Abstract
It is widely recognized that ADMET (Adsorption, Distribution, Metabolism, Excretion - Toxicology) liabilities kill the majority of drug candidates that progress to clinical trials. The development of computational models to predict small molecule membrane permeability is therefore of considerable scientific and public health interest. Empirical qualitative structure permeability relationship (QSPR) models of permeability have been a mainstay in industrial applications, but lack a deep understanding of the underlying biological physics. Others and we have shown that implicit solvent models to predict passive permeability for small molecules exhibit mediocre predictive performance when validated across experimental test sets. Given the vast increase in computer power, more efficient parallelization schemes, and extension of current atomistic simulation codes to general use graphical processing units (GPUs), the development and application of physical models based on all-atom simulations may now be feasible. Preliminary results from rigorous free energy calculations using all-atom simulations indicate that performance relative to implicit solvent models may be improved, but many outstanding questions remain. Here we review the current state of the art physical models for passive membrane permeability prediction, and present a prospective look at promising new directions for all-atom approaches.
Keywords: Passive membrane permeability, Implicit solvent, Molecular Dynamics, Homogenous solubility model
1. Introduction
A major challenge facing the drug discovery community is the high rate of attrition of potential small molecule drug candidates due to poor adsorption, distribution, metabolism, excretion, and toxicology (ADME-Tox, or ADMET) properties (1). Bioavailability and pharmacokinetics are two components of ADMET that have received increased attention since the 1990s, when failures in the clinic were predominantly due to these two factors (2). Subsequently, the pharmaceutical industry has committed significant efforts to understand the molecular basis of bioavailability and pharmacokinetics; and, as a result, the rate of clinical compound failure attributed to these two factors has decreased substantially (2). Yet, the persistence of candidate compound attrition is representative of a larger inability, as a field, to understand the many components controlling ADMET properties.
Successful models describing ADMET properties hold the promise to: i) identify compound liabilities early-on in the development phase, before hundreds of millions of dollars have been invested in clinical trials (3), ii) identify liabilities to be “designed out” of the candidate compounds, iii) identify where, and through which mechanism, these liabilities are operating in the host, iv) flag the liabilities for close monitoring using in vivo assays, and v) allow practitioners to manipulate and control these properties to their advantage. The ultimate goal is to not only identify the liability, but to also understand their causative physical and chemical processes.
Achieving a more comprehensive understanding of ADMET processes in vivo presents a substantial challenge, and certain pharmacokinetic properties are relevant to multiple aspects of attrition due to poor ADMET profiles. In particular, passive membrane permeability plays an essential role in the rational design of a drug from a small molecule candidate compound. For example, while a compound may be delivered orally, topically, by inhalation, or by injection, once it is in the body, it must pass an epithelial membrane barrier to exit systematic circulation and gain access to its target; passively diffusing through a membrane is, perhaps, the most common way this occurs. Moreover, diffusion of metabolites, produced largely by cytochrome P450 enzymes in the liver, must pass through renal membranes before clearing the body (4). Therefore, passive membrane permeability of small molecules is a critical component of overall ADMET issues, and correspondingly, it remains an area with significant ramifications to rational drug design and, by extension, to public health.
Experimentally, permeability can be measured using a number of different assays. In the drug discovery community, the most popular approaches have been parallel artificial membrane (PAMPA) (5, 6) and carcinoma colorectal cell based (CaCo-2) assays (7, 8). PAMPA assays are an economical high-throughput passive permeability construct that uses an artificial membrane of lipids (typically egg lecithin), dissolved in an organic solvent, to monitor the transport of solutes between wells. CaCo-2 assays, on the other hand, are biochemically more similar to in vivo intestinal absorption processes because they have intracellular junctions, hydrolase activity, and transport proteins. Thus, CaCo-2 measurements account for active and passive permeability, but they suffer from being less amenable to a high-throughput approach.
Varying degrees of complexity have been incorporated in models aimed to predict membrane permeation. In this perspective, we summarize the current state-of-the-art in terms of permeability prediction methods, with an emphasis on physics-based models. In particular, we revisit the all-atom approaches first developed in the early 1990s, and take a fresh look at their unique potential to combine the thermodynamic and kinetic components of passive membrane permeability in a comprehensive in silico framework.
2. Quantitative Structure-Permeability Relationship Models
The simplest and most utilized models are quantitative structure-permeability relationship (QSPR) methods. These models are built upon theories and ideas established in the seminal quantitative structure-activity relationship studies pioneered by Hansch in the early 1970s (9, 10). QSPR models are economical methods that utilize a set of molecular descriptors, such as molecular weight, hydrogen bond donors/acceptors, and polar surface area, in order to predict permeability constants based on statistical relationships among a set of compounds (11). Other considerations, such as molecular topology, may also make important contributions (12). These methods rely on training sets of molecules, which may vary widely depending on the practitioner’s environment or needs. For example, QSPR models have been developed to predict drug absorption (13), Caco-2 permeability (14), blood-brain barrier penetration (15), among others (for comprehensive methodological reviews, see (16, 17). It is widely recognized that the success or failure of these models is highly dependent on the training set, and therefore, the universality of the approach is limited, and transferability is a major issue (18). Even successful, predictive QSPR models are limited, as they provide essentially no insight to the actual atomic level mechanisms governing the assessed properties.
3. Physical Models of Passive Membrane Permeability
A fairly sophisticated understanding of the membrane diffusion process has been achieved for simple membrane bilayers. The permeation of small molecules through a membrane can be most simply understood in terms of the solubility-diffusion model of permeation. This model is represented by a three-step process, which includes the partitioning of the small molecule from the aqueous phase (bulk solvent, or cytoplasm) into the lipid bilayer, diffusion of the small molecule across the lipid bilayer, and the partitioning of the solute out of the bilayer and into the aqueous solvent (again, bulk solvent, or periplasm) (19, 20). The resistance, R, of solute permeation to a depth, z, is inversely related to its permeability, P, which can be defined in terms of the “local” solute partition coefficient between bulk water and the membrane at depth z, K(z), and the local diffusion constant, D(z), as:
| 1 |
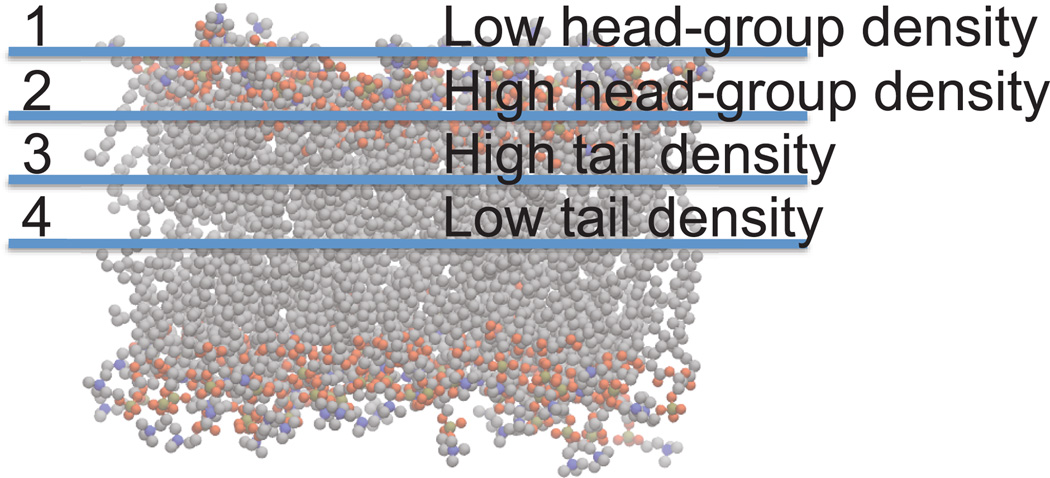
The membrane itself is comprised of a bilayer of lipid molecules, each with a polar headgroup and an aliphatic tail. As one would expect, the polar headgroups form a different physiochemical environment than the greasy, hydrophobic tails that constitute the interior region of the membrane. Moreover, the center of the bilayer itself is expected to contain voids that promote a physiochemical phenomena known as solute hopping (21, 22). Pioneering theoretical (23) work by Marrink and Berendsen described the spatial heterogeneity of biological lipid bilayers in terms of a so-called 4-region model (Figure 1). These physical underpinnings clearly indicate that small molecule transport through the biological membrane is anisotropic in nature.
Figure 1.
Anisotropy of membrane permeation can be understood in terms of the 4-region model. Carbon atoms are grey, oxygen atoms are red, phosphorous atoms are orange, and nitrogen atoms are blue. Hydrogen atoms have been omitted for the sake of clarity.
3.1 Implicit solvent models
Implicit solvent models assume that a single low-dielectric slab can represent the anisotropic complexity illustrated in figure 1. This removes the position dependence of the solute partition function and diffusion coefficient in equation 1, which may be simplified and solved for the permeability coefficient, P:
| 2 |
In equation 2, Kp is the solute partition coefficient between bulk water and the low-dielectric membrane slab, D is the solute diffusion coefficient through the membrane, and d is the membrane thickness. As it follows from a model in which the membrane is treated as a homogenous slab, equation 2 is consistent with the homogenous solubility diffusion model, which itself is closely linked to the barrier domain model (24). The barrier domain model posits that the rate of polar-compound membrane passage is limited by the highly ordered region of the membrane immediately behind the acyl-chain headgroup linkages, or region 3 in figure 1. Two factors contribute to reduced transport through this region. The ordered side chain structure in this region results in low diffusivity, while the apolar quality of the region disfavors solubility. The solute partition coefficient in equation 2 can be expressed in terms of the free energy difference of the solute dissolved in bulk water and in the low-dielectric membrane slab, or ΔG. Assuming that the membrane slab is a good approximation of the rate-limiting region of the membrane, or the barrier domain, then this leads to a permeability estimate analogous to transition state theory or, with R the universal gas constant and T the temperature. Moreover, if the differences in diffusion coefficients are negligible across similar sets of compounds, then the logged permeability of one compound relative to another is,
| 3 |
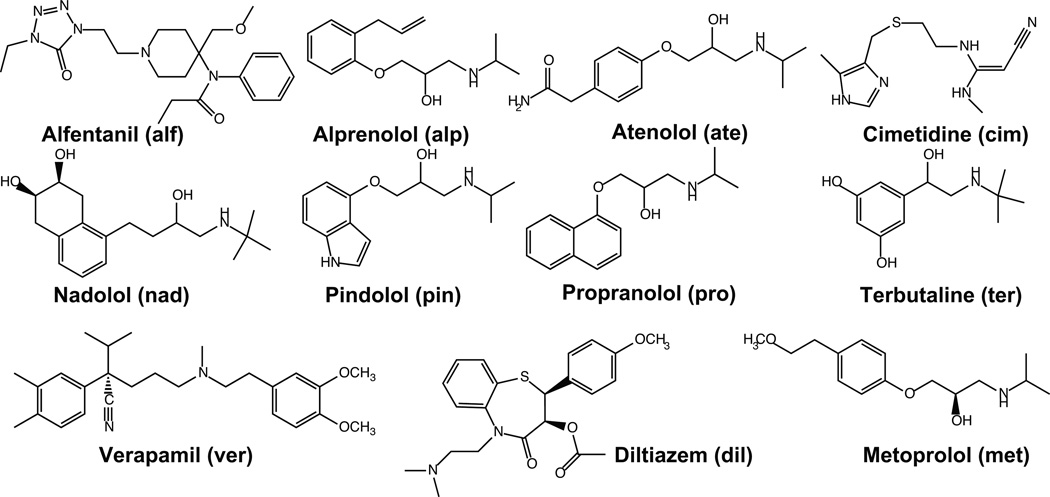
Jacobson et al. developed an implicit solvent model, based on equation 3, in which the global minimum conformation for a set of small molecules and cyclic peptides was determined in a Generalized Born (GB) implicit chloroform solvent, which replaced the non-uniform interactions between the solute and bilayer with an averaged, uniform interaction potential (25). The energy of this global minimum was then evaluated in GB implicit water and chloroform solvent models, and the difference was considered to be the free energy change of barrier crossing, or ΔG in equation 3 (26–28). They performed their initial experiments on a set of cyclic peptides, as well as a set of fluoroquinolines and a set of FDA-approved drugs, depicted in Figure 2. Our work in this area extended this approach by determining the global minimum conformation in each solvent environment and using those conformations in the evaluation of the free energy difference (29). Furthermore, we addressed the question of whether more exhaustive conformational sampling (performed through mode integration, or MINTA, importance sampling (30, 31), and the incorporation of multiple predominant states into the configuration integrals, would improve accuracy. Moreover, we extended the compound set to include the compounds in Figure 2 as well as a set of thirteen benzene congeners, intended to mimic more subtle structural differences. Our permeability estimates were compared to available PAMPA assay data (6). This work indicated that more rigorous treatments of the conformational distributions did not significantly improve correlation with experiment; R2 values never exceeded more than 0.75 (29), a limitation we attributed to a global minimum dominated free energy landscape and the severe simplifications of the model.
Figure 2.
11 small molecule, FDA approved, test compounds used to explore the accuracy of the implicit solvent and all-atom simulations. Three letter abbreviations, which appear in figure 6, are included.
Recently, Jacobson and colleagues presented a more extensive performance evaluation of the ability of these implicit solvent models to reproduce relative permeability rates using 9 different small molecule datasets against a variety of experimental assays (including PAMPA and cell-based assays) and QSPR predictions (32). The average R2 performance for log Pm against the PAMPA data was 0.71, whereas against the cell-based assays it was 0.64. This performance seems reasonable, given that no training data sets are required, and at the same time mediocre, both because the R2 values are not higher, and because the absolute permeabilities of the models underestimate experimental values by at least 5 orders of magnitude. Shortcomings could be attributed to many aspects of the simplified model, the most likely being the omission of entropic contributions and membrane anisotropy.
3.2 Explicit solvent models
In contrast to implicit solvent models, explicit-solvent, all-atom molecular dynamics (MD) simulations do not rely on extreme, simplifying assumptions and so, in principal, provide the level of detail necessary to describe membrane anisotropy (Figure 1). The development of MD-based models of small molecule membrane permeability through explicit biological lipid bilayers was initiated with the pioneering work of Bassolino-Klimas, Alper, and Stouch (33), who simulated benzene diffusion through a dimyristoylphosphatidylcholine (DMPC) lipid bilayer. Complementary efforts by Bemporad and Essex showed that these methods were able to correctly rank order the permeability rates for a set of eight organic solute molecules, but that the absolute permeabilities were an order of magnitude larger than experiment, which the authors attributed to insufficient sampling, ligand parameters inappropriately suited to the bilayer, and subtle issues surrounding ensemble choice (34). Since then, a number of studies have been carried out utilizing similar approaches, most recently see refs (34–39), and we explore these methods in more methodological detail below.
To determine whether the more detailed models are sufficiently accurate to capture the origins underlying permeability differences among a diverse small molecule set, exploratory molecular dynamics umbrella sampling simulations (40) were used to estimate the passive-permeability of the 11 small molecules shown in figure 2. To carry out the MD simulations, DMPC bilayers were modeled with 36 lipids per leaflet and a 20Å pad of water was positioned on either side of the membrane. TIP3P water (41) and CHARMM 36 parameters (42) were used. Following minimization, 10 ns of NPT equilibration, with an anisotropic barostat at 1 atm and a Langevin thermostat at 310.15 K, were performed in the NAMD molecular dynamics program (43). A more detailed description of the simulation protocol can be found in the supporting information. The “health” of the lipid bilayer models was determined by comparison to available experimental parameters, such as area per lipid headgroup, tail order parameters, and surface tension (data not shown).
3.2.a Thermodynamic influences: constructing potentials of mean force
Formally, the potential of mean force at z, or W(z), is the free energy at z relative to some arbitrary reference state, and it is directly connected to the local probability density, or ρ(z), through equation 4. The PMF gives rise to a mean force that causes the basins of the PMF to be more populated than other regions. It is this mean force, and the underlying PMF, that result in the equilibrium tendency of a molecule to partition to various depths within the lipid bilayer, facilitating the transport process. Additionally, coupling the PMF with an atomic-level description of the entire system entails the spatial entropic dependence that is neglected in the implicit solvent models, resulting in a more physically accurate model of the passive transport process.
| 4 |
Umbrella sampling simulations are the most established technique to determine PMF values, along a reaction coordination, in a biased fashion. Umbrella sampling is a straightforward approach that confines the solute of interest to discretized sections along the reaction coordinate, a.k.a. “windows”, using a harmonic biasing potential. Done properly, this stratification procedure enables effective and sufficient sampling over every region along the reaction coordinate and results in a set of overlapping, biased probability distributions. Using either the weighted histogram analysis method (WHAM) (44), or the multistate Bennett acceptance ratio (MBAR) method (45), these distributions can be unweighted and combined to minimize statistical error, resulting in unbiased probability distribution, which gives the PMF according to equation 4.
Following membrane equilibration, umbrella simulations for each of the molecules shown in figure 2 were prepared. Small molecule parameters were assigned using the CHARMM general force field (CGenFF) (46) using the MATCH program (47). Small molecules were oriented along each of their 3 principal axes and displaced in 32 1 Å increments from the bilayer center to the bulk solvent, along 1 leaflet, resulting in a total of 96 umbrella windows. Umbrella simulations were carried out for 3 ns in each window; simulations of propranolol, terbutaline, and verapamil were extended to 10 ns in each window to test convergence properties. The PMF along the membrane normal were estimated independently using the WHAM equations (44) for each of the 3 small molecule orientations. Averages were calculated across the 3 simulations, and standard errors were estimated using the standard deviations determined across the 3 simulations. A more detailed description of the umbrella simulation protocol and standard error calculation can be found in the supporting information.
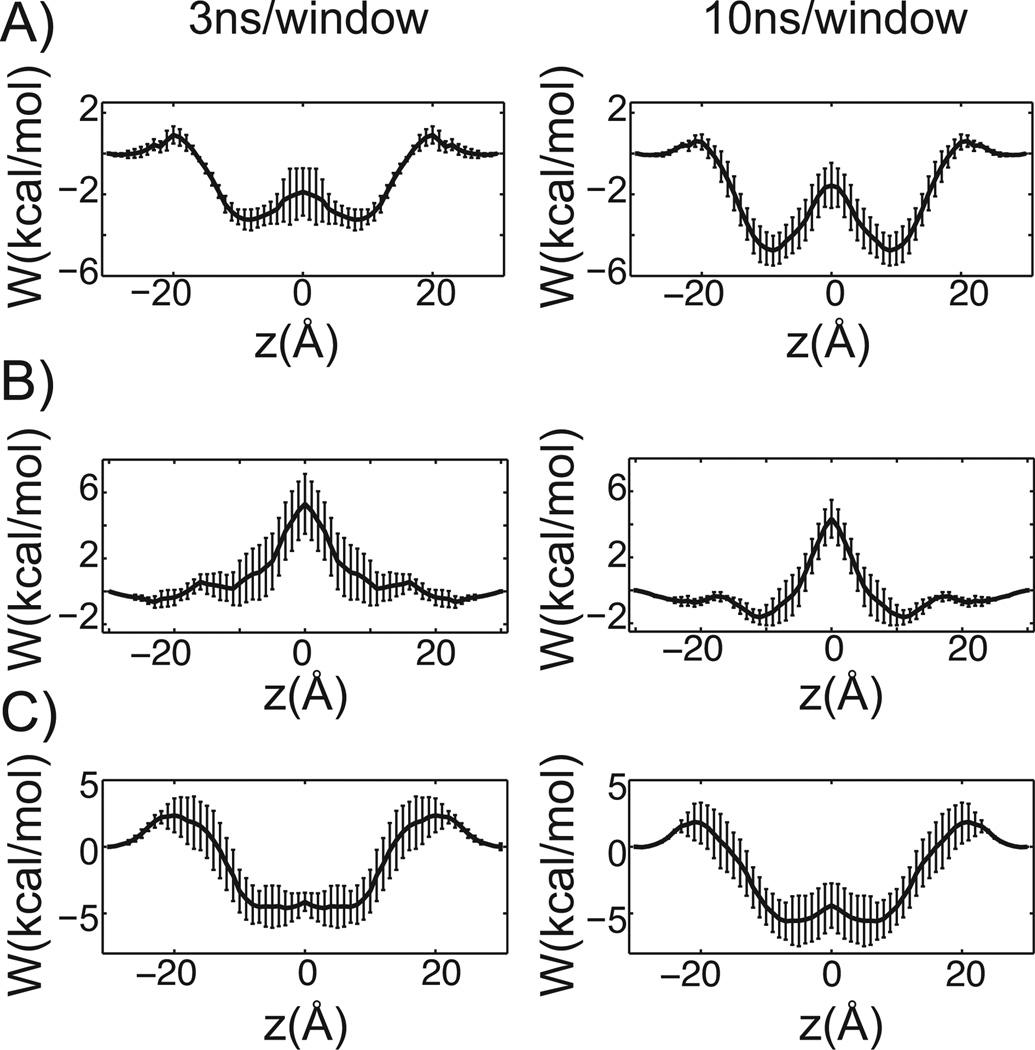
Results for propanolol, terbutaline and verapamil, are shown for 3 ns and 10 ns of sampling per window (Figure 3). While the general shapes of the PMFs are reasonably converged after 3ns of sampling per window, additional simulation time makes basins more pronounced and changes barrier heights, indicating that more than 10 ns of sampling per window may be necessary to properly average over the long-time conformational relaxation of the lipid tails and other slow system reorganization. To gain a better understanding of the accuracy of the PMF curves, we used them to estimate each small molecule’s water-DMPC membrane partition coefficient (38) and compared them to the experimental water-octanol partition coefficients. This comparison gave a correlation coefficient of 0.59, and both the experimental and predicted values spanned roughly the same orders of magnitude. Octanol has long been taken as an experimental proxy for lipid bilayers, however, as it is not an exact surrogate for a DMPC bilayer, perfect correlation can’t be expected due to fundamental structural and physicochemical differences between the environments (48). A correlation of 0.59 suggests that the estimated PMF curves reasonably describe the fundamental thermodynamics of small-molecule membrane partitioning. Nevertheless, lack of stronger correlation may hint at deeper force field deficiencies.
Figure 3.
Influence of sampling on PMF curves. A) through C), 3 ns of sampling per window is shown on the left, while 10 ns per window is on the right. The membrane center is positioned at 0 Å and the standard error is reported. A) Propranolol. B) Terbutaline. C) Verapamil.
Traditionally, biophysical simulations are carried out in aqueous solutions. Consistent with this, current state-of-the-art fixed charge force fields are parameterized to implicitly capture the polarization induced by a high dielectric solvent (49, 50). Such a parameterization strategy may lead to inappropriate interactions with apolar solvents such as the greasy region of the membrane bilayer. A strategy to optimize atomic partial charges to better reproduce experimental free energy differences between octanol and water may improve permeability accuracy estimates. Alternatively, polarizable force fields (51) may better capture multibody interactions whose description might be fundamentally important to model the thermodynamics and kinetics governing the passive transport process.
While umbrella simulations are the most widely used technique to estimate PMF curves, other, more contemporary, methods have also been advanced and may prove more efficient. For example, in the adaptive biasing force (ABF) method (52–55), an instantaneous biasing force that opposes the force acting on the solute along the reaction coordinate is estimated and applied. The estimate of this force improves with simulation length, and its application allows the system to diffuse along the reaction coordinate. Averaging the applied forces gives an estimate of the mean force, which can then be integrated along the reaction coordinate to yield the PMF. Other promising methods have also been developed (56) and may be applied. However, the most efficient methods for permeability estimates will yield not only a PMF, but also a local diffusion coefficient, which is discussed in the next section.
3.2.b Enter kinetics: estimating the local diffusion coefficient
While the PMF describes the propensity for a compound to partition from water into the membrane at a given depth, it is purely a thermodynamic quantity and says nothing about the rate of the process. Permeability is a transport process, and equilibrium thermodynamics must be integrated with a kinetic rate description. For example, if the diffusion constant is known along the reaction coordinate, then the mean first passage time can be estimated as a diffusive barrier crossing process (57). While monitoring the mean-square displacement (MSD) of a compound readily yields diffusion coefficients in the absence of substantial free energy barriers, it is impractical to estimate diffusion profiles along a rough free energy landscape. Instead, a variety of contemporary biased simulations can be paired with different diffusion estimators to arrive at a position dependent diffusion profile when substantial free energy barriers exist along a reaction coordinate, as for a compound moving through a lipid bilayer. For example, Marrink and Berendsen carried out equilibrium constrained simulations and a diffusion estimator that relates the autocorrelation of the constraint force to the local diffusion coefficient through the fluctuation-dissipation theorem (23). Extending work by Straub and Berne (58), Wolfe and Roux have applied an estimator that calculates a local diffusion coefficient from the velocity autocorrelation function extracted from a harmonically biased simulation (59), although it has not been applied to permeability predictions. More recently, Hummer extended the work of Wolfe and Roux to derive an estimator based on positional autocorrelation (60), whose use is reported here and described in greater detail below. Within non-equilibrium regimes, such as in steered molecular dynamics (SMD), Kosztin et al. have posited that one can use the slope of the mean dissipative work curve to estimate the local diffusion constant (61). Yet, this last method has been largely untested. Despite the variety of diffusion estimators, to the best of the authors’ knowledge, a detailed comparison of their performance has not been carried out, and selecting one estimator over another is still, unfortunately, driven by convenience. For example, when using a constrained simulation to calculate the PMF, the most convenient way to estimate the diffusion profile is through the autocorrelation of the constraint force, following the work of Marrink and Berendsen.
During harmonically restrained simulations, as for umbrella sampling PMF reconstruction, position dependent diffusion coefficients can be estimated within each window using theory formally derived by Hummer (60). If Di(z) is the local diffusion constant at the position of the harmonic potential minimum used to construct the ithwindow, its value can be estimated using equation 5:
| 5 |
where 〈δz2〉i is the mean square-fluctuation of the center of mass of the small molecule along the membrane normal from the ith umbrella window. Similarly, the autocorrelation time of the mean-square fluctuation of the center of mass of the small molecule from the ith umbrella window is given τi. The autocorrelation time is calculated by integrating the autocorrelation function of the mean fluctuations of the center of mass of the molecule along the membrane normal according to equation 6:
| 6 |
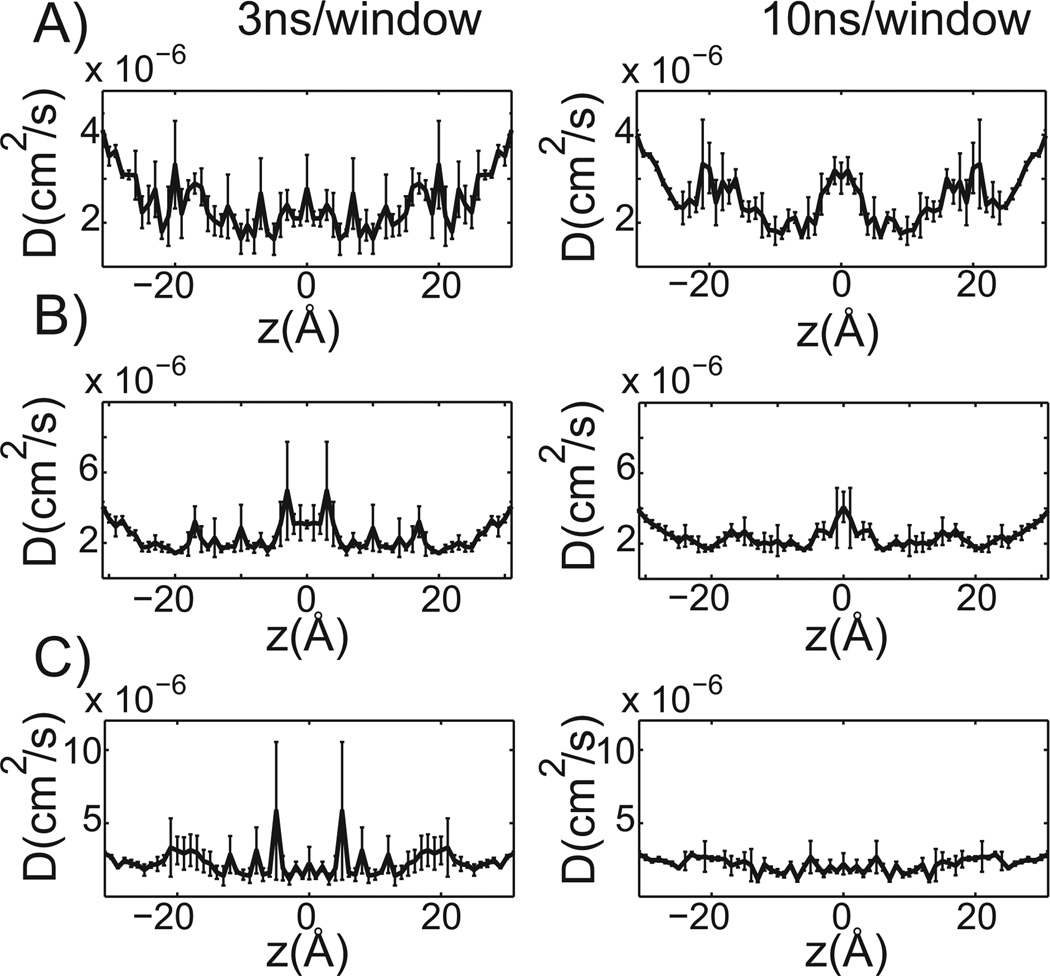
Unlike the PMF estimates, the diffusion profiles determined are noisy (Figure 4). Extending the simulation time modestly improves the results, as indicated by the position-dependent diffusion profiles for 3 ns and 10 ns of sampling per window for propranolol, terbutaline, and verapamil (Figure 4). All of the diffusion profiles are similarly shaped with slightly higher diffusivity at the bilayer center and lower diffusivity in hydrophobic tail region of the leaflets. While the estimated order of magnitude for the local diffusion profile agrees with values others have estimated for similarly sized molecules (37), the similarity and noisiness of the profiles may be insufficient to discriminate among structurally related small molecules as would be important in a lead optimization setting.
Figure 4.
Diffusion profiles at 3 ns and 10 ns of sampling per window. A) through C): 3 ns of sampling per window is shown on the left, while 10 ns per window is on the right. The membrane center is positioned at 0 Å and the standard error is reported. A) Propranolol. B) Terbutaline. C) Verapamil.
Looking forward prospectively, the validity of the estimators, as well as the affects of the biased simulation, need to be considered. For example, equation 5 assumes over-damped Langevin solute dynamics (23, 34, 59), which implies the solute undergoes Brownian motion (62). The extent to which this assumption holds at various points throughout the transport process has not been probed in detail. Additionally, simple unbiased positive controls have yet to be compared to estimates from biased simulations in regions where these comparisons are practical; e.g. at the membrane interface and in bulk water. While there are likely other outstanding issues that haven’t been mentioned here, careful study of the two issues raised may lead to long-term improvements in the accuracy of position dependent diffusion estimation. Such improvement will positively impact the reliability of transport process rate estimates.
3.2.c Uniting thermodynamics and kinetics to estimate passive-membrane permeability
The potentials of mean force and diffusion profiles described and reported in sections 3.2a and 3.2b can be incorporated to yield a permeability estimate. The local partition function, K(z), in equation 1, can be re-expressed in terms of the local solute concentration, at a depth z in the membrane, relative to the solute concentration in bulk water. Likewise, this can be related to the PMF given by equation 4, which allows equation 1 to be re-written as follows, in equation 7:
| 7 |
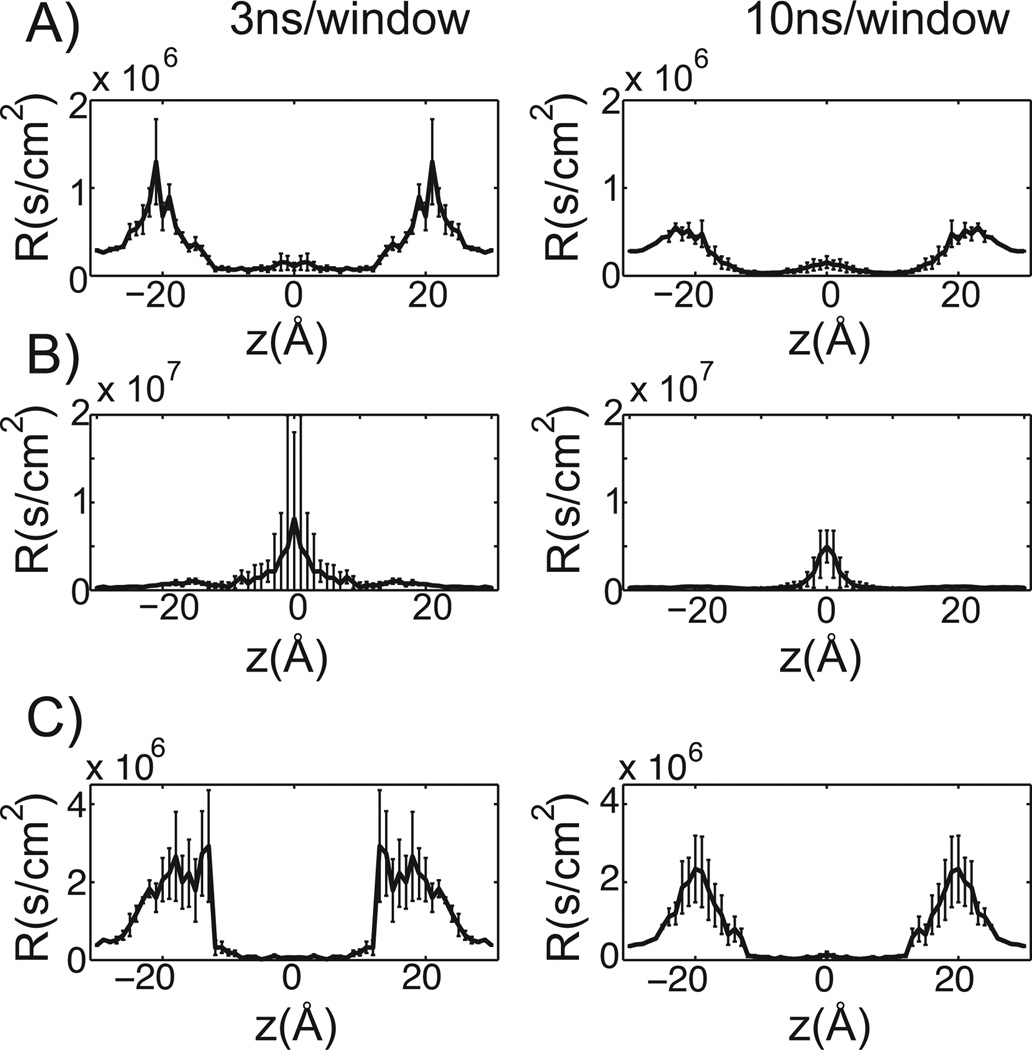
Equation 7 is then integrated across the bilayer and inverted to yield a permeability estimate. Regions where the PMF is much larger than RT, or the average thermal energy available to move the solute over the barrier, contribute exponentially to transport resistivity, while resistivity falls inversely with increasing diffusivity. Consequently, a solute is more likely to get “stuck” in a region bounded by large PMF barriers than a region bounded by low diffusivity. Figure 5 shows the predicted resistivity profiles for propranolol, terbutaline, and verapamil after 3 ns and 10ns of sampling per window. Standard errors are reported and were calculated across the 3 simulations corresponding to each solute starting orientation. Similar to the PMF estimates, the general shape of the resistivity curves are roughly converged after only 3 ns/window. While additional sampling decreases resistivity barrier heights and standard error, it has a modest affect on the estimated permeability (table 1)
Figure 5.
Resistivity profiles at 3 and 10ns of sampling per window. In figures A) through C), 3ns of sampling per window is shown on the left, while 10ns per window is on the right. The membrane center is positioned at 0 Å and the standard error is reported. A) Propranolol. B) Terbutaline. C) Verapamil.
Table 1.
The effect of simulation length on permeability estimates.
| Compound | log(P): 3 ns/window | Log(P): 10 ns/window | Log(P): experiment |
|---|---|---|---|
| Propanolol | −6.72 | −5.98 | 0.43 |
| Terbutaline | −7.94 | −6.96 | −7.25 |
| Verapamil | −7.57 | −7.01 | 0.26 |
The transport of propranolol and verapamil are most hindered at the membrane water interface, whereas terbutaline passage is significantly slowed at the center of the bilayer. This may be partly explained by considering the fraction of hydrophilic surface area for each compound (63). At roughly 31 percent, the hydrophilic component of the surface area of terbutaline is much larger than that of propranol (8 percent) and verapamil (5 percent). The larger hydrophilic surface area reduces the equilibrium tendency to partition into the hydrophobic center of the bilayer, as indicated by the roughly 5 kcal/mol PMF barrier for terbutaline (Figure 3B). The diffusion profile of terbutaline, on the other hand, is reasonably flat (Figure 4B). Consequently, the terbutaline resistivity barrier is largely a thermodynamic effect. This brief analysis speaks to the potential power of all atom permeability estimates. Macroscopically observable transport properties can be explained using a microscopic statistical mechanical description, which can be tied to physical molecular descriptors, like solvent accessible surface area. As physical molecular descriptors are systematically varied during lead optimization, QSPR can be mechanistically rationalized on a microscopic basis using accurate all atom permeability estimates, information that may prove invaluable during rational drug design efforts. A similar strategy was recently proposed to integrate information from implicit solvent models in ref (32).
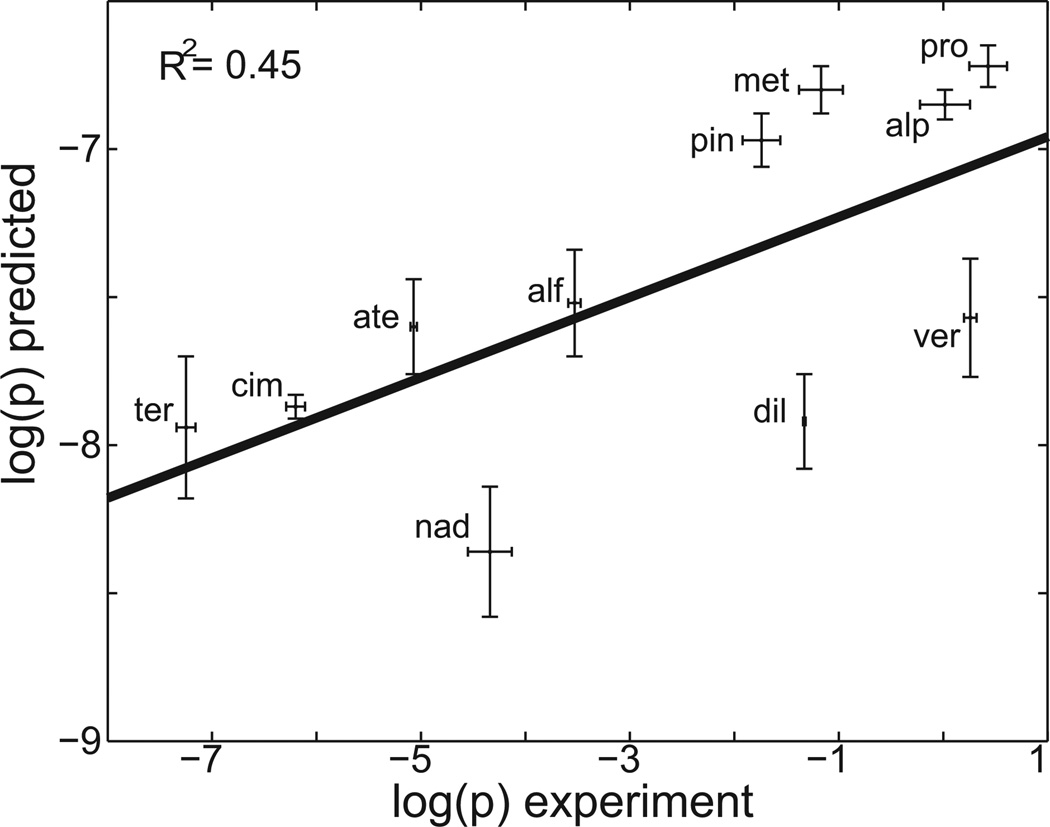
How accurate are all-atom permeability estimates? To answer this, in figure 6, the computationally predicted permeability, determined with 3 ns of sampling per window, is plotted as a function of the experimental permeability for the set of small molecules shown in figure 2. Using error propagation techniques, standard errors of the logged permeability estimates were calculated across the 3 simulations (see supporting information), corresponding to each solute starting orientation, and are shown as vertical error bars. Standard errors of the experimental permeabilities are shown as horizontal error bars and were calculated from (64). The experimental and computational values used to create the plot are provided in supporting information table S1. While the trend is promising, the coefficient of determination is 0.45, and the predicted values span only 1.5 orders of magnitude, while the experimental values range over 8 orders of magnitude. Although this is a modest improvement over a simple “null” model, which plots experimental permeability estimates versus molecular weight and gives a coefficient of determination of 0.33, stronger correlation is desirable. Table 1 shows that while increasing simulation time by more than 300 percent increases the estimated permeability rate by nearly an order of magnitude, estimates are still confined to a narrow range, relative to experiment. It may be that still more sampling is required. For example, when umbrella sampling with membrane-embedded leucine and arginine side chain analogs was carried out for over 200ns/window, the bilayer surface depressed or protruded to accommodate the side chain analog, leading to improved accuracy of the estimated water-membrane partition coefficient (38). However, because bilayer reorganization properties were different in harmonically restrained and unrestrained simulations, the relevance of the membrane structural reorganization observed during the restrained simulations is questionable (38). It may prove that non-equilibrium methods, such as SMD, carried out with a stiff spring and slow pulling speed (65–67), provide a better model of the naturally occurring transport process. Alternatively, promising emerging new methods, such as ABF, discussed in section 3.1, which would allow the solute to freely diffuse along the membrane normal, may prove more suitable.
Figure 6.
Computationally predicted vs. experimentally measured permeability values. Experimental values were measured in a PAMPA assay (63) and are shown along the horizontal axis. Computationally predicted values were calculated using equation 7 and are shown along the vertical axis. Reported estimates used 3 ns of sampling per window. Standard errors of the computed estimates are shown as vertical error bars, while standard errors of the experimental values are shown as horizontal error bars. Reported experimental standard errors were scaled by a factor of three to enhance their visibility. Data points are labeled with the corresponding three letter compound name abbreviations included in figure 2. The coefficient of determination, R2, is reported.
Neglect of alternative ligand protonation and tautomerization states are additional simplifying assumptions that may significantly impact the accuracy of passive membrane permeability estimates. For example, different tautomerization states may be thermodynamically favored, and can have higher or lower diffusivity, at different depths in the membrane, appreciably contributing to the experimental permeability measurement. Alternative protonation states, on the other hand, while seemingly more impactful because of larger columbic effects, may be less problematic, in some cases, than alternative tautomers. In part, this is because experimental permeability is often reported as an “intrinsic” permeability, or the permeability of the uncharged species alone (64), which was the case of the 11 compounds considered in this work. Additionally, charged species are less likely to dissolve to a significant extent in the greasy bilayer, so considering only electrically neutral compounds may be sufficient. However, charged compounds can be partially hydrated in the bilayer (68), and simply discounting their contribution might result in inaccurate permeability estimates.
4. Conclusions and future prospects
The limited accuracy of the exploratory explicit-solvent, all atom work presented here suggests that the answer to the question posed in this paper’s title (can physical models of passive membrane permeability help reduce drug candidate attrition and move us beyond QSPR?) is “no, at least not yet.” However, systematic exploration of PMF and diffusion estimators, refinement of ligand atomic partial charges to better match both the bilayer and aqueous environments, and inclusion of alternative tautomerization and protonation states, may facilitate more accurate permeability estimates. But does the promise of improved results justify the intellectual and computational investment that will be required to improve prediction accuracy? Critics will argue that the ends don’t justify the means. They will support their view by citing the strong experimental correlation that can be produced by carefully developed QSPR models (or even the implicit solvent models discussed here), which provide a satisfying connection between small molecule structure and experimentally observed membrane transport rates. While this is true, reliable QSPR models require a substantial front-end investment. Large sets of training molecules are necessary to elucidate the proper functional relationship between molecular descriptor values and experimentally determined membrane permeability rates. Additionally, for the best performance, QSPR models require that training molecules be closely related to test molecules, those whose permeability values will be predicted. Test molecules that are substantially different from training molecules may result in grossly inaccurate predictions. As a result, if a structurally homologous lead series is not available commercially or in-house, developing a successful QSPR model minimally requires the synthesis and testing of a series of related compounds, which may be impractical under budget and time constraints.
On the other hand, both the implicit solvent and explicit solvent models discussed rely on a molecular mechanics-based description of the system to estimate experimental permeability values, and so neither requires training sets. In addition to potentially significant experimental cost savings, the absence of training set requirements implies that these physical models won’t “break,” provided the underlying theory is correct, when molecules with very different structures are considered. However, because the implicit solvent models discard membrane anisotropy in favor of a highly simplified, homogenous low-dielectric slab, the level of mechanistic detail that can be extracted, even from a perfectly accurate calculation, is fundamentally limited. In contrast, because all-atom, physics-based models entail an atomistic description of the entire system, the relationship between small molecule structure and experimental measurements can be spatially resolved in terms of characteristic small molecule interactions with different parts of the system; e.g. interactions at the membrane water interface, with headgroups, or at the bilayer center. Such spatial resolution may be extremely valuable when rationalizing the permeability rates of a lead series or when developing a compound with desirable ADMET properties. Neither QSPR models, nor implicit solvent models, even with perfect accuracy, will ever yield such mechanistic detail, and are limited by comparison.
Still, critics will claim that despite the greater detail provided by explicit-solvent all-atom physics-based models, their computational requirements make them impractical. Their claim has (some) merit. For example, the exploratory work considered here required nearly 212,000 CPU hours. Assuming a devoted cluster of 560 processors running continuously, it would take nearly 16 days to complete these simulations. If two to four days are required for set up and analysis, a total of up to 20 days would elapse before the results were available. Even if the predictions were perfectly accurate, this may be too long to be practical (69) This may explain why a number of studies have investigated the transport of one or several molecules using one particular method (for examples, see refs (35–37), while systematic investigations comparing different methods (and their associated statistical and systematic errors) across larger sets of small molecules have not been reported. While the number of calculations that must be performed may seem limiting, contemporary software and hardware advances have enabled long MD simulations to be carried out on graphics processing units (GPUs), which has reduced the real-time requirement relative to that of a conventional CPU. Our own single-GPU MD benchmark calculations on so-called “gaming cards” indicate that standard codes routinely achieve benchmarks of ~30–50 ns / day for systems of appreciable size. With the promise of massively parallel GPU computing on the horizon (70, 71), in the very near future, the real-time requirements of these simulations will be routinely met under practical, fast-paced time constraints. Thus, despite the substantial intellectual and computational investment required to improve the accuracy and utility of all-atom, physics-based models, the return on investment to be significant and justifiable.
Supplementary Material
Acknowledgements
This work was funded in part by the National Institutes of Health through the NIH Director's New Innovator Award Program 1-DP2-OD007237. REA thanks the participants at the 2012 Free Energy Methods Workshop, especially Drs. Chris Chipot, Thomas Woolf, Jonathan Essex, and Pat Walters, for inspiring discussions. RVS thanks members of the Amaro lab for considerable discussion, in particular Drs. Vicki Feher, Özlem Demir, Luke Czapla, and Jesper SØrensen. RVS especially thanks Dr. Sara E. Nichols.
Footnotes
Supporting Information
Simulation Methods. A more detailed description of the reported simulation protocol.
Standard Error. Describes how standard error was calculated for the PMF, diffusion, and resistivity profiles, and logged peremeability estimates.
Table S1. Experimental and computationl permeability estimates and their standard errors.
References
- 1.Schuster D, Laggner C, Langer T. Why drugs fail--a study on side effects in new chemical entities. Curr Pharm Des. 2005;11:3545–3559. doi: 10.2174/138161205774414510. [DOI] [PubMed] [Google Scholar]
- 2.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–716. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 3.DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 4.Minneman K. Brody's Human Pharmacology: Molecular to Clinical. 5th Edition edn. Elsevier; 2010. [Google Scholar]
- 5.Kansy M, Senner F, Gubernator K. Physicochemical High Throughput Screening: Parallel Artificial Membrane Permeation Assay in the Description of Passive Absorption Processes. Journal of Medicinal Chemistry. 1998;41:1007–1010. doi: 10.1021/jm970530e. [DOI] [PubMed] [Google Scholar]
- 6.Avdeef A. The rise of PAMPA. Expert Opin Drug Metab Toxicol. 2005;1:325–342. doi: 10.1517/17425255.1.2.325. [DOI] [PubMed] [Google Scholar]
- 7.Artursson P, Palm K, Luthman K. Caco-2 monolayers in experimental and theoretical predictions of drug transport. Advanced Drug Delivery Reviews. 2001;46:27–43. doi: 10.1016/s0169-409x(00)00128-9. [DOI] [PubMed] [Google Scholar]
- 8.van Breemen RB, Li Y. Caco-2 cell permeability assays to measure drug absorption. Expert Opinion on Drug Metabolism & Toxicology. 2005;1:175–185. doi: 10.1517/17425255.1.2.175. [DOI] [PubMed] [Google Scholar]
- 9.Hansch C. Quantitative approach to biochemical structure-activity relationships. Accounts of Chemical Research. 1969;2:232–239. [Google Scholar]
- 10.Hansch C. Quantitative Relationships Between Lipophilic Character and Drug Metabolism. Drug Metabolism Reviews. 1972;1:1–13. [Google Scholar]
- 11.van de Waterbeemd H, Gifford E. ADMET in silico modelling: towards prediction paradise? Nat Rev Drug Discov. 2003;2:192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Engkvist O, Llinas A, Chen H. Beyond size, ionization state, and lipophilicity: influence of molecular topology on absorption, distribution, metabolism, excretion, and toxicity for druglike compounds. J Med Chem. 2012;55:3667–3677. doi: 10.1021/jm201548z. [DOI] [PubMed] [Google Scholar]
- 13.Egan WJ, Merz KM, Baldwin JJ. Prediction of Drug Absorption Using Multivariate Statistics. Journal of Medicinal Chemistry. 2000;43:3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 14.Ano R, Kimura Y, Shima M, Matsuno R, Ueno T, Akamatsu M. Relationships between structure and high-throughput screening permeability of peptide derivatives and related compounds with artificial membranes: application to prediction of Caco-2 cell permeability. Bioorganic & Medicinal Chemistry. 2004;12:257–264. doi: 10.1016/j.bmc.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Liu R, Sun H, So S-S. Development of Quantitative Structure?Property Relationship Models for Early ADME Evaluation in Drug Discovery. 2. Blood-Brain Barrier Penetration. Journal of Chemical Information and Computer Sciences. 2001;41:1623–1632. doi: 10.1021/ci010290i. [DOI] [PubMed] [Google Scholar]
- 16.Ekins S, Waller CL, Swaan PW, Cruciani G, Wrighton SA, Wikel JH. Progress in predicting human ADME parameters in silico. Journal of Pharmacological and Toxicological Methods. 2000;44:251–272. doi: 10.1016/s1056-8719(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 17.Katritzky AR, Kuanar M, Slavov S, Hall CD, Karelson M, Kahn I, et al. Quantitative Correlation of Physical and Chemical Properties with Chemical Structure: Utility for Prediction. Chemical Reviews. 2010;110:5714–5789. doi: 10.1021/cr900238d. [DOI] [PubMed] [Google Scholar]
- 18.Stouch TR, Kenyon JR, Johnson SR, Chen X-Q, Doweyko A, Li Y. In silico ADME/Tox: why models fail. Journal of Computer-Aided Molecular Design. 2003;17:83–92. doi: 10.1023/a:1025358319677. [DOI] [PubMed] [Google Scholar]
- 19.Finkelstein A. Water and nonelectrolyte permeability of lipid bilayer membranes. J Gen Physiol. 1976;68:127–135. doi: 10.1085/jgp.68.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolosin JM, Ginsburg H. The permeation of organic acids through lecithin bilayers Resemblance to diffusion in polymers. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1975;389:20–33. doi: 10.1016/0005-2736(75)90382-x. [DOI] [PubMed] [Google Scholar]
- 21.Träuble H. The movement of molecules across lipid membranes: A molecular theory. Journal of Membrane Biology. 1971;4:193–208. doi: 10.1007/BF02431971. [DOI] [PubMed] [Google Scholar]
- 22.Potts RO, Francoeur ML. Lipid biophysics of water loss through the skin. Proceedings of the National Academy of Sciences. 1990;87:3871–3873. doi: 10.1073/pnas.87.10.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marrink S-J, Berendsen HJC. Simulation of water transport through a lipid membrane. The Journal of Physical Chemistry. 1994;98:4155–4168. [Google Scholar]
- 24.Xiang T, Xu Y, Anderson BD. The barrier domain for solute permeation varies with lipid bilayer phase structure. J Membr Biol. 1998;165:77–90. doi: 10.1007/s002329900422. [DOI] [PubMed] [Google Scholar]
- 25.Bashford D, Case DA. Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 26.Kalyanaraman C, Jacobson M. An atomistic model of passive membrane permeability: application to a series of FDA approved drugs. Journal of Computer-Aided Molecular Design. 2007;21:675–679. doi: 10.1007/s10822-007-9141-z. [DOI] [PubMed] [Google Scholar]
- 27.Rezai T, Bock JE, Zhou MV, Kalyanaraman C, Lokey RS, Jacobson MP. Conformational Flexibility, Internal Hydrogen Bonding, and Passive Membrane Permeability: successful in Silico Prediction of the Relative Permeabilities of Cyclic Peptides. Journal of the American Chemical Society. 2006;128:14073–14080. doi: 10.1021/ja063076p. [DOI] [PubMed] [Google Scholar]
- 28.Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. Testing the Conformational Hypothesis of Passive Membrane Permeability Using Synthetic Cyclic Peptide Diastereomers. Journal of the American Chemical Society. 2006;128:2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]
- 29.Swift RV, Amaro RE. Modeling the pharmacodynamics of passive membrane permeability. Journal of Computer-Aided Molecular Design. 2011;25:1007–1017. doi: 10.1007/s10822-011-9480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolossváry I. Evaluation of the Molecular Configuration Integral in All Degrees of Freedom for the Direct Calculation of Binding Free Energies: Application to the Enantioselective Binding of Amino Acid Derivatives to Synthetic Host Molecules. Journal of the American Chemical Society. 1997;119:10233–10234. [Google Scholar]
- 31.Potter MJ, Gilson MK. Coordinate Systems and the Calculation of Molecular Properties. The Journal of Physical Chemistry A. 2001;106:563–566. [Google Scholar]
- 32.Leung SSF, Mijalkovic J, Borrelli K, Jacobson MP. Testing Physical Models of Passive Membrane Permeation. Journal of Chemical Information and Modeling. 2012 doi: 10.1021/ci200583t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bassolino-Klimas D, Alper HE, Stouch TR. Solute diffusion in lipid bilayer membranes: An atomic level study by molecular dynamics simulation. Biochemistry. 1993;32:12624–12637. doi: 10.1021/bi00210a010. [DOI] [PubMed] [Google Scholar]
- 34.Bemporad D, Essex JW, Luttmann C. Permeation of Small Molecules through a Lipid Bilayer:Äâ A Computer Simulation Study. The Journal of Physical Chemistry B. 2004;108:4875–4884. [Google Scholar]
- 35.Paloncýová M, Berka K, Otyepka M. Convergence of Free Energy Profile of Coumarin in Lipid Bilayer. Journal of Chemical Theory and Computation. 2012;8:1200–1211. doi: 10.1021/ct2009208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulander J, Haymet ADJ. Permeation Across Hydrated DPPC Lipid Bilayers: Simulation of the Titrable Amphiphilic Drug Valproic Acid. Biophys J. 2003;85:3475–3484. doi: 10.1016/S0006-3495(03)74768-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boggara MB, Krishnamoorti R. Partitioning of Nonsteroidal Antiinflammatory Drugs in Lipid Membranes: A Molecular Dynamics Simulation Study. Biophys J. 2010;98:586–595. doi: 10.1016/j.bpj.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neale C, Bennett WFD, Tieleman DP, Pomès R. Statistical Convergence of Equilibrium Properties in Simulations of Molecular Solutes Embedded in Lipid Bilayers. Journal of Chemical Theory and Computation. 2011;7:4175–4188. doi: 10.1021/ct200316w. [DOI] [PubMed] [Google Scholar]
- 39.Ghaemi Z, Minozzi M, Carloni P, Laio A. A Novel Approach to the Investigation of Passive Molecular Permeation through Lipid Bilayers from Atomistic Simulations. The journal of physical chemistry B. 2012;116:8714–8721. doi: 10.1021/jp301083h. [DOI] [PubMed] [Google Scholar]
- 40.Torrie GM, Valleau JP. Monte Carlo free energy estimates using non-Boltzmann sampling: Application to the sub-critical Lennard-Jones fluid. Chemical Physics Letters. 1974;28:578–581. [Google Scholar]
- 41.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 42.Klauda JB, Venable RM, Freites JA, OConnor JW, Tobias DJ, Mondragon-Ramirez C, et al. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. The Journal of Physical Chemistry B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 45.Shirts MR, Chodera JD. Statistically optimal analysis of samples from multiple equilibrium states. The Journal of Chemical Physics. 2008;129:124105–124110. doi: 10.1063/1.2978177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yesselman JD, Price DJ, Knight JL, Brooks CL. MATCH: An atom-typing toolset for molecular mechanics force fields. J Comput Chem. 2012;33:189–202. doi: 10.1002/jcc.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Young LR, Dill KA. Solute partitioning into lipid bilayer membranes. Biochemistry. 1988;27:5281–5289. doi: 10.1021/bi00414a050. [DOI] [PubMed] [Google Scholar]
- 49.Cieplak P, Cornell W, Bayly C, Kollman P. Application of the Multimolecule and Multiconformational RESP Methodology to Biopolymers: Charge Derivation for DNA, RNA, and Proteins. J Comput Chem. 1995;16:1357–1377. [Google Scholar]
- 50.Mackerell AD., Jr Empirical force fields for biological macromolecules: overview and issues. J Comput Chem. 2004;25:1584–1604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 51.Lopes PE, Roux B, Mackerell AD., Jr Molecular modeling and dynamics studies with explicit inclusion of electronic polarizability. Theory and applications. Theor Chem Acc. 2009;124:11–28. doi: 10.1007/s00214-009-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Gomez D, Darve E, Pohorille A. Assessing the efficiency of free energy calculation methods. The Journal of Chemical Physics. 2004;120:3563–3578. doi: 10.1063/1.1642607. [DOI] [PubMed] [Google Scholar]
- 53.Darve E, Pohorille A. Calculating free energies using average force. The Journal of Chemical Physics. 2001;115:9169–9183. [Google Scholar]
- 54.Darve E, Wilson MA, Pohorille A. Calculating free energies using a scaled-force molecular dynamics algorithm. Mol Simul. 2002;28:113–144. [Google Scholar]
- 55.Henin J, Chipot C. Overcoming free energy barriers using unconstrained molecular dynamics simulations. J Chem Phys. 2004;121:2904–2914. doi: 10.1063/1.1773132. [DOI] [PubMed] [Google Scholar]
- 56.Darve E, Chipot C, Pohorille A. Free Energy Calculations. 2007 [Google Scholar]
- 57.Schulten K, Schulten Z, Szabo A. Dynamics of reactions involving diffusive barrier crossing. The Journal of Chemical Physics. 1981;74:4426–4432. [Google Scholar]
- 58.Straub JE, Borkovec M, Berne BJ. Calculation of dynamic friction on intramolecular degrees of freedom. The Journal of Physical Chemistry. 1987;91:4995–4998. [Google Scholar]
- 59.Woolf TB, Roux B. Conformational Flexibility of o-Phosphorylcholine and o-Phosphorylethanolamine: A Molecular Dynamics Study of Solvation Effects. Journal of the American Chemical Society. 1994;116:5916–5926. [Google Scholar]
- 60.Hummer G. Position-dependent diffusion coefficients and free energies from Bayesian analysis of equilibrium and replica molecular dynamics simulations. New Journal of Physics. 2005;7:34. [Google Scholar]
- 61.Kosztin I, Barz B, Janosi L. Calculating potentials of mean force and diffusion coefficients from nonequilibrium processes without Jarzynski's equality. The Journal of Chemical Physics. 2006;124:064106–064111. doi: 10.1063/1.2166379. [DOI] [PubMed] [Google Scholar]
- 62.Zwanzig R. Nonequilibrium Statistical Mechanics. Oxford: Oxford University Press; 2001. [Google Scholar]
- 63.QikProp v, Schrodinger LLC. New York, NY: 2009. [Google Scholar]
- 64.Avdeef A, Artursson P, Neuhoff S, Lazorova L, Gråsjö J, Tavelin S. Caco-2 permeability of weakly basic drugs predicted with the Double-Sink PAMPA method. European Journal of Pharmaceutical Sciences. 2005;24:333–349. doi: 10.1016/j.ejps.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 65.Jensen MØ, Park S, Tajkhorshid E, Schulten K. Energetics of glycerol conduction through aquaglyceroporin GlpF. Proceedings of the National Academy of Sciences. 2002;99:6731–6736. doi: 10.1073/pnas.102649299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park S, Schulten K. Calculating potentials of mean force from steered molecular dynamics simulations. The Journal of Chemical Physics. 2004;120:5946–5961. doi: 10.1063/1.1651473. [DOI] [PubMed] [Google Scholar]
- 67.Amaro RE, Tajkhorshid E, Luthey-Schulten Z. Developing an energy landscape for the novel function of a (β/α)8 barrel: Ammonia conduction through HisF. Proceedings of the National Academy of Sciences. 2003;100:7599–7604. doi: 10.1073/pnas.1331150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schow EV, Freites JA, Cheng P, Bernsel A, von Heijne G, White SH, et al. Arginine in membranes: the connection between molecular dynamics simulations and translocon-mediated insertion experiments. J Membr Biol. 2011;239:35–48. doi: 10.1007/s00232-010-9330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drug Design: Structure- and Ligand-Based Approaches. Cambridge, New York, Melbourne, Madrid, Cape Town, Singapore, Sao Paulo, Delhi, Dubai, Tokyo: Cambridge University Press; 2010. [Google Scholar]
- 70.Harvey MJ, De Fabritiis G. High-throughput molecular dynamics: the powerful new tool for drug discovery. Drug Discovery Today. 2012 doi: 10.1016/j.drudis.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 71.Baker JA, Hirst JD. Molecular Dynamics Simulations Using Graphics Processing Units. Molecular Informatics. 2011;30:498–504. doi: 10.1002/minf.201100042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.