Abstract
Background
The UBE4B gene, located on chromosome 1p36, encodes a ubiquitin ligase that interacts with Hrs, a protein involved in EGFR trafficking, suggesting a link between EGFR trafficking and neuroblastoma pathogenesis. We have analyzed the roles of UBE4B in the outcomes of neuroblastoma patients and in neuroblastoma tumor cell proliferation, EGFR trafficking, and response to EGFR inhibition.
Methods
We examined the association of UBE4B expression with neuroblastoma patient survival using available microarray datasets. We measured UBE4B and EGFR protein levels in patient tumor samples and EGFR degradation rates in neuroblastoma cell lines and analyzed the effects of UBE4B on neuroblastoma tumor cell growth. The effects of the EGFR inhibitor cetuximab were examined in neuroblastoma cells expressing wild-type and mutant UBE4B.
Results
Low UBE4B gene expression is associated with poor outcomes in patients with neuroblastoma. UBE4B overexpression reduced neuroblastoma tumor cell proliferation, and UBE4B expression was inversely related to EGFR expression in patient tumor samples. EGFR degradation rates correlated with cellular UBE4B levels. Enhanced expression of catalytically active UBE4B resulted in reduced sensitivity to EGFR inhibition.
Conclusions
We have demonstrated associations between UBE4B expression and neuroblastoma patient outcomes and between UBE4B and EGFR expression in neuroblastoma tumor samples. Moreover, levels of UBE4B influenced neuroblastoma tumor cell proliferation, EGFR degradation, and response to EGFR inhibition. These results suggest UBE4B-mediated GFR trafficking may contribute to the poor prognosis of neuroblastoma tumors with 1p36 deletions, and that UBE4B expression may be a marker that can predict responses of neuroblastoma tumors to treatment.
Keywords: neuroblastoma, UBE4B, Hrs, EGFR, cetuximab
Introduction
Overall survival rates for children with high-risk neuroblastoma remain approximately 30% with current treatment regimens1,2. Cases of high-risk neuroblastoma are associated with frequent relapses and tumors that are resistant to treatment1,2, and children with refractory or recurrent neuroblastoma have poor responses to salvage therapy and very poor survival rates3,4. A better understanding of the mechanisms of neuroblastoma tumorigenesis will provide improved treatment options for these children.
Growth factors and their receptors play a critical role in the survival, growth, and differentiation of normal and malignant cells, and aberrant GFR expression and function are common features of neuroblastoma tumors. Ubiquitin-mediated sorting of activated growth factor receptors (GFRs) for lysosomal degradation can regulate the amplitude and kinetics of GFR signaling5,6, and defects in the pathways for intracellular GFR sorting and degradation leading to altered GFR expression and activity have been identified in many cancer types7. Appropriate trafficking of GFRs is critical for control of unrestrained signaling that is characteristic of transformed cells5,6,8,9.
Deletion of the chromosome 1p36 region is found in approximately one-third of neuroblastoma tumors and is associated with high-risk tumor features and a poor prognosis10-12. The UBE4B gene is located in the 1p36 region and encodes an E3/E4 ubiquitin ligase13,14. Recently, Martinsson and colleagues identified a mutation in the UBE4B gene in the tumor of a patient with neuroblastoma with a fatal outcome15. The expression of UBE4B was shown to be markedly diminished in a cohort of high-stage/poor-outcome tumors compared to low-stage/favorable-outcome tumors15,16, and UBE4B was therefore suggested to be a candidate tumor suppressor gene15.
We have observed that UBE4B interacts with hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs), a key regulator of the endosomal machinery for GFR trafficking, and that the UBE4B-Hrs interaction is critical for appropriate GFR trafficking and degradation14. Therefore, loss of UBE4B expression and function may be associated with aberrant GFR expression in neuroblastoma tumors. However, the roles of UBE4B in GFR trafficking in neuroblastoma tumor cells and of UBE4B-mediated GFR trafficking in the outcomes of neuroblastoma patients are unknown.
We hypothesized that UBE4B would be associated with neuroblastoma patient outcomes and neuroblastoma tumorigenesis. To explore the roles of UBE4B expression and function in the pathogenesis of neuroblastoma, we evaluated the association of UBE4B gene expression with neuroblastoma patient outcomes, and we investigated the roles of UBE4B in neuroblastoma tumor cell growth, in the regulation of EGFR expression, and in the responses of neuroblastoma tumor cells to EGFR inhibition. The results of these studies suggest UBE4B-mediated GFR trafficking may contribute to the poor prognosis of neuroblastoma tumors with 1p36 deletions and that UBE4B expression may be a marker that can predict responses of neuroblastoma tumors to treatment.
Methods
Cell culture
The characteristics of neuroblastoma cell lines SMS-KCNR, LA1-55N, NGP, SH-EP, SK-N-AS, SK-N-SH, and SH-SY5Y used in this study have been previously described17-20 and were generously provided by Susan Cohn (The University of Chicago Children’s Hospital, Chicago, IL) and John Maris (Children’s Hospital of Philadelphia, Philadelphia, PA). Cell lines used in these studies were authenticated by DNA profiling. Neuroblastoma cell lines were grown at 37°C in 5% CO2 in RPMI-1640 (Invitrogen, Carlsbad, CA) supplemented with 10% heat inactivated fetal bovine serum (USB, Minneapolis, MN), L-glutamine, sodium pyruvate, non-essential amino acids, and penicillin/streptomycin (Sigma Chemical Company, St. Louis, MO).
Neuroblastoma Patient Tumor Samples and Data
The patient tumor samples employed in these studies were provided by the Children’s Oncology Group Neuroblastoma Biology Committee and the Biopathology Center in Columbus, OH, as previously described21.
We obtained microarray analysis results of neuroblastoma patient tumor samples from the National Cancer Institute (NCI) Oncogenomics Data Center Section (http://pob.abcc.ncicrf.gov/cgi-bin/JK) from the databases “;Neuroblastoma Prognosis Database,” “Neuroblastoma Prognosis Database - Oberthuer Lab,” and “Exon Array Neuroblastoma Database.” These databases include patients with all tumor stages and included information regarding N-myc gene amplification status, and all patient data from these databases was included in our analysis. Additional studies were performed as detailed below using data from the “Neuroblastoma Prognosis Database - Seeger’s Lab” dataset.
Cell proliferation assay
SK-N-AS neuroblastoma tumor cells were infected with lentivirus constructs expressing GFP alone, wild-type UBE4B, or UBE4BP1140A, a mutant isoform with absent ubiquitin ligase activity13, as previously described22. 4,000 SK-N-AS neuroblastoma cells were plated in 96-well plates in 100 μL of culture media with serum or serum-free media supplemented with 50 ng/mL EGF. At baseline and after 24, 48, and 72 hours of incubation at 37°C, 10 μL of WST-1 reagent (Roche, Indianapolis, IN) was added to each well in each plate and absorbance at 450 nm was determined.
To evaluate proliferation in response to cetuximab, SK-N-AS cells were plated as above. After assessing baseline proliferation on day 1, existing media was discarded for all other plates, and 100μL of media supplemented with cetuximab (400nM, 1μM, or 4μM; provided by the M.D. Anderson Cancer Center pharmacy) was added to each well. Proliferation was assessed at 24, 48, and 72 hours as above and calculated at each time point as the percent change in absorbance compared to baseline absorbance. ANOVA analysis and post hoc Tukey tests were performed to establish significance.
EGFR degradation assay
For each experiment, cells at 80% confluency were incubated for 2 hours in DMEM supplemented with 1% BSA (Medium A). Media was then aspirated and cold medium A with EGF (50 ng/mL) was added. Plates were maintained at 4°C for one hour. Cells were washed three times with cold medium A and either kept on ice (0 min) or incubated with warm medium A for either 30 or 60 minutes. Following incubations, cells were rinsed with cold PBS and scraped into 1.5 mL PBS. Samples were centrifuged at 2,500g and cell pellets were resuspended in 30μL of lysis buffer (1μL each of 10mM leupeptin, 1μg/μL pepstatin, 0.3mM aprotinin, and 1.74μg/μL PMSF per 100 μL of M-PER (Pierce, Rockford, IL)). Samples were incubated for 1 hour at 4°C and then centrifuged at 15,000g.
Supernatant was collected and 50 μg of protein from each sample was separated by SDS-PAGE and transferred to nitrocellulose membranes as previously described21,23. Membranes were incubated in blocking buffer (5% nonfat dry milk in PBS) and then with a rabbit polyclonal anti-EGFR antibody (ABR, Golden, CO; 1:1000 dilution). Proteins were visualized with ECL and bands were quantified using ImageJ (NIH, ver 1.42). ANOVA analysis and post hoc Tukey tests were performed to establish significance.
Expression of UBE4B and EGFR in neuroblastoma tumor samples
Patient samples were homogenized using a Mikrodismembrator S (Sartorius, San Diego, CA), then incubated for 10 minutes in protein lysis buffer containing a phosphatase inhibitor (Sigma) and protease inhibitor cocktail (Roche). Lysates were centrifuged at 15,000g and supernatants collected. 30μg of protein was separated by SDS-PAGE and transferred to nitrocellulose membranes as above.
Membranes were blocked in 5% milk in TBST (100mM tris, 1.5M NaCl, 0.1% Tween-20, pH 7.9) and probed with antibodies directed against UBE4B, EGFR, and actin. Proteins were visualized and quantified as above. Spearman’s correlation analysis was performed on levels of UBE4B and EGFR.
Statistical analysis
Using gene expression results from the databases described above, patients were divided into high and low UBE4B gene expression groups by median-centered log2 ratios as detailed in the NCI Oncogenomics database website. The high-expression group contained 177 patients and the low-expression group contained 176 patients. Kaplan-Meier survival curves were plotted using the open-source statistical packages in R (http://www.r-project.org). We compared the survival curves by UBE4B gene expression groups using the log-rank test to examine the association between expression and patient survival outcomes in the whole cohort and in patients with stage 4 neuroblastoma and in those with stage 1, 2, 3, or 4S neuroblastoma. The Fisher-exact test was performed to determine whether the proportion of tumors with N-myc gene amplification was associated with UBE4B gene expression. For separate analyses of the Neuroblastoma Prognosis Database - Seeger’s Lab dataset, the median-centered log2 ratios for each of seven probes for UBE4B were standardized to have a mean of zero and unit length by location and scale transformation. The standardized log2 ratios were averaged across all seven probes. Based on the average of log2 ratios for each patient, 50 and 52 patients were in low and high UBE4B expression groups, respectively.
Results
Association of UBE4B Gene Expression with Neuroblastoma Patient Outcomes
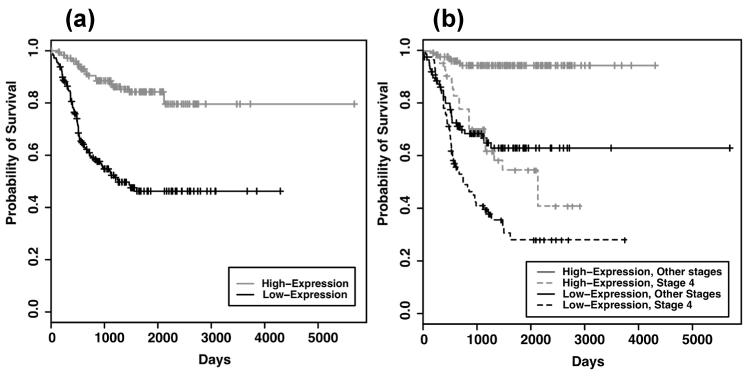
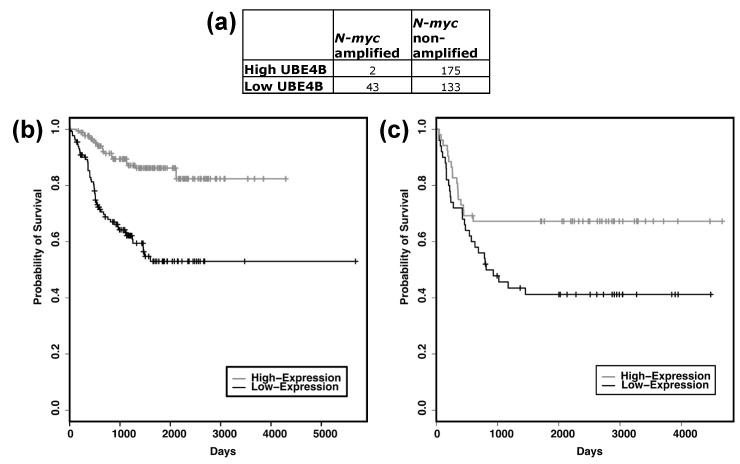
Due to the location of the UBE4B gene and its low expression in advanced neuroblastoma tumors16, loss of UBE4B expression and function may be critical for neuroblastoma tumorigenesis. Therefore, we evaluated the association of UBE4B gene expression with neuroblastoma patient outcomes, using results from microarray analyses of 353 neuroblastoma tumors obtained from the NCI as detailed above. Low expression of UBE4B was significantly associated with poor neuroblastoma patient outcomes (p<0.00001, Fig. 1A) and with worse outcomes in patients with either stage 4 disease (p=0.007) or with stage 1, 2, 3, and 4S disease (p<0.00001, Fig. 1B). Additionally, amplification of the N-myc oncogene was only seen in 2 of 177 patients with high UBE4B gene expression, as opposed to 43 of 176 with low UBE4B expression (p<0.00001, Fig. 2A), indicating an association of N-myc gene amplification with low UBE4B expression. These results demonstrate that UBE4B expression is associated with the outcomes of neuroblastoma patients.
Figure 1. Outcomes of neuroblastoma patients based on UBE4B expression.
The NCI Oncogenomics gene expression databases were evaluated for outcomes of neuroblastoma patients and Kaplan-Meier survival curves were generated. (A) Estimated overall survival for neuroblastoma patients with tumors having high UBE4B gene expression (grey, n=177) and low UBE4B gene expression (black, n=176) (log-rank test, p=3.02e-13) (B) Estimated overall survival for neuroblastoma patients with stage 4 tumors with high (grey, dashed; n=40) and low (black, dashed; n=87) UBE4B gene expression (log-rank test, p=0.007) and for patients with tumors of all other stages with high (grey, solid; n=137) and low (black, solid; n=89) UBE4B gene expression (log-rank test, p=7.59e−09).
Figure 2. Outcomes of neuroblastoma patients with non-amplified N-myc based on UBE4B expression.
The NCI Oncogenomics gene expression databases were evaluated for outcomes of neuroblastoma patients and Kaplan-Meier survival curves were generated. (A) Numbers of patients with high or low UBE4B gene expression compared to presence or absence of N-myc gene amplification (Fisher-exact test, p=2.87e-14). (B) Estimated overall survival for neuroblastoma patients with N-myc non-amplified tumors having high UBE4B gene expression (grey, n=175) and low UBE4B gene expression (black, n=133) (log-rank test, p=9.0e-9). (C) Estimated overall survival for neuroblastoma patients from the Neuroblastoma Prognosis Database-Seeger Lab dataset with tumors having high UBE4B gene expression (grey, n=52) and low UBE4B gene expression (black, n=50) (log-rank test, p=0.021).
In order to evaluate the prognostic relevance of UBE4B expression in patients without N-myc gene amplification, we separately analyzed the outcomes of the 308 patients without N-myc gene amplification in these databases. Low UBE4B expression was significantly associated with lower survival rates in these neuroblastoma patients (p<0.00001, Fig. 2B), suggesting that UBE4B expression is prognostically relevant in this subset of patients. For further confirmation of the prognostic value of UBE4B gene expression in neuroblastoma patients without N-myc amplification, we independently evaluated the association of UBE4B expression with patient survival in the Neuroblastoma Prognosis Database - Seeger Lab, a database with results from patients with neuroblastoma without N-myc gene amplification24. In these 102 patients, low UBE4B expression was also significantly associated with lower survival rates (p<0.05, Fig. 2C), confirming the association of UBE4B gene expression with overall survival in neuroblastoma patients without N-myc amplification.
Effects of UBE4B on Neuroblastoma Cell Proliferation
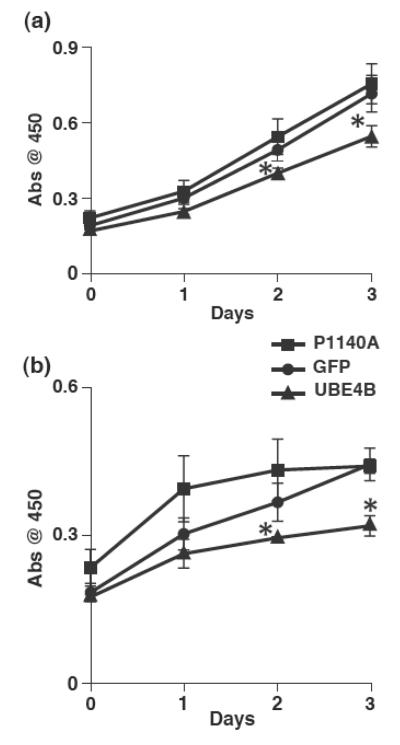
We then explored the mechanisms by which UBE4B might affect neuroblastoma patient outcomes. To investigate the effects of UBE4B on neuroblastoma tumor cell proliferation, SK-N-AS neuroblastoma tumor cells infected with lentivirus constructs expressing GFP alone, wild-type UBE4B, or UBE4BP1140A, a mutant isoform with absent ubiquitin ligase activity13, were grown in DMEM supplemented with either 10% fetal bovine serum or 50ng/mL EGF. Wild-type UBE4B expression, but not expression of the UBE4BP1140A mutant, resulted in reduced neuroblastoma tumor cell proliferation under both conditions (p<0.05, Fig. 3), suggesting that UBE4B expression and, more specifically, UBE4B ubiquitin ligase function, can reduce neuroblastoma tumor cell proliferation.
Figure 3. UBE4B effects on growth of neuroblastoma cells.
SK-N-AS neuroblastoma cells expressing either GFP (circles), wild type UBE4B (triangles), or UBE4BP1140A (squares) were grown in media containing 10% FBS (A) or 50ng/ml EGF (B). Cell growth was assessed using the WST-1 based spectrophotometric assay. *p<0.05.
Association of UBE4B and EGFR Protein Levels
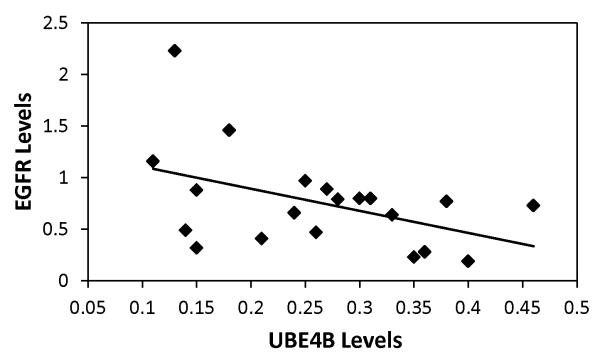
Aberrant GFR expression and function have been shown to be important in the pathogenesis of neuroblastoma25-30. We have observed that the UBE4B protein interacts with Hrs, a key regulator of EGFR endosomal trafficking and degradation31-34, and that the Hrs-UBE4B interaction is critical for efficient GFR trafficking and degradation14, suggesting that UBE4B expression and function may be associated with EGFR expression in neuroblastoma tumor cells. Therefore, we evaluated a cohort of 20 neuroblastoma tumor samples from the Children’s Oncology Group (COG) for expression of UBE4B and EGFR proteins by quantitative Western blotting. UBE4B protein expression was inversely correlated with EGFR protein expression in these patient samples (Spearman’s correlation Rho = −0.4545; p<0.05, Fig. 4), suggesting a possible role for UBE4B in regulation of EGFR levels in neuroblastoma tumors.
Figure 4. UBE4B and EGFR expression levels in tumors.
Human neuroblastoma tumor samples were obtained from the Children’s Oncology Group (COG). Tumor sample lysates were separated on SDS-PAGE gels. Immunoblots for UBE4B and EGFR were performed and relative protein levels were determined. A Spearman’s correlation was performed with UBE4B and EGFR protein levels as shown (Rho = −0.4545; p<0.05).
UBE4B-mediated EGFR Degradation in Neuroblastoma Cells
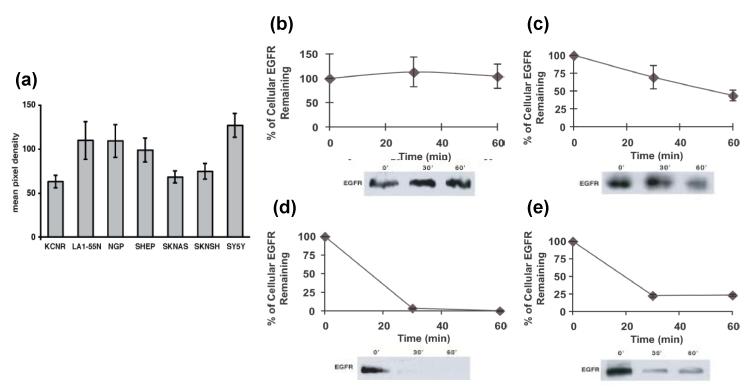
In order to explore the mechanisms by which UBE4B regulates EGFR expression in neuroblastoma tumor cells, we investigated the expression levels of UBE4B and the degradation rates of EGFR in neuroblastoma cell lines. Total UBE4B protein levels were measured in a panel of neuroblastoma tumor cells at baseline by quantitative Western blotting (Fig. 5A). In order to measure EGFR degradation rates in neuroblastoma tumor cells, cells with high (LA1-55N, SH-SY5Y) or low (SK-N-SH, SK-N-AS) UBE4B levels were briefly starved in serum-free media and then stimulated with EGF. Total cellular EGFR levels were then measured over the subsequent 60 minutes. Neuroblastoma tumor cells with higher levels of UBE4B had faster rates of EGFR degradation compared to those with lower UBE4B levels (Fig. 5B-E), suggesting that UBE4B affects EGFR levels through regulation of EGFR degradation.
Figure 5. UBE4B expression and EGFR degradation rates in neuroblastoma cell lines.
A. UBE4B protein levels were determined in neuroblastoma cell lines by immunoblot analysis of cell lysates. SK-N-SH (B), SK-N-AS (C), SH-SY5Y (D), and LA1-55N (E) neuroblastoma tumor cells were then starved and stimulated with EGF for 0, 30, or 60 minutes. Cell lysates were examined by immunoblotting for EGFR levels and levels were plotted over time.
Association of UBE4B with Neuroblastoma Tumor Cell Response to EGFR Inhibition
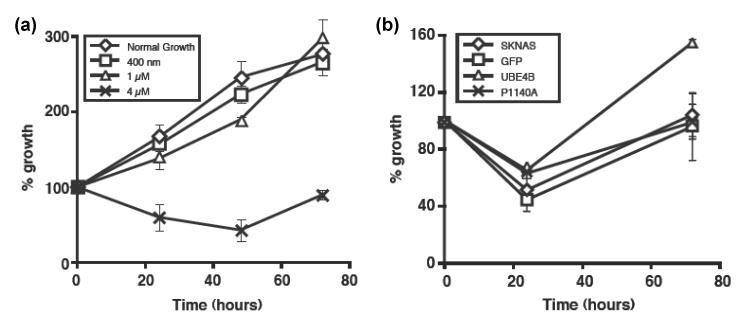
The association of UBE4B levels with EGFR degradation and with total EGFR levels suggested that UBE4B levels may also be associated with the response of neuroblastoma tumor cells to EGFR inhibitors. To investigate this possibility, neuroblastoma tumor cells were treated with a range of concentrations of cetuximab, an FDA-approved inhibitory antibody to EGFR. Treatment with 4μM cetuximab resulted in significant inhibition of neuroblastoma tumor cell proliferation (Fig. 6A, p<0.01). To investigate the role of UBE4B in the cetuximab-mediated inhibition of cell proliferation, parental neuroblastoma tumor cells or cells infected with lentivirus vectors expressing wild-type UBE4B, the UBE4BP1140A mutant, or GFP were grown as above and treated with 4μM cetuximab. Overexpression of UBE4B, but not the ligase-deficient mutant, resulted in loss of the cetuximab-mediated inhibition of proliferation (Fig. 6B), suggesting a role for UBE4B expression and its ubiquitin ligase activity in the responses of neuroblastoma tumor cells to EGFR inhibition.
Figure 6. UBE4B effects on growth of neuroblastoma cells and response to EGFR inhibitors.
A. SK-N-AS neuroblastoma tumor cells were treated with escalating concentrations of cetuximab. Cell growth was assessed at the indicated time points using the WST-1 based spectrophotometric assay. B. Parental SK-N-AS cells and SK-N-AS cells expressing either GFP (control), UBE4B, or UBE4BP1140A were grown in the presence of 4μM cetuximab (Erbitux) with growth assessed as above.
Discussion
Children with high-risk neuroblastoma have extremely poor outcomes, and additional understanding of the pathways involved in neuroblastoma pathogenesis will assist in the development of improved therapies in the future. The UBE4B gene is located in chromosome 1p36, a commonly deleted chromosomal region in neuroblastoma tumors, and UBE4B is involved in the trafficking of GFRs that ultimately results in their lysosomal degradation, suggesting a role for UBE4B-mediated GFR trafficking in neuroblastoma pathogenesis. Our results demonstrate an association of UBE4B gene expression with neuroblastoma patient outcomes and of UBE4B function with neuroblastoma tumor cell proliferation and response to EGFR inhibitors.
A large number of genes located within the 1p36 region have been proposed as tumor suppressors in neuroblastoma and other tumors, including CHD535, CAMTA136,37, miRNA-34a38,39, CASZ140, KIF1B41,42, and the HES gene family43. Studies are ongoing to determine the role of UBE4B expression and function in other tumors with 1p36 aberrations. Alterations of the chromosome 1p36 region are found in a wide range of tumors, including brain tumors, breast cancers, and leukemias, among others, and are often associated with aggressive tumor features and a poor prognosis44. In medulloblastoma tumor cells, UBE4B has been shown to induce degradation of p5345, suggesting a potential role for UBE4B in tumorigenesis independent of its effect on EGFR trafficking, and studies are ongoing to investigate the role of UBE4B in the trafficking of other transmembrane proteins in neuroblastoma.
EGF-induced signaling has been shown to be important in the pathogenesis of a variety of malignancies, and EGFR inhibitors are currently in use for treatment of a variety of malignancies46. EGFR expression has been documented in neuroblastoma tumors and cell lines, and inhibition of EGFR signaling has been shown to reduce growth of neuroblastoma tumor cells in vitro and in vivo21,30,47. Cetuximab has been shown to be effective against a variety of tumor cell lines, with IC50 values between 10nM and 10μM48. Therefore, our identified effective dose of 4μM fits within the observed cancer cell sensitivity range.
Previous studies have demonstrated that EGFR expression is associated with chemotherapy resistance in neuroblastoma cells and that chemotherapy is synergistic with EGFR inhibitors47,49. Additional studies have identified EGFR ubiquitination and degradation as the mechanism for colon cancer cell resistance to cetuximab50. The etiology of this EGFR-associated resistance and the mechanisms responsible for this association of EGFR with resistance to chemotherapy in neuroblastoma tumor cells are unknown. We have demonstrated that overexpression of UBE4B resulted in reduced sensitivity to cetuximab (Fig. 6), implicating UBE4B and the GFR trafficking pathway in the sensitivity of neuroblastoma tumor cells to treatment with EGFR inhibitors. These data also suggest that UBE4B-mediated GFR trafficking, through regulation of EGFR expression, may be responsible for the association of EGFR expression with treatment resistance. However, the mechanisms by which alterations in UBE4B expression and GFR trafficking might affect neuroblastoma tumor cell responses to therapy are unknown, and a better understanding of these mechanisms will likely lead to improved efficacy of current therapy and allow for identification of patient populations that might be more or less sensitive to GFR inhibitor treatment.
The results of these studies provide important information about the previously undescribed roles and mechanisms of GFR trafficking in neuroblastoma tumor cells and of the UBE4B protein in neuroblastoma tumor proliferation and response to treatment. Our results have identified a previously undiscovered link between GFR trafficking and neuroblastoma pathogenesis, and the correlation between UBE4B and EGFR levels we describe in neuroblastoma tumors may be relevant for multiple tumor types with known 1p36 alterations where EGFR inhibition is therapeutically effective. We have demonstrated associations between UBE4B expression and neuroblastoma patient outcomes, and we have shown that levels of UBE4B influence neuroblastoma tumor cell proliferation, EGFR trafficking, and response to EGFR inhibition. Our results suggest that inhibition of UBE4B-mediated GFR degradation may contribute to the poor prognosis of neuroblastoma tumors with 1p36 deletions, and that UBE4B expression may be a marker that can predict responses of neuroblastoma tumors to treatment. The roles of UBE4B levels as a prognostic marker for treatment response and of UBE4B-mediated EGFR degradation as a mechanism for resistance to EGFR inhibition are currently being explored. These results demonstrate a novel pathway with the potential for targeted drug development and a novel mechanism for tumor treatment sensitivity that may provide a prognostic marker to identify a patient population most likely to respond to inhibition of GFRs.
Acknowledgments
Grant support - NIH R01 MH58920 to Andrew Bean The authors report no conflicts of interest
References
- 1.Matthay KK, Reynolds CP, Seeger RC, et al. Long-Term Results for Children with High-Risk Neuroblastoma Treated on a Randomized Trial of Myeloablative Therapy Followed by 13-cis-Retinoic Acid: A Children’s Oncology Group Study. J Clin Oncol. 2009;27:1007–13. doi: 10.1200/JCO.2007.13.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zage PE, Kletzel M, Murray K, et al. Outcomes of the POG 9340/9341/9342 Trial for Children with High-Risk Neuroblastoma: a Report from the Children’s Oncology Group. Pediatr Blood Cancer. 2008;51:747–53. doi: 10.1002/pbc.21713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau L, Tai D, Weitzman S, Grant R, Baruchel S, Malkin D. Factors influencing survival in children with recurrent neuroblastoma. J Pediatr Hematol Oncol. 2004;26:227–32. doi: 10.1097/00043426-200404000-00003. [DOI] [PubMed] [Google Scholar]
- 4.London WB, Castel V, Monclair T, et al. Clinical and Biologic Features Predictive of Survival After Relapse of Neuroblastoma: A Report From the International Neuroblastoma Risk Group Project. J Clin Oncol. 2011;29:3286–92. doi: 10.1200/JCO.2010.34.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlessinger J. Cell Signaling by Receptor Tyrosine Kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 6.Burke P, Schooler K, Wiley HS. Regulation of Epidermal Growth Factor Receptor Signaling by Endocytosis and Intracellular Trafficking. Mol Biol Cell. 2001;12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosesson Y, Mills GB, Yarden Y. Derailed Endocytosis: An Emerging Feature of Cancer. Nat Rev Cancer. 2008;8:835–50. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 8.El-Rayes BF, LoRusso PM. Targeting the Epidermal Growth Factor Receptor. Br J Cancer. 2004;91:418–424. doi: 10.1038/sj.bjc.6601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marmor MD, Yarden Y. Role of Protein Ubiquitylation in Regulating Endocytosis of Receptor Tyrosine Kinases. Oncogene. 2004;23:2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- 10.Caron H, van Sluis P, de Kraker J, et al. Allelic Loss of Chromosome 1p as a Predictor of Unfavorable Outcome in Patients with Neuroblastoma. N Engl J Med. 1996;334:225–30. doi: 10.1056/NEJM199601253340404. [DOI] [PubMed] [Google Scholar]
- 11.Attiyeh AF, London WB, Mossé YP, et al. Chromosome 1p and 11q Deletions and Outcome in Neuroblastoma. N Engl J Med. 2005;353:2243–53. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 12.Maris JM, Guo C, Blake D, White PS, Hogarty MD, et al. Comprehensive analysis of chromosome 1p deletions in neuroblastoma. Med Pediatr Oncol. 2001;36:32–6. doi: 10.1002/1096-911X(20010101)36:1<32::AID-MPO1009>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 13.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U Box Proteins as a New Family of Ubiquitin-Protein Ligases. J Biol Chem. 2001;276:33111–20. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 14.Zage PE, Bean AJ. Growth Factor Receptor Trafficking as a Potential Therapeutic Target in Pediatric Cancer. Front Biol. 2012;7:1–13. [Google Scholar]
- 15.Krona C, Ejeskar K, Abel F, et al. Screening for Gene Mutations in a 500kb Neuroblastoma Tumor Suppressor Candidate Region in Chromosome 1p; Mutation and Stage-Specific Expression in UBE4B/UFD2. Oncogene. 2003;22:2343–51. doi: 10.1038/sj.onc.1206324. [DOI] [PubMed] [Google Scholar]
- 16.Caren H, Ejeskar K, Fransson S, Hesson L, Latif F, Sjoberg R-M, Krona C, Martinsson T. A Cluster of Genes Located in 1p36 are Down-Regulated in Neuroblastomas with Poor Prognosis, But Not Due to CpG Island Methylation. Mol Cancer. 2005;4:10. doi: 10.1186/1476-4598-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biedler JL, Helson L, Spengler BA. Morphology and Growth, Tumorigenicity, and Cytogenetics of Human Neuroblastoma Cells in Continuous Culture. Cancer Res. 1973;33:2643–2652. [PubMed] [Google Scholar]
- 18.Brodeur GM, Green AA, Hayes FA, Williams KJ, Williams DL, Tsiatis AA. Cytogenetic Features of Human Neuroblastomas and Cell Lines. Cancer Res. 1981;41:4678–4686. [PubMed] [Google Scholar]
- 19.Reynolds CP, Tomayko MM, Donner L, et al. Biological Classification of Cell Lines Derived From Human Extra-Cranial Neural Tumors. Prog Clin Biol Res. 1988;2:291–306. [PubMed] [Google Scholar]
- 20.Foley J, Cohn SL, Salwen HR, et al. Differential Expression of N-myc in Phenotypically Distinct Subclones of Human Neuroblastoma Cell Lines. Cancer Res. 1991;51:6338–6345. [PubMed] [Google Scholar]
- 21.Richards KN, Zweidler-McKay PA, Van Roy N, et al. Signaling of ERBB Receptor Tyrosine Kinases Promotes Neuroblastoma Growth In Vitro and In Vivo. Cancer. 2010;116:3233–43. doi: 10.1002/cncr.25073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutner RH, Zhang X-Y, Reiser J. Production, Concentration and Titration of Pseudotyped HIV-1-Based Lentiviral Vectors. Nat Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli UK. Cleavage of Structural Proteins During the Assembly of the Head of Bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Asgharzadeh S, Pique-Regi R, Sposto R, et al. Prognostic Significance of Gene Expression Profiles of Metastatic Neuroblastomas Lacking MYCN Gene Amplification. J Natl Cancer Inst. 2006;98:1193–203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- 25.Brodeur GM, Minturn JE, Ho R, et al. Trk Receptor Expression and Inhibition in Neuroblastoma. Clin Cancer Res. 2009;15:3244–50. doi: 10.1158/1078-0432.CCR-08-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsui T, Sano K, Tsukamoto T, et al. Human Neuroblastoma Cells Express Alpha and Beta Platelet-Derived Growth Factor Receptors Coupling with Neurotrophic and Chemotactic Signaling. J Clin Invest. 1993;92:1153–60. doi: 10.1172/JCI116684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimada A, Hirato J, Kuroiwa M, et al. Expression of KIT and PDGFR is Associated with a Good Prognosis in Neuroblastoma. Pediatr Blood Cancer. 2008;50:213–7. doi: 10.1002/pbc.21288. [DOI] [PubMed] [Google Scholar]
- 28.Meister B, Grunebach F, Bautz F, et al. Expression of Vascular Endothelial Growth Factor (VEGF) and its Receptors in Human Neuroblastoma. Eur J Cancer. 1999;35:445–9. doi: 10.1016/s0959-8049(98)00387-6. [DOI] [PubMed] [Google Scholar]
- 29.Hecht M, Papoutsi M, Tran HD, Wilting J, Schweigerer L. Hepatocyte Growth Factor/c-Met Signaling Promotes the Progression of Experimental Human Neuroblastomas. Cancer Res. 2004;64:6109–18. doi: 10.1158/0008-5472.CAN-04-1014. [DOI] [PubMed] [Google Scholar]
- 30.Ho R, Minturn JE, Hishiki T, et al. Proliferation of Human Neuroblastomas Mediated by the Epidermal Growth Factor Receptor. Cancer Res. 2005;65:9868–75. doi: 10.1158/0008-5472.CAN-04-2426. [DOI] [PubMed] [Google Scholar]
- 31.Sun W, Yan Q, Vida TA, Bean AJ. Hrs Regulates Early Endosome Fusion by Inhibiting Formation of an Endosomal SNARE Complex. J Cell Biol. 2003;162:125–137. doi: 10.1083/jcb.200302083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan Q, Sun W, Kujala P, Lotfi Y, Vida TA, Bean AJ. CART: An Hrs/Actinin-4/BERP/Myosin V Protein Complex Required for Efficient Receptor Recycling. Mol Biol Cell. 2005;16:2470–82. doi: 10.1091/mbc.E04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katzmann DJ, Babst M, Emr SD. Ubiquitin-Dependent Sorting Into the Multivesicular Body Pathway Requires the Function of a Conserved Endosomal Protein Sorting Complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 34.Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p Hse1p Complex Binds Ubiquitin and Mediates Endosomal Protein Sorting. Nat Cell Biol. 2002;4:534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- 35.Fujita T, Igarashi J, Okawa ER, et al. CHD5, A Tumor Suppressor Gene Deleted From 1p36.31 in Neuroblastomas. J Natl Cancer Inst. 2008;100:940–9. doi: 10.1093/jnci/djn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henrich KO, Bauer T, Schulte J, et al. CAMTA1, a 1p36 Tumor Suppressor Candidate, Inhibits Growth and Activates Differentiation Programs in Neuroblastoma Cells. Cancer Res. 2011;71:3142–51. doi: 10.1158/0008-5472.CAN-10-3014. [DOI] [PubMed] [Google Scholar]
- 37.Henrich KO, Fischer M, Mertens D, et al. Reduced Expression of CAMTA1 Correlates with Adverse Outcome in Neuroblastoma Patients. Clin Cancer Res. 2006;12:131–8. doi: 10.1158/1078-0432.CCR-05-1431. [DOI] [PubMed] [Google Scholar]
- 38.Tivnan A, Tracey L, Buckley PG, Alcock LC, Davidoff AM, Stallings RL. MicroRNA-34a is a Potent Tumor Suppressor Molecule In Vivo in Neuroblastoma. BMC Cancer. 2011;11:33. doi: 10.1186/1471-2407-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole KA, Attiyeh EF, Mosse YP, et al. A Functional Screen Identifies miR-34a as a Candidate Neuroblastoma Tumor Suppressor Gene. Mol Cancer Res. 2008;6:735–42. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, Yang X, Li Z, et al. CASZ1, a Candidate Tumor Suppressor Gene, Suppresses Neuroblastoma Tumor Growth Through Reprogramming Gene Expression. Cell Death Differ. 2011;18:1174–83. doi: 10.1038/cdd.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munirajan AK, Ando K, Mukai A, et al. KIF1Bbeta Functions as a Haploinsufficient Tumor Suppressor Gene Mapped to Chromosome 1p36. 2 by Inducing Apoptotic Cell Death. J Biol Chem. 2008;283:24426–34. doi: 10.1074/jbc.M802316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlisio S, Kenchappa RS, Vredeveld LC, et al. The Kinesin KIF1Bbeta Acts Downstream from EglN3 to Induce Apoptosis and Is a Potential 1p36 Tumor Suppressor. Genes Dev. 2008;22:884–93. doi: 10.1101/gad.1648608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zage PE, Nolo R, Fang W, Stewart J, Garcia-Manero G, Zweidler-McKay PA. Notch Pathway Activation Induces Neuroblastoma Tumor Cell Growth Arrest. Pediatr Blood Cancer. 2012;58:682–9. doi: 10.1002/pbc.23202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bagchi A, Mills AA. The Quest for the 1p36 Tumor Suppressor. Cancer Res. 2008;68:2551–6. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu H, Pomeroy SL, Ferreira M, et al. UBE4B Promotes Hdm2-Mediated Degradation of the Tumor Suppressor p53. Nat Med. 2011;17:347–55. doi: 10.1038/nm.2283. [DOI] [PubMed] [Google Scholar]
- 46.Mendelsohn J, Baselga J. Epidermal Growth Factor Receptor Targeting in Cancer. Semin Oncol. 2006;33:369–85. doi: 10.1053/j.seminoncol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Meyers MB, Shen WP, Spengler BA, et al. Increased Epidermal Growth Factor Receptor in Multidrug-Resistant Human Neuroblastoma Cells. J Cell Biochem. 1988;38:87–97. doi: 10.1002/jcb.240380203. [DOI] [PubMed] [Google Scholar]
- 48.Matar P, Rojo F, Cassia R, et al. Combined Epidermal Growth Factor Receptor Targeting with the Tyrosine Kinase Inhibitor Gefitinib (ZD1839) and the Monoclonal Antibody Cetuximab (IMC-C225): Superiority Over Single-Agent Receptor Targeting. Clin Cancer Res. 2004;10:6487–501. doi: 10.1158/1078-0432.CCR-04-0870. [DOI] [PubMed] [Google Scholar]
- 49.Michaelis M, Bliss J, Arnold SC, et al. Cisplatin-Resistant Neuroblastoma Cells Express Enhanced Levels of Epidermal Growth Factor Receptor (EGFR) and Are Sensitive to Treatment with EGFR-Specific Toxins. Clin Cancer Res. 2008;14:6531–7. doi: 10.1158/1078-0432.CCR-08-0821. [DOI] [PubMed] [Google Scholar]
- 50.Lu Y, Li X, Liang K, et al. Epidermal Growth Factor Receptor (EGFR) Ubiquitination as a Mechanism of Acquired Resistance Escaping Treatment by the Anti-EGFR Monoclonal Antibody Cetuximab. Cancer Res. 2007;67:8240–7. doi: 10.1158/0008-5472.CAN-07-0589. [DOI] [PubMed] [Google Scholar]