Abstract
The Smc5-Smc6 complex in Saccharomyces cerevisiae is both essential for growth and important for coping with genotoxic stress. While it facilitates damage tolerance throughout the genome under genotoxin treatment, its function during unperturbed growth is mainly documented for repetitive DNA sequence maintenance. Here we provide physical and genetic evidence showing that the Smc5–Smc6 complex regulates recombination at non-repetitive loci such as centromeres in the absence of DNA damaging agents. Mutating Smc6 results in the accumulation of recombination intermediates at centromeres and other unique sequences as assayed by 2D gel analysis. In addition, smc6 mutant cells exhibit increased levels of Rad52 foci that co-localize with centromere markers. A rad52 mutation that decreases centromeric, but not overall, levels of Rad52 foci in smc6 mutants suppresses the nocodazole sensitivity of these cells, suggesting that the Smc6-mediated regulation of recombination at centromeric regions impacts centromere-related functions. In addition to influencing recombination, the SUMO ligase subunit of the Smc5–Smc6 complex promotes the sumoylation of two kinetochore proteins and affects mitotic spindles. These results suggest that the Smc5–Smc6 complex regulates both recombination and kinetochore sumoylation to facilitate chromosomal maintenance during growth.
Introduction
The evolutionarily conserved Smc5 and Smc6 proteins are members of the Structural Maintenance of Chromosomes (SMC) family [1]–[3]. In budding yeast, they bind six other proteins, termed non-SMC elements (Nse1–6), to form the Smc5–Smc6 complex [4], [5]. This complex is required for both normal growth and resistance to DNA damaging agents [5]–[9]. Under DNA damaging conditions, it facilitates replication throughout the genome [10], [11]. One of its functions is to prevent the accumulation of recombination intermediates generated during perturbed replication, presumably through replication restart or ssDNA gap filling [12]–[16]. These recombination intermediates are detected as X-shaped DNA structures by 2-dimensional gel (2D gel) analysis, and can be toxic to the cell. Indeed, the removal of several recombination proteins, such as the strand exchange protein Rad51, improves the survival of mutants of the Smc5–Smc6 complex under genotoxic stress [14], [15]. It has been proposed that this complex can promote the resolution of recombination structures under these conditions [10], [12]–[17]. In addition, the Smc5–Smc6 complex also influences early steps of recombination, such as regulating the DNA association of the key recombination mediator protein Rad52 at stalled replication forks [18].
How the Smc5–Smc6 complex contributes to genome maintenance pathways during normal growth is less clear. In budding yeast, several studies have implicated this complex in the maintenance of repetitive DNA sequences, particularly the rDNA locus where it is enriched [5], [8], [16], [19]–[21]. In specific mutants of this complex, replication and segregation of rDNA or its neighboring DNA, but not other genomic loci, are particularly defective [8], [20]. Because these defects are largely unaffected by the removal of Rad52 [8], [20], the main function of the Smc5–Smc6 complex at this repetitive locus during growth appears largely independent of recombination, unlike the situation under replication stress. On the other hand, removal of Rad52, Rad51, and other recombination proteins can improve the growth of smc5 and smc6 mutants and even suppress the lethality of their null alleles [9], [14], [15]. These results suggest that the Smc5–Smc6 complex likely regulates recombination at other genomic loci during growth.
To test this idea, we examined whether the Smc5–Smc6 complex affects recombination at centromeric regions. Similar to rDNA, centromeres and the surrounding ∼25 kb regions on all chromosomes are enriched with this complex, as assayed by genome-wide ChIP analysis [19]. Unlike rDNA, the association of the Smc5–Smc6 complex with these centromeric regions requires Scc2–Scc4, similar to other regions on chromosomal arms [19]. Centromeric regions are critical for chromosomal stability and inheritance. They provide special chromatid structures, such as intra-chromatid cohesion, and allow the assembly of kinetochores, which are large protein complexes that connect to microtubules and enable chromosome segregation in mitosis [22], [23]. While a great deal is known about these aspects of centromeric regions, how recombination influences these loci has rarely been addressed. As DNA replication is often stalled at centromeric regions due to kinetochore blockage, proper regulation of recombination during these events could be important for the stability of these regions [24], [25]. Here, we examine how the Smc5–Smc6 complex influences these putative recombinational events at centromeres, using both 2D gel analysis to detect recombination intermediates, and live cell imaging to reveal the localization of recombination foci at centromeric regions. Results obtained using these two methods show that Smc6 is required to suppress recombination intermediates and modulate recombination events at centromeric regions. We also provide genetic evidence suggesting that these roles impact centromere related functions. In addition, our data show that a similar type of regulation may also occur at other unique, non-centromeric regions.
The stability of many genomic loci is influenced not only by DNA metabolism but also by the functions of associated protein factors. In this regard, centromeric sequences are uniquely complex due to the binding of more than 60 kinetochore proteins. The assembly and dynamics of kinetochore proteins are highly regulated by protein modifications, including sumoylation [26], [27]. Because the Nse2/Mms21 subunit (referred to as Mms21 hereafter) of the Smc5–Smc6 complex is a SUMO (small ubiquitin like modifier) ligase that promotes the addition of SUMO to substrates [5], [28], [29], we tested the potential role of this complex in the sumoylation of kinetochore proteins. We show that Mms21 regulates the sumoylation of specific kinetochore proteins and affects spindle function. These results thus suggest for the first time that the Smc5–Smc6 complex can combine recombinational repair with kinetochore protein regulation to promote chromosomal maintenance during growth.
Results
smc6–56 mutant cells contain increased levels of recombination intermediates at centromeric and other non-repetitive sequences
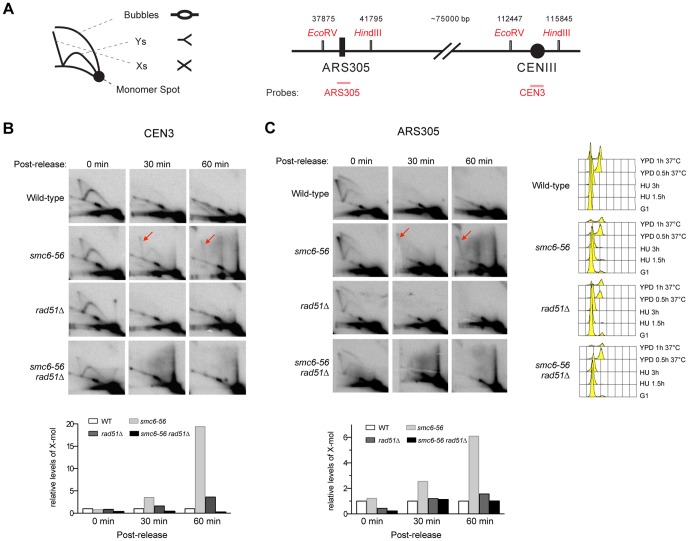
To address the question of whether the Smc5–Smc6 complex affects the metabolism of recombination intermediates in the absence of DNA damaging agents, we used 2D gel electrophoresis to examine these structures in wild-type cells and cells harboring a chromosome-integrated, temperature sensitive, smc6-56 allele [6]. We arrested both types of cells in G1 phase at permissive temperatures (of smc6-56) and further synchronized them in early S phase by HU (hydroxyurea) treatment, which inhibits dNTP production. Then the cells were released into normal media, allowing for replication at the non-permissive temperature of 37°C. Under this condition, wild-type and smc6-56 cells resumed replication as determined by the appearance of 2N DNA peaks in the FACS profiles and the disappearance of replication intermediates on 2D gels (Figs. 1A, 1B, 1C and data not shown). Using a probe recognizing the centromeric sequence on chromosome III, we detected increased levels of X-shaped DNA molecules in smc6-56 cells compared with wild-type cells at 30 and 60 minutes after release (Figs. 1A, 1B). These molecules were not observed in the absence of Rad51, confirming that they represent recombination intermediates likely formed between sister chromatids (Fig. 1B). Thus, smc6-56 cells accumulate recombination intermediates at centromeric regions in the absence of exogenous genotoxins.
Figure 1. smc6-56 cells accumulate recombination intermediates at centromeric and ARS305 sequences.
(A) Schematics of 2D gel and genomic regions containing ARS305 and CEN3 sequences. The numbers above the genomic region are base pair coordinates from the left end of chromosome III. (B–C) Cells were arrested in G1 using alpha-factor, and synchronized in S phase using 0.2 M HU for 3 hours at 25°C. Cells were then washed and released into YPD medium at 37°C. Samples before and after release at indicated time points were examined by 2D gel analysis. Membranes were hybridized to a probe specific for the centromeric sequence on chromosome III (B) and another specific for ARS305 (C). FACS analysis before and after release is presented on the right panel in (C). Quantification of X-molecules (red arrows) is shown in the bottom panels. For both loci, the level of X-molecules increases in smc6-56 cells compared with wild-type, and rad51Δ suppresses these increases.
To examine if increased levels of recombination intermediates under these conditions are specific to centromeric regions, we examined ARS305, an early replication origin located about 75 kb distal to the centromere on chromosome III (Fig. 1A). Increased levels of recombination intermediates in this region were also detected in smc6-56, but not in smc6-56 rad51Δ, cells at 30 and 60 minutes after release into normal media (Fig. 1C). These results demonstrate that smc6-56 cells contain increased levels of recombinational structures at both centromeric and other unique sequences during growth.
Rad52 foci levels increase at centromeric and non-centromeric regions in smc6-56 cells
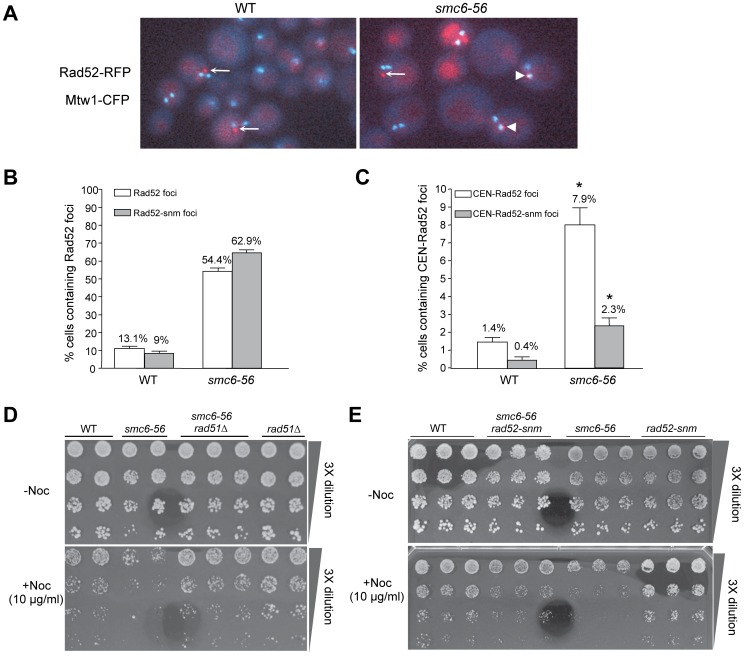
We next examined the levels of RFP-tagged Rad52 foci globally and at centromeric regions. This cytological approach assesses recombination events at a whole cell level [30] and complements the 2D gel analysis that assays specific sequences. Wild-type and smc6-56 cells were shifted to 37°C for 4 hours before examination. Consistent with previous reports, about 13% (46/351) of wild-type cells contained Rad52 foci ([30]; Fig. 2A and white bar in Fig. 2B). By contrast, 54% (86/203) of smc6-56 cells contained Rad52 foci, which is a 3.2-fold increase over wild-type levels (Fig. 2A and white bar in Fig. 2B).
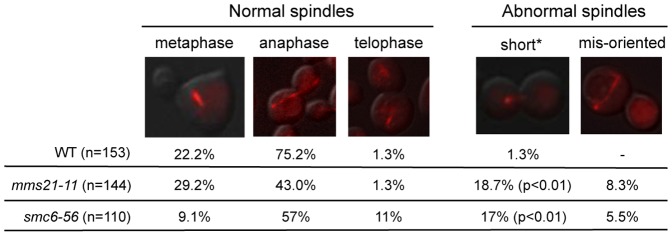
Figure 2. rad52-snm suppresses the increased repair foci at centromeric regions and nocodazole sensitivity of smc6-56 cells.
(A–C) smc6-56 displays increased levels of Rad52 foci at centromeric and non-centromeric regions. (A) Representative overlay images of Rad52-RFP and Mtw1-CFP for wild-type (WT) and smc6-56 cells. Arrowheads and arrows indicate the Rad52 foci that do and do not co-localize with Mtw1 foci, respectively. (B) Quantification of cells containing Rad52-RFP or Rad52-snm-RFP foci in wild-type and smc6-56 cells. (C) Quantification of cells containing Rad52-RFP or Rad52-snm-RFP foci that co-localize with Mtw1-CFP foci. The difference between the two numbers labeled by the asterisks is statistically significant (p<0.01). (D–E) rad51Δ and rad52-snm rescue the nocodazole sensitivity of smc6-56. 2–3 different spores for each genotype were examined.
We then examined how often Rad52 foci co-localized with centromere markers. Budding yeast centromeres and kinetochore proteins form one to two foci due to the clustering of centromeres [22]. Thus, live cell imaging of CFP-tagged kinetochore proteins, such as Mtw1, permits the assessment of colocalization of Rad52 foci with centromeric regions. We found that 1.4% (5/351) of wild-type cells contained Mtw1-colocalized Rad52 foci, which are referred to as CEN-Rad52 foci (Fig. 2A and white bars in Fig. 2C). About 8% (16/203) of smc6-56 cells contained CEN-Rad52 foci, a 5.6-fold increase over wild-type levels (Fig. 2A and white bars in Fig. 2C). Taken together, the above cell biological results are consistent with 2D gel analysis (Fig. 1), and suggest that recombinational repair increases in frequency and/or takes longer at both centromeric and other chromosomal loci in smc6-56 cells.
rad52-snm suppresses nocodazole sensitivity and the increased level of recombination foci at centromeric regions in smc6-56 cells
To assess whether the Smc6 regulation of recombination is biologically important at centromeric regions, we first examined whether alleviating the burden of recombination intermediates in smc6-56 cells could suppress centromere-related defects. A surrogate readout of these defects is sensitivity to the microtubule and spindle destabilization drug nocodazole, which sensitizes mutants defective in centromere and kinetochore functions. Interestingly, we found that smc6-56 mutants exhibited nocodazole sensitivity and that rad51Δ, which prevents the formation of recombination intermediates in smc6-56 cells (Fig. 1B, 1C), suppressed this sensitivity (Fig. 2D).
As rad51Δ eliminates recombination throughout the genome, we next asked whether a recombination mutant that affects centromere recombination influences smc6-56 nocodazole sensitivity. Various alleles of the key recombination protein Rad52 exhibit specific effects on different recombination processes [31]. One allele, rad52-snm (defective in Rad52 sumoylation, K43R,K44R,K253R) generally supports recombination functions in wild-type cells, but rescues the lethality of cells lacking the DNA helicases Sgs1 and Rrm3 [32], [33]. sgs1Δ cells accumulate recombination intermediates in the replication-blocking agent MMS (methyl methane sulfonate) similar to smc6-56 [12], [34]. Though the mechanism of the rad52-snm suppression of sgs1Δ is not well understood, the similar defects of sgs1Δ and smc6-56 suggest that rad52-snm may also influence recombination in the latter. Thus, we tested whether rad52-snm affects the levels of Rad52 foci in smc6-56 cells grown at high temperatures.
As shown in Figure 2B, 9% (22/244) of SMC6 cells contain Rad52-snm foci, compared with 62.9% (161/256) of smc6-56 cells (grey bars in Fig. 2B). The increase in rad52-snm foci in smc6-56 cells is similar to (though stronger than) that of Rad52 foci (white bars in Fig. 2B). Next, we determined the number of cells that exhibited colocalization between Mtw1 and rad52-snm foci. We found that 0.4% (1/244) SMC6 and 2.3% (6/256) smc6-56 cells contained these foci (grey bars in Fig. 2C). Chi-square tests show that the difference in the percentage of cells containing CEN-Rad52 foci vs. CEN-Rad52-snm foci is statistically significant for smc6-56 cells (p<0.01). Taken together, these observations suggest that rad52-snm reduces the levels of Mtw1-colocalized, but not overall, Rad52 foci in smc6-56 cells.
If the Smc6 regulation of recombination is biologically relevant at centromeres, rad52-snm, which specifically reduces the levels of recombination foci at centromeres in smc6-56 cells, may influence the nocodazole sensitivity of these cells as seen for rad51Δ. Indeed, rad52-snm improved the growth of smc6-56 cells on medium containing nocodazole (Fig. 2E). We interpret these results to mean that nocodazole sensitivity in smc6-56 cells is partly due to impairment of recombination at centromeric regions, and that these recombination events are regulated by Rad52 sumoylation.
rad52-snm does not affect recombination intermediate levels in smc6-56 cells
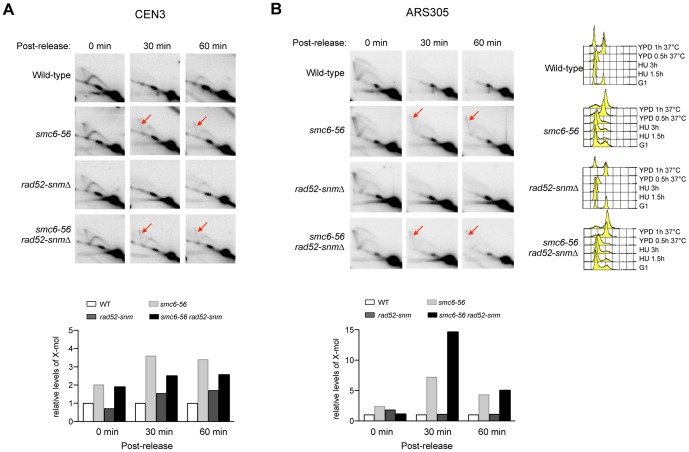
As mentioned above, how rad52-snm affects recombination is not fully understood. Its similarity with rad51Δ in suppressing the nocodazole sensitivity of smc6-56 raised the possibility that rad52-snm may affect recombination intermediate levels. To test this idea, we performed 2D gel analysis using the same experimental scheme as described above (Fig. 1). rad52-snm cells behaved like wild-type, consistent with the notion that this mutation does not grossly affect recombination (Fig. 3). rad52-snm did not suppress the increased recombination intermediate levels in smc6-56 cells at both CEN III and ARS305 loci (Fig. 3). These results suggest that rad52-snm affects recombination in a different manner than rad51Δ. As both suppressed the nocodazole sensitivity of smc6-56, rad52-snm likely alleviates a recombination defect distinct from recombination intermediate resolution. Considering the additional role of Smc5 and 6 in early recombination steps [18], it is possible that their regulation of both early and late steps of recombination contributes to nocodazole resistance.
Figure 3. rad52-snm does not affect recombination intermediate levels.
(A–B) Experiments were performed and analyzed as described in Figure 1. rad52-snm does not influence the levels of recombination intermediates (red arrows) in either wild-type or smc6-56 cells at both CEN III (A) or ARS305 (B).
mms21-11 and smc6-56 decrease the sumoylation of two kinetochore proteins
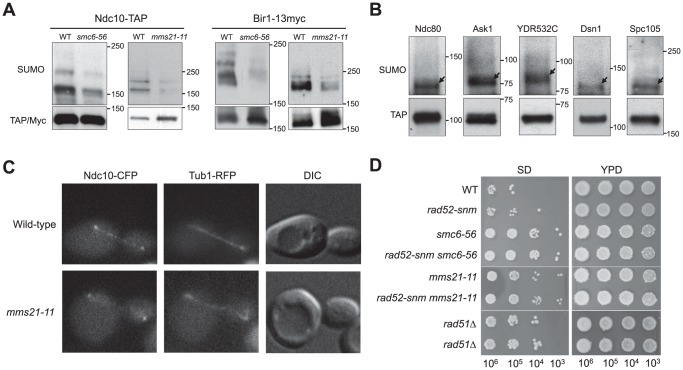
The results so far show that the Smc5–Smc6 complex regulates the levels of recombination intermediates and Rad52 foci at centromeres and other genomic loci, and that these effects likely pertain to centromere-related functions. As the Smc5–Smc6 complex contains a sumoylation enzyme subunit, Mms21, we tested whether this complex can also affect the protein components of the centromeres. Recent work has shown that sumoylation regulates kinetochore and spindle functions. Specifically, Ndc10 and Cep3, subunits of the Centromere Binding Factor 3 (CBF3), and Bir1 and Sli15, subunits of the chromosome passenger complex (CPC) were found to be sumoylated [26], [35]. We first examined whether their sumoylation levels were affected in mms21-11, a mutant lacking the SUMO ligase domain of Mms21 [5]. We found that Ndc10 and Bir1 sumoylation was diminished in mms21-11 cells, while Cep3 and Sli15 sumoylation levels showed no changes (Fig. 4A and data not shown). The sumoylation of Ndc10 and Bir1 was similarly decreased in smc6-56 cells (Fig. 4A). The co-depletion of sumoylation for these two proteins is consistent with a previous report showing that Bir1 sumoylation depends on Ndc10 sumoylation [26].
Figure 4. mms21-11 and smc6-56 affect sumoylation of specific kinetochore proteins, Ndc10 localization, and chromosome loss.
(A) Sumoylation of Ndc10 and Bir1 is reduced in mms21-11 and smc6-56 cells. The indicated proteins are tagged at their own chromosomal loci. Sumoylation of these proteins was examined using an anti-SUMO antibody (top panel). The unmodified proteins were detected by anti-TAP or anti-Myc antibody (bottom panel). The slightly different appearance of the SUMO bands is due to different gel percentages. (B) Sumoylation of kinetochore proteins. Sumoylation of each indicated kinetochore protein was examined as in (A). SUMO forms of the proteins are indicated by arrows and migrate at positions approximately 20 kD above the unmodified proteins. (C) Ndc10 spindle localization is defective in mms21-11 cells. Representative anaphase cells containing chromosomally tagged Ndc10-CFP and Tub1-RFP are shown. Note that Ndc10 is found in kinetochores and along spindles in wild-type cells. The spindle localization of Ndc10, but not the kinetochore localization, is defective in mms21-11. (D) mms21-11 and smc6-56 cells exhibit increased loss of chromosomes. Independent isolates of diploid strains were mated with haploid tester strains, and mating products from the indicated number of cells were selected on the SD (synthetic depleted) medium. YPD plates permit the growth of all cells regardless of mating status.
To test if additional kinetochore proteins are sumoylated in an Mms21-dependent manner, we examined sixty other kinetochore and spindle proteins tagged with the TAP module (Table 1; [36], [37]). We confirmed the modification of two linker kinetochore proteins (Ndc80 and Mcm21 [26]), and identified four new substrates: the outer kinetochore protein Ask1, and three linker kinetochore proteins, YDR532C, Dsn1 and Spc105 (Fig. 4B). As ∼17% (10/64) of tested kinetochore and spindle proteins are sumoylated, higher than the overall 8% of yeast proteins estimated to be sumoylated by proteomic studies [35], [38]–[42], there appears to be an enrichment of sumoylated substrates in these functional categories. Among all the sumoylated proteins identified here, mms21-11 strongly affected the sumoylation of only Ndc10 and Bir1 (Fig. 4A and data not shown).
Table 1. Proteins examined for sumoylation.
| Subcomplexes/functions | Sumoylated proteins | Non-sumoylated proteins |
| DAM1/DASH | Ask1 | Dam1, Duo1, Dad1–4, Spc19, Spc34, Hsk3 |
| CPC | Sli15, Bir1 | Ipl1 |
| CTF19/COMA | Mcm21 | Ctf3, Ctf19, Okp1, Ame1, Mcm16, Mcm22, Mcm19, Chl4, Nkp1–2 |
| SPC105 | Spc105, YDR532C | |
| MTW1/MIND | Dsn1 | Mtw1, Nnf1, Nsl1 |
| NDC80 | Ndc80 | Spc24, Nuf2, Cnn1 |
| CBF3 | Ndc10, Cep3 | Skp1, Ctf13 |
| Motor proteins and Microtubules | Stu2, Kip3, Cin8, Bim1, Bik1, Kip1, Kar3, Tub1, Tub3, Tub4 | |
| Checkpoint | Mad1–3, Bub1–3, Dbf2, Mob1, Slk19, Sgo1 | |
| Others | Cbf1, Cse4, Scm3, Mif2, Rdh54 |
mms21-11 impairs the spindle localization of Ndc10
Ndc10 sumoylation has been shown to promote its localization to spindles without affecting its kinetochore localization [26]. Considering the effect of mms21-11 on Ndc10 sumoylation, we tested whether this mutation affected Ndc10 spindle localization. While Ndc10 localized along the spindle in 95% (92/97) of wild-type anaphase cells, this localization was only observed in 69% (62/92, p<0.005) of mms21-11 anaphase cells (e.g. Fig. 4C). mms21-11 did not affect the kinetochore localization of Ndc10 (e.g. Fig. 4C). These results are consistent with the reported defect of a non-sumoylatable ndc10 mutant [26], and provide evidence linking the sumoylation function of Mms21 to Ndc10 spindle localization.
mms21-11 and smc6-56 lead to abnormal spindles and increased chromosome loss
The above results prompted us to examine whether mms21-11 also affects mitotic spindle morphology. As shown in Figure 5, mms21-11 cells contained spindles with abnormal morphologies, including mis-oriented (8.3%) and short spindles (18.7%), both of which were rarely seen in wild-type cells (Fig. 5). A similar defect was also observed in smc6-56 cells. This result suggests that sumoylation by the Smc5-Smc6 complex influences spindle properties.
Figure 5. Spindle morphology in wild-type, mms21-11, and smc6-56 cells.
Spindle morphology was examined 75 minutes after cells were released from G1 arrest when the majority of cells were at anaphase. Only medium to large budded cells were counted. A representative picture is shown for each spindle category. Similar results were obtained for two strains of each genotype and the results of one pair are shown. Asterisk denotes large budded cells with short spindles. p value indicates that there is a statistically significant difference between wild-type and mutants.
Next, we examined whether mms21-11 elevates chromosome loss, which is often linked to spindle defects. Although increased loss of heterozygosity has been reported for smc5 alleles [9], whether the sumoylation function of the Smc5–Smc6 complex affects chromosome loss has not been measured. We used a diploid bimating assay in which the loss of one copy of chromosome III (2N-1) allows cells to mate despite their diploid state [26], [43]. We spotted serial dilutions of mated cells to estimate the frequency of chromosome loss (see Methods). The chromosome loss frequency of wild-type diploids in this assay is similar to previous findings [26], [43]. mms21-11 homozygous diploid strains exhibited about a 100-fold increase in this assay (Fig. 4D). A similar increase was seen for smc6-56 cells at permissive temperature (Fig. 4D). We infer that sumoylation-dependent functions of the Smc5-Smc6 complex prevent chromosome loss. We observed that rad52-snm exhibited a normal level of chromosome loss and did not affect the levels in smc6-56 or mms21-11 cells (Fig. 4D). rad51Δ resulted in a similar level of chromosome loss as those of mms21-11 and smc6-56 (Fig. 4D), precluding the assessment of its possible suppression effect in this assay. While these results do not exclude the possibility that recombinational roles of the Smc5–Smc6 complex prevent chromosome loss, they raise the possibility that other functions of this complex, such as those involving kinetochore and spindle regulation, are important.
Discussion
Here we examine how mutations of the Smc5–Smc6 complex affect recombination at centromeric sequences, kinetochore protein modification, spindle properties, and chromosome loss. 2D gel analysis provides physical evidence of increased levels of recombination intermediates at centromeric regions in smc6-56 mutant cells (Fig. 1B). Consistent with this, live cell imaging shows that smc6-56 cells contain increased levels of CEN-Rad52 foci (Fig. 2C). These two pieces of evidence suggest that Smc6 is required to regulate recombination at centromeric DNA and surrounding regions during growth. Since a sumoylation-defective rad52 mutant that generates fewer centromeric foci can partially suppress the nocodazole sensitivity of smc6-56 (Figs. 2C, 2E), recombinational roles of Smc6 at centromeric regions likely affect centromere-related functions. These data also suggest that recombinational repair at centromere regions involves a subpathway that entails both sumoylated Rad52 and the Smc5–Smc6 complex. A requirement for both has also been found in double strand break repair in rDNA, though mms21-11 does not affect Rad52 sumoylation [32], [44]. These results provide the genetic bases for further examination of how the two collaborate in recombinational repair at both loci.
The notion that recombinational roles of Smc6 are relevant to centromere function is also consistent with the previous observation that removal of the recombination protein Mph1 suppresses the centromeric separation defect of smc6-56 [14]. This defect was initially thought to be unrelated to recombination, as rad51Δ did not suppress it [19]. However, we found that rad51Δ exhibited similar centromeric separation defects as smc6-56 (Fig. 6A–6B). The reason for this defect is unclear, but may be due to the deleterious effect of eliminating multiple recombination sub-pathways. Since rad51Δ moderately suppresses the nocodazole sensitivity of smc6-56 cells (Fig. 2D), the accumulation of recombination intermediates appears to be more deleterious than the lack of recombination.
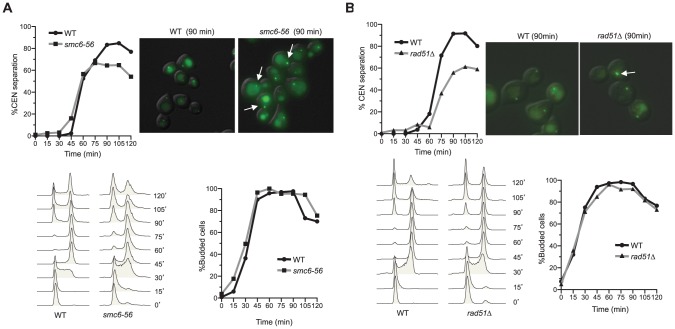
Figure 6. smc6-56 and rad51Δ cells are defective in centromeric LacO array separation.
(A–B) smc6-56 and rad51Δ cells exhibit defects in centromere separation. Cells contain a LacO array integrated 12 kb distal to the centromere on chromosome IV. Cells were arrested in G1 at 23°C and then shifted to 37°C for 1 hour before release into the cell cycle at 37°C. Samples were taken every 15 minutes to examine cell cycle progression by FACS analysis (bottom left) and budding index (bottom right). These time points were also examined for centromere separation by microscopy (top left). The difference between the percentage of wild-type and rad51Δ or between that of wild-type and smc6-56 cells containing separated GFP foci at 90, 105, and 120 min after release is statistically significant (p<0.01). Representative pictures at 90 minutes are shown (top right); arrows indicate cells with unseparated centromeres. Note that the background signals of LacI-GFP represent vacuolar staining.
Regulation of recombination by Smc6 is not restricted to centromeric regions. Increased levels of recombination intermediates and Rad52 foci were also detected in smc6-56 cells at non-centromeric regions (Figs. 1C and 2B). Such a general role fits with the presence of the Smc5–Smc6 complex at many chromosomal arm regions [19]. Although probing the physiological importance of such a role at non-centromeric regions is not the focus here, we speculate that other replication blockage sites likely require proper regulation of recombination by this complex. The effects on recombination by Smc6 during growth described here are reminiscent of those under replication stress caused by exogenous DNA damage [11]–[16]. This suggests that preventing the accumulation of toxic recombination structures is a crucial function of the Smc5–Smc6 complex both during growth and under DNA damage conditions. We therefore propose that the Smc5–Smc6 complex responds in a similar manner to multiple situations that create additional burden on the replication machinery, whether drug-induced or intrinsic to the nature of the locus. Thus, its function is not restricted to exogenous causes of replicative stress, but is likely to be an important component of the replication program. In agreement with this notion, the Smc5–Smc6 complex prevents sequence loss at telomeres and break-induced replication during growth, both of which can result from a failure to proper regulate recombination during replication [21], [45]. As rad51Δ and rad52-snm reduced the nocodazole sensitivity of smc6-56 cells, but differently affected recombination intermediate levels on 2D gel, it appears that the Smc5–Smc6 complex can affect more than one recombination steps as previously proposed.
This work also suggests that the Smc5–Smc6 complex affects kinetochore protein function. Our survey of 64 kinetochore and spindle proteins revealed ten SUMO substrates, four of which were not previously known (Table 1 and Figs. 4A–4B). Only Ndc10 and Bir1 sumoylation was decreased by mms21-11, an allele lacking the SUMO E3 ligase domain of Mms21 (Fig. 4A and data not shown). In addition, mms21-11 reduced the spindle localization of Ndc10 (Fig. 4C), consistent with the finding that non-sumoylatable ndc10 eliminates this localization [26]. The less penetrating defect of mms21-11 is likely due to its partial effect on Ndc10 sumoylation. As the Siz SUMO ligases also contribute to Ndc10 and Bir1 sumoylation, Mms21 may collaborate with them, though an indirect effect can not be excluded [26]. We do not expect that mms21-11 and non-sumoylatable ndc10 exhibit the same set of defects, because mms21-11 affects sumoylation of other proteins [5], [46]. Short and mis-oriented spindles seen in mms21-11 anaphase cells (Fig. 5) were not observed in non-sumoylatable ndc10 cells [26], and are likely due to a combined defect in the sumoylation of multiple Mms21 substrates. As smc6-56 exhibited similar defects as mms21-11 in spindle morphology and Ndc10 and Bir1 sumoylation (Figs. 4A and 5), the smc6-56 allele likely impairs the sumoylation function of the Smc5–Smc6 complex.
In summary, our results suggest that the Smc5–Smc6 complex affects both recombination and kinetochore protein function during growth. While this work does not delineate the molecular connections between these two effects, results here provide bases for further study of their interplay. As mutants of the Smc5–Smc6 complex in fission yeast also exhibit chromosomal loss and an increased sensitivity to a microtubule destabilization drug, the roles of this complex at centromeric regions may be conserved [47], [48]. It is highly plausible that the integration of recombinational regulation and sumoylation via the Smc5–Smc6 complex contributes to the maintenance of other genomic regions during normal growth and under genotoxic stress. Future work will be needed to elucidate the interplay as well as the biological influence of these dual functions in genome maintenance.
Materials and Methods
Yeast strains, plasmids, primers and genetic manipulations
Strains containing chromosomal TAP-tagged (Table 1) kinetochore proteins were obtained from Open Biosystems [37]. These tagged strains were crossed to smc6-56 or mms21-11 strains to generate the corresponding mutant derivatives. Other yeast strains are listed in Table 2; where applicable, a single representative of each genotype is listed. The LacO-array assay strain, rad52-snm strain, and the RFP-Tub1 strain were kind gifts from Andrew Murray, Michael Lisby, and Kerry Bloom, respectively. To construct Mtw1-CFP, the following primers were used to amplify the CFP region on the pFA6a-CFP plasmid: tag-mtw1-cfp-F2 (ATTGAAGAGCCTCAATTGGATTTACTTGATGATGTGTTACGGATCCCCGGGTTAATTAA) and tag-mtw1-cfp-R1 (AAGGTTGGCTGGCTACAGGATTCGAATTTTACGAAGTACTGAATTCGAGCTCGTTTAAAC). The PCR products were then used to tag Mtw1 at its own chromosomal locus using standard yeast protocols.
Table 2. Yeast strains used in this study.
| Name | Relevant Genotype | Source |
| X2123-2A | smc6-56-13myc::HIS3 W303 | [14] |
| X2761-3C | rad51Δ::LEU2 W303 | This work |
| X2761-2B | rad51Δ::LEU2 smc6-56-13myc::HIS3 W303 | This work |
| X1432-5a | RAD52-YFP MTW1-CFP W303 | This work |
| X1432-5b | RAD52-YFP MTW1-CFP smc6-56-13myc::HIS3 W303 | This work |
| X1429-7d | rad52-K43R,K44R,K253R-YFP MTW1-CFP smc6-56-13myc::HIS3 W303 | This work |
| X1429-13d | rad52-K43R,K44R,K253R-YFP MTW1-CFP W303 | This work |
| X2050-13B | rad52-K43R,K44R,K253R smc6-56-13myc::HIS3 W303 | This work |
| X1465-5D | NDC10-TAP::HIS3 mms21-11::LEU2 S288C | This work |
| X1711-8C | NDC10-TAP::HIS3 smc65-56 S288C | |
| T658-3 | NDC10-CFP::HPH ura3-1::mCherry-TUB1::URA3 W303 | This work |
| T657-1 | NDC10-CFP::HPH ura3-1::mCherry-TUB1::URA3 mms21-11::LEU2 W303 | This work |
| PWY93-3B | BIR1-13Myc::KAN W303 | [52] |
| X1508-2A | BIR1-13Myc::KAN mms21-11::LEU2 W303 | This work |
| X1740-6D | BIR1-13Myc::KAN smc6-56 W303 | This work |
| X3049-1 | mms21-11::KAN/mms21-11::URA W303 | This work |
| X3042-2 | smc6-56-13myc::HIS/smc6-56-13myc::KAN W303 | This work |
| X4573-6 | smc6-56-13myc::HIS/smc6-56-13myc::KAN rad52-K43R,K44R,K253R/rad52-K43R,K44R,K253R W303 | This work |
| X4204-1 | rad52-K43R,K44R,K253R/rad52-K43R,K44R,K253R W303 | This work |
| X4571-7 | mms21-11::KAN/mms21-11::HIS rad52-K43R,K44R,K253R/rad52-K43R,K44R,K253R W303 | This work |
| X4209-6 | rad51Δ::LEU2/rad51Δ::LEU2 W303 | This work |
| X2133-18c | his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::LacO::TRP1 W303 | This work |
| X2133-14b | his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::LacO::TRP1 smc6-56-13myc::HIS3 W303 | This work |
| X1991-1A | his3-11::pCUP1-GFP12-lacI12::HIS trp1-1::LacO::TRP1 W303 | This work |
| X2133-15D | his3-11::pCUP1-GFP12-lacI12::HIS3 trp1-1::LacO::TRP1 rad51Δ::LEU2 W303 | This work |
| X2065-28B | rad51Δ::LEU2 smc6-56-13myc::HIS3 W303 | This work |
Strains in this study are either derivatives of W1588-4C, a RAD5 derivative of W303 (MAT a ade2-1 can1-100 ura3-1 his3-11,15 leu2-3,112 trp1-1 rad5-535 [53]), or in S288C background. Strain backgrounds are indicated.
Cell imaging
All imaging was performed on an Axioimager microscope with a 100× objective lens (NA = 1.4). Cells were processed for microscopy as described previously [30], except that the exposure times used for each fusion protein are as follows: Rad52-RFP, 1s; Tub1-RFP, 1s; Mtw1-CFP, 1s; and LacO assay, 1s. In all cases, 14–18 Z-sections with a 0.5 µm step size were taken to cover the whole yeast cell and maximal projections are shown for all figures. For experiments examining asynchronous smc6-56 cells, both mutant and wild-type cells were incubated at 37°C for 4 hours before imaging. For each measurement, at least two strains per genotype were tested. Statistical analysis of focus frequency was carried out using a Chi-Square test, and the p-values for each relevant comparison are indicated in the figure legends and tables.
2D gel analysis to examine recombination intermediates
Purification of DNA intermediates, 2D gel analysis, and quantification of replication intermediates were carried out as previously described [49]. The DNA samples were digested with EcoRV and HindIII and analyzed with probes recognizing CEN3 and ARS305. The following oligos were used to amplify ARS305 and CEN3 probes: ARS305-FW: GTTCCGAAACAGGACACTTAGC, ARS305-RV: ATCCAGGAGGGACTCAATGTAG, CEN3-FW: CCGAGAGAGCTGCAAAATTAG, CEN3-RV: GATTCTCACCGCATGACAAGTG. The PCR-generated CEN3 probe is ∼1 kb that contains the 118 bp centromeric sequence and ∼900 bp of surrounding sequence.
Detection of sumoylated proteins
Strains containing TAP-tagged proteins involved in kinetochore and spindle functions (Table 1) were obtained from the collection of Open Biosystems [37]. Each strain was inoculated in YPD to mid-log phase and cell lysates were made under denaturing conditions to preserve sumoylated proteins as described [50]. Immunoprecipitation of the TAP tagged protein and detection of the sumoylated forms have been described previously [50]. Western blotting was performed according to standard procedures using the following primary antibodies: anti-Myc (9E10; Sigma), anti-TAP (P1291, Sigma) and anti-SUMO [5]. Detection was performed using Enhanced Chemiluminescence (GE Healthcare). Note that sumoylated forms normally comprise a small percentage of the proteins and are not detected by the anti-TAP antibody on the exposures shown.
Centromere separation assays
This assay was performed as described in [51]. In brief, wild-type, smc6-56 and rad51Δ cells were grown at 23°C to early log phase. Cells were arrested at G1 phase by the addition of alpha-factor at 23°C. Cells were then shifted to 37°C for 1 hour to inactivate the Smc5–Smc6 complex in the presence of alpha-factor and then released into normal media at 37°C. Samples were taken every 15 minutes to examine cell cycle progression by FACS analysis and budding index, and to examine centromere separation by microscopy. Alpha-factor was added back to the culture 45 minutes after release to minimize the number of cells entering the next cell cycle. For each measurement, at least two strains of each genotype were analyzed and produced consistent results; thus, results from one trial are presented. Statistical analysis of focus frequency was carried out using Chi-square tests and the p-values for relevant comparisons are indicated in the figure legends.
Chromosomal loss assays
This assay was performed as described previously, with minor modifications [43]. In brief, 1 OD of diploid cells were mated with the same number of MAT a tester cells for 24 hours, and serial dilutions of mated cells were spotted onto YPD and SD (synthetic deplete) plates and incubated at 30°C for 36 hours before being photographed. When diploid cells lose one of the two copies of chromosome III, they gain the ability to mate with Mat a cells, resulting in growth on SD plates. More than 3 independent diploid clones were tested for each genotype.
Acknowledgments
We thank Michael Lisby, Andrew Murray, Kerry Bloom, and Eric Muller for providing strains; and Zhao lab members, particularly Catherine Cremona, Prabha Sarangi, and Xiao Peng, for comments on the paper.
Funding Statement
This work was supported by National Institute of General Medical Science grant R01 GM080670 and American Cancer Society Research Scholar Grant RSG-12-013-01-CCG to X. Zhao and by the Associazione Italiana per la Ricerca sul Cancro grant IG 10637 and the ERC grant 242928 to D. Branzei. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lehmann AR, Walicka M, Griffiths DJ, Murray JM, Watts FZ, et al. (1995) The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol Cell Biol 15: 7067–7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Verkade HM, Bugg SJ, Lindsay HD, Carr AM, O'Connell MJ (1999) Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol Biol Cell 10: 2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fousteri MI, Lehmann AR (2000) A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J 19: 1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hazbun TR, Malmstrom L, Anderson S, Graczyk BJ, Fox B, et al. (2003) Assigning function to yeast proteins by integration of technologies. Mol Cell 12: 1353–1365. [DOI] [PubMed] [Google Scholar]
- 5. Zhao X, Blobel G (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA 102: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Onoda F, Takeda M, Seki M, Maeda D, Tajima J, et al. (2004) SMC6 is required for MMS-induced interchromosomal and sister chromatid recombinations in S. cerevisiae . DNA Repair 3: 429–439. [DOI] [PubMed] [Google Scholar]
- 7. Hu B, Liao C, Millson SH, Mollapour M, Prodromou C, et al. (2005) Qri2/Nse4, a component of the essential Smc5/6 DNA repair complex. Mol Microbiol 55: 1735–1750. [DOI] [PubMed] [Google Scholar]
- 8. Torres-Rosell J, Machin F, Farmer S, Jarmuz A, Eydmann T, et al. (2005) SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat Cell Biol 7: 412–419. [DOI] [PubMed] [Google Scholar]
- 9. Cost GJ, Cozzarelli NR (2006) Smc5p promotes faithful chromosome transmission and DNA repair in Saccharomyces cerevisiae . Genetics 172: 2185–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bermudez-Lopez M, Ceschia A, de Piccoli G, Colomina N, Pasero P, et al. (2010) The Smc5/6 complex is required for dissolution of DNA-mediated sister chromatid linkages. Nucleic Acids Res 38: 6502–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bustard DE, Menolfi D, Jeppsson K, Ball LG, Dewey SC, et al. (2012) During replication stress Non-Smc-Element 5 is required for Smc5/6 complex functionality at stalled forks. J Biol Chem 287: 11374–11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, et al. (2006) Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522. [DOI] [PubMed] [Google Scholar]
- 13. Sollier J, Driscoll R, Castellucci F, Foiani M, Jackson SP, et al. (2009) The S. cerevisiae Esc2 and Smc5–6 proteins promote sister chromatid junction mediated intra-S repair. Mol Biol Cell 20: 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Y-H, Choi K, Szakal B, Arenz J, Duan X, et al. (2009) Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc Natl Acad Sci USA 106: 21252–21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi K, Szakal B, Chen YH, Branzei D, Zhao X (2010) The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae . Mol Biol Cell 21: 2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chavez A, Agrawal V, Johnson FB (2010) Sumoylation and the structural maintenance of chromosomes (SMC) 5/6 complex slow senescence through recombination intermediate resolution. J Biol Chem 285: 11922–11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wehrkamp-Richter S, Hyppa RW, Prudden J, Smith GR, Boddy MN (2012) Meiotic DNA joint molecule resolution depends on Nse5–Nse6 of the Smc5-Smc6 holocomplex. Nucleic Acids Res [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Irmisch A, Ampatzidou E, Mizuno K, O'Connell MJ, Murray JM (2009) Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J 28: 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindroos HB, Strom L, Itoh T, Katou Y, Shirahige K, et al. (2006) Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell 22: 755–767. [DOI] [PubMed] [Google Scholar]
- 20. Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, et al. (2007) Anaphase onset before complete DNA replication with intact checkpoint responses. Science 315: 1411–1415. [DOI] [PubMed] [Google Scholar]
- 21. Noël J-F, Wellinger RJ (2011) Abrupt telomere losses and reduced end-resection can explain accelerated senescence of Smc5/6 mutants lacking telomerase. DNA Repair 10: 271–282. [DOI] [PubMed] [Google Scholar]
- 22. Verdaasdonk JS, Bloom K (2011) Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol 12: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kotwaliwale C, Biggins S (2006) Microtubule capture: a concerted effort. Cell 127: 1105–1108. [DOI] [PubMed] [Google Scholar]
- 24. Greenfeder SA, Newlon CS (1992) Replication forks pause at yeast centromeres. Mol Cell Biol 12: 4056–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, et al. (2003) The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell 12: 1525–1536. [DOI] [PubMed] [Google Scholar]
- 26. Montpetit B, Hazbun TR, Fields S, Hieter P (2006) Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol 174: 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mukhopadhyay D, Dasso M (2010) The fate of metaphase kinetochores is weighed in the balance of SUMOylation during S phase. Cell Cycle 9: 3194–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, et al. (2005) Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol 25: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Potts PR, Yu H (2005) Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol 25: 7021–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- 31. Mortensen UH, Erdeniz N, Feng Q, Rothstein R (2002) A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. Genetics 161: 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sacher M, Pfander B, Hoege C, Jentsch S (2006) Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol 8: 1284–1290. [DOI] [PubMed] [Google Scholar]
- 33. Altmannova V, Eckert-Boulet N, Arneric M, Kolesar P, Chaloupkova R, et al. (2010) Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res 38: 4708–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, et al. (2005) Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 19: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wohlschlegel JA, Johnson ES, Reed SI, Yates JR 3rd (2004) Global analysis of protein sumoylation in Saccharomyces cerevisiae . J Biol Chem 279: 45662–45668. [DOI] [PubMed] [Google Scholar]
- 36. Westermann S, Drubin DG, Barnes G (2007) Structures and functions of yeast kinetochore complexes. Annu Rev Biochem 76: 563–591. [DOI] [PubMed] [Google Scholar]
- 37. Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, et al. (2003) Global analysis of protein expression in yeast. Nature 425: 737–741. [DOI] [PubMed] [Google Scholar]
- 38. Panse VG, Hardeland U, Werner T, Kuster B, Hurt E (2004) A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem 279: 41346–41351. [DOI] [PubMed] [Google Scholar]
- 39. Zhou W, Ryan JJ, Zhou H (2004) Global analyses of sumoylated proteins in Saccharomyces cerevisiae - Induction of protein sumoylation by cellular stresses. J Biol Chem 279: 32262–32268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, et al. (2005) A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics 4: 246–254. [DOI] [PubMed] [Google Scholar]
- 41. Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, et al. (2005) Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae . J Biol Chem 280: 4102–4110. [DOI] [PubMed] [Google Scholar]
- 42. Wykoff DD, O'Shea EK (2005) Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol Cell Proteomics 4: 73–83. [DOI] [PubMed] [Google Scholar]
- 43. Spencer F, Gerring SL, Connelly C, Hieter P (1990) Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124: 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, et al. (2007) The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9: 923–931. [DOI] [PubMed] [Google Scholar]
- 45. Hwang J-Y, Smith S, Ceschia A, Torres-Rosell J, Aragón L, et al. (2008) Smc5-Smc6 complex suppresses gross chromosomal rearrangements mediated by break-induced replications. DNA Repair 7: 1426–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takahashi Y, Dulev S, Liu X, Hiller NJ, Zhao X, et al. (2008) Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet 4: e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pebernard S, Schaffer L, Campbell D, Head SR, Boddy MN (2008) Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J 27: 3011–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murray JM, Tavassoli M, al-Harithy R, Sheldrick KS, Lehmann AR, et al. (1994) Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol Cell Biol 14: 4878–4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D (2010) Replication and Recombination Factors Contributing to Recombination-Dependent Bypass of DNA Lesions by Template Switch. PLoS Genetics 6: e1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, et al. (2012) Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the Mec1 checkpoint. Mol Cell 45: 422–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Straight AF, Belmont AS, Robinett CC, Murray AW (1996) GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- 52. Widlund PO, Lyssand JS, Anderson S, Niessen S, Yates JR 3rd, et al. (2006) Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol Biol Cell 17: 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao X, Muller EG, Rothstein R (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2: 329–340. [DOI] [PubMed] [Google Scholar]