Abstract
The paired box homeotic gene 3 (PAX3) is a crucial regulator for the maintenance of melanocytic progenitor cells and has a poorly defined role in melanoma. To understand how PAX3 affects melanocyte and melanoma proliferation, we identified potential PAX3 downstream targets through gene expression profiling. Here we identify TBX2, a key developmental regulator of cell identity and an anti-senescence factor in melanoma, as a directly regulated PAX3 target. We also found that TBX2 is involved in the survival of melanoma cells, and is overexpressed in some melanoma specimens. The identification of TBX2 as a target for PAX3 provides a key insight into how PAX3 may contribute to melanoma evolution and may provide opportunities for pro-senescence therapeutic intervention aimed at disrupting the ability of PAX3 to regulate TBX2.
Introduction
A number of pathways involved in the development and survival of the melanocyte lineage have been identified. The paired box homeotic gene 3 (PAX3) is a key factor in some of these pathways and is essential for maintaining melanocytic progenitor cells (1–3). A chromosomal deletion, a splice-site mutation, or an amino acid substitution of PAX3 can cause Splotch-retarded, Splotch and Splotch-delayed mice, respectively (4). Splotch-delayed homozygous embryos survive to birth, compared to Splotch mutant embryos, which die at E13 due to neural tube defects (5). Heterozygous Splotch-delayed (Splotch/+) have pigmentation abnormalities characterized by a white patch on the belly due to defective neural crest-derived melanocyte development (6). PAX3 mutations in humans produce type I and type III Waardenburg syndrome (7, 8), a condition characterized by melanocyte deficiencies in the skin and inner ear. Collectively, these studies indicate important roles for PAX3 both in the proliferation and/or survival of developing melanocytes, as well as differentiation pathways such as pigmentation. PAX3 overexpression is also frequently detected in melanomas (3, 9–14). Approximately 30–70% of primary melanoma specimens and 77% of cultured primary melanoma cells are known to have PAX3 overexpression (3, 12, 14, 15).
The upstream regulators of PAX3 have been thoroughly investigated. Several transcription factors, which regulate the transcription of PAX3 in a tissue-specific manner, have been identified, including POU transcription factors (Oct-1, Brn1, and Brn-2) (16, 17), tale homeodomain-containing proteins (Pbx and Prep1) (18), myostatin (19), HoxA1 (16), Tead2 (20), and N-Myc (21). All these regulators of PAX3 modulate cell proliferation. These findings further suggest that PAX3 is an important factor in regulating cell proliferation and survival. In addition, our recent study demonstrated that fibroblast growth factor basic (FGF2)-signal transducer and activator of transcription 3 (STAT3) signaling modulates PAX3 transcription in melanocytes (22). Upon activation by FGF2, STAT3 binds PAX3 upstream regulatory sequences to transactivate its promoter. We also showed that transforming growth factor beta (TGF-β) represses PAX3 expression and that ultraviolet (UV) B irradiation downregulates TGF-β activity (15). This suggests that de-repression of PAX3 transcription upon UVB irradiation might be important in pigment induction.
PAX3 is a transcription factor (23) and exerts its functions in melanocyte development and maturation through its transcriptional modulation of key melanocyte regulators, including the microphthalmia-associated transcription factor (MITF) gene (24, 25). Other crucial downstream targets of PAX3 include tyrosinase-related protein-1 (Tyrp-1) (26), c-Ret (27), TGF-β2 (28), and Wnt1 (29–31). However, the spectrum of the direct transcriptional targets of PAX3 has not been fully investigated. Moreover, while its role in development has been characterized, it is widely speculated that PAX3 will play a key role in melanoma progression. However, how PAX3 might impact on melanoma is poorly understood. Here, we sought to better understand the downstream effectors in melanocytes and melanoma in order to characterize their role in regulating melanocyte growth and melanoma evolution.
To this end, we systematically identified other downstream candidates of PAX3 by microarray analysis and identified 91 candidates as PAX3-dependent downstream targets. Since it has been shown that T-box 2 (TBX2) regulates the proliferation of melanocytes and enhances melanoma invasiveness (9–11), we chose to study TBX2 and investigate whether it is directly regulated by PAX3 at the transcriptional level. We found that TBX2 is up-regulated in PAX3-overexpressed melanoma tissues. Our biochemical studies also demonstrated that PAX3 protein binds to the TBX2 promoter and that TBX2 is a PAX3-responsive gene. This data indicates that TBX2 is directly regulated by PAX3 at the transcriptional level and confirm that TBX2 is a critical factor in the regulation of melanocyte growth.
Results
Microarray screen for PAX3 target genes
To identify PAX3 transcriptional targets, global mRNA expression levels were profiled in low and high PAX3-expressing human primary melanocytes (HPMs) with or without cycloheximide, a RNA-protein translation inhibitor (32). The pooled HPMs (2nd passage) (n=5) were either infected with lenti-PAX3 virus to generate melanocytes with a high level of PAX3 expression (H-HPMs), or with pRetro-shPAX3 virus to generate melanocytes with a low level of PAX3 expression (L-HPMs) (Figure 1A). Using this system, expression profiling was performed with Human Genome U133 Plus 2.0 Array Chips (Affymetrix Inc., Santa Clara, CA).
Figure1. Microarray screen for PAX3-dependent target genes.
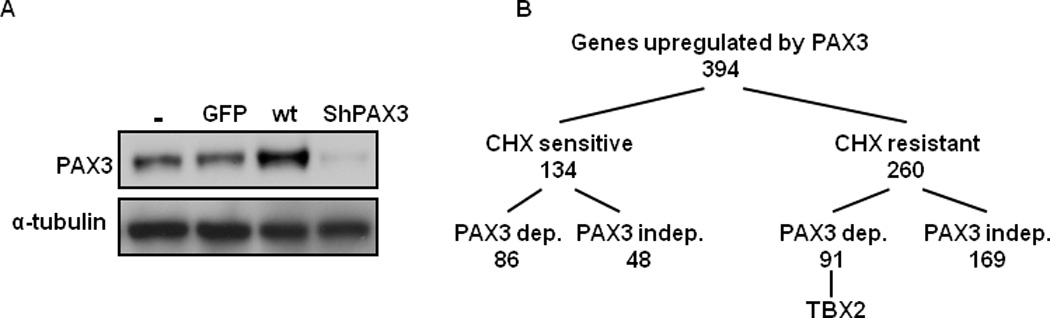
(A) Expression of virus-encoded proteins in primary human melanocytes infected for 48 hr with empty control (−), negative (GFP), wild-type (wt), or shRNA (shPAX3) PAX3 virus. (B) Summary of screen results from expression profiling of melanocytes overexpressed with PAX3 or knockdown PAX3 ± cycloheximide (CHX).
As summarized in Figure 1B, a total 394 genes were upregulated at multiple time points by PAX3 overexpression, with 217 upregulated independently of PAX3 and 177 displaying PAX3 dependency. A target gene was considered PAX3-dependent if the ratio of its maximum induction in the presence of PAX3 overexpression, to the maximum repression in the presence of PAX3 knockdown was ≥4. Of the PAX3-dependent targets, 169 were sensitive to cycloheximide, and 91 were resistant. Table 1 shows selected sets of cycloheximide-resistant PAX3-dependent targets identified by this screen. Among these potential targets, TBX2 was noteworthy because TBX2 is overexpressed in melanomas, regulates the proliferation of melanoma cells, and enhances melanoma invasiveness (9–11). Furthermore, TBX2 regulates the promoter of the familial melanoma genes, CDKN2A (p19ARF) (33) and p21CIP1 (34), and attenuates the induction of CDKN2A mediated by E2F, MYC, or H-RAS (35). Therefore, a possible relationship between PAX3 and TBX2 was further investigated.
Table 1.
PAX3 Target Genes (Select CHX-Resistant Targets)
TBX2 is upregulated in PAX3-overexpressed melanoma tissues
To identify whether PAX3 protein co-localizes with TBX2 protein in melanoma cells, we performed immunofluorescence staining of PAX3 protein and TBX2 protein in WM293 melanoma cells. Specifically, we demonstrated that both PAX3 and TBX2 proteins are nuclear (Figure 2A). Furthermore, nuclear foci of TBX2 and PAX3 were detected (Figure 2A).
Figure2. Correlation between TBX2 expression and PAX3 expression in melanomas.
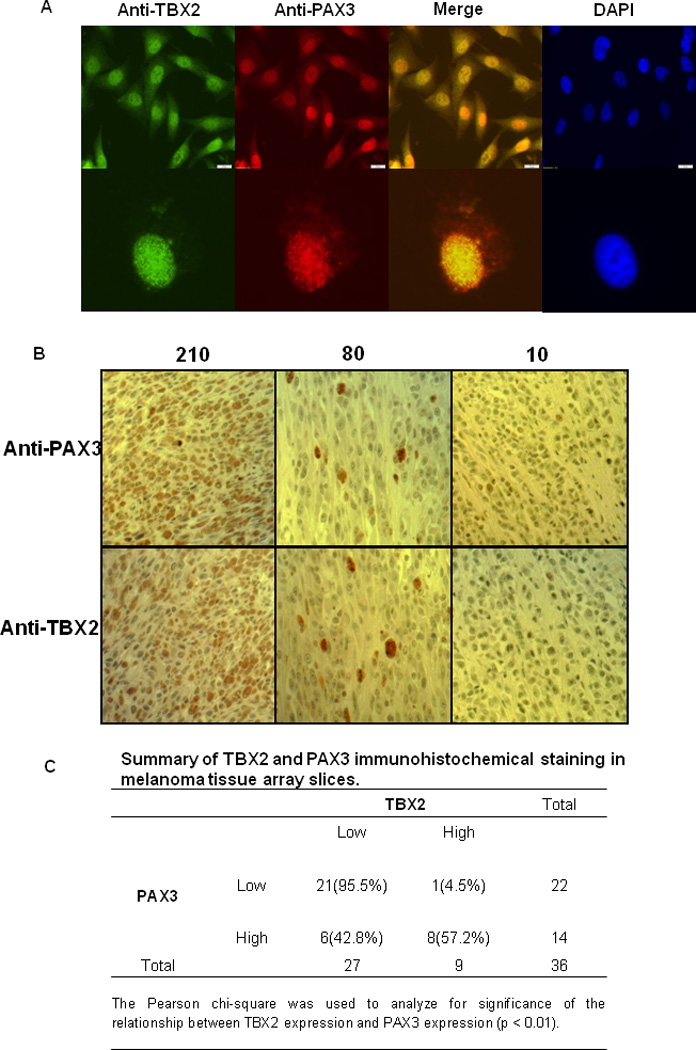
(A) TBX2 protein and PAX3 protein are co-localized in melanocyte nuclei. Melanoma cells were stained with anti-PAX3 and anti-TBX2 antibodies by Immunofluorescence. (B) Different H score of TBX2 and PAX3 staining of melanoma specimens. (C) Summary of TBX2 and PAX3 immunohistochemical staining in melanoma tissue array slices.
Next, we preformed immunohistochemical staining and scoring analysis (H score) of melanoma specimens for PAX3 (15). Immunohistochemical scoring (H score) was determined by multiplying the staining intensity by the percentage of positive tumor cells (15, 36). To determine whether PAX3-enhanced TBX2 expression was also observed in melanoma, we performed immunohistochemical staining and scoring analysis of adjacent melanoma sections from the same source, for TBX2 using a validated anti-TBX2 antibody (see below). As determined by the bimodal H score distribution, tissues with scores <80 were designated as TBX2 low, and scores >80 were designated as TBX2 high (Figure 2B). We found a significantly positive correlation between TBX2 and PAX3 expression (Figure 2C). Note that in the panels with an H-score of 80 (Figure 2B), the pattern of strong positive staining for Tbx2 and Pax3 is very similar, suggesting that these represent the same cells on adjacent sections.
TBX2 is a PAX3 responsive gene
To validate the TBX2 antibody used, TBX2 expression was determined in B16, A375 and melc cells by Western blot using Abcam rabbit polyclonal TBX2 antibodies. As shown in Figure 3A, TBX2 protein is robust in B16 cells and undetectable in melc cells, consistent with the results from previous work using a mouse monoclonal antibody (37). To test whether TBX2 is a PAX3-responsive gene in melanoma cells, we introduced PECE-PAX3 or empty vectors into the MeWo melanoma cells, and assessed TBX2 expression by quantitative RT-PCR and Western blot (Figure 3B). This melanoma cell line was used because MeWo cells carry neither Braf nor N-Ras mutations (38). The PECE-PAX3 construct, generously provided by Dr. Goossens, has been described elsewhere (39). As shown in Figure 3B, TBX2 expression was significantly induced in response to PAX3 at both the mRNA and protein levels.
Figure3. TBX2 is induced by PAX3 overexpression.
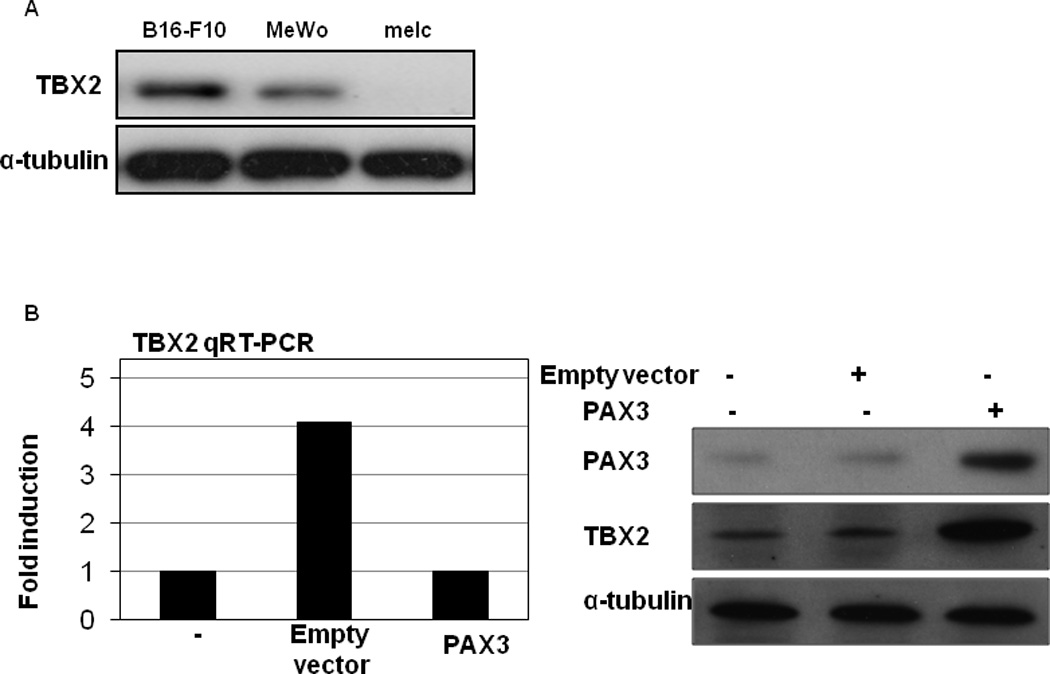
(A) TBX2 antibody validation. TBX2 expression was determined in B16, A375 and melc cells by Western blot using Abcam rabbit polyclonal TBX2 antibodies. (B) B16 melanoma cells were transfected with empty pECE plasmid, pECE-PAX3 plasmid, or no plasmid. TBX2 RNA levels were measured by quantitative RT-PCR and normalized to GAPDH (left panel). Results are expressed as the mean of the experiment done in triplicate ± the SEM. TBX2 and PAX3 protein expression were analyzed by Western blot and α-tubulin was used as a loading control (right panel).
We recently demonstrated that FGF2 stimulation up-regulates the expression of PAX3 in melanocytes (22). To determine whether TBX2 expression is regulated by PAX3 in the context of FGF2 stimulation in melanocytes, HPMs at 50–70% confluency were treated with FGF2 (10nmol/L). As shown in Figure 4A, the expression of both TBX2 mRNA and protein was upregulated by 6 hr. Importantly, inhibition STAT3 activation abrogated FGF2-induced TBX2 up-regulation (Figure 4A). Furthermore, overexpression of PAX3 reverses the effects of a STAT3 inhibitor on FGF2-induced TBX2 upregulation (Figure 4B). To identify whether PAX3 is required in FGF2-inudced TBX2 up-regulation, we generated B16 cells with stable reduction of PAX3 expression by specific shRNA knockdown, and examined PAX3 expression after FGF2 stimulation. We found that the FGF2-stimulated TBX2 overexpression was abolished following shPAX3 knockdown (Figure 4C), indicating that PAX3 is required for FGF2-stimulated TBX2 up-regulation. These experiments suggest that TBX2 is transcriptionally regulated by PAX3 in melanocytes and that this regulation occurs in the physiological context of FGF2 stimulation.
Figure4. TBX2 is a PAX3 responsive gene.
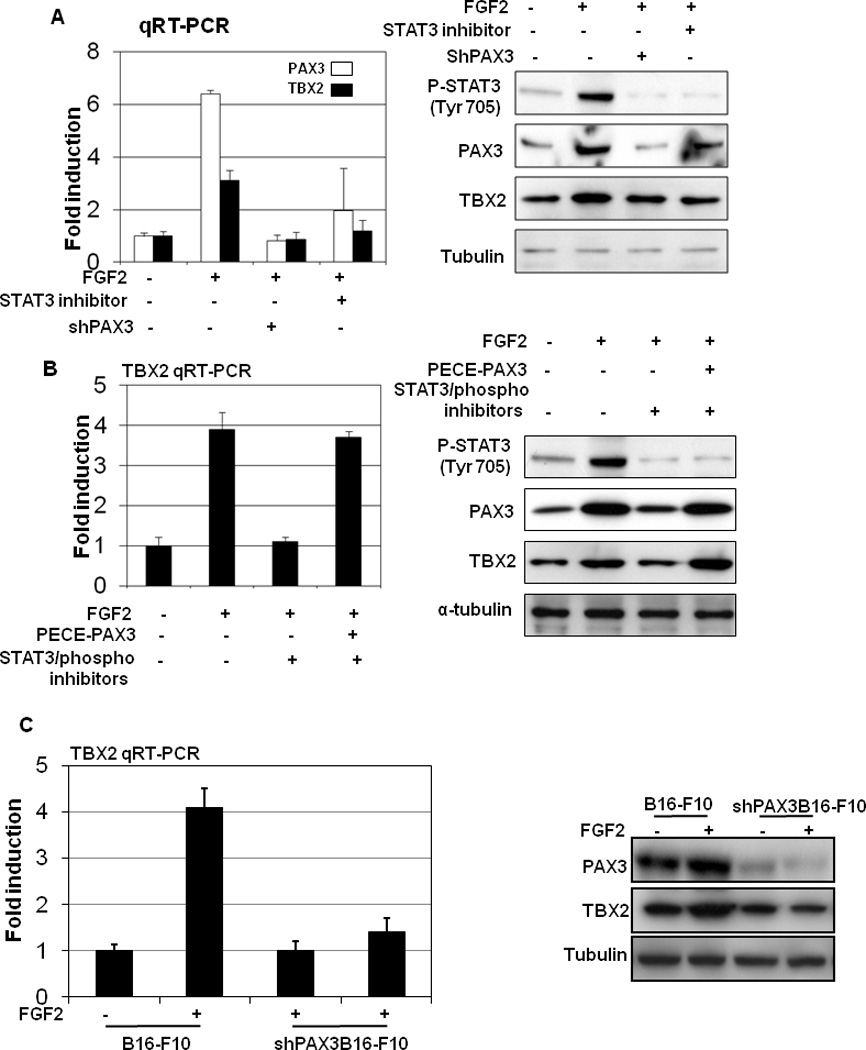
(A) Human primary melanocytes (HPMs) at 75% of confluency were stimulated with FGF2 (10nmol/L). RNA and protein were collected at time 0 and at different time points after stimulation. The left panels represent TBX2 RNA levels as measured by quantitative RT-PCR and normalized to GAPDH. Results are expressed as the mean of the experiment done in triplicate ± SEM. Induction is calculated relative to TBX2 levels in vehicle-treated cells. Protein levels of TBX2, which were analyzed by Western blot, are shown on the right, along with α-tubulin, which served as loading control. (B) Overexpression of PAX3 rescues STAT3 inhibitor inhibited FGF2-induced TBX2 upregulation. B16 melanoma cells were transfected with pECE-PAX3 plasmid for 24 hr and then treated with a specific STAT3 inhibitor, 5, 15-DPP (20umol/L), and a STAT3 phosphorate inhibitor, FLLL31 (5umol/L), together with FGF2 (10nmol/L) for 6 hr. TBX2 RNA levels were measured by quantitative RT-PCR and normalized to GAPDH (left panel). Results are expressed as the mean of the experiment done in triplicate ± the SEM. TBX2 and PAX3 protein expression were analyzed by Western blot, and α-tubulin was used as a loading control (right panel). (C) PAX3 is required in FGF2-stimulated TBX2 repression. B16 cells with stable siPAX3 were treatment with FGF2 (10nmol/L). RNA and protein were collected 6 hr following FGF2 stimulation. The left panels represent TBX2 RNA levels as measured by quantitative RT-PCR and normalized to GAPDH. Results are expressed as the mean of the experiment done in triplicate ± the standard error of the mean (SEM). Repression is calculated relative to TBX2 levels in vehicle (DMSO)-treated cells. Protein levels of TBX2 were analyzed by Western blot.
PAX3 protein binds the TBX2 promoter
PAX3 is a transcription factor containing a paired domain (PD) and a paired-type homeodomain (HD). The HD binds with a consensus sequence harboring an ATTA motif, and the PD recognizes consensus sequences harboring a GTTCC motif, though frequently Pax3 may bind variant sequences that are difficult to predict. Complementary to the gene expression based approach, we used a web tool known as TESS (Transcription Element Search System, http://www.cbil.upenn.edu/tess) to detect probable PAX3 target genes by analyzing gene promoter sequences within 1.0 kb upstream of the transcription start site of genes listed in Table 1. Consistent with our results using PAX3-expressing and knock-down cell lines, we identified a PD binding site in the TBX2 promoter (−781 to −777) (Figure 5A).
Figure5. In vivo association of PAX3 with the TBX2 promoter by chromatin immunoprecipitation (ChIP).
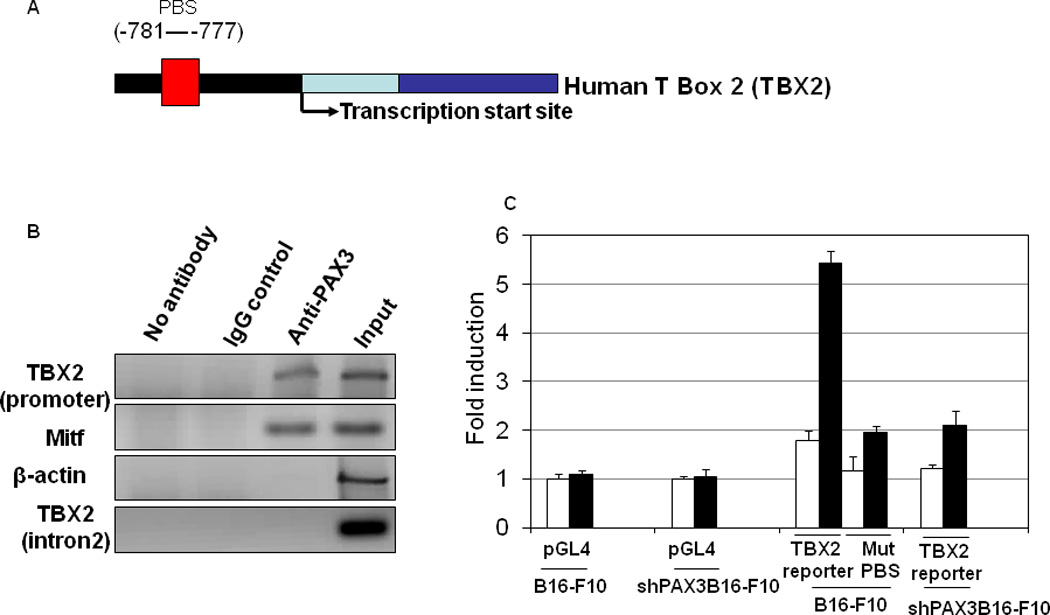
(A) Schematic representation of the human TBX2 locus. (B) B16 melanoma cells were subjected to ChIP assays with the indicated antibodies and PCR primers. (C) Promoter activity was studied using reporter assays with different promoter portions (including mutated PD), which drive expression of firefly luciferase. Relative luciferase activity was normalized to empty pGL4-basic plasmid using the dual-luciferase assay system (Promega). Results are expressed as the mean of the experiment done in triplicate ± SEM. Analysis of TBX2 promoter transactivation capacity by FGF2 stimulation of B16 cells or B16 cells with stable shPAX3 transiently transfected with TBX2 reporter, employed mutations at PAX3 binding sites (PBS).
Chromatin immunoprecipitation revealed that the PD binding site of the TBX2 promoter was amplified in crosslinked chromatin isolated from B16-F10 mouse melanoma cells (40) and immunoprecipitated with a specific anti-PAX3 antibody. However, no amplification occurred in the multiple controls, including IgG antibody, the same PAX3 antibody tested for binding to an intronic sequence in TBX2, and the same PAX3 antibody tested for binding to the β-actin promoter. As a positive control, MITF, a well-documented PAX3 target gene (15, 27, 39, 41, 42) was also examined and PAX3 binding confirmed (Figure 5B). B16-F10 cells were used here because these cells are more easily transfected in comparison to other melanoma cell lines.
To further identify whether the PD binding site in the TBX2 promoter (−781 to −777) is essential for FGF2-induced activation of TBX2 transcription, a series of TBX2-promoter-luciferase reporters were tested for FGF2 responsiveness in B16 mouse melanoma cells. Site-specific mutation at the evolutionarily conserved PAX3 binding site (−781 to −777) abolished the FGF2-induced activation of the TBX2 promoter (Figure 5C). These data indicate that the PD of TBX2 promoter from −781 to −777 is required for FGF2-induced TBX2 transcription. Next, TBX2-promoter-luciferase reporters were tested for FGF2 responsiveness in B16 mouse melanoma cells with stable PAX3 silencing. PAX3 silencing abolished the FGF2-induced activation of PAX3 promoter (Figure 5C). Together, these results suggest that the PD of TBX2 promoter from −781 to −777 represents an important PAX3 binding site that mediates FGF2 responsiveness.
TBX2 contributes to the melanoma cell proliferation
We next sought to determine if interference of TBX2 activity, regulated by PAX3, affected the proliferation and survival of melanocytes, since PAX3 is funciontally relevant for melanocyte growth. Previous work has established that TBX2 overexpression promotes melanocyte proliferation and transformation, and is required by primary melanoma cells for immortality (43). Furthermore, TBX2 activity is regulated by direct interaction with the retinoblastoma (Rb1) protein (44) and is critically required to suppress senescence in melanoma (37). To further confirm these previous reports, we tested whether knockdown of TBX2 affected the viability of melanoma cells.
We first observed that knockdown of TBX2 (Figure 6A), using the ShTBX2 vector, significantly inhibited proliferation of melanoma cells and induced cell cycle arrest in the G1 phase, compared to control cells transfected with an empty vector or the ShLuc vector (Figure 6A). These data strongly suggest that TBX2 has an important role in promoting growth of melanocytes and melanoma cells.
Figure6. TBX2 contributes to the melanoma cell survival.
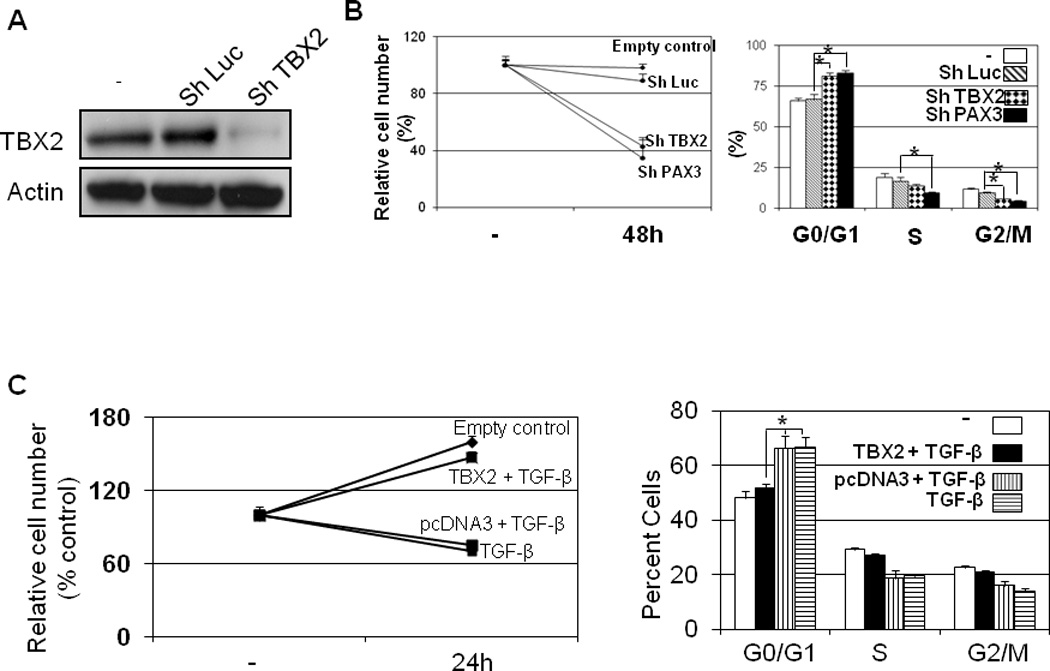
(A) Knockdown of TBX2 in B16 cells. (B) Knockdown of TBX2 induced cell cycle arrest and the inhibition of proliferation B16 melanoma cells were transfected with ShTBX2 or control vector. After 48 hr, cells were fixed. Samples were subjected to MTT assays to evaluate the relative cell numbers (left panel) and to flow cytometry assays to determine the cell-cycle distribution (right panel). (*P<0.01 by t test). (B) Overexpression of TBX2 rescued the TGF-β-induced growth inhibition. B16 mouse melanoma cells were transfected with TBX2 vector and then treated with TGF-β 24 hr later. Samples were subjected to MTT assays to evaluate the relative cell numbers (left panel) and to flow cytometry assays to determine the cell-cycle distribution (right panel) 24 hr after TGF-β treatment.
To determine whether overexpression of TBX2 could rescue TGF-β-induced growth inhibition in melanocytes, B16 mouse melanoma cells, which are sensitive to TGF-β treatment (45), were transfected with the TBX2 expression vector and then treated with TGF-β. Overexpression of TBX2 increased the expression of its target gene, MITF, and reversed TGF-β-induced cell cycle arrest, thereby preventing a decrease in cell number (Figure 6B).
Discussion
Identifying the program of transcription driving melanoma progression is a key issue. PAX3 is widely recognized an important regulator of MITF, a transcription factor that plays a key role in controlling melanoma sub-population identity (46), but few other targets for PAX3 have been identified to date. Here we found among the repertoire of genes regulated by PAX3 identified using gene array analysis, that PAX3 regulates the expression of TBX2, a transcription factor identified as key suppressor of melanoma senescence (34, 37). The regulation of TBX2 by PAX3 is direct, and mediates the induction of TBX2 by FGF2, and we both confirm that TBX2 plays a key role in the survival of melanoma cells and demonstrate that TBX2 and PAX3 are co-expressed in melanoma specimens.
In addition to the direct regulation of TBX2 transcription by PAX3 in melanocytes and melanoma cells identified here, PAX3 also regulates TBX2 transcription indirectly via by MITF (47). It is well known that the transcription of, and therefore the activity of MITF, the central transcriptional modulator in these cells, is regulated by an array of trancription factors including PAX3, CREB, SOX10 and LEF1 (15, 48–51). In turn, MITF regulates a network of targets with diverse functions in differentiation, survival, DNA damage repair and the cell cycle, including TBX2, which is a key transcription factor during development (26, 47). Importantly, MITF expression is also required to suppress senescence (52). Thus PAX3 is a key regulator of two anti-senescence factors in melanoma, MITF and TBX2 and targetting PAX3 expression or activity may therefore be a potential route to pro-senescence therapy for melanoma.
The role of TBX2 in melanocytes is tightly interwoven with MITF, with TBX2 acting to repress MITF activated genes such as TYRP1 (10, 47, 53–55) and CDKN1a (p21) (34, 37, 56). The ability of PAX3 to coordinately up-regulate both genes therefore will thererfore enable cells to control the kinetic of activation of their common downstream targets; if TBX2 is active as a transcription repressor MITF will only transiently up-regulate their common targets in response to elevated Pax3 activity for example in response to FGF2 or TGFβ signalling, but if TBX2 is inactivated bye iether post-translational modification or gene silencing, MITF may promote sustained transcription activation of genes such as CDKN1a and TYRP1. The cooordinate regulation of these two genes by PAX3 therfore provides cells with opportunities to fine tune the expression of tehir target genes.
MITF and TBX2 both function in melanocyte proliferation and senescence (34, 35, 50, 57, 58). Given the overlap in MITF and TBX2 target genes, the fact that MITF directly regulates a set of genes required for DNA replication, repair, and mitosis (59), functions in which TBX2 also plays a key role, may indicate that their repertoire of shared targets may be extensive, though validation of this idea will await genome-wide analysis of TBX2 (and TBX3) binding sites.
The molecular mechanisms of MITF and TBX2-regulated melanocyte proliferation aslo overlap. First, upstream regulators and interactors of MITF and TBX2 are similar. Rb1 protein can interact with MITF to regulate the expression of p21 (56). At the same time, hypo-phosphorylated Rb1 protein interacts directly with TBX2 to regulate melanocyte proliferation (44). This suggests that the activity of both proteins may be coordinately regulated via Rb1 phosphorylation during the cell cycle.
Besides TBX2, we also observed a large overlap between the direct transcriptional targets of MITF and PAX3 in primary melanocytes (60). This observation supports the notion that besides being a direct upstream regulator of MITF, PAX3 may cooperate with MITF to modulate gene expression, and thereby melanocyte functions. Further experiments are underway to determine the extent of the cooperation between these two transcription factors and their impact on various cellular processes in melanocytes. However, Slug and Myg1, both of which function in melanocyte development (61–64), were identified as potential targets of PAX3 in our microarray studies (Figure 1 and Table 1). In addition, we found PAX3 binding sequences in each promoter (for Slug, −294 to −299; for Myg1, −541 to −546) suggesting that these genes will be directly regulated by PAX3 directly at the transcriptional level.
Materials and Methods
Cell lines and reagents
Primary melanocytes were isolated from normal discarded foreskins as described (65, 66). Human primary melanocytes were cultured in Medium 254 (Cascade Biology) with 5ml human melanocyte growth supplement. The final concentrations of the components in the supplemented medium are bovine pituitary extract (BPE) (0.2% v/v), fetal bovine serum (0.5% v/v), bovine insulin (5 µg/ml), bovine transferrin (5 µg/ml), basic fibroblast growth factor (3 ng/ml), hydrocortisone (0.18 µg/ml), heparin (3 µg/ml) and phorbol 12-myristate 13-acetate (PMA) (10ng/ml) (http://products.invitrogen.com/ivgn/product/S0025). B16-F10 cells were generously provided by Dr. David Fisher at MGH, Harvard Medical School. Melanoma cells were maintained in DMEM plus 10% FBS (GIBCO/BRL). Adherent cells were irradiated as described elsewhere (67). FGFb (FGF2) was purchased from Sigma (St. Louis, MO). Short hairpin RNA duplexes that target TBX2 were purchased from Origene (Rockville, MD). Short hairpin RNA duplexes that target PAX3 were purchased from Openbiosystem (Lafayette, CO).
MTT Assay of Cell Numbers
Ten thousand cells were plated per well in a 96-well plate. FGF2 (10nmol/L) was added following overnight culture. MTT solution was added 24 hr later. Optical density was read at 560 nm, and background was subtracted at 670 nm.
Histology
Immunohistochemical studies were performed on melanoma specimens in tissue slices. Primary antibodies included polyclonal anti-PAX3 antibody (C-20, Santa Cruz Biotec.) and polyclonal anti-TBX2 antibody (Abcam). Staining was performed with Dako DAB or AEC Detection Kit ((DAKO EnVision + System HRP, DAKO). Tissues known to express the antigen of interest were used as positive controls, whereas removal of the primary antibodies in the test tissues was used as the negative control.
Sections (5 µm) of paraffin-embedded tissues were analyzed by Immunofluorescence according to standard protocols with the following antibodies: anti-PAX3 (C-20, Santa Cruz Biotec.) and anti-TBX2 (C-17, Santa Cruz Biotec.).
Flow cytometry
Cells were fixed in ice-cold 70% ethanol before DNA staining with 50 µg/ml propidium iodide (Sigma Aldrich) in PBS containing 0.5 mg/ml RNase (Amersham). DNA content was analyzed by flow cytometry (Becton Dickinson FACSCalibur).
Chromatin immunoprecipitation
Chromatin immunoprecipitation of the human TBX2 sequence from human primary melanocytes was performed as described elsewhere (15, 68). PCR was carried out using primers specific to the promoter region of human TBX2 (forward, 5’-TGCGGAGAACTCCAGGTT-3’ reverse, 5’-AGCTGGTGAGTGGAGCCA-3’).
Real-Time RT-PCR, Western Blotting, Immunoprecipitation, and Enzyme Immunoassay
For quantitative RT-PCR, total RNA was converted into cDNA using the SuperScript™ III reverse transcriptase kit (Invitrogen). cDNA expression was quantified using the QuantiTect Probe RT-PCR kit (Qiagen, Valencia, CA) and an ICycler machine (BioRad, Hercules, CA). Gene-specific primer sets were used as reported for human TBX2: forward: 5’-TAGAGCCTCGTCTGGCAAGT -3’; reverse: 5’-ACCAGAGACACCTTCGGATG -3’; mouse TBX2: forward: 5’-GGCTTCAGCGGTCAGAATAG -3’; reverse: 5’-AAGGAGGTGCCCTGGTAGTT-3’. Taqman PCR reactions were done in triplicate for each sample and normalized to GAPDH.
Western blotting was performed using the following antibodies: anti-PAX3 (The Developmental Studies Hybridoma Bank at the University of Iowa); anti-MITF (C5, generously provided by Dr. David E. Fisher at Massachusetts General Hospital); and anti-TBX2 (Abcam, ab33298.).
Gene profiling analysis
To identify PAX3 transcriptional targets, global mRNA expression levels were profiled in low and high PAX3 expressing human primary melanocytes (HPMs) with or without cycloheximide, a RNA-protein translation inhibitor. The pooled HPMs (2nd passage) (n=5) were either infected with lenti-PAX3 virus to generate melanocytes with a high level of PAX3 expression (H-HPMs), or with pRetro-shPAX3 virus to generate melanocytes with a low level of PAX3 expression (L-HPMs) (Supplementary Figure 3A). Using this system, expression profiling was undertaken with Human Genome U133 Plus 2.0 Array Chips (Affymetrix Inc., Santa Clara, CA). Significant induction of transcription was defined as: (1) a 2-fold or greater increase relative to the HPMs baseline after PAX3 overexpression; (2) a target gene was considered PAX3-dependent if the ratio of its maximum induction in the presence of PAX3 overexpression to the maximum repression in the presence of PAX3 knockdown was ≥4; (3) a target gene was excluded as a transcriptional target if the ratio of its maximal induction in the absence of cycloheximide relative to the presence of cycloheximide was ≥2. Cycloheximide (CHX) was used at a final concentration of 100µg/ml for 24 hours.
Luciferase Reporter Assay
In a fragment of the human TBX2 promoter (−889/+112, relative to the transcription start site), a series mutation of TBX2 binding elements (GTTCC to GGTCC) were generated by PCR and were inserted into the pGL-4 basic vector (Promega). Promoter constructs were co-transfected with the pRL-TK plasmids (Promega). Twenty-four hours after transfection, cells were treated with FGF2 (10nM), and another 24 hr later were lysed and assayed using dual luciferase reagents (Promega). Promoter activity was measured by comparing luciferase levels, normalized to the constitutively expressed Renilla.
Supplementary Material
Significance.
While PAX3 is a well-established regulator of melanocyte development, its role in melanoma is unclear. TBX2 has been identified as a key regulator of epigenetic inheritance in development, and disruption of TBX2 expression or function can restore senescence in melanoma cell lines, raising the possibility of pro-senescence therapy for this disease. However little is known as to how TBX2 is regulated. This study identifies a direct link between PAX3 and TBX2 that suggests that signals regulating PAX3 expression and activity will also control TBX2 expression with implications for both melanocyte development and melanoma.
Acknowledgments
The PAX3 monoclonal antibody developed by Dr. Charles P Ordahl was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biology, Iowa City, IA, USA. This work was supported by National Institutes of Health (7RO1CA137098 for RC), American Cancer Society (RSG-09-022-01-CNE), and The Harry J. Lloyd Charitable Trust (RC). RC is an American Cancer Society Research Scholar.
Footnotes
No potential conflicts of interest were disclosed.
References
- 1.Blake JA, Ziman MR. Dev Growth Differ. 2005 Dec;47:627. doi: 10.1111/j.1440-169X.2005.00835.x. [DOI] [PubMed] [Google Scholar]
- 2.Steingrimsson E, Copeland NG, Jenkins NA. Cell. 2005 Apr 8;121:9. doi: 10.1016/j.cell.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 3.Scholl FA, et al. Cancer Res. 2001 Feb 1;61:823. [PubMed] [Google Scholar]
- 4.Tassabehji M, et al. Hum Mol Genet. 1994 Jul;3:1069. doi: 10.1093/hmg/3.7.1069. [DOI] [PubMed] [Google Scholar]
- 5.Moase CE, Trasler DG. J Med Genet. 1992 Mar;29:145. doi: 10.1136/jmg.29.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein DJ, Vogan KJ, Trasler DG, Gros P. Proc Natl Acad Sci U S A. 1993 Jan 15;90:532. doi: 10.1073/pnas.90.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Read AP, Newton VE. J Med Genet. 1997 Aug;34:656. doi: 10.1136/jmg.34.8.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tassabehji M, et al. Nature. 1992 Feb 13;355:635. doi: 10.1038/355635a0. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez M, Aladowicz E, Lanfrancone L, Goding CR. Cancer Res. 2008 Oct 1;68:7872. doi: 10.1158/0008-5472.CAN-08-0301. [DOI] [PubMed] [Google Scholar]
- 10.Carreira S, Dexter TJ, Yavuzer U, Easty DJ, Goding CR. Mol Cell Biol. 1998 Sep;18:5099. doi: 10.1128/mcb.18.9.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abrahams A, Mowla S, Parker MI, Goding CR, Prince S. J Biol Chem. 2008 Jan 25;283:2223. doi: 10.1074/jbc.M705651200. [DOI] [PubMed] [Google Scholar]
- 12.Barr FG, et al. Cancer Res. 1999 Nov 1;59:5443. [PubMed] [Google Scholar]
- 13.Vachtenheim J, Novotna H. Cell Mol Biol (Noisy-le-grand) 1999 Nov;45:1075. [PubMed] [Google Scholar]
- 14.Plummer RS, et al. Mod Pathol. 2008 Mar 7; [Google Scholar]
- 15.Yang G, et al. Mol Cell. 2008 Nov 21;32:554. [Google Scholar]
- 16.Pruitt SC, Bussman A, Maslov AY, Natoli TA, Heinaman R. Gene Expr Patterns. 2004 Oct;4:671. doi: 10.1016/j.modgep.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Smit DJ, Smith AG, Parsons PG, Muscat GE, Sturm RA. Eur J Biochem. 2000 Nov;267:6413. doi: 10.1046/j.1432-1327.2000.01737.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhu BK, Pruitt SC. Melanoma Res. 2005 Oct;15:363. doi: 10.1097/00008390-200510000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Amthor H, et al. Dev Biol. 2002 Nov 15;251:241. doi: 10.1006/dbio.2002.0812. [DOI] [PubMed] [Google Scholar]
- 20.Milewski RC, et al. Development. 2004 Feb;131:829. doi: 10.1242/dev.00975. [DOI] [PubMed] [Google Scholar]
- 21.Harris RG, White E, Phillips ES, Lillycrop KA. J Biol Chem. 2002 Sep 20;277:34815. doi: 10.1074/jbc.M109609200. [DOI] [PubMed] [Google Scholar]
- 22.Dong L, et al. Cell Death Differ. Oct 14; [Google Scholar]
- 23.Medic S, Ziman M. Crit Rev Biochem Mol Biol. 2009 Jun;44:85. doi: 10.1080/10409230902755056. [DOI] [PubMed] [Google Scholar]
- 24.Lang D, et al. Nature. 2005 Feb 24;433:884. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe A, Takeda K, Ploplis B, Tachibana M. Nat Genet. 1998 Mar;18:283. doi: 10.1038/ng0398-283. [DOI] [PubMed] [Google Scholar]
- 26.Galibert MD, Yavuzer U, Dexter TJ, Goding CR. J Biol Chem. 1999 Sep 17;274:26894. doi: 10.1074/jbc.274.38.26894. [DOI] [PubMed] [Google Scholar]
- 27.Lang D, Epstein JA. Hum Mol Genet. 2003 Apr 15;12:937. doi: 10.1093/hmg/ddg107. [DOI] [PubMed] [Google Scholar]
- 28.Mayanil CS, et al. J Biol Chem. 2006 Aug 25;281:24544. doi: 10.1074/jbc.M512449200. [DOI] [PubMed] [Google Scholar]
- 29.Conway SJ, et al. Cardiovasc Res. 2000 Aug;47:314. doi: 10.1016/s0008-6363(00)00098-5. [DOI] [PubMed] [Google Scholar]
- 30.Fenby BT, Fotaki V, Mason JO. Biochim Biophys Acta. 2008 Feb;1779:115. doi: 10.1016/j.bbagrm.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Kubic JD, Young KP, Plummer RS, Ludvik AE, Lang D. Pigment Cell Melanoma Res. 2008 Dec;21:627. doi: 10.1111/j.1755-148X.2008.00514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martegani E, Popolo L, Alberghina L, Sturani E. Biochim Biophys Acta. 1980 Dec 11;610:318. doi: 10.1016/0005-2787(80)90013-1. [DOI] [PubMed] [Google Scholar]
- 33.Gandini S, et al. Eur J Cancer. 2005 Sep;41:2040. doi: 10.1016/j.ejca.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 34.Prince S, Carreira S, Vance KW, Abrahams A, Goding CR. Cancer Res. 2004 Mar 1;64:1669. doi: 10.1158/0008-5472.can-03-3286. [DOI] [PubMed] [Google Scholar]
- 35.Jacobs JJ, et al. Nat Genet. 2000 Nov;26:291. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 36.Camp RL, Rimm EB, Rimm DL. Cancer. 1999 Dec 1;86:2259. doi: 10.1002/(sici)1097-0142(19991201)86:11<2259::aid-cncr13>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Vance KW, Carreira S, Brosch G, Goding CR. Cancer Res. 2005 Mar 15;65:2260. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- 38.Gao L, et al. Cancer Res. 2006 Aug 15;66:7880. doi: 10.1158/0008-5472.CAN-06-0254. [DOI] [PubMed] [Google Scholar]
- 39.Bondurand N, et al. Hum Mol Genet. 2000 Aug 12;9:1907. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- 40.Firnberg N, Neubuser A. Dev Biol. 2002 Jul 15;247:237. doi: 10.1006/dbio.2002.0696. [DOI] [PubMed] [Google Scholar]
- 41.Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Hum Genet. 2000 Jul;107:1. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- 42.Verastegui C, Bille K, Ortonne JP, Ballotti R. J Biol Chem. 2000 Oct 6;275:30757. doi: 10.1074/jbc.C000445200. [DOI] [PubMed] [Google Scholar]
- 43.Peres J, et al. Genes Cancer. Mar;1:272. doi: 10.1177/1947601910365160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vance KW, Shaw HM, Rodriguez M, Ott S, Goding CR. Mol Biol Cell. Aug 1;21:2770. doi: 10.1091/mbc.E09-12-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan D, et al. J Exp Ther Oncol. 2002 Sep-Oct;2:286. [Google Scholar]
- 46.Hoek KS, et al. Pigment Cell Melanoma Res. 2008 Dec;21:665. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 47.Carreira S, Liu B, Goding CR. J Biol Chem. 2000 Jul 21;275:21920. doi: 10.1074/jbc.M000035200. [DOI] [PubMed] [Google Scholar]
- 48.Dorsky RI, Raible DW, Moon RT. Genes Dev. 2000 Jan 15;14:158. [PMC free article] [PubMed] [Google Scholar]
- 49.Yasumoto K, et al. EMBO J. 2002 Jun 3;21:2703. doi: 10.1093/emboj/21.11.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goding CR. Genes Dev. 2000 Jul 15;14:1712. [PubMed] [Google Scholar]
- 51.Busca R, Ballotti R. Pigment Cell Res. 2000 Apr;13:60. doi: 10.1034/j.1600-0749.2000.130203.x. [DOI] [PubMed] [Google Scholar]
- 52.Ohanna M, et al. Genes Dev. Jun 15;25:1245. doi: 10.1101/gad.625811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yavuzer U, Goding CR. Mol Cell Biol. 1994 May;14:3494. doi: 10.1128/mcb.14.5.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yavuzer U, et al. Oncogene. 1995 Jan 5;10:123. [PubMed] [Google Scholar]
- 55.Bertolotto C, et al. Mol Cell Biol. 1998 Feb;18:694. doi: 10.1128/mcb.18.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carreira S, et al. Nature. 2005 Feb 17;433:764. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- 57.Hoogaars WM, et al. Pigment Cell Melanoma Res. 2008 Jun;21:379. doi: 10.1111/j.1755-148X.2008.00461.x. [DOI] [PubMed] [Google Scholar]
- 58.Giuliano S, et al. Cancer Res. May 1;70:3813. doi: 10.1158/0008-5472.CAN-09-2913. [DOI] [PubMed] [Google Scholar]
- 59.Strub T, et al. Oncogene. May 19;30:2319. doi: 10.1038/onc.2010.612. [DOI] [PubMed] [Google Scholar]
- 60.McGill GG, et al. Cell. 2002 Jun 14;109:707. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 61.Gupta PB, et al. Nat Genet. 2005 Oct;37:1047. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanchez-Martin M, et al. Am J Med Genet A. 2003 Oct 1;122A:125. doi: 10.1002/ajmg.a.20345. [DOI] [PubMed] [Google Scholar]
- 63.Philips MA, et al. Biol Cell. 2009 Jun;101:361. doi: 10.1042/BC20080086. [DOI] [PubMed] [Google Scholar]
- 64.Kingo K, et al. J Dermatol Sci. 2006 Nov;44:119. doi: 10.1016/j.jdermsci.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 65.Horikawa T, Norris DA, Zekman T, Morelli JG. Pigment Cell Res. 1996 Apr;9:58. doi: 10.1111/j.1600-0749.1996.tb00089.x. [DOI] [PubMed] [Google Scholar]
- 66.Dunham WR, Klein SB, Rhodes LM, Marcelo CL. J Invest Dermatol. 1996 Sep;107:332. doi: 10.1111/1523-1747.ep12363174. [DOI] [PubMed] [Google Scholar]
- 67.Cui R, et al. Cell. 2007 Mar 9;128:853. [Google Scholar]
- 68.Cui R, et al. J Biol Chem. 2005 Nov 25;280:39152. doi: 10.1074/jbc.M504655200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


