Abstract
Integration of cellular signaling pathways with androgen receptor (AR) signaling can be achieved through phosphorylation of AR by cellular kinases. However, the kinases responsible for phosphorylating the androgen receptor at numerous sites and the functional consequences of AR phosphorylation are only partially understood. Bioinformatic analysis revealed AR serine 213 (S213) as a putative substrate for PIM1, a kinase overexpressed in prostate cancer. Therefore, phosphorylation of AR serine 213 by PIM1 was examined using a phosphorylation site-specific antibody. Wild type PIM1, but not catalytically inactive PIM1, specifically phosphorylated AR but not an AR serine to alanine mutant (S213A). In vitro kinase assays confirmed that PIM1 can phosphorylate AR S213 in a ligand independent manner and cell type specific phosphorylation was observed in prostate cancer cell lines. Upon PIM1 overexpression AR phosphorylation was observed in the absence of hormone and was further increased in the presence of hormone in LNCaP, LNCaP-abl, and VCaP cells. Moreover, phosphorylation of AR was reduced in the presence of PIM kinase inhibitors. An examination of AR mediated transcription showed that reporter gene activity was reduced in the presence of PIM1 and wild type AR, but not S213A mutant AR. Androgen mediated transcription of endogenous PSA, Nkx3.1, and IGFBP5 was also decreased in the presence of PIM1 whereas IL6, cyclin A1, and caveolin 2 were increased. Immunohistochemical analysis of prostate cancer tissue microarrays showed significant P-AR S213 expression that was associated with hormone refractory prostate cancers, likely identifying cells with catalytically active PIM1. In addition, prostate cancers expressing a high level of P-AR S213 were twice as likely to be from biochemically recurrent cancers. Thus, AR phosphorylation by PIM1 at S213 impacts gene transcription and is highly prevalent in aggressive prostate cancer.
Keywords: PIM1, AR, phosphorylation, prostate cancer, hormone refractory
Introduction
The androgen receptor (AR), a phospho-protein (1), must respond to carefully timed developmental and extracellular signals to direct differentiation and proliferation of the prostate but the impact of AR phosphorylation on AR function and cancer progression is not well understood. Studies using pharmacological inhibitors and kinase overexpression have shown that Akt can phosphorylate the AR on serines 213 and 791 depending on cell type (2–4). Moreover, our previous studies show that AR is rapidly phosphorylated at S213 in response to dihydrotestosterone (DHT) or the synthetic androgen, R1881 and is tightly regulated in prostate epithelial cells and tissues (5). While AR S213 is embedded in a putative Akt consensus site, recent bioinformatic analysis (http://www.netphorest.info) indicates that it is also a consensus site for PIM1 kinase. Using the phosphorylation site-specific antibody against AR phospho-serine 213 (P-AR S213) developed in our laboratory, we examined whether PIM1 could phosphorylate AR S213.
PIM1 is expressed as two isoforms, a longer form (44 kDa) resulting from an alternative translation initiation site (6) and localized to the plasma membrane and a shorter form (33 kDa) that is localized to the cytoplasm and the nucleus (7–8). PIM1 promotes cell cycle progression and cell survival by phosphorylation of Cdc25A (9), downregulation of the cyclin-dependent kinase inhibitor, p27 (10) and inactivation of the pro-apoptotic pathway by phosphorylating BAD protein on the regulatory serine 112 site (11).
While PIM1 has been more extensively studied in lymphoma, there is increasing evidence to suggest that PIM1 overexpression plays a role in prostate cancer (12–13). Consistent with the synergy between c-myc and PIM1 in promoting leukemia (14–15), a mouse model of c-myc-driven prostate cancer shows that PIM1 is upregulated (16) and in a tissue recombination model, PIM1 synergizes with c-myc to induce carcinoma (17). In addition, a metastatic mouse model of prostate specific p53 and Rb deficiencies demonstrate increased levels of PIM1 protein (18).
Several substrates of PIM1 have been identified: Cdc25A, p27, BAD, HP1γ, 4EBP1, and p21, (9–11, 19–21). Here we identify AR as a novel PIM1 substrate. In the context of prostate cancer, the proto-oncogene (22) PIM1 can phosphorylate AR S213 in a ligand independent manner. Moreover, AR S213 phosphorylation is prevalent in recurring prostate cancers, suggesting possible upregulation of a phosphorylating kinase and the marking of cells with functionally active PIM1 in castration resistant prostate cancer.
Results
PIM1 Phosphorylates the Androgen Receptor at Serine 213
PIM1 and Akt kinases were assessed for their impact on AR phosphorylation at serine 213. PIM1 (isoform 2, 33kDa) kinase was expressed in human embryonic kidney (HEK) 293 cells with either wild type AR or an AR serine to alanine (S213A) mutant that cannot be phosphorylated (Figure 1A). Figure 1A indicates that expression of PIM1 kinase results in robust phosphorylation at AR S213 (lanes 2 and 8), without detectable phosphorylation of the AR S213A mutant (lanes 5 and 11) indicating that PIM1 specifically phosphorylates S213. While phosphorylation is more prevalent in the presence of ligand (10nM R1881, lane 8), there is also marked phosphorylation in the absence of exogenously added ligand (0nM R1881, lane 2), suggesting that in the presence of PIM1 phosphorylation can occur under low hormone conditions.
Figure 1. Phosphorylation of AR WT by PIM1 Kinase.
A) 293 cells were transiently transfected with either AR WT or AR mutant S213A and vector only, PIM1, or HA-myr-Akt. Cells were steroid starved in 10% cFBS overnight and then treated with vehicle or 10nM R1881 for 2 hours. Total protein lysates were subjected to Western blots against P-AR S213, AR (total), PIM1, HA (HA-epitope tagged myristoylated (Myr)-Akt), and tubulin (loading control). B 293 cells were transiently transfected with AR and PIM1 WT or PIM1 K67M and treated as in A. Protein lysates were subjected to Western blot analysis using antibodies against P-AR S213, AR, endogenous PIM1, exogenous PIM1, and tubulin.
Since AR S213 may also be phosphorylated by Akt (2–4), we examined the impact of constitutively active Akt (Myr-Akt) on AR S213 phosphorylation. The results indicate that Akt also appears to increase AR S213 phosphorylation (Figure 1A). However, consistent with previous results (23–24), the total AR protein levels are increased in the presence of Akt, indicating that the apparent increase in phosphorylation is likely due to upregulation of AR protein levels (compare AR panel, lane 7 versus 9 to P-AR S213 panel, lane 7 versus 9). Upregulation of AR through Akt seems to be independent of AR phosphorylation since the AR mutant S213A is also increased. However we cannot exclude the possibility that Akt makes a contribution to AR phosphorylation under these conditions. To determine if the catalytic activity of PIM1 kinase was necessary for AR S213 phosphorylation, a kinase deficient PIM1 mutant (K67M) was co-expressed with AR and compared to PIM1 WT. As shown in Figure 1B, co-expression of PIM1 K67M and AR did not increase P-AR S213 over the basal level of phosphorylation whereas the PIM1 WT increased phosphorylation of AR S213.
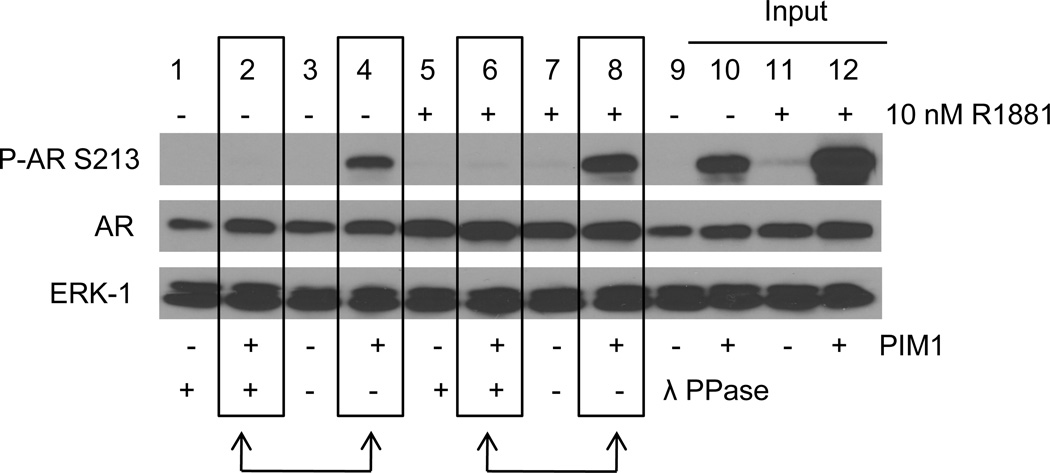
Our previous work indicated that the ligand-dependent modification detected by the P-AR S213 antibody, was phosphorylation (5) as demonstrated by the absence of antigen detection following treatment with lambda phosphatase. The apparent increase in AR S213 phosphorylation in the presence of PIM1 was also sensitive to lambda phosphatase treatment (Figure 2). Lambda phosphatase (λ PPase) treatment abolished AR S213 phosphorylation in response to PIM1 in the absence of R1881 (Figure 2, lane 2 vs lane 4) and the presence of R1881 (Figure 2, lane 6 vs 8), indicating that overexpression of PIM1 in fact, results in phosphorylation on AR S213.
Figure 2. Lambda Phosphatase Treatment Abolishes Phosphorylation of AR by PIM1.
293 cells were transiently transfected with AR and vector only or PIM1. Cells were steroid starved overnight and then treated with vehicle or 10nM R1881 for 2 hours. Total protein lysates were mock treated or lambda phosphatase treated for 30 minutes. Lanes 9–12 (Inputs) are protein lysates prior to lambda phosphatase treatment. Protein lysates were subjected to Western blots against P-AR S213, AR, and ERK-1 (loading control).
PIM1 Phosphorylates AR in vitro
Results shown above indicate that AR is phosphorylated at S213 in the presence of overexpressed PIM1, but do not prove that PIM1 can directly phosphorylate AR. To determine if AR is a PIM1 substrate we performed in vitro kinase assays. As a comparison, the ability of Akt to phosphorylate AR S213 was also examined in these assays since Akt has been reported to phosphorylate S213 (2–4). As shown in Figure 3A, P-AR S213 was detected in the presence of GST-PIM1, but not His-tagged Akt (His-Akt) or GST-GSK3, included as a negative control. In addition, AR S213 phosphorylation occurred in both the presence and absence of ligand (compare 0nM R1881 to 10nM R1881). Interestingly, active Akt was not able to phosphorylate AR under the conditions of these kinase reactions. To ensure the His-tagged Akt was catalytically active, we incubated Akt with GST- GSK3 α/β, a well-established substrate of Akt. As shown in Figure 3B, Akt was able to phosphorylate GSK3 α/β at S21/9 and as expected, active PIM1 phosphorylated Bad at S112 (Figure 3C).
Figure 3. PIM1 Phosphorylates AR in vitro.
A 293 cells were transiently transfected with FLAG-AR. Cells were steroid starved overnight and then treated with vehicle or 10nM R1881 for 2 hours. FLAG-AR was immunopurified and then subjected to kinase reactions in the presence of recombinant GSK3, PIM1, or Akt. The proteins were immunoblotted for P-AR S213, AR, PIM1, and P-Akt S473. B and C Kinase reactions using recombinant His-Akt or GST-PIM1 with their known substrates, GSK3 and Bad, respectively. Kinase activity was detected using antibodies against P-GSK3α/β S21/9, P-Akt S473, and P-Bad S112. D Recombinant AR and PIM1 were combined in a kinase reaction in the presence of R1881. Phosphorylation was detected by P-AR S213 antibody. AR and PIM1 antibodies were also used as controls for the presence of AR and PIM1.
In addition, recombinant full length His-AR, was used in the in vitro kinase assay to eliminate the possibility that phosphorylation was occurring because of proteins in complex with immunoprecipitated AR. An in vitro kinase reaction containing 10nM R1881 was performed using His-AR in the presence or absence of recombinant GST-PIM1. Phosphorylation of AR S213 was only observed in the presence of PIM1 (Figure 3D), once again indicating that AR is a substrate of PIM1 kinase.
PIM1 Expression in LNCaP and LNCaP-abl Cells Enhances AR Phosphorylation
Experiments shown above indicate that under conditions of AR and PIM1 overexpression, AR becomes phosphorylated on S213. To determine if increased levels of PIM1 result in phosphorylation of endogenous AR, PIM1 was overexpressed in prostate cancer cell lines including androgen dependent LNCaP and VCaP cells and an androgen independent LNCaP subline, LNCaP-abl (abl). Overexpression of PIM1 resulted in phosphorylation of AR S213 that was readily detectable in LNCaP and abl cell lines compared to the vector only conditions (Figure 4A). As in the 293 cell line experiments, phosphorylation occurred in both the presence and absence of hormone. As expected, expression of the kinase deficient mutant K67M did not result in P-AR S213 expression in LNCaP or abl cells (data not shown). In VCaP cells P-AR S213 was observed in a hormone-sensitive manner in the absence of PIM1 overexpression (see VCaP, vector only (v.o.) lanes, and compare minus versus plus R1881). This is in contrast to LNCaP and LNCaP-abl cells where little phosphorylation was observed in the absence of overexpressed PIM1. Moreover, phosphorylated AR S213 significantly increased in the presence of overexpressed PIM1 in VCaP cells. VCaP cells stably overexpressing vector only (v.o.) or PIM1 were generated to assess the effect of PIM1 on cell proliferation (Figure 4B, right). PIM1 overexpression does not appear to effect cell proliferation in adherent culture conditions but under anchorage independent conditions using ultra-low attachment plates, VCaP cells overexpressing PIM1 formed more spheres than the vector control cell line (Figure 4B, left). The increased number of spheres formed in the VCaP-PIM1 cell line suggests that these cells are more transformed than the vector only cell line. A similar result has been reported for LNCaP cells overexpressing PIM1 grown in soft agar (25).
Figure 4. Phosphorylation of Endogenous AR in Response to PIM1 Overexpression.
A LNCaP, LNCaP-abl, and VCaP cells were transiently infected with either vector only or PIM1. Cells were steroid starved overnight and then treated with vehicle or 10nM R1881 for 2 hours. Protein lysates were subjected to Western blot analysis using antibodies against P-AR S213, AR, PIM1, and tubulin. B VCaP stable cell lines overexpressing vector only (vo) or PIM1 (right panel) were grown on ultra-low attachment plates. Spheres that grew in the anchorage independent condition were counted under a microscope. The graph represents three independent experiments performed in triplicate. Each point represents the number of spheres in a replicate and the horizontal bar denotes the mean (vo: 245.8 ± 21.4 and PIM1: 274.8 ± 22.0). The p-value was calculated using an unpaired t-test.
Inhibition of AR S213 Phosphorylation in the Presence of PIM kinase Inhibitors
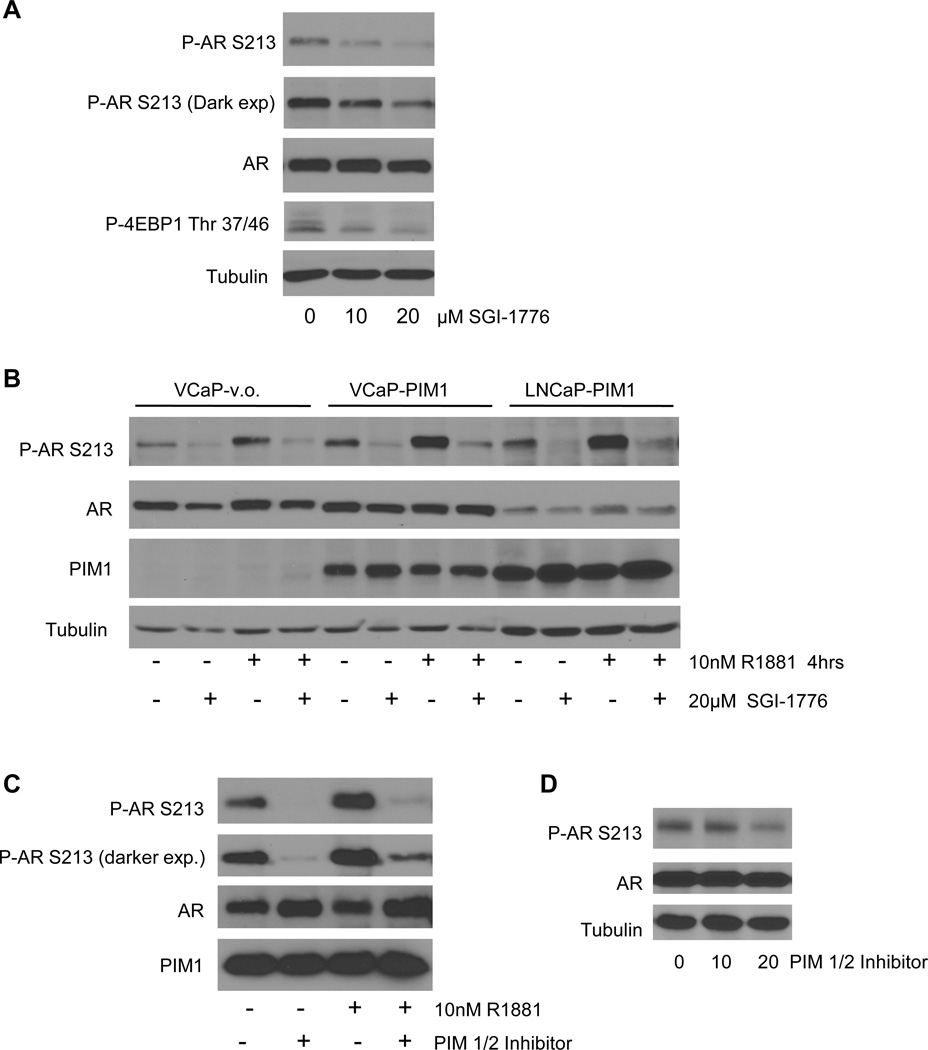
To determine if inhibition of PIM1 affected phosphorylation of AR S213, VCaP cells were treated with 10nM R1881 in the presence of a PIM kinase inhibitor, SGI-1776. Treatment with the inhibitor reduced phosphorylation of S213 and P-4EBP1 Thr37/46, an established substrate of PIM1 (21, 26–27), in a dose dependent manner (Figure 5A). SGI-1176 also significantly reduced the level of P-AR S213 in VCaP and LNCaP cells overexpressing PIM1 (Figure 5B), indicating that PIM1 inhibition of both endogenous and upregulated levels of PIM1 result in decreased expression of P-AR S213. In addition, a structurally different PIM1 inhibitor, PIM1/2 inhibitor V, reduced endogenous phosphorylation of S213 in VCaP cells (Figure 5D). PIM1/2 inhibitor V was able to dramatically inhibit S213 phosphorylation in in vitro kinase reactions where immunopurified AR was incubated with GST-PIM1 in the presence or absence of PIM 1/2 inhibitor V (Figure 5C). The robust phosphorylation of AR by GST-PIM1 was reduced in the presence of inhibitor, independent of the presence of hormone.
Figure 5. PIM1/2 Inhibition Decreases P-AR S213.
A) VCaP cells were steroid starved overnight then treated with 10nM R1881 and PIM kinase inhibitor SGI-1776 as indicated for 4 hours. Total protein lysates were analyzed by Western blot with antibodies against P-AR S213, AR (total), P-4EBP1 Thr 37/46, and tubulin. B) VCaP and LNCaP cells were transiently infected with either vector only or PIM1. Cells were steroid starved overnight and then treated with vehicle or 10nM R1881 in the presence of absence of 20µM SGI-1776 for 4 hours. Protein lysates were subjected to Western blot analysis using antibodies against P-AR S213, AR, PIM1, and tubulin. C) 293 cells were transiently transfected with FLAG-AR. Cells were steroid starved overnight and then treated with vehicle or 10nM R1881 for 2 hours. FLAG-AR was immunopurified and incubated with recombinant PIM1 in the presence and absence of PIM1/2 inhibitor V. Kinase reactions were subjected to Western blot analysis using antibodies against P-AR S213, AR, and PIM1. D) VCaP cells were steroid starved and then treated with 10 nM R1881 and PIM1/2 inhibitor V for 2 hours.
PIM1 Expression Affects AR Mediated Transcription
The impact of PIM1 phosphorylation of AR S213 on AR function is not known. To evaluate the effect of PIM1 phosphorylation of AR on gene transcription we conducted luciferase assays using the AR reporter gene ARR3-luciferase (28) in the presence of either WT AR or AR S213A. As shown in Figure 6A, co-expression of WT AR and PIM1 repressed AR mediated transcription in a dose dependent manner. In contrast, PIM1 co-expression with AR S213A had no effect on the same reporter gene (Figure 6B). The effect of PIM1 overexpression on AR WT and AR S213A transcriptional activity is not due to AR protein levels, as AR levels are equivalent or increased slightly in the presence of PIM1 and 24 hours exposure to R1881 (Figure 6A and 6B, insets). In support of these studies, examination of increased PIM1 expression in VCaP and LNCaP cells with endogenous levels of AR showed diminished luciferase reporter gene activity in the presence of PIM1 (Figure 6C and D). In LNCaP cells stably expressing vector only or PIM1, we next examined androgen mediated transcription of endogenous target genes PSA and Nkx3.1. Androgen treatment increased mRNA transcript levels of PSA and Nkx3.1 in the vector only cells as expected. However, in cells overexpressing PIM1, levels of PSA and Nkx3.1 mRNA were reduced 1.9 to 3.7 fold (Figure 6E). Repression of androgen mediated transcription of PSA and Nkx3.1 was not due to decreased levels of AR protein. Figure 6F shows that AR protein levels were equivalent between vector only and PIM1 expressing cells and as expected AR levels were increased in the presence of androgen. Thus, PIM1 appears to decrease androgen mediated transcription of PSA and Nkx3.1, genes expressed by differentiated prostate epithelial cells.
Figure 6. Regulation of AR mediated Transcription by PIM1.
A and B) 293 cells were transiently transfected with AR WT or AR mutant S213A, lacZ reporter, ARR3-Luciferase reporter, and increasing amounts of PIM1 in the presence and absence of 10nM R1881. Insets show AR and PIM1 protein levels for A and B. Data was normalized to 10 nM R1881 in the absence of PIM1 and set to 100% activity. C) VCaP cells were transfected with lacZ reporter, ARR3-Luciferase reporter, and increasing amounts of PIM1 in the presence and absence of 10nM R1881. D) LNCaP cells were transfected as above in the presence of 10nM R1881. E and F LNCaP cells stably expressing vector only (vo), or PIM1 (pools#1 and #2) were steroid starved overnight and then treated in the presence and absence of 10nM R1881 for 24 hours. qPCR was performed to quantify transcript levels of PSA and Nkx3.1 (E) and AR and PIM1 protein expression was determined (F). Graphs are representative of at least three independent experiments. Error was calculated using the standard deviation.
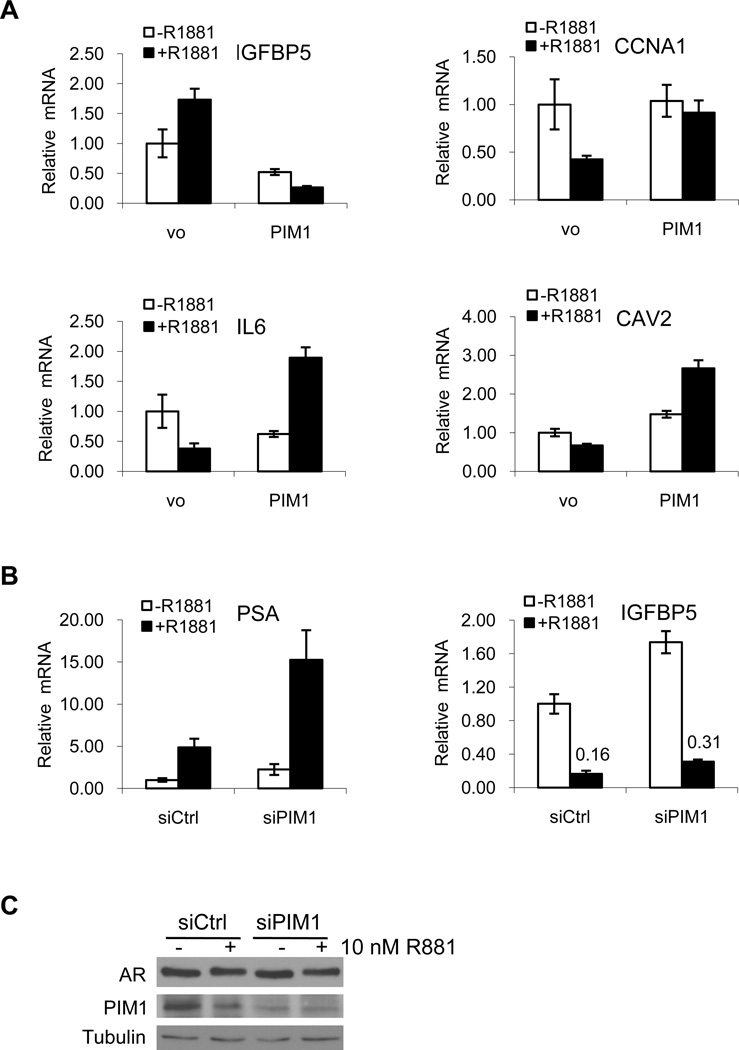
We also examined the impact of PIM1 expression on genes selected from the RT2 Profiler PCR Array, representing 84 genes regulated in prostate cancer. Of the 84 genes, 13 were upregulated greater than 2 fold and 7 were downregulated greater than 1.5 fold when comparing PIM1 and vector only in the presence of R1881 (data not shown). IL6, cyclin A1 (CCNA1), caveolin 2 (CAV2), and IGFBP5 were highly up- or downregulated and were chosen for validation based on androgen regulated expression, effect of PIM1 on transcription, and relevance to prostate cancer. Androgen regulated expression of IGFBP5, CCNA1, IL6, and CAV2 was observed (vo controls (Figure 7A)). In the presence of PIM1 and R1881, IL6, cyclin A1, and caveolin 2 were upregulated 5, 2.2, and 4 fold, respectively while IGFBP5 was downregulated almost 7 fold (Figure 7A, compare black bars). In addition, siRNA knockdown of PIM1 in VCaP cells resulted in increased PSA and IGFBP5 mRNA compared to non-silencing controls (Figure 7B, compare black bars). Reduced protein expression of PIM1 in siRNA treated cells and equivalent AR levels are shown in Figure 7C.
Figure 7. PIM1 Affects AR Target Gene Expression.
A) LNCaP stable cell lines overexpressing vector only (vo pool #1) or PIM1 (pool #2) were treated as in Figure 6E and the gene expression of IGFBP5, CCNA1, IL6, and CAV2 was quantified by qPCR. B) VCaP cells were transfected with siRNA against PIM1 and treated as above. Gene expression of PSA and IGFBP5 was quantified by qPCR and protein expression of AR and PIM1 determined C. Graphs are representative of at least three independent experiments. Error was calculated using the standard deviation.
AR S213 and PIM1 Expression in Human Prostate Cancer
To investigate the possible clinical relevance of AR S213 phosphorylation, we examined the expression pattern of antigen detected by antibody against P-AR S213 compared to antigen detected by antibody that recognizes both phosphorylated and non-phosphoryated AR (total AR). The studies were conducted on two different sets of tissue microarrays (TMAs) obtained from the New York University (NYU) Prostate Cancer Biorepository Network and Memorial Sloan Kettering Cancer Center (MSKCC). Disease outcome data was available for both tissue microarrays. Immunohistochemistry was performed on both TMAs examining the expression of P-ARS213 (Figure 8A, panels A–C) and AR (not shown). The NYU TMA was also analyzed for PIM1 expression (Figure 8A, panels D–F). Histoscore (H-score) analysis of the TMAs indicates that nuclear P-AR S213 expression was significantly increased in hormone refractory cancers versus non-hormone refractory cancers (H-score of 64.28 ± 55.59 vs 13.41 ± 35.80, p<0.0001) (Figure 8B). In addition, expression of nuclear P-AR S213 was increased in cases with high Gleason scores (Gleason 8–10) vs low Gleason scores (Gleason 5–7) (H-score 43.07 ± 54.27 vs 0.90 ± 3.51, p<0.001) (Figure 8B). Expression of nuclear P-AR S213 and PIM1 was much more prevalent in hormone refractory cancer (Figure 8A, panels A–B and panels D–E) compared to non-hormone refractory cancer (Figures 8A, panels C and F). PIM1 was expressed in 50% (32/64) of all cases, of which 14 were hormone refractory. 88.9% (16/18) of hormone refractory cancers were positive for P-AR S213 and 72.2% (12/18) of hormone refractory cases were positive for P-AR S213 and PIM1.
Figure 8. Expression of Phosphorylated AR S213 in Hormone Refractory Prostate Cancers.
A) Immunohistochemistry on prostate tissues using antibodies against P-AR S213 (A–C) or PIM1 (D–F). For P-AR S213, a separate histoscore was given for nuclear and cytoplasmic staining positivity while PIM1 was present only in the nucleus. The total histoscore ranges from 0 for a completely negative sample to a maximum score of 300. Magnification is 10X with insets of the same tissue at a 40X magnification. B) Histoscores (H-Scores) of hormone refractory (HR) vs non- hormone refractory prostate cancers and Gleason score 5–7 vs Gleason 8–10 were compared. The distribution of H-Scores is shown with the mean (horizontal bar). The differences between the means were determined to be statistically significant by t-test.
In the MSKCC TMA, nuclear P-AR S213 expression was detected in a wide range of cases. A far greater proportion of biochemically recurrent cancers have an H-score >100 (62.5% recurrent vs 31.4% non-recurrent). If the H-score is dichotomized to 125, 40.6% of P-AR S213 positive cases were recurrent vs 17.1% non-recurrent. While a high P-AR S213 H-score was twice as likely to be from recurrent cancer, the H-scores of recurrent vs non-recurrent cancer did not reach statistical significance. Overall, P-AR S213 expression did not correlate to clinical parameters available with the MSKCC TMA, including PSA levels, Gleason score, stage, recurrence, disease free interval or capsular penetration.
While the results from the microarrays have a similar trend, differences may be ascribed to the tissue type represented on the TMAs. The NYU TMA consists of tissue from clinically advanced prostate cancer obtained from transurethral resection of the prostate (TURP). Samples designated as hormone refractory were from patients that had failed hormone ablation therapies. The MSKCC TMA consists of tissue from patients that have undergone radical prostatectomy. Recurrent and non-recurrent cases were determined by biochemical recurrence defined as three consecutive rises in PSA levels, as opposed to cancer progression after hormone ablation.
Interestingly, a small number of cases had cytoplasmic staining in addition to nuclear staining of P-AR S213 (21.5% NYU; 14.9% MSKCC). Positive P-AR S213 staining in the cytoplasm was associated with a shorter disease-free interval (30 months vs 55 months). All cases of cytoplasmic P-AR S213 in the NYU TMA were Gleason 9 and 10.
Discussion
The AR possesses consensus sequences for the kinases Akt (RXRXXS/T) and PIM1 (RXRHXS) that are highly similar, suggesting that either kinase may phosphorylate AR depending on the cellular context. Our studies indicate that in prostate epithelial cells PIM1 robustly phosphorylates AR at S213 and AR is likely a direct substrate of PIM1. While overexpression of Akt does result in slightly increased AR S213 phosphorylation in 293 cells (Figure 1) and VCaP cells (29), this more likely reflects the increase in AR protein levels in the presence of Akt. However, it is possible that the kinase activity of the myristoylated, constitutively active Akt construct used was not robust enough to elicit phosphorylation of AR S213 and that more than one kinase can phosphorylate AR S213 depending on the cellular context.
Our studies indicate that AR is phosphorylated at S213 by endogenous PIM1 in VCaP cells. This is in contrast to LNCaP cells, where very little phosphorylation is observed in the absence of PIM1 overexpression. Since levels of PIM1 appear similar between all the cell lines, this may reflect higher catalytic activity of PIM1 in VCaP cells than LNCaP cells. Alternatively, it may simply be easier to detect S213 phosphorylation in VCaP cells since AR levels are much higher than in LNCaP cells (Figure 4A).
We observe that overexpression of PIM1 results in decreased expression of an androgen-responsive reporter gene; consistent with a previous study that showed repression of a probasin-luciferase reporter gene (30). However, PIM1 expression appears to have differing affects on endogenous AR target gene transcription increasing proliferative and decreasing differentiation signals. PIM1 overexpressing cells exhibit decreased androgen mediated transcription of PSA, Nkx3.1, and IGFBP5 compared to vector only containing cells. Repression or loss of the tumor suppressor, Nkx3.1, has been reported as an early event in PTEN models of prostate cancer and correlates with cancer progression (31–32). Nkx3.1 and PSA are also markers of differentiated prostate epithelium (reviewed in (33)) suggesting that PIM1 overexpression may cause de-differentiation of prostate tissue. IGFBP5 has also been shown to be downregulated in metastases (34–35). In addition to repressing some AR target genes, we also show that PIM1 is involved in activation of AR target genes important in cell proliferation and signaling such as cyclin A1, IL6, and caveolin 2. Cyclin A1 has been shown to be involved in prostate cancer invasion and metastasis (36–37). IL6 has been implicated in androgen independent activation of AR (38), is elevated in the sera of patients with advanced disease (39–41), and may be involved in intracrine production of androgens (42). CAV2 expression was demonstrated to be increased in poorly differentiated prostate tissues (43). We speculate that activation of PIM1 in prostate cancer cells may repress genes involved in tumor suppression and differentiation and activate genes important in cellular transformation.
While we have not demonstrated the mechanism by which PIM1 phosphorylation of AR S213 changes AR target gene expression, we speculate that AR phosphorylation likely alters interaction with chromatin, as recently demonstrated for AR S81 (44), as well as interaction with cofactors resulting in differential target gene expression as demonstrated for phosphorylated glucocorticoid receptor interaction with Mediator (MED) 14 (45). Understanding the impact of AR S213 phosphorylation by PIM1 is also complicated by the fact that PIM1 is likely to have AR independent effects on chromatin since PIM1 phosphorylates HP1γ (19) and histone H3 (46). These findings are consistent with the fact that our data demonstrates synergistic effects of PIM1 overexpression with hormone treatment.
Hormone refractory prostate cancer still relies on the AR, but the impact on AR-mediated gene transcription is pleiotropic during cancer progression. AR pathways associated with proliferation, de-differentiation, and decreased apoptosis are upregulated while AR pathways that inhibit proliferation, induce differentiation, or promote apoptosis are downregulated. Gene profiling of normal prostatic epithelium, low to high grade prostate cancers, and metastatic prostate cancers revealed that androgen receptor signaling and certain androgen receptor pathways that inhibit proliferation, increase apoptosis, or induce differentiation are downregulated in high grade prostate cancers and metastatic disease (47–48). Consistent with PSA mRNA downregulation in metastases (48), a recent study demonstrated that the proportion of tumor cells expressing low amounts of PSA increase in high grade cancer and hormone refractory cancers (49). Thus, although the finding that PSA is repressed in the presence of PIM1 appears to be paradoxical considering that PSA is associated with tumor growth, studies have shown that the utility of PSA as a surrogate marker for disease progression decreases as tumors become more de-differentiated in high grade and hormone refractory tumors (49–50). It is possible that P-AR S213 expression marks cells that express low amounts of PSA and are resistant to androgen ablative treatments.
While PIM1 overexpression in prostate cancer (12–13, 25) is detectable by immunohistochemistry, there is no easy method to determine when PIM1 is active in vivo since its activation does not correlate with phosphorylation detectable by an antibody. While not predictive of disease outcome, detection of AR phosphorylated at S213 is much more prevalent in aggressive, hormone refractory prostate cancer (Figure 8). Our studies indicate that AR is a direct target of PIM1 and as such, detection of P-AR S213 can serve as an in vivo marker for activated PIM1. This may be important to stratify patient populations who might benefit from small molecule PIM1 inhibitors in development. In fact, a PIM1 inhibitor was in a trial for lymphoma and prostate cancer (see clinical trials.gov Identifier NCT00848601, Safety of SGI-1776, A PIM kinase inhibitor to treat docetaxol refractory prostate cancer and relapsed/refractory non Hodgkin’s lymphoma). There is also some evidence that PIM1 inhibitors may enhance chemotherapy because treatment of prostate cancer cells with PIM1 inhibitors enhances sensitivity to taxane-based chemotherapy (51). Thus our studies indicate that AR is a PIM1 substrate and phosphorylated AR may serve as a biomarker for activated PIM1.
Materials and Methods
Cell Culture and Reagents
LNCaP, VCaP, and HEK-293 cell lines were obtained and cultured as recommended by the ATCC (Manassas, VA) LNCaP cells used in this study were from passages 23–30. Androgen independent LNCaP-abl cells (gift from Z. Culig) were maintained in RPMI-1640 supplemented with 10% charcoal stripped serum (cFBS) (52). R1881 (methyltrienolone) was purchased from Perkin Elmer (Waltham, MA) and reconstituted in ethanol. PIM kinase inhibitors SGI-1776 and PIM1/2 kinase inhibitor V were purchased from Selleck Chemicals (Houston, TX) and EMD Chemicals (Gibbstown, NJ), respectively. Both inhibitors were reconstituted in DMSO.
PIM1 and mutant PIM1 K67M cDNA was purchased from Origene Technologies (Rockville, MD). The open reading frame (ORF) for the shorter, isoform 2 of PIM1 and mutant PIM1 K67M was subcloned into the EcoRI/ XbaI sites of pcDNA3.1(+) with a N-terminal FLAG tag and into the HpaI site of the pLB(N)CX retroviral vector (neomycin resistance cassette replaced with blasticidin resistance; gift from Greg David, NYU School of Medicine).
LNCaP and VCaP stable cell lines expressing vector only or PIM1 were generated after retroviral infection with the above construct and selection in blasticidin (Invitrogen; Carlsbad, CA). Pools were created and screened for expression of PIM1 via Western blot.
Transient Transfections and Infections
HEK-293 cells were transiently transfected with pcDNA3-HA-AR WT or pcDNA3-HA-AR S213A and vector only, pCMV6-myr-Akt1-HA pcDNA3.1(+)-FLAG-PIM1 WT or pcDNA3.1(+)-FLAG-PIM1 K67M using Lipofectamine (Invitrogen) according to the manufacturer’s recommendations. VCaP cells were transfected with siRNA directed against PIM1 (siGENOME SMARTpool; Dharmacon, Lafayette, CO), steroid starved and treated with R1881 for 24 hours.
Retroviral particles were produced in the 293T/17 cell line (ATCC). LNCaP, LNCaP-abl, and VCaP cell lines were infected on two consecutive days with vector only or PIM1 retroviral particles and polybrene. After the 2nd infection, cells were steroid starved overnight and then treated with R1881.
Protein Extraction and Western blot Analysis
Protein extraction and Western blot analysis was performed as previously described (29). The following antibodies were used: phospho-AR S213 (5), AR (441), PIM1 (12H8), ERK-1 (K-23) (Santa Cruz Biotechnology; Santa Cruz, CA); phospho-Akt S473, phospho-Bad S112, Bad (total), phospho-GSK3 α/β S21/9, phospho-4EBP1 Thr 37/46 (Cell Signaling Technology; Danvers, MA); Tubulin, HA-epitope tag (Covance; Denver, PA); and PIM1 (Abcam; Cambridge, MA).
Lambda Phosphatase
HEK-293 cells were transiently transfected as described above with pcDNA3-HA-AR WT and vector only or pcDNA3.1(+)-Flag-PIM1) and assayed as previously described (53).
Kinase Reactions
pcDNA3-FLAG-AR WT was transiently transfected into HEK-293 cells as described above. Cells were lysed in 1X Cell Lysis Buffer (Cell Signaling Technology) and then incubated with Anti-FLAG M2 Affinity Gel (Sigma-Aldrich; St. Louis, MO). Immunoprecipitates were extensively washed in lysis buffer and eluted from the agarose beads with 3X FLAG peptide (Sigma-Aldrich). Immunopurified FLAG-AR was incubated with 1 µg of recombinant protein GST-GSK3 (Cell Signaling), GST-PIM1, active (Millipore; Billerica, MA), or His-Akt, active (Millipore) in kinase buffer (Cell Signaling) supplemented with 0.2mM ATP. The reaction proceeded for 30 minutes at 30°C. In vitro kinase reactions were conducted in Assay Dilution Buffer I and Magnesium/ATP cocktail (Millipore). 3µg of His-Bad (Millipore) or GST-GSK3 and 250ng of His-Akt or GST-PIM1 kinases were incubated for 10 minutes at 30°C. 1µg of full length His-Androgen Receptor (Abcam) and 250ng GST-PIM1 were incubated for 30 minutes at 30°C in the presence of 10nM R1881. Inhibition of phosphorylated AR in the presence of PIM kinase inhibitor was performed as previously described (27) with some modifications. Immunopurified FLAG-AR and 1µg of GST-PIM1 kinase reactions proceeded in the presence and absence of 100µM Pim kinase 1/2 inhibitor V for 30 minutes at 30°C. Kinase reactions were terminated with the addition of 3X SDS sample buffer.
Sphere Formation
VCaP stables lines were plated at a low density on ultra-low attachment plates (Corning, Corning, NY) and cultured the absence of serum and hormone as previously described (54), with the addition of N2 (Invitrogen).
Quantitative RT-PCR
Total RNA was extracted and quantitative RT-PCR was performed as previously described for PSA, Nkx 3.1, and RPL19 (55) with the following modifications. RNA was reverse transcribed using First Strand cDNA Synthesis for Real-Time PCR (USB; Cleveland, OH) and quantitative PCR (qPCR) was performed with HotStart-IT SYBR green qPCR Master Mix (2X), (USB). The RT2 Profiler PCR Array (PAHS-135) and primers for CCNA1, IL6, and CAV2 were purchased from SABiosciences (Valencia, CA). The primers sequences used to quantify IGFBP5 were F 5’-ATTGTGACCGCAAAGGATTC and R 5’-AGGTGTGGCACTGAAAGTCC.
Human Prostate Cancer Tissue Microarrays, Immunohistochemistry, and Scoring
Human prostate cancer tissue microarrays were obtained from the New York University (NYU) Prostate Cancer Biorepository Network and Memorial Sloan-Kettering Cancer Center. Tissues were acquired according to each institution’s institutional review board’s policies. The NYU tissue microarray contains 65 cases represented as 4 cores per case. The samples are designated as being either hormone refractory (n=18) or non-hormone refractory (n=47), which includes hormone naïve (n=18) samples. The samples range from Gleason score 5 through 10. The Memorial Sloan Kettering Cancer Center (MSKCC) tissue microarray contains 68 cases represented as 3 cores per case. The cases are designated as being either biochemically recurrent (n=33) or non-recurrent (n=35). The samples range from Gleason score 4 through 9. Other clinical parameters include PSA levels, Gleason score, stage, recurrence, disease free interval or capsular penetration.
Immunohistochemistry and image acquisition was conducted as previously described (29) on the NYU and MSKCC tissue microarrays for P-AR S213. In addition, the NYU microarray was immunostained for PIM1 (Abcam). Protein expression in each core was assessed using the semi-quantitative weighted histoscore method, representing staining intensity (negative (0), weak (1), moderate (2), and strong (3) and the percentage of positive cells within each intensity category. The final histoscore was determined for each case by taking the average histoscore of all the present cores. Both TMAs were scored by two independent observers. Statistical significance (p<0.05) was determined by t-test.
Acknowledgments
We acknowledge funding from the New York University School of Medicine Department of Urology at New York University School of Medicine.
This work was supported by NIH R01CA112226 (S.L.), funds from the NYU Department of Urology, DOD grant PC094786 for the Prostate Cancer Biorepository Network (PCBN, J.M), DOD grant PC080010 (PL) and NIH UO1 1U01CA149556-01(PL).
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Ward RD, Weigel NL. Steroid receptor phosphorylation: Assigning function to site-specific phosphorylation. Biofactors. 2009 Nov-Dec;35(6):528–536. doi: 10.1002/biof.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. Embo. J. 2002 Aug 1;21(15):4037–4048. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin HK, Yeh S, Kang HY, Chang C. Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7200–7205. doi: 10.1073/pnas.121173298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen Y, Hu MC, Makino K, Spohn B, Bartholomeusz G, Yan DH, et al. HER-2/neu promotes androgen-independent survival and growth of prostate cancer cells through the Akt pathway. Cancer Res. 2000 Dec 15;60(24):6841–6845. [PubMed] [Google Scholar]
- 5.Taneja SS, Ha S, Swenson NK, Huang HY, Lee P, Melamed J, et al. Cell-specific regulation of androgen receptor phosphorylation in vivo. J Biol Chem. 2005 Dec 9;280(49):40916–40924. doi: 10.1074/jbc.M508442200. [DOI] [PubMed] [Google Scholar]
- 6.Saris CJ, Domen J, Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 1991 Mar;10(3):655–664. doi: 10.1002/j.1460-2075.1991.tb07994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Y, Xu K, Dai B, Guo Z, Jiang T, Chen H, et al. The 44 kDa Pim-1 kinase directly interacts with tyrosine kinase Etk/BMX and protects human prostate cancer cells from apoptosis induced by chemotherapeutic drugs. Oncogene. 2006 Jan 5;25(1):70–78. doi: 10.1038/sj.onc.1209058. [DOI] [PubMed] [Google Scholar]
- 8.Xie Y, Xu K, Linn DE, Yang X, Guo Z, Shimelis H, et al. The 44-kDa Pim-1 kinase phosphorylates BCRP/ABCG2 and thereby promotes its multimerization and drug-resistant activity in human prostate cancer cells. J Biol Chem. 2008 Feb 8;283(6):3349–3356. doi: 10.1074/jbc.M707773200. [DOI] [PubMed] [Google Scholar]
- 9.Mochizuki T, Kitanaka C, Noguchi K, Muramatsu T, Asai A, Kuchino Y. Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J Biol Chem. 1999 Jun 25;274(26):18659–18666. doi: 10.1074/jbc.274.26.18659. [DOI] [PubMed] [Google Scholar]
- 10.Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008 Jul 1;68(13):5076–5085. doi: 10.1158/0008-5472.CAN-08-0634. [DOI] [PubMed] [Google Scholar]
- 11.Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004 Jul 30;571(1–3):43–49. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 12.Cibull TL, Jones TD, Li L, Eble JN, Ann Baldridge L, Malott SR, et al. Overexpression of Pim-1 during progression of prostatic adenocarcinoma. J Clin Pathol. 2006 Mar;59(3):285–288. doi: 10.1136/jcp.2005.027672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valdman A, Fang X, Pang ST, Ekman P, Egevad L. Pim-1 expression in prostatic intraepithelial neoplasia and human prostate cancer. Prostate. 2004 Sep 1;60(4):367–371. doi: 10.1002/pros.20064. [DOI] [PubMed] [Google Scholar]
- 14.van Lohuizen M, Verbeek S, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989 Feb 24;56(4):673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 15.Verbeek S, van Lohuizen M, van der Valk M, Domen J, Kraal G, Berns A. Mice bearing the E mu-myc and E mu-pim-1 transgenes develop pre-B-cell leukemia prenatally. Mol Cell Biol. 1991 Feb;11(2):1176–1179. doi: 10.1128/mcb.11.2.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003 Sep;4(3):223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Kim J, Roh M, Franco OE, Hayward SW, Wills ML, et al. Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene. 2010 Apr 29;29(17):2477–2487. doi: 10.1038/onc.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Z, Flesken-Nikitin A, Corney DC, Wang W, Goodrich DW, Roy-Burman P, et al. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 2006 Aug 15;66(16):7889–7898. doi: 10.1158/0008-5472.CAN-06-0486. [DOI] [PubMed] [Google Scholar]
- 19.Koike N, Maita H, Taira T, Ariga H, Iguchi-Ariga SM. Identification of heterochromatin protein 1 (HP1) as a phosphorylation target by Pim-1 kinase and the effect of phosphorylation on the transcriptional repression function of HP1(1) FEBS Lett. 2000 Feb 4;467(1):17–21. doi: 10.1016/s0014-5793(00)01105-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Bhattacharya N, Mixter PF, Wei W, Sedivy J, Magnuson NS. Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochim Biophys Acta. 2002 Dec 16;1593(1):45–55. doi: 10.1016/s0167-4889(02)00347-6. [DOI] [PubMed] [Google Scholar]
- 21.Chen WW, Chan DC, Donald C, Lilly MB, Kraft AS. Pim family kinases enhance tumor growth of prostate cancer cells. Mol Cancer Res. 2005 Aug;3(8):443–451. doi: 10.1158/1541-7786.MCR-05-0007. [DOI] [PubMed] [Google Scholar]
- 22.Cuypers HT, Selten G, Quint W, Zijlstra M, Maandag ER, Boelens W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984 May;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 23.Ha S, Ruoff R, Kahoud N, Franke TF, Logan SK. Androgen receptor levels are upregulated by Akt in prostate cancer. Endocrine-Related Cancer. 2011 Apr 1;18(2):245–255. doi: 10.1530/ERC-10-0204. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xin L, Teitell MA, Lawson DA, Kwon A, Mellinghoff IK, Witte ON. Progression of prostate cancer by synergy of AKT with genotropic and nongenotropic actions of the androgen receptor. Proc Natl Acad Sci U S A. 2006 May 16;103(20):7789–7794. doi: 10.1073/pnas.0602567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Roh M, Abdulkadir SA. Pim1 promotes human prostate cancer cell tumorigenicity and c-MYC transcriptional activity. BMC Cancer. 2010;10:248. doi: 10.1186/1471-2407-10-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beharry Z, Mahajan S, Zemskova M, Lin Y-W, Tholanikunnel BG, Xia Z, et al. The Pim protein kinases regulate energy metabolism and cell growth. Proceedings of the National Academy of Sciences. 2011 Jan 11;108(2):528–533. doi: 10.1073/pnas.1013214108. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia Z, Knaak C, Ma J, Beharry ZM, McInnes C, Wang W, et al. Synthesis and evaluation of novel inhibitors of Pim-1 and Pim-2 protein kinases. J Med Chem. 2009 Jan 8;52(1):74–86. doi: 10.1021/jm800937p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rennie PS, Bruchovsky N, Leco KJ, Sheppard PC, McQueen SA, Cheng H, et al. Characterization of two cis-acting DNA elements involved in the androgen regulation of the probasin gene. Molecular Endocrinology. 1993 Jan 1;7(1):23–36. doi: 10.1210/mend.7.1.8446105. 1993. [DOI] [PubMed] [Google Scholar]
- 29.Ha S, Ruoff R, Kahoud N, Logan SK, Franke TF. Androgen receptor levels are upregulated by Akt in prostate cancer. Endocr Relat Cancer. 2011 Feb 11; doi: 10.1530/ERC-10-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson J, Peltola KJ, Koskinen PJ, Janne OA, Palvimo JJ. Attenuation of androgen receptor-dependent transcription by the serine/threonine kinase Pim-1. Lab Invest. 2003 Sep;83(9):1301–1309. doi: 10.1097/01.lab.0000087585.03162.a3. [DOI] [PubMed] [Google Scholar]
- 31.Song H, Zhang B, Watson MA, Humphrey PA, Lim H, Milbrandt J. Loss of Nkx3.1 leads to the activation of discrete downstream target genes during prostate tumorigenesis. Oncogene. 2009;28(37):3307–3319. doi: 10.1038/onc.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowen C, Bubendorf L, Voeller HJ, Slack R, Willi N, Sauter G, et al. Loss of NKX3.1 Expression in Human Prostate Cancers Correlates with Tumor Progression1,2. Cancer Research. 2000 Nov 11;60(21):6111–6115. 2000. [PubMed] [Google Scholar]
- 33.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes & Development. 2010 Sep 15;24(18):1967–2000. doi: 10.1101/gad.1965810. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001 Aug 23;412(6849):822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 35.Su Y, Wagner ER, Luo Q, Huang J, Chen L, He BC, et al. Insulin-like growth factor binding protein 5 suppresses tumor growth and metastasis of human osteosarcoma. Oncogene. 2011 Sep 15;30(37):3907–3917. doi: 10.1038/onc.2011.97. [DOI] [PubMed] [Google Scholar]
- 36.Wegiel B, Bjartell A, Ekberg J, Gadaleanu V, Brunhoff C, Persson JL. A role for cyclin A1 in mediating the autocrine expression of vascular endothelial growth factor in prostate cancer. Oncogene. 2005;24(42):6385–6393. doi: 10.1038/sj.onc.1208795. [DOI] [PubMed] [Google Scholar]
- 37.Wegiel B, Bjartell A, Tuomela J, Dizeyi N, Tinzl M, Helczynski L, et al. Multiple cellular mechanisms related to cyclin A1 in prostate cancer invasion and metastasis. J Natl Cancer Inst. 2008 Jul 16;100(14):1022–1036. doi: 10.1093/jnci/djn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim O, Jiang T, Xie Y, Guo Z, Chen H, Qiu Y. Synergism of cytoplasmic kinases in IL6-induced ligand-independent activation of androgen receptor in prostate cancer cells. Oncogene. 2004 Mar 11;23(10):1838–1844. doi: 10.1038/sj.onc.1207304. [DOI] [PubMed] [Google Scholar]
- 39.Twillie DA, Eisenberger MA, Carducci MA, Hseih W-S, Kim WY, Simons JW. Interleukin-6: A candidate mediator of human prostate cancer morbidity. Urology. 1995;45(3):542–549. doi: 10.1016/S0090-4295(99)80034-X. [DOI] [PubMed] [Google Scholar]
- 40.Michalaki V, Syrigos K, Charles P, Waxman J. Serum levels of IL-6 and TNF-[alpha] correlate with clinicopathological features and patient survival in patients with prostate cancer. Br J Cancer. 2004;90(12):2312–2316. doi: 10.1038/sj.bjc.6601814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drachenberg DE, Elgamal A-AA, Rowbotham R, Peterson M, Murphy GP. Circulating levels of interleukin-6 in patients with hormone refractory prostate cancer. The Prostate. 1999;41(2):127–133. doi: 10.1002/(sici)1097-0045(19991001)41:2<127::aid-pros7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 42.Chun JY, Nadiminty N, Dutt S, Lou W, Yang JC, Kung H-J, et al. Interleukin-6 Regulates Androgen Synthesis in Prostate Cancer Cells. Clinical Cancer Research. 2009 2009 Aug 1;15(15):4815–4822. doi: 10.1158/1078-0432.CCR-09-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gould ML, Williams G, Nicholson HD. Changes in caveolae, caveolin, and polymerase 1 and transcript release factor (PTRF) expression in prostate cancer progression. Prostate. 2010 Nov 1;70(15):1609–1621. doi: 10.1002/pros.21195. [DOI] [PubMed] [Google Scholar]
- 44.Chen S, Gulla S, Cai C, Balk SP. Androgen receptor serine 81 phosphorylation mediates chromatin binding and transcriptional activation. J Biol Chem. 2012 Mar 9;287(11):8571–8583. doi: 10.1074/jbc.M111.325290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen W, Dang T, Blind RD, Wang Z, Cavasotto CN, Hittelman AB, et al. Glucocorticoid receptor phosphorylation differentially affects target gene expression. Mol Endocrinol. 2008 Aug;22(8):1754–1766. doi: 10.1210/me.2007-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007 Aug;9(8):932–944. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
- 47.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39(1):41–51. doi: 10.1038/ng1935. [10.1038/ng1935]. [DOI] [PubMed] [Google Scholar]
- 48.Hendriksen PJ, Dits NF, Kokame K, Veldhoven A, van Weerden WM, Bangma CH, et al. Evolution of the androgen receptor pathway during progression of prostate cancer. Cancer Res. 2006 May 15;66(10):5012–5020. doi: 10.1158/0008-5472.CAN-05-3082. [DOI] [PubMed] [Google Scholar]
- 49.Qin J, Liu X, Laffin B, Chen X, Choy G, Jeter CR, et al. The PSA(-/lo) Prostate Cancer Cell Population Harbors Self-Renewing Long-Term Tumor-Propagating Cells that Resist Castration. Cell Stem Cell. 2012 May 4;10(5):556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Payne H, Cornford P. Prostate-specific antigen: An evolving role in diagnosis, monitoring, and treatment evaluation in prostate cancer. Urologic Oncology: Seminars and Original Investigations. 2011;29(6):593–601. doi: 10.1016/j.urolonc.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Mumenthaler SM, Ng PY, Hodge A, Bearss D, Berk G, Kanekal S, et al. Pharmacologic inhibition of Pim kinases alters prostate cancer cell growth and resensitizes chemoresistant cells to taxanes. Mol Cancer Ther. 2009 Oct;8(10):2882–2893. doi: 10.1158/1535-7163.MCT-09-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Culig Z, Hoffmann J, Erdel M, Eder IE, Hobisch A, Hittmair A, et al. Switch from antagonist to agonist of the androgen receptor bicalutamide is associated with prostate tumour progression in a new model system. Br J Cancer. 1999 Sep;81(2):242–251. doi: 10.1038/sj.bjc.6690684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M, et al. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003 Nov 14;302(5648):1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- 54.Garraway IP, Sun W, Tran CP, Perner S, Zhang B, Goldstein AS, et al. Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. The Prostate. 2010;70(5):491–501. doi: 10.1002/pros.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nwachukwu JC, Mita P, Ruoff R, Ha S, Wang Q, Huang SJ, et al. Genome-wide impact of androgen receptor trapped clone-27 loss on androgen-regulated transcription in prostate cancer cells. Cancer Res. 2009 Apr 1;69(7):3140–3147. doi: 10.1158/0008-5472.CAN-08-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]