Abstract
Since protein/protein interactions usually trigger signaling processes, inhibitors of those interactions must preclude protein binding without eliciting the signaling process themselves. To accomplish those goals, small molecules need to target those protein residues that contribute the most to binding (binding hotspots) without disturbing those residues that initiate signaling processes (allosteric hotspots). The availability of a blueprint identifying binding and allosteric hotspots will significantly aid inhibitor design and optimization. In this paper, we show that in some situations the blueprint can be constructed by combining the standard technique of alanine scanning mutagenesis with isothermal titration calorimetry (ITC). We demonstrate the approach by developing the combined binding and allosteric hotspots blueprint for CD4/gp120, the initial interaction leading to HIV-1 cell infection. A major finding of these studies is that not all binding hotspots are allosteric hotspots opening the possibility for the rational design of inhibitors and antagonist or agonist modulators.
Keywords: Binding Affinity, Enthalpy, Entropy, Thermodynamic Optimization, Isothermal Titration Calorimetry, Alanine Scanning Mutagenesis
The development of small molecule inhibitors of protein/protein interactions has attracted significant attention as a new frontier in drug design (1–3). Two important issues hinder the design of these inhibitors. First is the large difference in size between the small molecule and the protein/protein binding interface. A small molecule only covers a small fraction of the protein binding surface, which forces the designer to identify and target only a cluster of residues; hopefully the selected cluster contributes significantly to binding and the presence of the inhibitor effectively dissociates the proteins. Since, very often, the binding of a protein to another (e.g. protein ligand to cell surface receptor) initiates a signaling process, there is always the risk that the inhibitor itself may act as a surrogate protein ligand and trigger the signal that is supposed to be inhibited. In fact, those unwanted effects have been reported for HIV-1 cell entry inhibitors (4). It has been recognized for many years that the interaction energy between two proteins is not evenly distributed between the residues in the binding surface but localized to only a few residues, so-called hotspots (5). The binding affinity consequences of mutating protein interface residues to Ala have provided the experimental basis for those conclusions and define the approach of Ala scanning mutagenesis (3, 5). Ala scanning mutagenesis allows identification of the residues that contribute the most to binding affinity (binding hotspots). Targeting binding hotspots has been a major goal in the design of drugs that can disrupt protein-protein interactions (3, 6–8). Ideally, one would like to target those residues that contribute the most to binding (binding hotspots) while simultaneously avoiding the residues that trigger the signaling process (allosteric hotspots). In this paper we present the technique of Thermodynamic Guided Alanine Scanning Mutagenesis aimed at accomplishing those goals. This technique is an extension of the traditional Ala scanning mutagenesis approach, made it possible by taking advantage of the additional information provided by microcalorimetry. We demonstrate the technique with the optimization of cell entry HIV-1 inhibitors.
The first event in HIV-1 infection is the binding of the virus envelope glycoprotein gp120 to the cell surface receptor CD4 (6, 9). In its unliganded state, gp120 is characterized by the presence of intrinsically disordered domains. In particular, the residues that define the coreceptor binding epitope are disordered and only become binding competent when CD4 binds gp120, a process that triggers a large allosteric structuring in gp120. This conformational change is reflected in a very large favorable binding enthalpy and very large unfavorable binding entropy. These enthalpy and entropy values are similar to those observed in protein folding; in fact, it has been estimated that they correspond to the folding of about 130 residues (10). The large favorable enthalpy reflects the formation of hydrogen bonds, van der Waals and other interactions associated with folding, while the unfavorable entropy reflects the large folding ordering effect, which overcomes the favorable entropy associated with desolvation. The binding of CD4 triggers the structuring of the coreceptor binding site in gp120 allowing it to bind to the chemokine coreceptor and initiate the sequence of events that lead to fusion of the viral and host cell membranes (11).
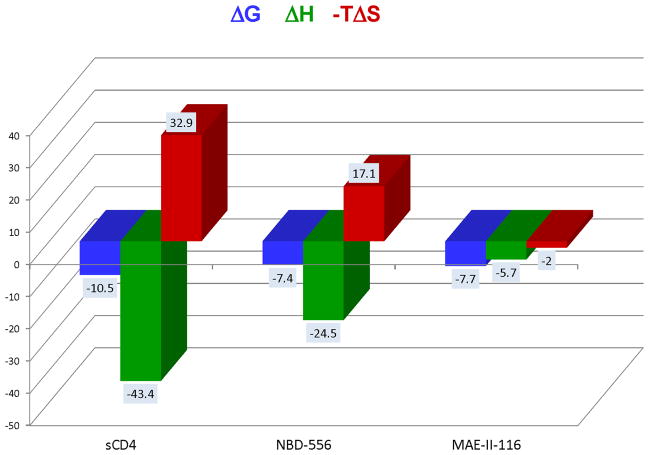
The development of small molecule inhibitors of CD4/gp120 binding has been hindered by low potency and by triggering the unwanted activation of gp120. In fact, the initially promising small molecular weight inhibitor NBD-556, which competes with CD4, activated the coreceptor site in gp120, making the virus infective to CD4-negative cells (4). NBD-556 has a thermodynamic signature that closely resembles that of CD4 (Figure 1) indicating that it also triggers the structuring of gp120. A surprising observation during the optimization of NBD-556 was that not all analogs triggered the structuring of gp120 and that this effect was independent of their binding affinity. An example is shown in Figure 1. In this figure, it is shown that the thermodynamic signatures of NBD-556 and its analog MAE-II-116 are very different despite having similar binding affinities. MAE-II-116 binds with a slight favorable enthalpy and also favorable entropy indicating that it does not trigger the large structuring process in gp120 (4, 12). These observations prompted the hypothesis that binding hotspots and allosteric hotspots did not completely overlap making it possible to develop competitive inhibitors that do not trigger the unwanted signaling process (12, 13). In this paper, we demonstrate that this is indeed the case and develop a method to identify binding and allosteric hotspots for sCD4/gp120. This technology can be extended to other proteins and provides a blueprint for the optimization of inhibitors of protein/protein interactions.
Figure 1.
Thermodynamic signatures for sCD4, NBD-556 and MAE-II-116. The data for NBD-556 and MAE-II-116 were published previously (4, 12). The values for sCD4 were determined in this study.
METHODS AND MATERIALS
Expression and Purification of gp120
Expression and purification of gp120 protein from the B subtype YU2 HIV-1 isolate (GenBank accession no. M93258) were carried out as reported previously with minor modifications (14). Briefly, the pcDNA3.1/Zeo (−) plasmid with the full-length YU2 gp120 gene insertion was used to transfect 293F cells for protein expression. Filtered cell culture supernatant was loaded directly onto an antibody affinity column equilibrated with PBS, pH 7.4. After washing with five column volumes of PBS, protein was eluted off the column and equilibrated with PBS, pH 7.4 as described previously (14).
Expression and Purification of Soluble CD4 Proteins
The expression plasmid for soluble wild type four-domain CD4 (sCD4wt) was generously provided by I. Chaiken and M. Contarino (Drexel University College of Medicine, Philadelphia, PA). Point mutations were generated using the QuikChange Site-Directed Mutagenesis Kit (Catalog # 200523) from Agilent Technologies according to the manufacturer’s instructions. The DNA sequence of the whole coding region of each sCD4 mutant was confirmed by sequencing to be as expected. Automated DNA sequencing was provided by The Synthesis & Sequencing Facility at Johns Hopkins University School of Medicine. Transfection grade large scale plasmid preparation was carried out with the QIAGEN Plasmid Plus Maxi Kit (Catalog # 12963) from QIAGEN (Germantown, MD) according to the manufacturer’s instructions.
The plasmids for wild-type sCD4 and the Ala mutants were transfected into 293F cells for protein expression using the same procedure as that for gp120 described above. After incubation, cell cultures were centrifuged at 10,000g at 4 °C for 12 minutes. The supernatant was filtered and loaded onto a HisTrap HP 5 mL column (GE Healthcare) equilibrated with 20 mM Tris and 150 mM NaCl, pH 7.4. All column chromatography was performed on an ÄKTA FPLC system (Amersham Biosciences, Uppsala, Sweden) at 4 °C. After washing with 10 column volumes of buffer A (20 mM Tris, 20 mM imidazole, and 150 mM NaCl, pH 7.4), protein was eluted with a linear gradient to buffer B (20 mM Tris, 300 mM imidazole, and 150 mM NaCl, pH 7.4) in five column volumes. Fractions containing sCD4 were pooled, concentrated, and loaded onto a HiPrep 26/60 Sephacryl S-200 gel filtration column (GE Healthcare) equilibrated with PBS, pH 7.4 (Roche Diagnostics GmbH, Mannheim, Germany). Fractions spanning the peak of sCD4 were pooled and concentrated. Aliquots were frozen in dry ice and stored at −80 °C. The structural integrity of the Ala mutants was evaluated by differential scanning calorimetry (DSC) as described before (15). Wild type and mutants exhibited Tm’s bracketed between 58 and 61°C and all of them were completely folded at 25°C.
Differential Scanning Calorimetry
Differential Scanning Calorimetry (DSC) experiments of wild-type and mutant sCD4 proteins were performed using VP-DSC microcalorimeters from MicroCal/GE Healthcare (Northampton, MA, USA). The sCD4 proteins were diluted with PBS, pH 7.4, to 0.5–1 mg/mL. All solutions were degassed for 15 minutes immediately before loading into the calorimetric cells. The protein solutions were scanned from 15 to 75 °C at a rate of 1 °C/min. Data were analyzed using the CpPlus9 program developed in this laboratory.
Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) was carried out using VP- ITC microcalorimeters from MicroCal/GE Healthcare (Northampton, MA, USA). In all titration experiments, the two binding partners were equilibrated with PBS, pH 7.4, and titrations were performed at 25 °C by injecting 10 μL aliquots of the sCD4 protein into the calorimetric cell containing gp120 (volume ~ 1.4 mL). The concentrations of CD4 and gp120 were 25 and 2 μM, respectively. The heat evolved upon each injection of inhibitor was obtained from the integral of the calorimetric signal. The heat associated with binding to gp120 in the cell was obtained by subtracting the heat of dilution from the heat of reaction. The individual heats were plotted against the molar ratio, and the enthalpy change (ΔH) and association constant (Ka = 1/Kd) were obtained by nonlinear regression of the data.
RESULTS AND DISCUSSION
Alanine Scanning Mutagenesis
Traditionally, alanine scanning mutagenesis has been utilized to assess the contributions of interface residues to binding affinity and to identify binding hotspots (5). Since protein/protein interactions usually initiate an allosteric signaling process, a similar approach can be used to identify allosteric hotspots (i.e. residues that initiate the signaling process) provided that a technique is available to quantitate signaling. For the sCD4/gp120 interaction, the allosteric signaling is defined by a structuring process that manifests itself in terms of large enthalpy and entropy changes. Consequently, isothermal titration calorimetry (ITC) becomes an ideal technique due to its ability to simultaneously measuring ΔG, ΔH and −TΔS, and assess residue contributions to binding affinity and to allosteric structuring. Contributions to binding affinity will be revealed in changes in Gibbs energy, ΔG, while contributions to allosteric structuring will be revealed in the enthalpy and entropy changes. For example, the mutation to alanine of a residue that contributes significantly to binding and structuring will be reflected in a decrease in ΔG and a large decrease in ΔH. On the other hand, the Ala mutation of a residue that contributes to binding and not to structuring will be reflected in a decrease in ΔG and a small decrease in ΔH.
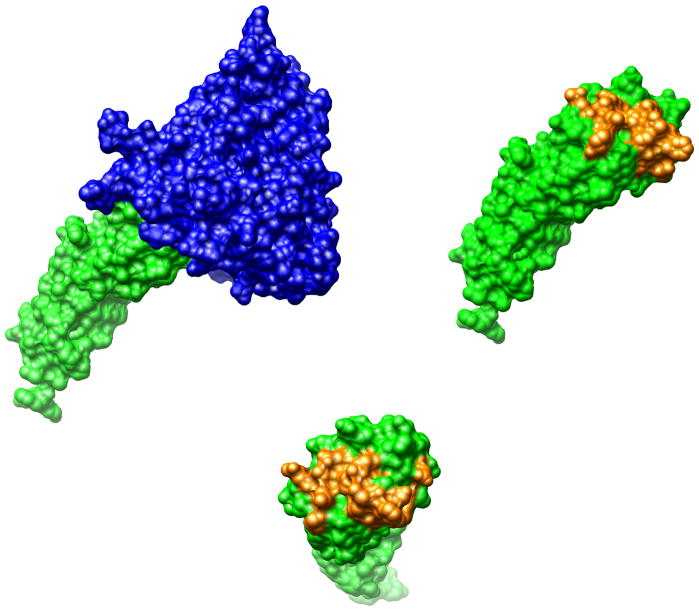
Several crystallographic structures containing the gp120-CD4 complex have been determined (16–19). According to these structures (Figure 2), the binding interface between CD4 and gp120 consists of about 20 residues in both proteins (18) comprising a total area close to 2000 Å2. The crystal structure of unliganded sCD4 (20, 21) indicates that it does not undergo any change in conformation upon binding to gp120, being therefore ideally suited for alanine scanning mutagenesis. A total of fifteen residues in CD4 were mutated to alanine one at a time (Gln 25, Lys 29, His 27, Gln 33, Lys 35, Asn 39, Gln 40, Ser 42, Phe 43, Leu 44, Thr 45, Pro 48, Arg 59, Asp 63 and Glu 85). As shown in Figure 2, these residues define most of the interacting surface of CD4 with gp120 and their Ala mutations can be used to assess if the influence of each residue to the allosteric structuring of gp120 is proportional to its contribution to binding affinity.
Figure 2.
The structure of gp120 (blue) in complex with CD4 (green) extracted from pdb file 1G9N. The protein/protein interaction surface extends to about 2,000 Å2. The residues in CD4 that comprise most of the interaction with gp120 (Gln 25, His 27, Lys 29, Gln 33, Lys 35, Asn 39, Gln 40, Ser 42, Phe 43, Leu 44, Thr 45, Pro 48, Arg 59, Asp 63 and Glu 85) are shown in yellow. Single amino acid sCD4 mutants in which these amino acids were individually mutated to alanine were used in the analysis presented in this paper.
ITC Experiments
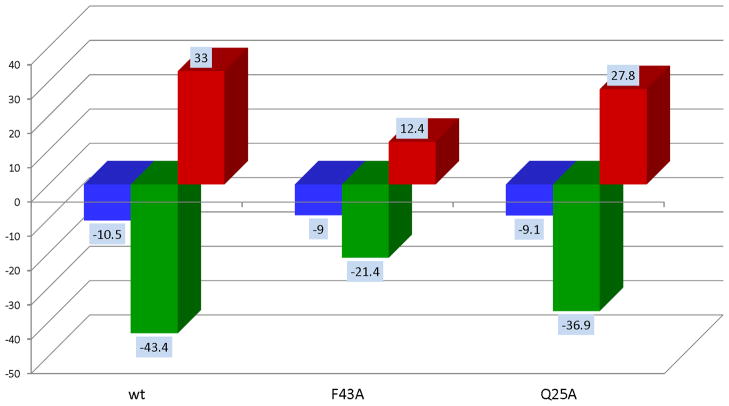
The binding of each Ala mutant of sCD4 to gp120 was measured by ITC. Several situations were observed indicating that not all binding hotspots elicit the same degree of allosteric structuring. As an example, Figure 3 illustrates two types of results obtained for two different Ala mutations, F43A and Q25A. Both Ala mutations lower the binding affinity by similar amounts (about one order of magnitude) but they do so by different enthalpy/entropy combinations. The F43A mutation is characterized by an enthalpy change which is 22 kcal/mol lower than the wild type whereas the Q25A mutation is only 6.5 kcal/mol lower than the wild type. The same behavior is observed with the entropy change, −TΔS, which decreases 20.6 kcal/mol for F43A and only 5.2 kcal/mol for Q25A. These results indicate that F43 contributes strongly to binding and also to the allosteric structuring of gp120 since its Ala mutation results in a large drop in binding enthalpy. Q25, on the other hand, contributes about the same as F43 to binding but much less to the allosteric structuring of gp120 as indicated by the smaller drop in ΔH or −TΔS. This result is very important because it indicates that residues that contribute the same to binding affinity do not necessarily contribute the same to the allosteric effect, opening the possibility of selectively targeting residues for affinity or allosteric inhibition. For competitive inhibitors, in which inhibiting the allosteric signaling is the main objective, residues that contribute to binding and not structuring become the target for optimization.
Figure 3.
Thermodynamic signatures for gp120 binding to wild type sCD4 and the Ala mutants F43A and Q25A. Mutations at specific locations may affect binding affinities (ΔG) differently indicating the contribution of specific residues to binding. Some mutations like F43A and Q25A reduce affinity by similar amounts but through different enthalpy/entropy combinations. F43A reduces the binding enthalpy (ΔH) by about half indicating that F43 is not only a hotspot for binding affinity but also for the allosteric structuring of gp120. Mutation of Q25, on the other hand lowers the affinity by a similar amount to that of F43A but with a much smaller reduction in binding enthalpy indicating that Q25 does not contribute significantly to allosteric structuring of gp120 even though it contributes significantly to binding. The magnitude of the allosteric structuring effect is also appreciated in the entropy changes. F43A does not cause as much structuring (smaller −TΔS) as wt or Q25A.
Binding and Allosteric Hotspots
The results for fifteen Ala mutations are summarized in Figure 4 and depicted in the structure of CD4 (pdb file 1G9N). The residue contributions to the Gibbs energy of binding of sCD4 for gp120 exhibit a distribution pattern resembling those observed for other proteins (3, 5). As observed for other proteins, the binding energy is not uniformly distributed throughout the surface but concentrated in a few hotspots in a non-random fashion. In CD4 the binding hotspots appear to be clustered in two different areas, the area surrounding Phe 43 and the area surrounding Gln 25 and Lys 35. These areas contribute strongly to binding affinity. The surprising result is that the residue contributions to allosteric structuring are not proportional to the contributions to binding. The cluster around Phe 43 contributes significantly to binding and allosteric structuring whereas the cluster around Gln 25 contributes significantly to affinity but not to allosteric structuring. Since the allosteric structuring occurs in gp120, the results indicate that the gp120 residues that interact with Phe 43 and immediate neighbors are strong initiators the allosteric cascade whereas those that interact with Gln 25 and Lys 35 are not. The most fundamental conclusion from these experiments is that not all the residues that contribute to binding affinity contribute equally to allosteric structuring. All residues that contribute to allosteric structuring also contribute to binding but not vice versa. These observations have important consequences for the design of inhibitors of protein/protein interactions.
Figure 4.
Thermodynamic guided Ala mutagenesis results. In the left panel, residues have been colored according to their contributions to binding affinity and on the right panel according to their contributions to allosteric structuring. Colored in red are the residues that contribute the most (ΔΔG > 0.8 kcal/mol, ΔΔH > 20 kcal/mol), in pink are those in the middle (0.1 kcal/mol < ΔΔG < 0.8 kcal/mol, 1 kcal/mol < ΔΔH < 20 kcal/mol) and in white those residues that do not contribute ΔΔG < 0.1 kcal/mol, ΔΔH < 1 kcal/mol). Colored in green are residues that have not been mutated.
The results of the thermodynamic guided alanine scanning mutagenesis indicate that the triggers for the allosteric structuring of gp120 are localized to specific regions. This conclusion agrees with previous observations obtained with single amino acid mutations in gp120. It has been known for several years that some mutations in gp120 can shift the unliganded state to a conformation resembling that induced by CD4 binding. Most notably, the S375W mutation fills the top of the Phe43 cavity of gp120 inducing a conformation closer to that of the CD4-bound state in unliganded gp120 (22). As a result, CD4 binding becomes more favorable and characterized by smaller enthalpy and entropy changes (22). The introduction of a second mutation in the Phe43 cavity, T257S, stabilizes the CD4-bound conformation even further (23). It is clear that allosteric properties are localized in discrete regions and can be modulated by single amino acid mutations and, consequently they are also susceptible to be affected by small molecules that bind at specific sites.
Inhibitor Optimization
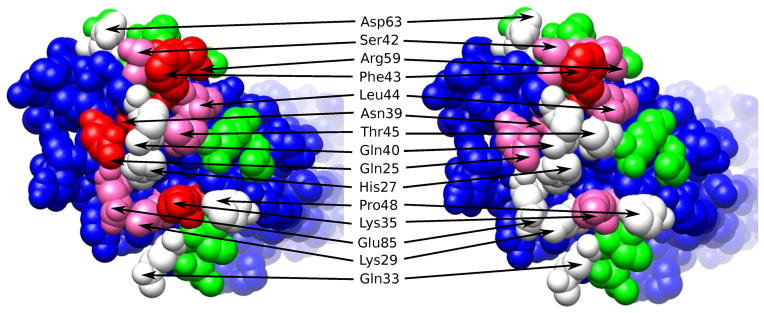
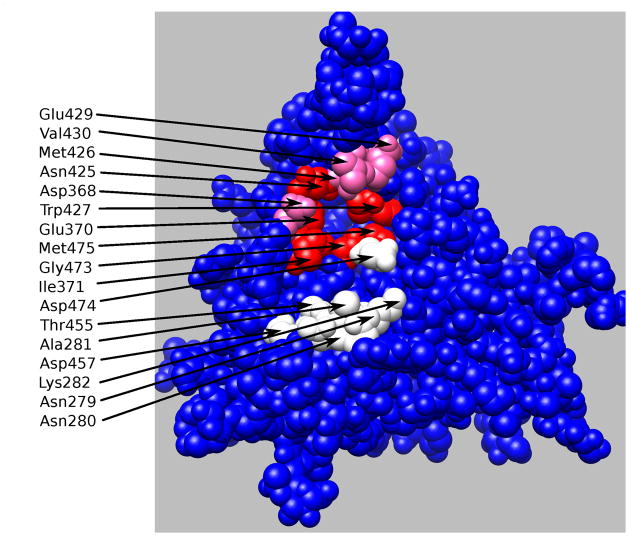
The thermodynamic guided alanine mutagenesis results shown in Figure 4 can be mapped into the gp120 binding surface in order to identify the CD4 partners responsible for binding and allosteric effects. The results are shown in Figure 5. In preparing this figure, gp120 residues that were at less than 4Å from the mutated CD4 residues (pdb file 1G9N) were labeled according to the measured CD4 contribution to binding affinity and allosteric effect. There are clearly two different clusters of residues that contribute to affinity but only one of them contributing significantly to the allosteric structuring effect. These results provide a blue print for the optimization of CD4/gp120 inhibitors. First, the competitive efficiency of an inhibitor is related to the degree with which the inhibitor disrupts critical interactions between the two proteins (i.e. preferentially targeting residues that contribute significantly to binding, white, pink and red colored residues in Figure 5); second, if the goal is to inhibit the signaling process, targeting should select those residues that contribute significantly to binding but not to the allosteric effect (white colored residues in Figure 5).
Figure 5.
Residues at the gp120 interface with CD4 contributing to binding affinity and allosteric effect are colored red, pink and white. The red residues contribute to strongly both affinity and allosteric effects. Residues colored in pink contribute strongly to affinity and only moderately to the allosteric effect. The white residues contribute only to affinity.
The results obtained with the thermodynamic guided alanine scanning mutagenesis agree with the structural characteristics of recently published CD4/gp120 inhibitors that do not trigger the unwanted allosteric structuring effect (24). This improved set of analogs make a strong hydrogen bond with Asp 368 in gp120, an amino acid that does not trigger a strong structuring effect (Figure 5). Since Asp 368 is only moderately non-allosteric (pink colored in Figure 5), the results suggest the existence of an activation threshold and, consequently, that even better inhibitors could be developed by targeting even less allosteric residues. The availability of the design blueprint will accelerate the development of better HIV-1 cell entry inhibitors.
CONCLUSIONS
Proteins often interact with other proteins with affinities in the nanomolar and high picomolar levels (25–35). The development of protein/protein interaction inhibitors requires harnessing all the binding potency of the small molecule into critical spots that will disrupt protein/protein binding without triggering the signaling cascade that needs to be blocked. Thermodynamic guided alanine scanning mutagenesis provides an experimental way to simultaneously identify binding and allosteric hotspots. Binding hotspots can be identified in a straightforward fashion from the measured binding affinities of the Ala mutants. Allosteric hotspots, on the other hand, require investigation of the thermodynamic signature of the proteins, which in many situations like CD4/gp120, will reflect the triggering of the allosteric signaling. In those cases, ITC provides a unique technique for identifying binding and allosteric hotspots in proteins.
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB0641252) and the National Institutes of Health (GM56550 and GM57144).
References
- 1.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 2.Berg T. Small-molecule inhibitors of protein-protein interactions. Curr Opin Drug Discov Devel. 2008;11:666–674. [PubMed] [Google Scholar]
- 3.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 4.Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, et al. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry. 2006;45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 6.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 7.Keskin O, Ma B, Nussinov R. Hot regions in protein--protein interactions: the organization and contribution of structurally conserved hot spot residues. Journal of molecular biology. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 8.Keskin O, Ma B, Rogale K, Gunasekaran K, Nussinov R. Protein-protein interactions: organization, cooperativity and mapping in a bottom-up Systems Biology approach. Physical biology. 2005;2:S24–35. doi: 10.1088/1478-3975/2/2/S03. [DOI] [PubMed] [Google Scholar]
- 9.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, et al. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 10.Leavitt SA, SchOn A, Klein JC, Manjappara U, Chaiken IM, Freire E. Interactions of HIV-1 proteins gp120 and Nef with cellular partners define a novel allosteric paradigm. Curr Protein Pept Sci. 2004;5:1–8. doi: 10.2174/1389203043486955. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 12.Schon A, Madani N, Smith AB, Lalonde JM, Freire E. Some binding-related drug properties are dependent on thermodynamic signature. Chemical biology & drug design. 2011;77:161–165. doi: 10.1111/j.1747-0285.2010.01075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schon A, Lam SY, Freire E. Thermodynamics-based drug design: strategies for inhibiting protein-protein interactions. Future medicinal chemistry. 2011;3:1129–1137. doi: 10.4155/fmc.11.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brower ET, Schon A, Freire E. Naturally occurring variability in the envelope glycoprotein of HIV-1 and development of cell entry inhibitors. Biochemistry. 2010;49:2359–2367. doi: 10.1021/bi1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schon A, Velazquez-Campoy A. Calorimetry. In: Jiskoot W, Crommelin DJA, editors. Methods for Structural Analysis of Protein Pharmaceuticals. Arlington, VA: AAPS Press; 2005. pp. 573–589. [Google Scholar]
- 16.Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong PD, Ryu SE, Hendrickson WA, Axel R, Sweet RM, Folena-Wasserman G, et al. Molecular characteristics of recombinant human CD4 as deduced from polymorphic crystals. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:6423–6427. doi: 10.1073/pnas.87.16.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu SE, Kwong PD, Truneh A, Porter TG, Arthos J, Rosenberg M, et al. Crystal structure of an HIV-binding recombinant fragment of human CD4. Nature. 1990;348:419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, et al. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 2002;76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dey B, Pancera M, Svehla K, Shu Y, Xiang SH, Vainshtein J, et al. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J Virol. 2007;81:5579–5593. doi: 10.1128/JVI.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalonde JM, Kwon YD, Jones DM, Sun AW, Courter JR, Soeta T, et al. Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors. Journal of medicinal chemistry. 2012;55:4382–4396. doi: 10.1021/jm300265j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbate EA, Berger JM, Botchan MR. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes & development. 2004;18:1981–1996. doi: 10.1101/gad.1220104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banner DW, D’Arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, et al. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 27.Hage T, Sebald W, Reinemer P. Crystal structure of the interleukin-4/receptor alpha chain complex reveals a mosaic binding interface. Cell. 1999;97:271–281. doi: 10.1016/s0092-8674(00)80736-9. [DOI] [PubMed] [Google Scholar]
- 28.Kelekar A, Chang BS, Harlan JE, Fesik SW, Thompson CB. Bad is a BH3 domain-containing protein that forms an inactivating dimer with Bcl-XL. Mol Cell Biol. 1997;17:7040–7046. doi: 10.1128/mcb.17.12.7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 30.Philo JS, Aoki KH, Arakawa T, Narhi LO, Wen J. Dimerization of the extracellular domain of the erythropoietin (EPO) receptor by EPO: one high-affinity and one low-affinity interaction. Biochemistry. 1996;35:1681–1691. doi: 10.1021/bi9524272. [DOI] [PubMed] [Google Scholar]
- 31.Rickert M, Wang X, Boulanger MJ, Goriatcheva N, Garcia KC. The structure of interleukin-2 complexed with its alpha receptor. Science. 2005;308:1477–1480. doi: 10.1126/science.1109745. [DOI] [PubMed] [Google Scholar]
- 32.Shibata H, Yoshioka Y, Ohkawa A, Abe Y, Nomura T, Mukai Y, et al. The therapeutic effect of TNFR1-selective antagonistic mutant TNF-alpha in murine hepatitis models. Cytokine. 2008;44:229–233. doi: 10.1016/j.cyto.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Sundstrom M, Lundqvist T, Rodin J, Giebel LB, Milligan D, Norstedt G. Crystal structure of an antagonist mutant of human growth hormone, G120R, in complex with its receptor at 2.9 A resolution. The Journal of biological chemistry. 1996;271:32197–32203. doi: 10.1074/jbc.271.50.32197. [DOI] [PubMed] [Google Scholar]
- 34.Syed RS, Reid SW, Li C, Cheetham JC, Aoki KH, Liu B, et al. Efficiency of signalling through cytokine receptors depends critically on receptor orientation. Nature. 1998;395:511–516. doi: 10.1038/26773. [DOI] [PubMed] [Google Scholar]
- 35.Walsh ST, Jevitts LM, Sylvester JE, Kossiakoff AA. Site2 binding energetics of the regulatory step of growth hormone-induced receptor homodimerization. Protein science : a publication of the Protein Society. 2003;12:1960–1970. doi: 10.1110/ps.03133903. [DOI] [PMC free article] [PubMed] [Google Scholar]