Abstract
Investigating the phylogenetic relationships within physiologically essential gene families across a broad range of taxa can reveal the key gene duplication events underlying their family expansion and is thus important to functional genomics studies. P-Type II ATPases represent a large family of ATP powered transporters that move ions across cellular membranes and includes Na+/K+ transporters, H+/K+ transporters, and plasma membrane Ca2+ pumps. Here, we examine the evolutionary history of one such transporter, the Sarco(endo)plasmic reticulum calcium ATPase (SERCA), which maintains calcium homeostasis in the cell by actively pumping Ca2+ into the sarco(endo)plasmic reticulum. Our protein-based phylogenetic analyses across Eukaryotes revealed two monophyletic clades of SERCA proteins, one containing animals, fungi, and plants, and the other consisting of plants and protists. Our analyses suggest that the three known SERCA proteins in vertebrates arose through two major gene duplication events after the divergence from tunicates, but before the separation of fishes and tetrapods. In plants, we recovered two SERCA clades, one being the sister group to Metazoa and the other to Apicomplexa clade, suggesting an ancient duplication in an early eukaryotic ancestor, followed by subsequent loss of one copy in Opisthokonta, the other in protists, and retention of both in plants. We also report relatively recent and independent gene duplication events within invertebrate taxa including tunicates and the leech Helobdella robusta. Thus, it appears that both ancient and recent gene duplication events have played an important role in the evolution of this ubiquitous gene family across the eukaryotic domain.
Introduction
A clear understanding of the evolutionary history of gene families is essential for studying their function, expression, and the evolutionary forces responsible for their diversification. Many evolutionary events such as gene duplication are important drivers of the expansion of gene families but can also confound functional genomics and gene expression studies that focus on orthologous genes. Consequently, a solid phylogeny of the genes of interest, especially multiple-copy genes, is needed before performing gene expression or comparative studies. The availability of many sequenced genomes greatly facilitates the investigation of the evolutionary history of many environmentally relevant gene families, such as the P-type II ATPases. This family of cation transporters plays a key role in the adaptation of organisms to variable environments, including variation in cation concentrations, due to their shared specificities for Ca2+, K+ and Na+ [1]. Although the nomenclature of this gene family has been revisited, it is generally accepted that P-type II ATPases include five closely related sub-families (SERCA, PMCA, NK/HK, ENA, and ACU) [1], [2], [3]. This study focuses on investigating the key evolutionary events that have led to the extensive diversification of sarco(endo)plasmic calcium ATPases (SERCA) across the major domains of eukaryotes.
Scarco(endo)plasmic Reticulum Calcium-ATPase (SERCA) is a key player in calcium signalling [4], which is involved in many aspects of cellular function [5], including transcription [6], cell motility [7], apoptosis, exocytosis, and signal transduction [8]. For example, during calcium-mediated signal transduction, the depolarization of the cell membrane in active cells causes an extensive influx of calcium into the cytoplasm. However, this influx of calcium needs to be reversed for proper cellular function [5]. To reduce cytoplasmic Ca2+ concentrations, SERCA uses ATP to actively pump calcium into the sarco(endo)plasmic reticulum for storage [4], [9]. The essential cellular function of SERCA makes it an interesting target for evolutionary studies as it is ubiquitous and indispensable across eukaryotic taxa.
Given the importance of the SERCA proteins to both cellular and organismal physiology, changes in the function, location, and expression of SERCA constitute significant evolutionary events. Previous genetic studies revealed that several gene duplication events occurred in the evolution of the SERCA. Three genes are present in vertebrates (ATP2A1-3), coding for three SERCA isoforms, SERCA 1-3 [9], while only one gene has been described in invertebrates, with the exception of the human parasitic blood fluke, Schistosoma mansoni, which has at least two [9], [10]. Interestingly, each of the vertebrate genes undergoes alternative splicing, resulting in ten SERCA proteins: SERCA 1a/b, SERCA 2a/b and SERCA 3a/b/c/d/e/f [11], [12]. These isoforms and their splice variants show a range of tissue specific expression patterns. For example, SERCA 1a is expressed in fast twitch muscles of adults and SERCA 1b in neonates [13]. SERCA 2a is expressed primarily in cardiac and slow-twitch skeletal muscles, whereas its splice variant, SERCA 2b, is expressed in almost all non-muscle cells and is often considered the house keeping variant [9], [12]. Furthermore, SERCAs 3 and 2b are found in a wide range of cells including lymphocytes, epithelial, endothelial, and mast cells, as well as Purkinje neurons of the cerebellum [9], [14]. The efficiency of the pump varies among the isoforms with SERCA 1a/b having a higher turnover rate than SERCA 2b and a higher affinity for calcium than SERCA 3 [14], [15]. Between the two SERCA 2 isoforms, SERCA 2b has a 2-fold higher calcium binding ability but a 2-fold lower turnover rate [14], [16]. The single SERCA gene in invertebrates also undergoes alternative splicing and shows tissue specific expression in a similar way to the vertebrate SERCA 2 [17], [18], [19]. SERCA 2 can be alternatively spliced to make a SERCA 2b (1042aa) protein or the shorter variant SERCA 2a (997aa), which has exons 22 and 24 spliced out compared with the longer protein [9], [20]. A similar pattern of splicing has been demonstrated in invertebrates such as Artemia franciscana and Caenorhabditis elegans [9], [17]. The single SERCA protein in invertebrates has been suggested to be most closely related to the vertebrate SERCA 2 based on the observation that SERCA2b is the housekeeping variant [9].
The P-type ATPases show a complex history of gene duplication events. For example, Arabidopsis has 46 known P-type ATPase genes with multiple isoforms in each family; comparatively humans have about 36 genes. Several hypotheses have been proposed to explain the evolution of this complex gene family. First, multiple isoforms could have evolved to be expressed in different cell types, and would have tissue specific regulation [21]. In this scenario, mutations in the promoter are important for isoforms to be expressed in distinctive amounts in different tissues or at different developmental stages [22]. Second, different isoforms could have evolved to function optimally under different cellular conditions or stressors (e.g., toxic cations) [23], allowing the organism to inhabit a wide variety of habitats and niches. In this case, mutations in coding sequence of the genes are most important, as they can alter the biochemical properties of the protein that are advantageous in specific environments. Lastly, it has been suggested that a fraction of isoforms are functionally redundant duplicates [21]. These alternative hypotheses are consistent with the theoretical predictions that the evolutionary fate of gene duplicates is difficult to distinguish [24]. To address this issue, an initial step is to characterize the historical gene duplication events that have occurred during the evolution of such gene families. SERCA is the most well characterized P-type ATPase, having X-ray crystallographic structures of its different conformational states and domains [25]. Despite the extensive knowledge and interest in its structure, function, and expression, little is known about SERCA’s evolutionary history. Here, we use protein sequences and phylogenetic reconstruction to examine the relationship among SERCA homologues across eukaryotic taxa. Specifically, we assess the role of gene duplication in the evolution of vertebrate SERCA isoforms and test previous hypotheses regarding the phylogenetic relationships of three vertebrate SERCA isoforms with invertebrate SERCA. Furthermore, we explore the protein-based eukaryotic phylogeny of SERCA to examine various likely gene duplication events in other phylogenetic lineages and their evolutionary implications.
Materials and Methods
Sequence Retrieval
A total of 81 SERCA amino acid sequences of vertebrate, invertebrate, plant, fungi, and other unicellular eukaryotes such as protists and ciliated protozoans were retrieved from Genbank, Uniprot, Ensembl, and JGI (Table S1). Sequences were chosen to span the known and confirmed SERCA genes across the eukaryotic kingdom. Preference was given to sequences that had protein level and transcript level evidence for the sequence over those inferred only from homology (Table S1). Searches were conducted using the key terms “Sarcoplasmic/endoplasmic calcium ATPase”, “SR Ca2+ ATPase” and “SERCA”. In addition, BLAST searches were conducted based on annotated sequences. If multiple splice variants of the protein were reported, the canonical sequence was used.
Sequence Alignment and Phylogenetic Analyses
The amino acid sequence alignment was performed using ClustalW [26] with a gap opening penalty of 10 and gap extension penalty of 0.2. The highly variable 3′ terminal ends of the sequences were trimmed to avoid ambiguity. No other variable regions were excluded from the alignment. Phylogenetic analyses were performed using neighbour-joining (NJ) and Bayesian inference (BI) approaches, in MEGA [27] and MrBayes [28] respectively. Members of other P-type II ATPases sub-families were used as outgroups (Table S1) in all phylogenetic reconstructions. Outgroup sequences included the Plasma Membrane Ca2+ ATPase (ATP2B1), Secretory Pathway Ca2 + ATPase (ATP2C1 and ATP2C2), Na+/K+-transporting ATPase (ATP4A1), K+-transporting ATPase (ATP4A), as well as the fungi-specific Na+/K+ ATPase (ACU1) and Na+ transport ATPase (ENA1) [29]. We used a Bayesian inference method for tree construction with the WAG model of amino acid substitution. This model was selected prior to the final analysis using a model jumping algorithm implemented in MrBayes [28], [30]. This algorithm regularly swaps between 9 different fixed-rate amino acid models throughout the analysis and selects the model with the highest contribution to posterior probability density [28], [30]. The WAG model can handle a large number of sequences and is applicable to a wide range of protein families, but retains the advantages of a maximum-likelihood approach and accounts for multiple substitutions at the same site [31]. Three independent runs of 4 Markov chains were conducted for 1,000,000 generations with a sampling frequency of 10 and the first 25% of sampled trees discarded as burn-in. We confirmed the topology of the Bayesian tree with a cluster based neighbour-joining tree using pairwise deletion and the Jones-Taylor-Thornton (JTT) amino acid model [32]. Node support was analyzed using 1000 bootstrap replicates.
Results and Discussion
Overall Phylogenetic Pattern
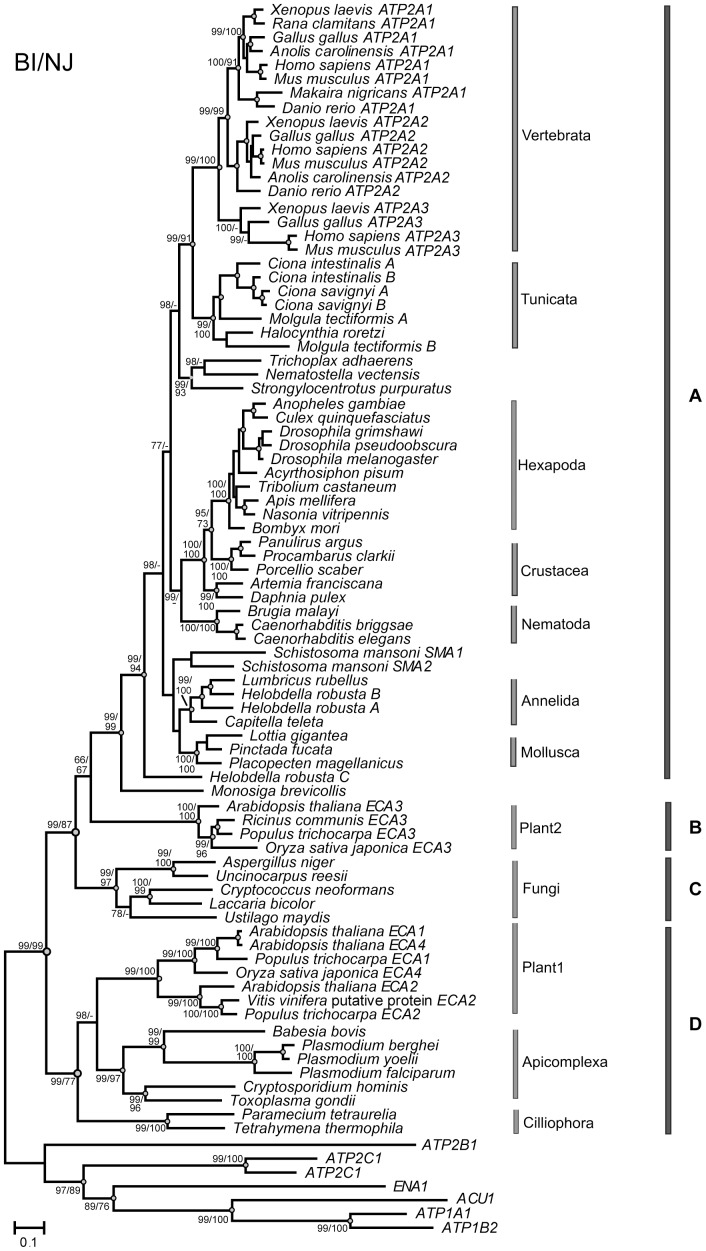
The SERCA alignment consisted of 81 sequences (61 unique taxa) spanning 1575 amino acids and contained 220 conserved and 818 parsimony-informative sites. Both the BI and NJ analyses returned highly congruent tree topologies. There were two major monophyletic clades. The first group contains clades A, B and C, which consist of metazoan, fungal, and plant sequences, respectively (Fig. 1). The second group contains clade D that encompasses plant and protist sequences (Fig. 1). Within clade A, the chordates are monophyletic and contain two reciprocally monophyletic clades corresponding to vertebrates and tunicates.
Figure 1. Bayesian phylogenetic reconstruction of SERCA amino acid sequences from 57 taxa.
The numbers at the nodes indicate posterior probabilities/bootstrap supports. Nodes highlighted with gray circles represent consensus neighbouring-joining (NJ) and Bayesian Inference (BI) analyses with bootstrap support higher than 70%.
SERCA Gene Duplication and Evolution
Within metazoans, the SERCA sequences of the chordates form a well supported monophyletic group that includes two sister clades, corresponding to the vertebrates and tunicates. In vertebrates, each of the three SERCA isoforms (i.e. SERCA1-3; coded by ATP2A1-3, respectively) form highly supported monophyletic groups. This pattern indicates that the three SERCA isoforms arose through two rounds of gene (or whole genome) duplication events after divergence from the tunicates but before the separation of fishes and tetrapods. Furthermore, ATP2A1 and 2 are sister groups and likely arose from a more recent duplication event than the one leading to ATP2A3. Our findings do not support the previous suggestion that SERCA2 is the most ancestral of the three isoforms [9]. Furthermore, tunicates show evidence of several independent and relatively recent gene duplication events (Fig. 1).
Our study reveals that invertebrates also have experienced several instances of gene duplication events leading to multiple SERCA paralogs. Previous studies identified that the parasite trematode Schistosoma mansoni has two SERCA proteins that are coded by the genes SMA1 and SMA2 [10]. Based on our analysis, the two S. mansoni calcium ATPases group together, despite the low bootstrap support and high sequence divergence (29.5%). This suggests that SMA1 and 2 are likely paralogs that arose from an ancient gene duplication event. Furthermore, Helobdella robusta also possesses three SERCA genes. While SERCA A and B group with other annelid sequences SERCA C is highly divergent and basal to the animal clade. Thus, SERCA C likely arose due to an ancient gene duplication event, while SERCA A and B are the result of a relatively recent gene duplication event. This is consistent with findings of several other gene duplications within the H. robusta genome, across a variety of unrelated gene families [33], [34], [35].
Plants are particularly reliant on calcium signalling to control many vital physiological processes and stress responses [36], [37]. Because plants use the Ca2+ to respond to different stimuli, the temporal and spatial patterns of the signals must vary [38]. Thus, the regulation of proteins involved in calcium signalling needs to be fine-tuned in order to induce a stimulus specific response [39]. While Arabidopsis is known to have four Endoplasmic Ca2+ ATPases (coded by genes ECA 1-4) [40], most plants have only three [21]. Based on our analysis, the plant sequences form two monophyletic clades, Plant1 and Plant2 (Fig. 1), consistent with previous findings in Viridiplantae [3]. Plant1 clade consists of the ECA 1, 2, and Arabidopsis ECA4 genes and is sister to the clade formed by apicomplexan sequences. However, Plant2, which contains the ECA 3 genes, is sister to metazoan sequences. The presence of two divergent clades of SERCA genes in plants indicates a possible ancient duplication early in eukaryotic evolution followed by the loss of ECA 3-like protein from the protists and the loss of the other duplicate in the lineage leading to animals and fungi, whereas the plants retained both copies. Alternatively, lateral gene transfer (LGT) between protists and plants could have resulted in the Plant1 group. However, LGT events are rare and their prevalence in eukaryotes is unclear [41], [42]. Interestingly, the ECA 3 encoded protein serves to pump calcium into both the Golgi and endoplasmic reticulum [43]. However, animals and fungi use a separate set of proteins (SPCA) to pump calcium into the Golgi that appear to be absent in plants [43]. The altered function and localization of the ECA 3 encoded protein to the Golgi may have played a role in its evolution and maintenance in plant genomes and warrants further investigation. The evolution of complex calcium signalling in plants was likely facilitated by duplication of Ca2+ ATPase genes which diverged in their patterns of regulation and localization.
Gene duplication events are an essential part of the evolutionary process as they generate novel gene functions and families. The initial increased dosage of gene products resulting from a gene duplication event may be beneficial or detrimental to the organism. The function of the new gene will be retained through stabilizing selection if the increased dosage is beneficial or lost through purifying selection if it is detrimental [24]. However, if increased dosage has no effect, the gene is no longer under selective pressure and is free to accumulate mutations. Therefore, the duplicated gene can either become a pseudogene, or gain novel function through changes in the protein structure or expression pattern [44], [45]. Duplicated genes may also gain novel function by translocation into different regulatory regions. Such events can drastically alter the location, timing, and conditions of their expression. It appears that duplicated SERCA genes gain novel functions; this is especially apparent in the three vertebrate SERCA genes that exhibit tissue specific expression patterns likely resulting from divergence in regulation of these genes following the duplication events. Evidence of both ancient and recent gene duplication events in many taxa demonstrates the capacity of SERCA genes to multiply and retain functional significance.
From Gene Tree to Species Tree: Paraphyly of Crustacea
If we ignore the major ancient gene duplication events, the overall phylogenetic pattern recovered was consistent with that of the combined protein data of α-tubulin, β-tubulin, actin, and elongation factor 1–alpha [46]. Moreover, the recovered phylogeny provides valuable information about the evolutionary path of crustaceans. The phylogenetic relationships among arthropod taxa, especially those within Pancrustacea, remain unclear in many phylogenetic studies [47]. Here, the monophyly of the Pancrustacea SERCA proteins is highly supported. The SERCAs of hexapods form a monophyletic group. However, crustaceans appear to be paraphyletic; Panulirus argus, Procambarus clarkii, and Porcellio scaber are sister to the hexapods, and not to the clade formed by the branchiopods Daphnia pulex and Artemia franciscana. This observation is consistent with other molecular and morphological based studies that support the monophyly of Pancrustacea, including all members Crustacea and Hexapoda [48], [49], [50], [51], [52], [53]. To date there is no consensus regarding the placement of Hexapoda within the paraphyletic crustacean group. Proposed sister clades include Branchiopoda [50], [51], Malacostraca [48], [49], and Copepoda [52]. Our SERCA protein-based phylogeny supports Malacostraca (lobsters, shrimp, woodlice) as the sister group to Hexapoda. However, future detailed studies based on a combination of morphological and molecular data are still necessary to elucidate the phylogenetic relationships within Pancrustacea.
Conclusion
Overall, our phylogenetic analyses reveal several recent and ancient gene duplication events across different taxonomic levels during the evolution of SERCA genes. Notably, gene duplication events have resulted in proteins with new function and expression patterns in plants and vertebrates. Our results have refined the understanding of the complex evolutionary history of this gene family and will greatly facilitate gene expression and comparative studies that focus on SERCA genes.
Supporting Information
List of protein sequences used for phylogenetic analyses.
(DOCX)
Acknowledgments
We thank Bora Demiri, John Colbourne, Norman Yan, and two anonymous reviewers for their careful and insightful comments on the manuscript.
Funding Statement
This study was funded by the Natural Sciences and Engineering Council (NSERC) of Canada grant to Melania E. Cristescu. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Corradi N, Sanders IR (2006) Evolution of the P-type II ATPase gene family in the fungi and presence of structural genomic changes among isolates of Glomus intraradices . BMC Evol Biol 6: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benito B, Garciadeblas B, Schreier P, Rodriguez-Navarro A (2004) Novel P-type ATPases mediate high-affinity potassium or sodium uptake in fungi. Eukar Cell 3: 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pedersen CNS, Axelsen KB, Harper JF, Palmgren MG (2012) Evolution of plant P-type ATPases. Front Plant Sci 3: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berridge MJ, Bootman MD, Roderick HL (2003) Calcium signaling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol 4: 517–529. [DOI] [PubMed] [Google Scholar]
- 5. Clapham DE (2007) Calcium signaling. Cell 131: 1047–1058. [DOI] [PubMed] [Google Scholar]
- 6. Flavell SW, Greenberg ME (2008) Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci 31: 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qi H, Moran MM, Navarro B, Chong JA, Krapivinsky G, et al. (2007) All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc Natl Acad Sci USA 104: 1219–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kudla J, Batistic O, Hashimoto K (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wuytack F, Raeymaekers L, Missiaen L (2002) Molecular physiology of the SERCA and SPCA pumps. Cell Calcium 32: 279–305. [DOI] [PubMed] [Google Scholar]
- 10. Talla E, de Mendonça RL, Degand I, Goffeau A, Ghislain M (1998) Schistosoma mansoni Ca2+-ATPase SMA2 restores viability to yeast Ca2+-ATPase-deficient strains and functions in calcineurin-mediated Ca2+tolerance. J Biol Chem 273: 27831–27840. [DOI] [PubMed] [Google Scholar]
- 11. Martin V, Bredoux R, Corvazier E, Van Gorp R, Kovacs T, et al. (2002) Three novel sarco/endoplasmic reticulum Ca2+ATPase (SERCA) 3 isoforms. Expression, regulation, and function of the membranes of the SERCA3 family. J Biol Chem 277: 24442–24452. [DOI] [PubMed] [Google Scholar]
- 12.Hovnanian A (2007) SERCA pumps and human diseases. In: Carafoli, E, Brini, M, editors. Calcium Signaling and Disease: Molecular Pathology of Calcium. New York: Springer. 337–363.
- 13. Brandl CJ, deLeon S, Martin DR, MacLennan DH (1987) Adult forms of the Ca2+ ATPase of sarcoplasmic reticulum. Expression in developing skeletal muscle. J Biol Chem 262: 3768–3774. [PubMed] [Google Scholar]
- 14. East JM (2000) Sarco(endo)plasmic reticulum calcium pumps: recent advances in our understanding of structure/function and biology. Mol Membr Biol 17: 189–200. [DOI] [PubMed] [Google Scholar]
- 15. Lytton J, Westlin M, Burk SE, Shull GE, MacLennan DH (1992) Functional comparisons between isoforms of the sarcoplasmic or endoplasmic reticulum family of calcium pumps. J Biol Chem 267: 14483–14489. [PubMed] [Google Scholar]
- 16. Verboomen H, Wuytack F, De Smedt H, Himpens B, Casteels R (1992) Functional difference between SERCA2a and SERCA2b Ca2+pumps and their modulation by phospholamban. Biochem J 286: 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Escalante R, Sastre L (1993) Similar alternative splicing events generate two sarcoplasmic or endoplasmic reticulum Ca-ATPase isoforms in the crustacean Artemia franciscana and in vertebrates. J Biol Chem 268: 14090–14095. [PubMed] [Google Scholar]
- 18. Fan W, Li C, Li S, Feng Q, Xie L, et al. (2007) Cloning, characterization, and expression patterns of three Sarco/Endoplasmic Reticulum Ca +2 -ATPase isoforms from pearl oyster (Pinctada fucata). Acta Biochim Biophys Sin 39: 722–730. [DOI] [PubMed] [Google Scholar]
- 19. Mandal A, Arunachalam SC, Meleshkevitch EA, Mandal PK, Boudko DY, et al. (2009) Cloning of sarco-endoplasmic reticulum Ca2+ -ATPase (SERCA) from Caribbean spiny lobster Panulirus argus . J Comp Physiol B 179: 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campbell AM, Kessler PD, Sagara Y, Inesi G, Fambrough DM (1991) Nucleotide sequences of avian cardiac and brain SR/ER Ca(+2)-ATPases and functional comparisons with fast twitch Ca(+2)-ATPase. Calcium affinities and inhibitor effects. J Biol Chem 266: 16050–16055. [PubMed] [Google Scholar]
- 21. Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, et al. (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132: 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lingrel JB (1992) Na, K-ATPase: Isoform structure, function, and expression. J Bioenerg Biomembr 24: 263–270. [DOI] [PubMed] [Google Scholar]
- 23. Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, et al. (2001) Phylogenetic relationships within cation transporter families of Arabidopsis . Plant Phisiol 26: 1646–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Innan H, Kondrashov F (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108. [DOI] [PubMed] [Google Scholar]
- 25. Kühlbrandt W (2004) Biology, structure and mechanism of P-Type ATPases. Nat Rev Mol Cell Biol 5: 282–295. [DOI] [PubMed] [Google Scholar]
- 26. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 28. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 29. Axelsen KB, Palmgren MG (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol 46: 84–101. [DOI] [PubMed] [Google Scholar]
- 30.Ronquist F, Huelsenbeck JP, van der Mark P (2005) MrBayes 3.1 Manual. Published online at: http://mrbayes.csit.fsu.edu/manual.php.
- 31. Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18: 691–699. [DOI] [PubMed] [Google Scholar]
- 32. Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282. [DOI] [PubMed] [Google Scholar]
- 33. Kourakis MJ, Martindale MQ (2001) Hox gene duplication and deployment in the annelid leech Helobdella. Evol Dev 3: 145–153. [DOI] [PubMed] [Google Scholar]
- 34. Cristescu ME, Innes DJ, Stillman JH, Crease TJ (2008) D- and L-lactate dehydrogenases during invertebrate evolution. BMC Evol Biol 8: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cho SJ, Vallès Y, Giani VC, Seaver EC, Weisblat DA (2010) Evolutionary dynamics of the wnt gene family: a lophotrochozoan perspective. Mol Biol Evol 27: 1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reddy ASN, Ali GS, Celesnik H, Day IS (2011) Coping with stresses: Roles of Calcium- and Calcium/Calmodulin-regulated gene expression. Plant Cell 23: 2010–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Conde A, Manuela Chaves M, Gerós H (2011) Membrane transport, sensing and signalling in plant adaptation to environmental stress. Plant Cell Physiol 52: 1583–1602. [DOI] [PubMed] [Google Scholar]
- 38. Kudla J, Batistič O, Hashimoto K (2010) Calcium signals: The lead currency of plant information processing. Plant Cell 22: 541–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signalling. Plant Cell 19: S401–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kabala K, Klobus G (2005) Plant Ca2+ -ATPases. Acta Physiol Plant 27: 559–574. [Google Scholar]
- 41. Andersson JO (2005) Lateral gene transfer in eukaryotes. Cell Mol Life Sci 62: 1182–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, et al. (2005) The tree of eukaryotes. Trends Ecol Evol 20: 670–676. [DOI] [PubMed] [Google Scholar]
- 43. Mills RF, Doherty ML, López-Marqués RL, Weimar T, Dupree P, et al. (2008) ECA3, a Golgi-localized P2A-type ATPase, plays a crucial role in manganese nutrition in Arabidopsis. Plant Physiol 146: 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV (2002) Selection in the evolution of gene duplications. Genome Biol 3: research0008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, et al. (2011) The ecoresponsive genome of Daphnia pulex . Science 331: 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baldauf SL (2000) A kingdom-level phylogeny of Eukaryotes based on combined protein data. Science 290: 972–977. [DOI] [PubMed] [Google Scholar]
- 47. Jenner RA (2010) Higher-level crustacean phylogeny: Consensus and conflicting hypotheses. Arthropod Struct Dev 39: 143–153. [DOI] [PubMed] [Google Scholar]
- 48. Harzsch S (2002) The phylogenetic significance of crustacean optic neuropils and chiasmata: a re-examination. J Comp Neurol 453: 10–21. [DOI] [PubMed] [Google Scholar]
- 49. Sinakevitch I, Douglass JK, Scholtz G, Loesel R, Strausfeld NJ (2003) Conserved and convergent organization in the optic lobes of insects and isopods, with reference to other crustacean taxa. J Comp Neurol 467: 150–172. [DOI] [PubMed] [Google Scholar]
- 50. Regier JC, Shultz JW, Kambic RE (2005) Pancrustacean phylogeny: hexapods are terrestrial crustaceans and maxillopods are not monophyletic. Proc R Soc B 272: 395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mallatt J, Giribet G (2006) Further use of nearly complete 28S and 18S rRNA genes to classify Ecdysozoa: 37 more arthropods and a kinorhynch. Mol Phylogenet Evol 40: 772–794. [DOI] [PubMed] [Google Scholar]
- 52. von Reumont BM, Meusemann K, Szucsich NU, Dell’Ampio E, Gowri-Shankar V, et al. (2009) Can comprehensive background knowledge be incorporated into substitution models to improve phylogenetic analyses? A case study on major arthropod relationships. BMC Evol Biol 9: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Edgecombe GD (2010) Arthropod phylogeny: an overview from the perspectives of morphology, molecular data and the fossil record. Arthropod Struct Dev 39: 74–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of protein sequences used for phylogenetic analyses.
(DOCX)