bull 65: 167–181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
R. odorabile clones at the end of the recovery phase (12 days). Clones shown are (A) T = 12, ambient nutrient exposure and 31°C (B) T = 12, high nutrient exposure and 31°C. Large boxes show the entire sponge clone, smaller boxes show internal mesohyl tissue, from each respective sponge clone.
(TIF)
Bacterial diversity of all sponge samples amplified with 454 amplicon pyrosequencing. Rarefaction curves are based on OTUs at 97% sequence similarity. Calculations were performed in Mothur (Schloss et al., 2009)
(TIF)
Comparison of bacterial community composition of individual samples was tested, at the OTU level, using an nMDS plot. Lowest stress = 0.0904, R2 = 0.9711. Calculations were performed in Mothur (Schloss et al., 2009). Unweighted Unifrac analysis also revealed there were no significant differences between time points at the OTU level.
(TIF)
The relative abundance of the 50 most abundant OTUs (according to the sum of relative abundance across all samples). Samples are clustered according to phylogenetic affiliation. Scale is percentage of reads in each OTU (white = 0%). C = control with replicate A, B or C. A = ambient nutrient treatment with replicate A, B or C. M = medium nutrient treatment with replicate A, B or C
(TIF)
(a) The proportion of reads that were assigned to an SC or SCC per sample (b) The proportion of reads that were assigned to an SC per bacterial phylum and (c) The proportion of reads that were assigned to an SCC per bacterial phylum. The number of reads per phylum (b and c) is calculated as a percentage of the total reads that were assigned to a SC/SCC in each sample.
(TIF)
A. Average nutrient parameters measured in the experiment for ambient, low, medium and recovery nutrient exposure levels (n = 3 for each nutrient level). B. Average fold change in nutrient concentrations measured (for low, medium and recovery) during the course of the experiment from ambient values (n = 3 for each nutrient level).
(TIF)
Overview of the total number of sequence reads obtained per sample and the total numbers of OTUs obtained per sample are shown (after noise removal, quality filtering etc.) OTU information is reported at 97% similarity
(TIF)