Abstract
Background
Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) catalyses the key reaction in the photosynthetic assimilation of CO2. In C4 plants CO2 is supplied to Rubisco by an auxiliary CO2-concentrating pathway that helps to maximize the carboxylase activity of the enzyme while suppressing its oxygenase activity. As a consequence, C4 Rubisco exhibits a higher maximum velocity but lower substrate specificity compared with the C3 enzyme. Specific amino-acids in Rubisco are associated with C4 photosynthesis in monocots, but it is not known whether selection has acted on Rubisco in a similar way in eudicots.
Methodology/Principal Findings
We investigated Rubisco evolution in Amaranthaceae sensu lato (including Chenopodiaceae), the third-largest family of C4 plants, using phylogeny-based maximum likelihood and Bayesian methods to detect Darwinian selection on the chloroplast rbcL gene in a sample of 179 species. Two Rubisco residues, 281 and 309, were found to be under positive selection in C4 Amaranthaceae with multiple parallel replacements of alanine by serine at position 281 and methionine by isoleucine at position 309. Remarkably, both amino-acids have been detected in other C4 plant groups, such as C4 monocots, illustrating a striking parallelism in molecular evolution.
Conclusions/Significance
Our findings illustrate how simple genetic changes can contribute to the evolution of photosynthesis and strengthen the hypothesis that parallel amino-acid replacements are associated with adaptive changes in Rubisco.
Introduction
Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase, EC 4.1.1.39) serves as the main gateway for inorganic carbon to enter metabolic pathways in most ecosystems and hence is unique in its importance to support life. Observations of significant variation in Rubisco kinetics between plant species [1], [2],[3], the correlation of Rubisco kinetics with temperature [4] and CO2 availability [5], and positive selection on Rubisco at the molecular level in all principal lineages of land plants [6] support the hypothesis that all Rubiscos may be well adapted to their subcellular environment [7]. However, the molecular mechanisms responsible for optimizing the relationship between Rubisco specificity and its maximum rate of catalytic turnover in particular conditions are still open to debate [8]. Here we use a phylogeny-based approach to investigate how the occurrence of C4 photosynthesis has influenced Rubisco evolution at the molecular level in eudicots as represented by the family Amaranthaceae sensu lato.
Rubisco discriminates imperfectly between CO2 and O2 as substrates, and under present-day atmospheric conditions (385 p.p.m. CO2), the carboxylase activity of Rubisco is undersaturated in C3 plants, and the oxygenase activity gives rise directly to the competing process of photorespiration. Photorespiratory rates in C3 plants increase steeply with increasing temperature and give rise to a distinct temperature optimum for net photosynthesis, above which plant yields decline steeply. Increased carbon loss via photorespiration at higher temperatures is attributable mainly to the declining specificity of Rubisco for CO2 relative to O2 (S c/o). In fact, it has been proposed that the very slow turnover of Rubisco (k cat ≈3 s−1) is a direct consequence of the enzyme's particular reaction mechanism, in which S c/o is maximized by tight binding of the transition-state intermediate [7]. Land plants also depend on the enzyme rubisco activase which removes tightly binding inhibitors at the active site of Rubisco and thus prevents the loss of its catalytic activity. The cascade of side-reactions performed by Rubisco is yet to be fully understood although recent achievements in mathematical modelling of Rubisco reactions offer the theoretical background for predicting ‘side-effects’ by simulating the overall kinetic behaviour [9]. Another corollary of low k cat and of the large size of the holoenzyme (560 kDa) is that Rubisco comprises up to 50% of soluble protein in photosynthetic tissues and is probably the most abundant enzyme on Earth [10].
In terrestrial plants with C4 photosynthesis or crassulacean acid metabolism (CAM), and in many aquatic organisms, photorespiration is partially or completely suppressed by the operation of an auxiliary CO2-concentrating mechanism. C4 plants initially fix atmospheric carbon in the mesophyll cells using phosphoenolpyruvate carboxylase, an enzyme with a high effective affinity for CO2 (HCO3 − being the true substrate of the enzyme). Further four-carbon compounds (malate or aspartate) produced by this fixation are transported to the specialized bundle-sheath cells, where CO2 is released and fixed by Rubisco. Rubisco from C4 plants, which experiences ∼10-fold higher CO2 concentrations in bundle-sheath cells than does the enzyme in C3 plants [11], has a lower affinity for CO2 but a higher k cat (≈4 s−1). Having less specific but faster Rubisco and no photorespiration losses, C4 plants require 60 to 75% less Rubisco to match the photosynthetic capacity of C3 plants [12], [13]. In fact, many C4 plants such as maize, sugarcane and sorghum are among the most productive of all species cultivated agriculturally. Although C4 plants appeared relatively recently in evolutionary terms and constitute only 3% of terrestrial plant species, they are already among the most successful and abundant groups in warm climates and are responsible for about 20% of terrestrial gross primary productivity [14], [15].
C4 photosynthesis evolved independently in at least 62 recognizable lineages of angiosperms and represents one of the most striking examples of a convergent biochemical adaptation in plants [16]. However, since its discovery, most attention has been devoted to the more numerous and agriculturally important C4 monocots in the Poaceae, while C4 eudicots have been studied less intensively. The family Amaranthaceae sensu lato (i.e. including Chenopodiaceae) [17], [18] contains about 180 genera and 2500 species, of which approximately 750 are C4 species [16], making it by far the largest C4 family among eudicots and the third-largest among angiosperms (after Poaceae and Cyperaceae). C4 photosynthesis evolved at least 15 times within Amaranthaceae [16] making this family a good model to study coevolution of C4 photosynthesis and Rubisco. Notably, the Amaranthaceae exceed the Poaceae and Cyperaceae in the diversity of photosynthetic organ anatomy [19], and is the only angiosperm family containing terrestrial C4 plants that lack Kranz anatomy, with three species having a single-cell rather than the more usual dual-cell C4 system [20], [21]. The predominantly tropical Amaranthaceae sensu stricto and primarily temperate and subtropical Chenopodiaceae have long been treated as two closely related families (see review in [19]) until the formal proposal that Chenopodiaceae should be included within the expanded Amaranthaceae based on a lack of separation between the two families in sequence data [17]. Amaranthaceae sensu lato (henceforth referred to as Amaranthaceae) constitutes the most diverse lineage of the Caryophyllales. Both C3 and C4 species from this family are adapted to a range of conditions from temperate meadows to the tropics, hot deserts and salt marshes. However, it has been shown that the abundance of C4 Amaranthaceae is correlated with precipitation but not temperature, in contrast to the abundance of C4 Poaceae and Cyperaceae, which is correlated with temperature but not precipitation [22].
Despite C4 Amaranthaceae showing different suites of anatomical and biochemical adaptations as well as ecological preferences compared to C4 Poaceae and Cyperaceae, like C4 monocots they possess faster but less CO2-specific Rubiscos than their C3 relatives [3], [5], [23]. Thus, Rubisco of C4 eudicots and monocots represents a notable example of convergent evolution of enzyme properties in phylogenetically distant groups. However, it is not known whether this functional convergence in Rubisco kinetics evolved via similar or different structural changes in protein [24]. Molecular adaptation can be inferred from comparison of the rates of non-synonymous (changing amino-acid protein sequence, d N) and synonymous (resulting in no change at the protein level, d S) mutations along a phylogenetic tree using maximum likelihood and Bayesian frameworks [25]. Recently, such methodology has been applied to the chloroplast gene rbcL, which encodes the large subunit of Rubisco that forms the enzyme's active site, and showed that positive Darwinian selection is acting within most lineages of plants [6]. Only a small fraction of Rubisco residues appear to be under positive selection, while most residues have been under purifying selection [6]. Some of these residues have been shown to be under positive selection within C4 lineages of Poaceae and Cyperaceae [26] and in the small Asteraceae genus, Flaveria [27], which contains both C3 and C4 species. However, no specific analysis has yet been made of Rubisco sequence evolution in a large group of C4 eudicots. In this study, we investigate positive selection on the rbcL gene of plants from the Amaranthaceae family and, in particular, focus on coevolution of Rubisco and C4 photosynthesis asking whether positive selection on the rbcL gene occured on branches leading to C4 clades and/or within C4 clades. Finally, we address the following question: which amino-acid replacements were associated with transitions from C3 to C4 photosynthesis in Amaranthaceae, and are these replacements unique to this lineage or shared with C4 monocots and/or Flaveria?
Materials and Methods
Phylogenetic analysis
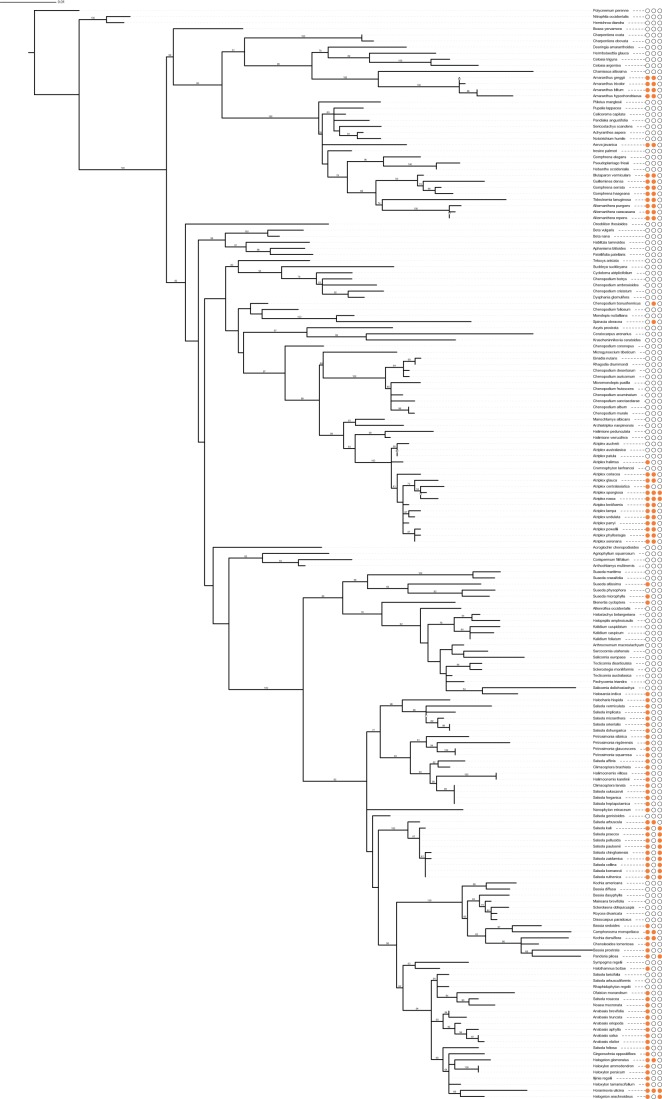
We obtained all Amaranthaceae rbcL nucleotide sequences available in GenBank and aligned them. Sequences shorter than 1341 base pairs and sequences with missing data were excluded. The resulting trimmed alignment consisted of 179 rbcL sequences of 1341 base pairs long which represented 94% of the rbcL coding region and corresponded to positions 64 to 1404 of the rbcL sequence of Spinacia oleracea (GenBank AJ400848). The analysed dataset consisted of 95 C3 and 84 C4 species (Table S1). Most of the included sequences came from four studies [19], [28], [29], [30] and evenly represented all main lineages within the family (Fig. 1). Phylogeny was reconstructed using a maximum-likelihood inference (ML) conducted with RAxML version 7.2.6 [31] using the raxmlGUI interface [32]. We conducted five independent runs from different starting points to assess convergence within two likelihood units of the best tree, which was consistently selected. The parameters of partition were allowed to vary independently under the GTRGAMMA model of evolution as implemented in RAxML. ML nodal support was calculated by analysing 1000 bootstrap replicates. The best-scoring ML tree was used for tests of positive selection (see below).
Figure 1. Maximum likelihood phylogram based on rbcL sequences of 179 Amaranthaceae species.
Numbers above the branches are ML bootstrap support percentages. Filled orange circles of the first, second and third columns after species names indicate presence of C4 photosynthesis, serine at the position 281 and isoleucine at the position 309, respectively. The figure was composed using iTOL program [62].
Tests for positive selection
Positive, neutral, or purifying selection at the molecular level can be inferred by comparing rates of non-synonymous (d N) and synonymous (d S) mutations along a phylogenetic tree [33]. Under neutrality, the two rates are expected to be equal (d N/d S = 1), while purifying (negative) or adaptive (positive) selection is expected to deflate (d N/d S<1) or inflate (d N/d S>1) this ratio, respectively. One can use likelihood ratio tests to detect positive selection that affects only a subset of codons in a protein-coding gene, with positive selection indicated by accelerated nonsynonymous substitutions. Models assuming positive selection along all phylogeny or prespecified branches only (e.g. C4 lineages in our case) can be employed within Phylogenetic Analysis by Maximum Likelihood (PAML) framework [33].
We used the codeml program in the PAML v.4.4 package [33] to estimate d N/d S ratio in the model M0, that allows for a single d N/d S value across the whole phylogenetic tree obtained previously (see Phylogenetic analyses section). Further, codeml was used to perform likelihood ratio tests (LRTs) for positive selection among amino acid sites. The tree length value obtained from the model M0 was compared with tree length values obtained from other models to control for consistency among models. We performed two LRTs to compare null models which assume the same selective pressure along all branches of a phylogeny and do not allow positive selection (d N/d S >1) with nested models which do allow it [33]. The first LRT, M1a-M2a, compares the M1a model (Nearly Neutral) which allows 0≤ d N/d S ≤1 with the M2a model (Selection model; same as the M1a model plus an extra class under positive selection with d N/d S >1). The second LRT, M8a-M8, compares the M8a model which assumes a discrete beta distribution for d N/d S, which is constrained between 0 and 1 including a class with d N/d S = 1 with the M8 model which allows the same distribution as M8a but an extra class under positive selection with d N/d S >1.
Finally, we performed two branch-site tests of positive selection along prespecified foreground branches [33], [34], [35]. The first was the A model for basal C4 branches only where positive selection was allowed only on branches leading to C4 clades. The second was the A model for all C4 branches where positive selection was allowed on branches leading to C4 clades and branches within C4 clades. The A1-A LRT compares the null model A1 with the nested model A. Both the A1 and A models allow d N/d S ratios to vary among sites and among lineages. The A1 model allows 0< d N/d S <1 and d N/d S = 1 for all branches, and also two additional classes of codons with fixed d N/d S = 1 along prespecified foreground branches while restricted as 0< d N/d S <1 and d N/d S = 1 on background branches. The alternative A model allows 0< d N/d S <1 and d N/d S = 1 for all branches, and also two additional classes of codons under positive selection with d N/d S >1 along prespecified foreground branches while restricted as 0< d N/d S <1 and d N/d S = 1 on background branches. C4 lineages were marked as foreground branches.
For all LRTs, the first model is a simplified version of the second, with fewer parameters, and is thus expected to provide a poorer fit to the data (lower maximum likelihood). The M1a, M8a and A1 models are null models which do not allow codons with d N/d S >1, whereas the M2a, M8 and A models are alternative models which do allow codons with d N/d S >1. The significance of the LRTs was calculated assuming that twice the difference in the log of maximum likelihood between the two models was distributed as a chi-square distribution with the degrees of freedom (df) given by the difference in the numbers of parameters in the two nested models [34], [36]. For the M1a-M2a comparison df = 2, and for M8a-M8, A1-A and M0 vs 2-rates model comparisons df = 1. Each LRT was run two times using different initial d N/d S values (0.1 and 0.4) to test for suboptimal local peaks. To identify amino acid sites potentially under positive selection, the parameter estimates from M2a, M8 and A models were used to calculate the posterior probabilities that an amino acid belongs to a class with d N/d S >1 using the Bayes Empirical Bayes (BEB) approaches implemented in PAML [37]. Independently from codeml we used the SLR program which implements “sitewise likelihood-ratio” (SLR) method for detecting non-neutral evolution, a statistical test that can identify sites under positive selection even when the strength of selection is low [38]. The SLR test [38] consists of performing a likelihood-ratio test on a sitewise basis, testing the null model (neutrality, d N/d S = 1) against an alternative model (d N/d S ≠1). SLR method is a test of whether a given site has undergone selection or not, and the test statistic summarizes the strength of the evidence for selection rather than the strength of the selection itself [38]. The same input files with sequence alignment and species phylogeny were used for both codeml and SLR.
Analysis of correlated evolution on phylogenies
Closely related taxa are not independent data points and they consequently violate the assumptions of conventional statistical methods [39]. Thus, we used analysis of correlated evolution on phylogenies to test the significance of correlation between pairs of discrete characters: (1) the presence/absence of C4 photosynthesis and (2) the presence/absence of particular amino-acid at sites found to be under positive selection along C4 branches in the A model of codeml. For this purpose, we used the phylogeny obtained using RAxML (see above) and performed Pagel's test of correlated (discrete) character evolution [40] implemented in the Mesquite package (version 2.72) [41]. Test was performed separately for each Rubisco residue under positive selection along C4 branches and Bonferroni correction was performed for simultaneous statistical testing.
Structural analysis of Rubisco
We used the published Rubisco protein structure from spinach (Spinacia oleracea, Amaranthaceae) from data file 1RBO [42] obtained from the RCSB Protein Data Bank. Throughout the paper, the numbering of Rubisco large subunit residues is based on the spinach sequence. The locations and properties of individual amino acids in the Rubisco structure were analysed using DeepView – Swiss-PdbViewer v.3.7 [43] and by CUPSAT [44].
Results
Phylogenetic analysis
The ML phylogenetic tree (Fig. 1) for rbcL sequences from 179 Amaranthaceae species was largely congruent with previously obtained phylogenies and accepted taxonomic subdivisions of the family [19], [28], [29], [30], [45], [46], [47], [48]; however no statistical tests for topological similarity between our tree and previously published trees were performed because of different sizes and species compositions of datasets. A minimum of 16 independent origins of C4 photosynthesis were represented in the Amaranthaceae phylogeny if conservative approach for observed polytomies had been taken (Fig. 1), which is consistent with the estimate by Sage et al. [16]. The other assumption of this estimate was that no reversals from C4 to C3 were allowed. Predominance of C4 gains over reversals to C3 is supported by both empirical data and theoretical work [49].
Tests for positive selection
Likelihood ratio tests (LRTs) for variation in d N/d S ratios and for positive selection [33] were applied to the dataset of rbcL sequences from 179 C3 and C4 Amaranthaceae species. LRTs that were run using two different initial d N/d S values (0.1 and 0.4) to test for suboptimal local peaks produced identical results. LRTs for positive selection [33] showed that the models assuming positive selection (M2a and M8) fit the data better than the nested models without positive selection (M1a and M8a; p-value <0.00001; Table 1). To test whether selection occurs specifically in C4 clades we used two branch site models (aka model A [33], [34]), one of which allowed positive selection only on branches leading to C4 clades and the other also allowed positive selection within the C4 clades. Each of these models was compared to an alternative model that allowed for no positive selection and only the latter of the two models demonstrated better fit to data than the model without positive selection (p-value <0.05; Table 1).
Table 1. Analysis of the Amaranthaceae rbcL genes for positively selected sites.
| Model with positive selection a | Null model a | LRT d | ||||||
| log-likelihood | Parameters b | Positively selected sites c | log-likelihood | Parameters b | 2l | P-value | ||
| Analysis for positively selected sites common for C3 and C4 clades | ||||||||
| M2a | −10711.44 | κ = 3.00, p 0 = 0.93, ω 0 = 0.02, p s = 0.01, ω s = 2.62 | 32, 145, 279, 439 | M1a | −10729.19 | κ = 2.94, p 0 = 0.93, ω 0 = 0.02 | 35.5 | <0.00001 |
| M8 | −10705.58 | κ = 2.94, p 0 = 0.96, p = 0.15, q = 3.04, ω s = 1.56 | 32, 43, 145, 225, 262, 279, 439, 443 | M8a | −10717.70 | κ = 2.90, p 0 = 0.94, p = 0.20, q = 5.42 | 24.2 | <0.00001 |
| SLR | NA | κ = 2.75, ω = 0.10 | 32, 145, 225, 279, 439 | NA | NA | NA | NA | NA |
| Analysis for positively selected sites specific for branches leading to C4 clades | ||||||||
| A | −10729.13 | κ = 2.94, p 0 = 0.93, ω 0 = 0.02, p s = 0.00, ω s = NA | no | A1 | −10729.13 | κ = 2.94, p 0 = 0.93, ω 0 = 0.02 | 0.0 | 1.00000 |
| Analysis for positively selected sites specific for C4 clades | ||||||||
| A | −10723.60 | κ = 2.94, p 0 = 0.92, ω 0 = 0.02, p s = 0.01, ω s = 3.15 | 281, 309 | A1 | −10726.15 | κ = 2.94, p 0 = 0.92, ω 0 = 0.02 | 5.1 | 0.02384 |
M1a (nearly neutral), M2a (positive selection), M8a (beta & ω = 1) and M8 (beta & ω) are PAML site models; A1 and A are PAML branch site models; SLR is “sitewise likelihood-ratio” method.
κ is transition/transversion rate ratio; ω is d N/d S ratio; ω s is d N/d S ratio in a class under putative positive selection; p 0 and p s are proportion of codons with ω<1 and ω>1, respectively; p and q are parameters of beta distribution in the range (0, 1); for the SLR test, the parameter values given are those optimal under M0.
The sites listed are those at which positive selection is detected with a cutoff (significance level or posterior probability, as appropriate to the method used) >95%; those >99% are in italics. For the SLR test, the italic underlined sites are those at which there is still evidence for positive selection after correcting for multiple comparisons.
LRT is likelihood ratio test, 2l is twice the difference of model log-likelihoods.
Sites under positive selection
Four sites were identified as evolving under positive selection with a posterior probability >0.95 by BEB [37] implemented in the M2a model (residues 32, 145, 279, 439), but eight sites when BEB was implemented in the M8 model (all the same that in M2a plus sites 43, 225, 262, 443). Independent SLR analysis showed five sites evolving under positive selection (32, 145, 225, 279, 439), but only for one of them (site 279) evidence for positive selection remained significant after correcting for multiple comparisons. Two sites (residues 281 and 309) were shown to be under positive selection within C4 clades while under relaxed or purifying selection within C3 clades with a posterior probability >0.99 by BEB in the A model for C4 branches. Both sites had only two alternative amino acids in this dataset (Table 2). One of the two alternative amino acids was more frequent among C4 species, while the other was more frequent among C3 species (Table 2), but there were no fixed differences between C4 and C3 species. We refer to amino acids more frequently associated with C4 taxa as the ‘C4’ amino acids, but only for the sake of brevity, as they are not invariantly associated with C4 photosynthesis. Pagel's test of correlated character evolution [40] on phylogeny showed significant positive associations (p-value <0.05) between the presence of C4 photosynthesis and the presence of ‘C4’ amino acids at sites 281 and 309, shown to be under positive selection along C4 branches.
Table 2. Characteristics of amino-acid replacements under positive selection in the C4 lineages of Amaranthaceae.
| AA No.a | AA changes ‘C3’→‘C4’ | Type of changes b | ΔHc | ΔPd | ΔVe | SAf (%) | ΔGg (kJ/mol) | RFPS (%) h | % C3/% C4 species i | Location of residue | Structural motifs within 5 Å | Inter-actions j | ||
| 281 | A | → | S | HN → UP | −2.6 | 1.1 | 0.4 | 0.00 | DS (−10.6) | 2.7 | 2.1/34.5 | Helix 4 | Helices 4, 5 | DD |
| 309 | M | → | I | HN → HN | 2.6 | −0.5 | 3.8 | 8.50 | S (−1.3) | 19.6 | 0.0/16.7 | Strand F | Strand E; Helices F, 5 | ID |
Amino acid (AA) numbering is based on the spinach sequence after [63].
Side chain type changes. Types abbreviations: H – hydrophobic; N – nonpolar aliphatic; P – polar uncharged; U – hydrophilic (after [64]).
Hydropathicity difference [65].
Polarity difference [66].
van der Waals volume difference [67].
Solvent accessibility calculated using the spinach structure (pdb file 1RBO) by CUPSAT [44].
Overall stability of the protein predicted using the spinach structure (pdb file 1RBO) by CUPSAT [44]. DS – destabilizing, S – stabilizing.
RFPS – relative frequency of the particular residue to be under positive selection in C3 plants. Data from 112 rbcL datasets with detected positive selection from [6].
Percentage of C3 and C4 species that have ‘C4’ amino acid among the 95 C3 species and 84 C4 species of Amaranthaceae analysed.
Interactions in which the selected residues and/or residues within 5 Å of them are involved. ID – intradimer interactions; DD – dimer-dimer interactions (after [63]).
Discussion
Widespread positive selection on Rubisco
As the performance of Rubisco can directly affect plant growth and crop yields, substantial efforts have been made to study its structure and function, with the ultimate aim of trying to improve Rubisco performance [50]. The last few years have brought new approaches to improving our understanding of Rubisco evolution and its genetic mechanisms. The initial molecular-phylogenetic analysis of rbcL showed that positive selection is widespread among all main lineages of land plants, but is restricted to a relatively small number of Rubisco amino acid residues within functionally important sites [6]. Following studies showed that rbcL is under positive selection in particular taxonomic groups [26], [27], [51], [52], [53], [54], [55], [56]. Coevolution of residues is common in Rubisco of land plants as well as positive selection and there is an overlap between coevolving and positively selected residues [57]. Hence, phylogeny-based genetic analyses suggest there has been a constant fine-tuning of Rubisco to optimize its performance in specific conditions, in agreement with empirical observations that Rubisco enzymes from different organisms show diversity of kinetics better related to species ecology than phylogeny [4].
All eight residues shown under selection in Amaranthaceae using SLR and PAML models M2 and M8 were already shown to be under Darwinian selection in other groups of plants [6]. Five of these residues (145, 225, 262, 279 and 439) were among twenty most commonly selected Rubisco large subunit residues [6]. Findings in Amaranthaceae are in agreement with the previously described uneven distribution of putative fine-tuning residues in Rubisco [6]. Residues 43, 145, 225, 262 and 279 had only two alternative amino acids in the analyzed dataset, while residues 32 and 439 had three and residue 443 had four alternative amino acids. Residue 145 is involved in dimer-dimer interactions, residue 225 is involved in interactions with small subunit, while residue 262 is involved in both [8]. C4 photosynthesis has increased the availability of CO2 for Rubisco in numerous independently evolved lineages of C4 plants, including Amaranthaceae, driving selection for less specific but faster enzymes which have both higher K M(CO2) and k cat values [3], [5], [23]. In the present study, we found that model A assuming positive selection on C4 branches provided a significantly better fit to the analysed Amaranthaceae dataset than the null model without selection (Table 1). We found no positive selection on branches which lead to C4 clades of Amaranthaceae, but we found positive selection specific for all C4 branches including branches which lead to C4 clades and branches within C4 clades (Table 1). This may be an argument in support of the hypothesis that C3 ancestors of C4 species, C3–C4 intermediates and C4 species at the dawn of their origin have Rubisco with C3 kinetics, but once C4 pump is fully functional it creates a strong selective pressure for acquiring Rubisco with C4 kinetics which then evolves during the stage of optimisation of C4 photosynthesis [58].
Parallel amino-acid replacements in Rubisco from phylogenetically distant lineages
Bayesian analyses of rbcL sequences in a phylogenetic framework allowed us to identify two residues under directional selection along C4 branches within Amaranthaceae (Table 2). There are no common trends in physicochemical properties of ‘C4’ amino acids with respect to properties such as residue hydrophobicity, solvent accessibility, or location within the tertiary structure of the enzyme (Table 2). Alanine at the position 281 was replaced by serine at least eleven times within the studied species with nine of replacements taking place within C4 clades and two replacements in C3 species Chenopodium bonus-henricus and Spinacia oleracea (Fig. 1). Methionine at the position 309 was replaced by isoleucine at least four times, all of which within C4 clades (Fig. 1). Only three C4 species, Atriplex spongiosa, A. rosea and Horaninovia ulicina, had both ‘C4’ amino acids simulteniously. Seven C4 clades of which one was monospecific had ‘C4’ amino acids, while nine C4 clades of which six consisted of only one species did not have ‘C4’ amino acids (Fig. 1). More frequent occurrence of ‘C4’ amino acids in clades consisting of many species compared to monospecific clades corresponds to our findings of stronger positive selection within C4 clades (Table 1).
Interestingly, both selected residues in C4 Amaranthaceae are among the eight residues selected in C4 Cyperaceae and Poaceae [26] and the ‘C4’ amino acid 309I is also among selected in C4 Flaveria [27]. None of the ‘C4’ amino acids is fixed among C4 species, but they are more frequent among C4 lineages, ranging from 17 to 35% in C4 Amaranthaceae, and from 14 to 87% in C4 Cyperaceae and Poaceae (Table 2; percentage for C4 Cyperaceae and Poaceae calculated using numbers from [26]). Although ‘C4’ amino acids are not fixed among all C4 species, there is a significant positive association between their presence and C4 photosynthetic type in Amaranthaceae. Given the existence of C4 species without ‘C4’ amino acids , it is likely that other as yet unidentified amino acids replacements may be involved in Rubisco adaptation. The model of sequence evolution used to identify Rubisco residues under positive selection within C4 lineages averages selective pressure among selected branches (C4 branches in our case) and hence allows detection only of the most typical substitutions, potentially missing ones that are unique for a particular branch. Other possible explanations are variation in Rubisco kinetic properties not only between C3 and C4 groups of species but also within these groups [3], [4], [5], [23] and putative differences in other proteins which form the Rubisco complex (small subunit, Rubisco activase). Although the large subunits contain active sites, changes in small subunits may make significant contribution to kinetic properties of plant and algal Rubiscos [59], including differences observed between C3 and C4 plants [60], and the rbcS genes encoding small subunits have been shown under positive selection in C4 Flaveria [27].
Identical amino-acids in Rubisco of C4 Amaranthaceae and C4 Cyperaceae and Poaceae, representing eudicots and monocots with significantly different anatomy and ecological preferences [22], constitute a remarkable example of parallel molecular evolution in phylogenetically distant groups. This example becomes even more interesting if C3 plants are considered as well. Various groups of C3 plants such as some aquatic species and C3 species from cold habitats have faster but less CO2-specific Rubisco compared with their C3 relatives from terrestrial and warm conditions, respectively [3], [23]. Hence, some groups of C3 plants can arrive at the same evolutionary solutions for Rubisco fine-tuning as C4 plants. Indeed, ‘C4’ amino acids shown for C4 Amaranthaceae in the present study and for C4 monocots and Flaveria previously [26], [27], have been reported to be under positive selection in various groups of C3 plants by Kapralov and Filatov [6]. Moreover, residue 309 is among the most frequently positively selected sites in land plants, and although residue 281 itself is not, its close neighbours, residues 279 and 282, are among the most often positively selected ones [6]. Thus, we can conclude that both ‘C4’ amino acids, 281S and 309I, evolved in parallel in various phylogenetically distant lineages of C3 and C4 plants in which faster but less specific Rubisco was needed.
The residue 309 is located on the interface of large subunits within a large subunit dimer, while the residue 281 is involved into dimer-dimer interactions (Table 2). Methionine at position 309 is replaced by the smaller and more hydrophobic isoleucine, which has a stabilising and favourable effect on overall molecule stability according to CUPSAT calculations using spinach pdb-structure [44], while A281S replacement decreases hydrophobicy and may be destabilising (Table 2).
Effects of A281S replacement on kinetics of land plants Rubisco has not been studied, while recent study by Whitney et al. [61] using mutagenic approach showed that M309I replacement in Flaveria changed Rubisco kinetics from “C3-like” to “C4-like” making the enzyme faster but less CO2-specific. Importance of M309I replacement for changes in kinetics of Flaveria Rubisco was predicted using in silico approach similar to one used in the present study [27] and confirmed in planta by the study of Whitney et al. [61] making it a good case in support of further application of phylogeny-based methods for detecting residues under positive selection in Rubisco and elsewhere.
Towards the periodic table of functional amino-acid replacements in Rubisco
Continuing population growth creating increasing demand for food, coupled with future climate change and its potentially dire consequences such as biome collapse and crop failure, both call for an improved understanding of mechanisms allowing plant species to adapt the photosynthetic process to a wide range of conditions. Hence, there is a necessity for more phylogeny-based studies of genes encoding Rubisco from various lineages of phototrophs established in different conditions to better understand Rubisco evolution at the molecular level. The integration of phylogenetic and biochemical research is required to study how Darwinian selection has created a range of enzymes with different kinetic and physical properties tailored to function in virtually all ecosystems on our planet. Knowledge of the role of specific residues in Rubisco adaptation to the particular conditions may provide clues for engineering better enzymes suited to contemporary agricultural needs as well as helping to understand what modifications in the enzyme may have been (and perhaps will be) driven by adaptation to different environmental conditions.
Supporting Information
List of studied species.
(XLSX)
Acknowledgments
We thank the Herbaria of the University of Oxford and the Curator, Dr Stephen Harris, for access to the collections, and Dr Tim Massingham (European Bioinformatics Institute) for help with the SLR program.
Funding Statement
This research was funded by NERC (http://www.nerc.ac.uk/; grant number NE/H007741/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jordan DB, Ogren WL (1981) Species variation in the specificity of ribulose biphosphate carboxylase/oxygenase. Nature 291: 513–515. [Google Scholar]
- 2. Jordan DB, Ogren WL (1983) Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase. Archives of Biochemistry and Biophysics 227: 425–433. [DOI] [PubMed] [Google Scholar]
- 3. Yeoh H-H, Badger MR, Watson L (1981) Variations in kinetic properties of ribulose-1,5-bisphosphate carboxylases among plants. Plant Physiology 67: 1151–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Galmés J, Flexas J, Keys AJ, Cifre J, Mitchell RAC, et al. (2005) Rubisco specificity factor tends to be larger in plant species from drier habitats and in species with persistent leaves. Plant, Cell and Environment 28: 571–579. [Google Scholar]
- 5. Kubien DS, Whitney SM, Moore PV, Jesson LK (2008) The biochemistry of Rubisco in Flaveria . Journal of Experimental Botany 59: 1767–1777. [DOI] [PubMed] [Google Scholar]
- 6. Kapralov MV, Filatov DA (2007) Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evolutionary Biology 7: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tcherkez GGB, Farquhar GD, Andrews TJ (2006) Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. PNAS 103: 7246–7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spreitzer RJ, Salvucci ME (2002) Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annual Review of Plant Biology 53: 449–475. [DOI] [PubMed] [Google Scholar]
- 9. Witzel F, Götze J, Ebenhöh O (2010) Slow deactivation of ribulose 1,5-bisphosphate carboxylase/oxygenase elucidated by mathematical models. FEBS Journal 277: 931–950. [DOI] [PubMed] [Google Scholar]
- 10. Ellis RJ (1979) The most abundant protein in the world. Trends in Biochemical Sciences 4: 241–244. [Google Scholar]
- 11. von Caemmerer S, Furbank R (2003) The C4 pathway: an efficient CO2 pump. Photosynthesis Research 77: 191–207. [DOI] [PubMed] [Google Scholar]
- 12.Long S (1999) Environmental responses. In: Sage RF, Monson RK, editors. C4 plant biology. San Diego: CA: Academic Press. 215–249.
- 13. Smith ME, Koteyeva NK, Voznesenskaya EV, Okita TW, Edwards GE (2009) Photosynthetic features of non-Kranz type C4 versus Kranz type C4 and C3 species in subfamily Suaedoideae (Chenopodiaceae). Functional Plant Biology 36: 770–782. [DOI] [PubMed] [Google Scholar]
- 14. Lloyd J, Farquhar GD (1994) 13C discrimination during CO2 assimilation by the terrestrial biosphere. Oecologia 99: 201–215. [DOI] [PubMed] [Google Scholar]
- 15. Still CJ, Berry JA, Collatz GJ, DeFries RS (2003) Global distribution of C3 and C4 vegetation: Carbon cycle implications. Global Biogeochemical Cycles 17: 1006. [Google Scholar]
- 16.Sage RF, Christin P-A, Edwards EJ (2011) The C4 plant lineages of planet Earth. Journal of Experimental Botany. [DOI] [PubMed]
- 17. The Angiosperm Phylogeny Group (1998) An ordinal classification for the families of flowering plants. Annals of the Missouri Botanical Garden 85: 531–553. [Google Scholar]
- 18. The Angiosperm Phylogeny Group (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society 161: 105–121. [Google Scholar]
- 19. Kadereit G, Borsch T, Weising K, Freitag H (2003) Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. International Journal of Plant Sciences 164: 959–986. [Google Scholar]
- 20. Voznesenskaya EV, Franceschi VR, Kiirats O, Freitag H, Edwards GE (2001) Kranz anatomy is not essential for terrestrial C4 plant photosynthesis. Nature 414: 543–546. [DOI] [PubMed] [Google Scholar]
- 21. Edwards GE, Franceschi VR, Voznesenskaya EV (2004) Single-cell C4 photosynthesis versus the dual-cell (Kranz) paradigm Annual Review of Plant Biology. 55: 173–196. [DOI] [PubMed] [Google Scholar]
- 22. Pyankov VI, Ziegler H, Akhani H, Deigele C, Lüttge U (2010) European plants with C4 photosynthesis: geographical and taxonomic distribution and relations to climate parameters. Botanical Journal of the Linnean Society 163: 283–304. [Google Scholar]
- 23. Sage RF (2002) Variation in the k cat of Rubisco in C3 and C4 plants and some implications for photosynthetic performance at high and low temperature. Journal of Experimental Botany 53: 609–620. [DOI] [PubMed] [Google Scholar]
- 24. Hudson G, Mahon J, Anderson P, Gibbs M, Badger M, et al. (1990) Comparisons of rbcL genes for the large subunit of ribulose- bisphosphate carboxylase from closely related C3 and C4 plant species. The Journal of Biological Chemistry 265: 808–814. [PubMed] [Google Scholar]
- 25.Yang Z (2006) Computational Molecular Evolution. Oxford: Oxford University Press. 376 p.
- 26. Christin PA, Salamin N, Muasya AM, Roalson EH, Russier F, et al. (2008) Evolutionary switch and genetic convergence on rbcL following the evolution of C4 photosynthesis. Molecular Biology and Evolution 25: 2361–2368. [DOI] [PubMed] [Google Scholar]
- 27. Kapralov MV, Kubien DS, Andersson I, Filatov DA (2011) Changes in Rubisco kinetics during the evolution of C4 photosynthesis in Flaveria (Asteraceae) are associated with positive selection on genes encoding the enzyme. Molecular Biology and Evolution 28: 1491–1503. [DOI] [PubMed] [Google Scholar]
- 28. Wen Z-B, Zhang M-L, Zhu G-L, Sanderson S (2010) Phylogeny of Salsoleae s.l. (Chenopodiaceae) based on DNA sequence data from ITS, psbB–psbH, and rbcL, with emphasis on taxa of northwestern China. Plant Systematics and Evolution 288: 25–42. [Google Scholar]
- 29. Kadereit G, Mavrodiev EV, Zacharias EH, Sukhorukov AP (2010) Molecular phylogeny of Atripliceae (Chenopodioideae, Chenopodiaceae): Implications for systematics, biogeography, flower and fruit evolution, and the origin of C4 photosynthesis. Am J Bot 97: 1664–1687. [DOI] [PubMed] [Google Scholar]
- 30. Kadereit G, Freitag H (2011) Molecular phylogeny of Camphorosmeae (Camphorosmoideae, Chenopodiaceae): Implications for biogeography, evolution of C4-photosynthesis and taxonomy. Taxon 60: 51–78. [Google Scholar]
- 31. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 32.Silvestro D, Michalak I (2011) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution: 1–3.
- 33. Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 34. Yang Z, Nielsen R (2002) Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Molecular Biology and Evolution 19: 908–917. [DOI] [PubMed] [Google Scholar]
- 35. Yang Z, dos Reis M (2011) Statistical properties of the branch-site test of positive selection. Molecular Biology and Evolution 28: 1217–1228. [DOI] [PubMed] [Google Scholar]
- 36. Yang Z, Swanson WJ (2002) Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Molecular Biology and Evolution 19: 49–57. [DOI] [PubMed] [Google Scholar]
- 37. Yang Z, Wong WS, Nielsen R (2005) Bayes empirical bayes inference of amino acid sites under positive selection. Molecular Biology and Evolution 22: 1107–1118. [DOI] [PubMed] [Google Scholar]
- 38. Massingham T, Goldman N (2005) Detecting amino acid sites under positive selection and purifying selection. Genetics 169: 1753–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Felsenstein J (1985) Phylogenies and the comparative method. The American Naturalist 125: 1–15. [DOI] [PubMed] [Google Scholar]
- 40. Pagel M (1994) Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proceedings of the Royal Society of London Series B: Biological Sciences 255: 37–45. [Google Scholar]
- 41.Maddison WP, Maddison DR (2010) Mesquite: a modular system for evolutionary analysis. Version 2.73.
- 42. Taylor TC, Fothergill MD, Andersson I (1996) A common structural basis for the inhibition of ribulose 1,5-bisphosphate carboxylase by 4-carboxyarabinitol 1,5-bisphosphate and xylulose 1,5-bisphosphate. Journal of Biological Chemistry 271: 32894–32899. [DOI] [PubMed] [Google Scholar]
- 43. Guex N, Peitsch M (1997) SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- 44. Parthiban V, Gromiha MM, Schomburg D (2006) CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Research 34: W239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Müller K, Borsch T (2005) Phylogenetics of Amaranthaceae based on matK/trnK sequence data: evidence from parsimony, likelihood, and Bayesian analyses. Annals of the Missouri Botanical Garden 92: 66–102. [Google Scholar]
- 46. Kapralov MV, Akhani H, Voznesenskaya EV, Edwards G, Franceschi V, et al. (2006) Phylogenetic relationships in the Salicornioideae/Suaedoideae/Salsoloideae s.l. (Chenopodiaceae) clade and a clarification of the phylogenetic position of Bienertia and Alexandra using multiple DNA sequence datasets. Systematic Botany 31: 571–585. [Google Scholar]
- 47. Akhani H, Edwards G, Roalson EH (2007) Diversification of the Old World Salsoleae s.l. (Chenopodiaceae): molecular phylogenetic analysis of nuclear and chloroplast data sets and a revised classification. International Journal of Plant Sciences 168: 931–956. [Google Scholar]
- 48. Kadereit G, Ackerly D, Pirie MD (2012) A broader model for C4 photosynthesis evolution in plants inferred from the goosefoot family (Chenopodiaceae s.s.). Proceedings of the Royal Society B: Biological Sciences 279: 3304–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Christin P-A, Freckleton RP, Osborne CP (2010) Can phylogenetics identify C4 origins and reversals? Trends in Ecology and Evolution 25: 403–409. [DOI] [PubMed] [Google Scholar]
- 50. Whitney SM, Houtz RL, Alonso H (2011) Advancing our understanding and capacity to engineer Nature’s CO2-sequestering enzyme, Rubisco. Plant Physiology 155: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kapralov MV, Filatov DA (2006) Molecular adaptation during adaptive radiation in the Hawaiian endemic genus Schiedea . PLoS One 1: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Iida S, Miyagi A, Aoki S, Ito M, Kadono Y, et al. (2009) Molecular adaptation of rbcL in the heterophyllous aquatic plant Potamogeton . PLoS One 4: e4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kato S, Misawa K, Takahashi F, Sakayama H, Sano S, et al. (2011) Aquatic plant speciation affected by diversifying selection of organelle DNA regions. Journal of Phycology 47: 999–1008. [DOI] [PubMed] [Google Scholar]
- 54. Miwa H, Odrzykoski IJ, Matsui A, Hasegawa M, Akiyama H, et al. (2009) Adaptive evolution of rbcL in Conocephalum (Hepaticae, bryophytes). Gene 441: 169–175. [DOI] [PubMed] [Google Scholar]
- 55. Sen L, Fares M, Liang B, Gao L, Wang B, et al. (2011) Molecular evolution of rbcL in three gymnosperm families: identifying adaptive and coevolutionary patterns. Biology Direct 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Young JN, Rickaby REM, Kapralov MV, Filatov DA (2012) Adaptive signals in algal Rubisco reveal a history of ancient atmospheric carbon dioxide. Philosophical Transactions of the Royal Society B: Biological Sciences 367: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang M, Kapralov M, Anisimova M (2011) Coevolution of amino acid residues in the key photosynthetic enzyme Rubisco. BMC Evolutionary Biology 11: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sage RF (2004) The evolution of C4 photosynthesis. New Phytologist 161: 341–370. [DOI] [PubMed] [Google Scholar]
- 59. Genkov T, Meyer M, Griffiths H, Spreitzer RJ (2010) Functional hybrid Rubisco enzymes with plant small subunits and algal large subunits. Journal of Biological Chemistry 285: 19833–19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ishikawa C, Hatanaka T, Misoo S, Miyake C, Fukayama H (2011) Functional incorporation of sorghum small subunit increases the catalytic turnover rate of Rubisco in transgenic rice. Plant Physiology 156: 1603–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Whitney SM, Sharwood RE, Orr D, White SJ, Alonso H, et al. (2011) Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) carboxylation rate in Flaveria . Proceedings of the National Academy of Sciences 108: 14688–14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Letunic I, Bork P (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Research 39: W475–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Knight S, Andersson I, Brändén C-I (1990) Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2·4 Å resolution: Subunit interactions and active site. Journal of Molecular Biology 215: 113–160. [DOI] [PubMed] [Google Scholar]
- 64.Nelson DL, Cox MM (2005) Lehninger principles of biochemistry. New York: WH Freeman and Company.
- 65. Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology 157: 105–132. [DOI] [PubMed] [Google Scholar]
- 66. Grantham R (1974) Amino acid difference formula to help explain protein evolution. Science 185: 862–864. [DOI] [PubMed] [Google Scholar]
- 67. Zamyatin AA (1972) Protein volume in solution. Progress in Biophysics and Molecular Biology 24: 107–123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of studied species.
(XLSX)