Abstract
Background
Cyclophilin A (CypA) is vital for HCV replication. Cyp inhibitors successfully decrease viral loads in HCV-infected patients. However, their mechanisms of action remain unknown. Since interferon (IFN) can also suppress HCV replication, we asked whether a link between CypA and the IFN response exists.
Methods
We used cellular and recombinant pulldown approaches to investigate the possibility of a specific association of CypA with host ligands.
Results
We found for the first time that CypA binds to a major component of the IFN response – the IFN regulatory factor 9 (IRF9). IRF9 is the DNA-binding component of the transcriptional IFN-stimulated gene factor 3 (ISGF3). CypA binds directly IRF9 via its peptidyl-prolyl isomerase (PPIase) pocket. Cyp inhibitors such as cyclosporine A (CsA) or non-immunosuppressive derivates such as alisporivir and SCY-635, prevent IRF9-CypA complex formation. CypA binds to the C-terminal IRF-association-domain (IAD), but not to the DNA-binding or linker domains of IRF9. Remarkably, CypA associates with the multimeric ISGF3 complex. We also obtained evidence that CypA neutralization enhances IFN-induced transcription. Interestingly, the hepatitis C virus (HCV) nonstructural 5A (NS5A) protein, which is known to modulate the IFN response, competes with IRF9 for CypA binding and can prevent the formation of IRF9-CypA complexes.
Conclusions
This study demonstrates for the first time that CypA binds specifically to a component of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway - IRF9. This study also reveals a novel opportunity of HCV to modulate the IFN response via NS5A.
INTRODUCTION
Following viral infection, IFN is produced in a biphasic fashion that involves a number of transcription factors, including the IFN regulatory factors (IRFs) 1, 3, 7, and 9 [1–5]. Because HCV replicates via a double-stranded RNA (dsRNA) intermediate, it activates protein kinase R (PKR) [6, 7], IRF-1 [6, 8], and IRF-3 [8, 9] and downstream antiviral genes that are activated by these factors [10, 11]. However, HCV persists in the liver despite strong ISG induction [12–15]. HCV evolved to develop multiple strategies to attenuate the IFN response. A key player is the HCV NS3/4A protein, which cleaves the adapter molecules TRIF and IPS-1 [16], and thereby, blocks TLR3 and RIG-I signaling [17]. Another key player is the core, which interferes with JAK/STAT signaling and ISG expression [18]. Another key protein, which attenuates the IFN response, is NS5A. NS5A interferes directly with the function of ISGs by i) inhibiting 2′–5′ oligoadenylate synthetase (2′–5′ OAS) [19]; ii) inducing IL-8, which inhibits overall ISG expression [20]; and iii) inhibiting PKR function [21].
NS5A was found to interact with CypA [22–29]. Importantly, Cyp inhibitors, which bind to the isomerase pocket of CypA, abrogate CypA-NS5A interactions and block HCV replication both in vitro and in vivo [23, 26, 30–33]. Interestingly, a recent study showed that the administration of the Cyp inhibitor SCY-635 to HCV patients modulates the IFN response [34]. Specifically, SCY-635 triggered rapid elevation of IFN, IFN 1, IFN 3, and 2′–5′ OAS plasma levels [34]. These findings suggested a link between HCV, CypA and the IFN response. We thus in this study investigated whether CypA associates directly or indirectly with components of the IFN response.
MATERIAL AND METHODS
Drugs and plasmids
Alisporivir was obtained from Novartis, SCY-635 from SCYNEXIS, sanglifehrin B from Biotica, and CsA, FK506 and juglone from Calbiochem. Human CypA-Strep and Flag-TLR3 were cloned into the pcDNA3 vector. Human IRF9-HA vector was purchased from InvivoGen, the C-terminal HA removed, an N-terminal Flag added and the Flag-IRF9 recloned into pcDNA3. All IRF9 truncations were created in the same Flag-IRF9 vector. Flag-STAT1 and -STAT2 plasmids were obtained from A. García-Sastre. Con1 NS5A-Flag plasmid was obtained from P. Targett-Adams.
Co-precipitations and liquid chromatography/tandem mass spectrometry (LC-MS/MS) studies
Cellular precipitations followed by LC-MS/MS analyses were conducted as described in Supplementary Material.
Interaction studies with recombinant proteins
Production of recombinant proteins, pulldown and ELISA studies were conducted as described in Supplementary Material.
IFN-induced transcriptional studies
HepG2 cells were pretreated with DMSO, IFN (10 U), CsA (2 µM) or a combination of IFN and CsA. Eight hours post-treatment, cells were transfected with plasmids encoding luciferase driven either by an ISRE (Stratagene), IFNA1 or IFNA4 promoter (10 µg). Luciferase activity was analyzed after 24 h. Relative luciferase activity was measured as fold activation (relative to the basal level for the reporter gene in the presence of DMSO after normalization to co-transfected relative light unit activity). The CypA knockdown was conducted 3 days before IFN treatment as we described previously [35]. To analyze the effect of NS5A on IFN-induced transcription, cells were transfected with NS5A plasmid (15 µg) 3 days before IFN treatment. Amounts of 2',5'-oligoadenylate synthetase (2’,5’-OAS) in Amicon filter-concentrated HepG2 cell lysates was quantified by ELISA (USCN Life Science Inc) following the manufacturer’s instructions.
RESULTS
CypA and IRF9 form complexes
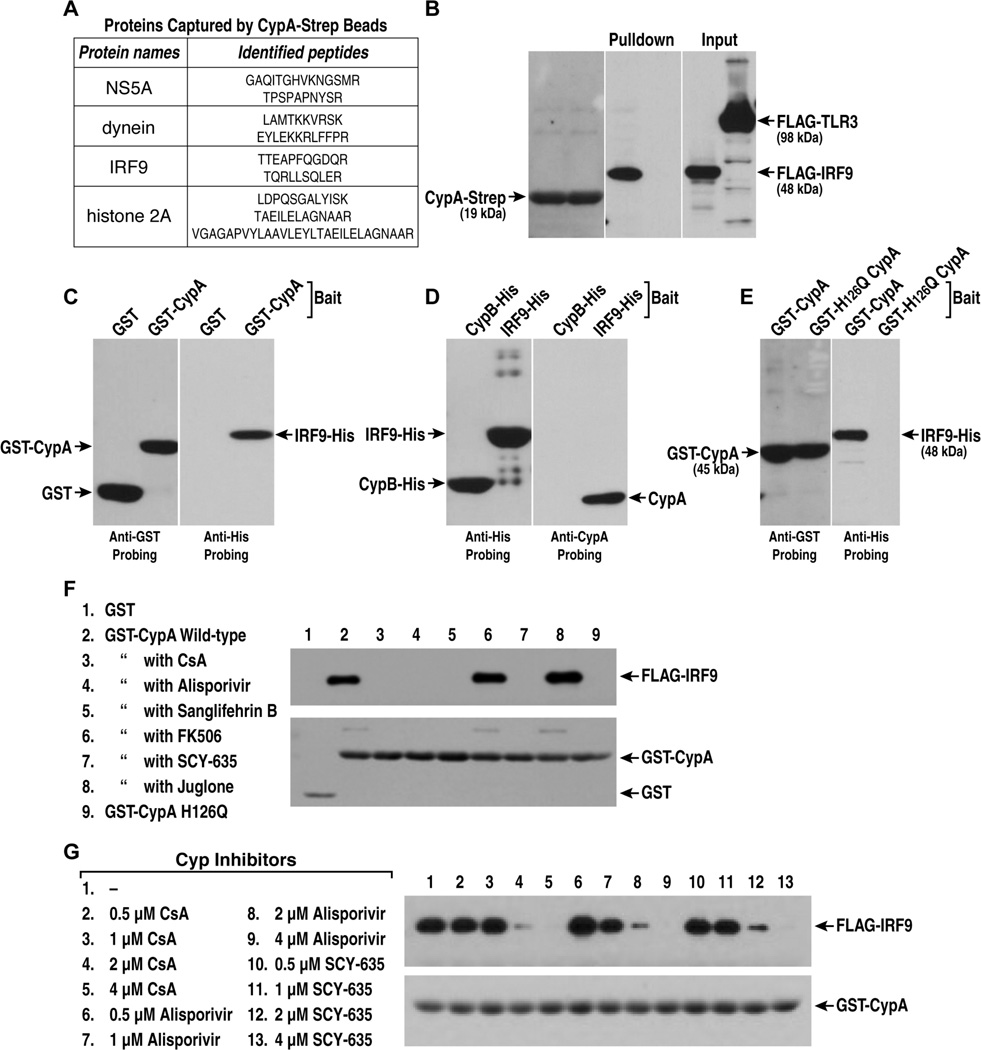
To investigate the possibility that CypA is linked to the IFN response, we asked whether CypA associates with elements of the innate response. To address this issue, we fused a Strep-tag peptide to the N-terminus of human CypA and used this fusion protein as bait to recover CypA-interacting proteins from cell lysates of HCV-infected hepatocytes treated with or without CsA. Bound material was analyzed by LC-MS/MS as we previously reported [36]. Figure 1 shows identified peptides of known proteins pulled down in the absence, but not in the presence of CsA (Figure 1A). Among them, we identified the HCV NS5A protein (Figure 1A), further confirming that NS5A is a viral ligand for CypA [22, 23, 25–30]. Host proteins were also identified such as dynein (Figure 1A). This is in accordance with a previous study that showed that CypA associates with the dynein/dynactin motor protein complex [37]. Interestingly, one component of the IFN response – IRF9 [38] - was pulled down by CypA in the absence of CsA, but not in the presence of the drug (Figure 1A). To examine whether the CypA-IRF9 association is genuine, we co-transfected 293T cells with Flag-IRF9 and CypA-Strep plasmids, and conducted pulldowns using CypA-Strep as bait. We used Flag-TLR3 - another component of the innate response - as negative control. Importantly, CypA efficiently pulled down IRF9, but not TLR3 (Figure 1B). Except IRF9, CypA-Strep was unable to pull down all exogenous tagged host proteins tested so far including CypB, CypC, TRIM5 alpha, TBC1D20 and c-Src (data now shown).
Fig. 1. CypA binds IRF9.
A. Proteins pulled down by CypA-Strep in the absence, but not in the presence of CsA. B. 293T cells were transfected with CypA-Strep, Flag-IRF9 or Flag-TLR3. Pulled down material analyzed with anti-Strep or -Flag IgG. C. GST or GST-CypA was mixed with IRF9-His. Bound material analyzed with anti-GST or -His IgG. D. CypA was mixed with IRF9-His or CypB-His. Bound material analyzed with anti-His or -CypA IgG. E. Same as C. F. GST-CypA was mixed with lysates containing Flag-IRF9 and inhibitors. Bound material analyzed with anti-Flag and -GST IgG. G. Same as F.
CypA directly binds IRF9 via its isomerase pocket
We then asked whether the association between CypA and IRF9 is direct. To test this hypothesis, we used recombinant CypA and IRF9 and asked whether they bind together. GST-CypA, but not GST, captures IRF9-His efficiently (Figure 1C). Similarly, IRF9-His captures CypA, whereas CypB-His used as a negative control bait does not (Figure 1D). We then asked whether CypA binds IRF9 by its enzymatic pocket [39, 40]. To address this issue, we introduced the H126Q mutation into CypA that abolishes its enzymatic activity [40]. Importantly, wild-type (GST-CypA), but not the isomerase-deficient CypA (GST-H126Q CypA) captures IRF9 (Figure 1E). Together these data suggest that CypA binds IRF9 specifically and directly by its isomerase pocket.
Cyp inhibitors prevent CypA-IRF9 complex formation
Since Cyp inhibitors neutralize the enzymatic activity of CypA [39, 40], we asked whether they prevent CypA-IRF9 contacts. We used GST-CypA as bait to capture Flag-IRF9 from cell lysates in the presence or absence of Cyp inhibitors including i) the immunosuppressive Cyp inhibitor - CsA; ii) two non-immunosuppressive CsA derivates - alisporivir [41] and SCY-635 [34]; and iii) the immunosuppressive macrolide – sanglifehrin B [42]. We also used two non-Cyp inhibitors: i) the immunophilin FKBP inhibitor – FK506 [43]; and ii) the peptidyl-prolyl isomerase Pin1 inhibitor – juglone [44]. As above, GST-CypA, but not GST, captures Flag-IRF9 (Figure 1F). Importantly, all Cyp inhibitors blocked CypA-IRF9 interactions, whereas the non-Cyp inhibitors do not (Figure 1F). Remarkably, the Cyp inhibitors blocked CypA-IRF9 interactions in a dose-dependent manner (Figure 1G).
CypA is a component of the ISGF3 complex
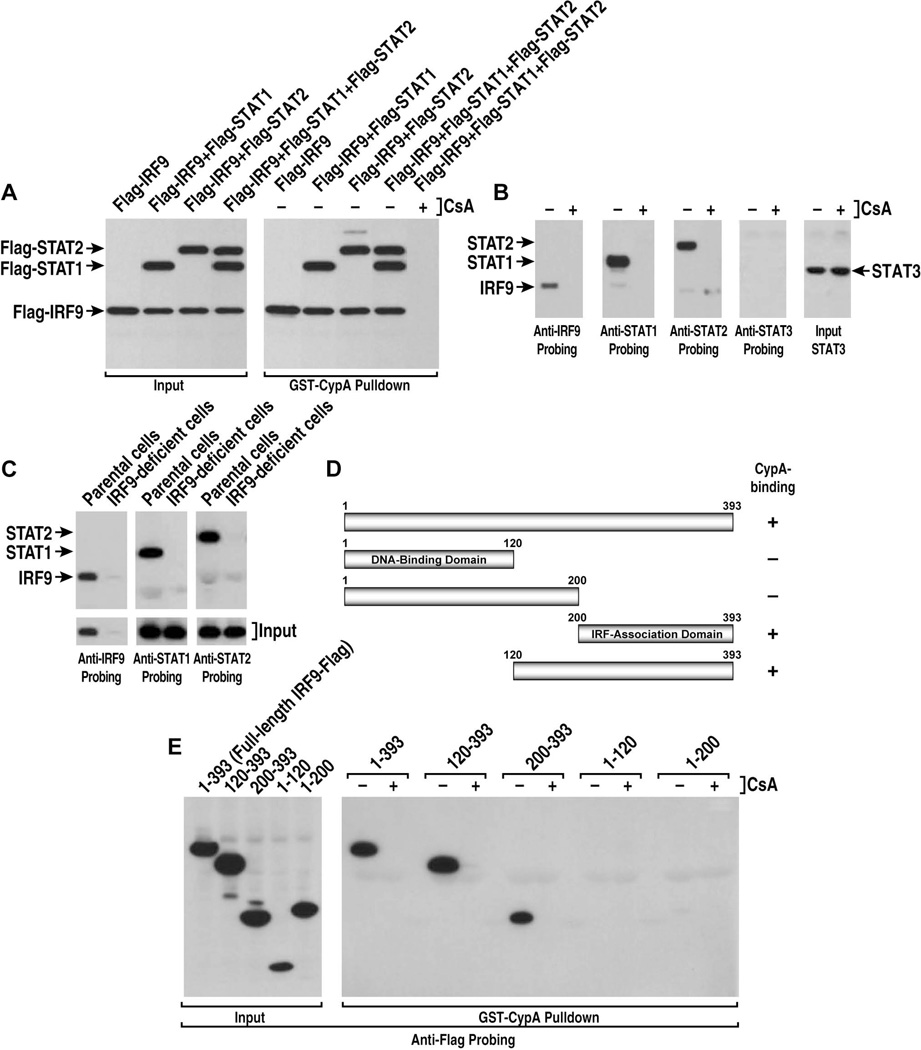
A critical event of the JAK/STAT pathway is the translocation of the ISGF3 complex to the nucleus to activate antiviral genes. The ISGF3 complex is composed of at least three proteins – STAT1, STAT2 and IRF9. We thus asked whether CypA, by interacting with IRF9, is part of this complex. To address this issue, Huh7 cells were co-transfected with Flag-IRF9, Flag-STAT1 and Flag-STAT2 and GST-CypA was used as bait to recover ISGF3 components from cell lysates. Cells were treated with IFN for 24 h before cell lysis. Importantly, GST-CypA captures all components of the ISG3 complex - IRF9, STAT1 and STAT2 (Figure 2A, right panel). All proteins were similarly expressed (Figure 2A, left panel). Note that cell lysates contain both exogenous and endogenous IRF9, STAT1 and STAT2. Remarkably, CsA prevents the capture of the trimolecular complex by GST-CypA (Figure 2A, right panel, last lane), suggesting that CypA is part of the ISGF3 complex. We obtained similar results using CypA-Strep as bait to pull down ISGF3-tagged complexes (data not shown). Rather than using exogenous proteins, we then used endogenous proteins. Importantly, anti-CypA antibodies pulled down CypA together with IRF9, STAT1 and STAT2 from lysates of IFN - treated cells (Figure 2B). CsA prevents the pulldown of IRF9, STAT1 and STAT2 by CypA (Figure 2B). In contrast to IRF9, STAT1 and STAT2, we found that STAT3, although well expressed in cell lysates, is not precipitated with CypA (Figure 2B). We then asked whether the expression of IRF9 is necessary for an interaction of CypA with STAT1 and STAT2. Specifically, we asked whether or not STAT1 and STAT2 are co-precipitated with CypA in parental (2fTGH) or IRF9-deficient (U2A) human cells. We found that in the absence of IRF9, endogenous CypA fails to precipitate both endogenous STAT1 and STAT2 (Figure 2C), suggesting that the presence of IRF9 facilitates/mediates the co-precipitation of STAT1 and STAT2 by CypA. These data suggest that CypA is part of the ISGF3 complex even under native conditions
Fig. 2. CypA is a component of the ISGF3 complex.
A. IFN -treated Huh7 cells were transfected with Flag-IRF9, Flag-STAT1 and Flag-STAT2. GST-CypA and glutathione beads were added to lysates. Bound material analyzed with anti-Flag IgG. B. IFN -treated Huh7 cell lysate was mixed with anti-CypA IgG and CsA. Bound material analyzed with anti-IRF9, -STAT1, -STAT2 and -STAT3 IgG. C. Same as B, except that parental and IRF9-deficient cells were used. D. IRF9 truncations. E. 293T cells were transfected with IRF9 truncations. GST-CypA and glutathione beads were added to cell lysates with or without CsA. Bound material analyzed with anti-Flag IgG.
The IAD of IRF9 contains a CypA-binding site
We then searched for the CypA-binding region within IRF9. Parental IRF9 (1–393) contains well-defined domains. Region 1–120 corresponds to the DNA-binding domain; region 120–200 corresponds to a linker region; and region 200–393 corresponds to the IRF association domain (IAD) [45]. We therefore created the following IRF9 truncated forms: 1–120, 1–200, 120–393 and 200–393 (Figure 2D) and examined their capacities to bind CypA. Importantly, 1–120 and 1–200 IRF9 failed to bind CypA (Figure 2E, right panel). In contrast, 120–393 and 200–393 bind CypA well (Figure 2E, right panel). All truncated forms were well expressed (Figure 2E, left panel). These data suggest that the IAD serves as a major CypA-binding site. The interaction is specific since CsA blocks IAD-CypA interactions (Figure 2E, right panel).
NS5A and IRF9 compete for CypA binding
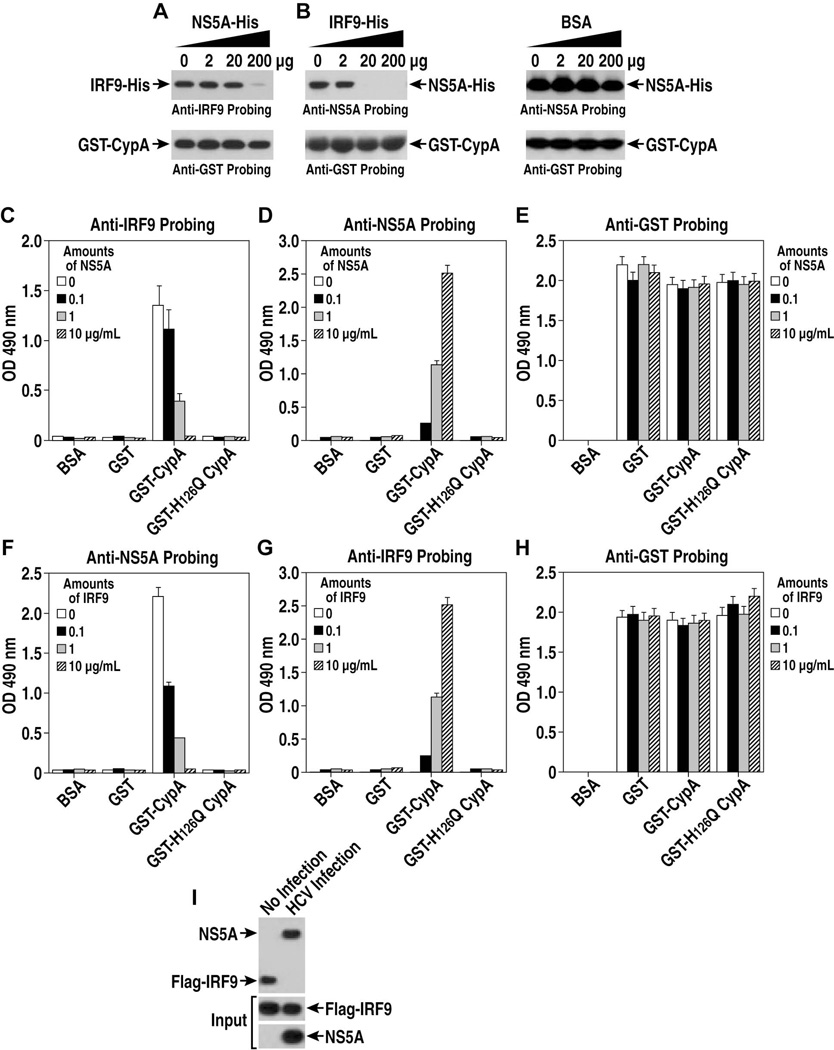
Since CypA binds HCV NS5A [22, 23, 26, 30], we asked whether NS5A and IRF9 compete for CypA binding. The addition of increasing concentrations of NS5A-His diminishes the capacity of GST-CypA to capture IRF9-His (Figure 3A). The reciprocal competition experiment led to similar results (Figure 3B). In contrast to IRF9, increasing concentrations of BSA do not inhibit CypA-NS5A interactions (Figure 3B), demonstrating the specificity of the NS5A/IRF9 competition. We confirmed the GST pulldown results by ELISA. Specifically, immobilized GST-CypA captures IRF9 in the absence, but not in the presence of increasing concentrations of NS5A (Figure 3C). Remarkably, NS5A dissociates IRF9 from CypA (Figure 3C). While amounts of IRF9 captured by CypA decreased (Figure 3C), amounts of NS5A captured by CypA increased (Figure 3D). The specificity of these captures was demonstrated by the lack of IRF9 and NS5A capture by GST and GST-H126Q CypA (Figure 3C and D). We were able to conduct a reciprocal competition (Figure 3F and G). Similar amounts of GST proteins were immobilized onto the plates (Figure 3E and H). We then asked whether NS5A competes with IRF9 for CypA binding during HCV infection. We found that JFH-1 infection of IRF9-expressing Huh7 cells significantly decreases levels of IRF9 co-precipitated with CypA (Figure 3I), suggesting that indeed NS5A and IRF9 can compete for CypA binding during infection, at least in vitro. Altogether these data demonstrate that NS5A and IRF9 compete for CypA contact.
Fig. 3. NS5A disrupts CypA-IRF9 complexes.
A. GST-CypA was mixed with IRF9 and NS5A. Bound material analyzed with anti-IRF9 and -GST IgG. B. Same as A. C. IRF9 and NS5A were added to BSA, GST, GST-CypA and GST-H126Q CypA plates. Captured IRF9 detected with anti-IRF9 IgG. D. Captured NS5A detected with anti-NS5A IgG. E. Same as C. F. Same a C. G. Same as D. H. Same as F. Errors bars of graphs represent standard errors of triplicates. I. Cells were electroporated with Flag-IRF9 and JFH-1, lysed and mixed with anti-CypA IgG. Bound material analyzed with anti-Flag and -NS5A IgG.
CypA inhibition enhances IFN-induced ISRE transcriptional activity
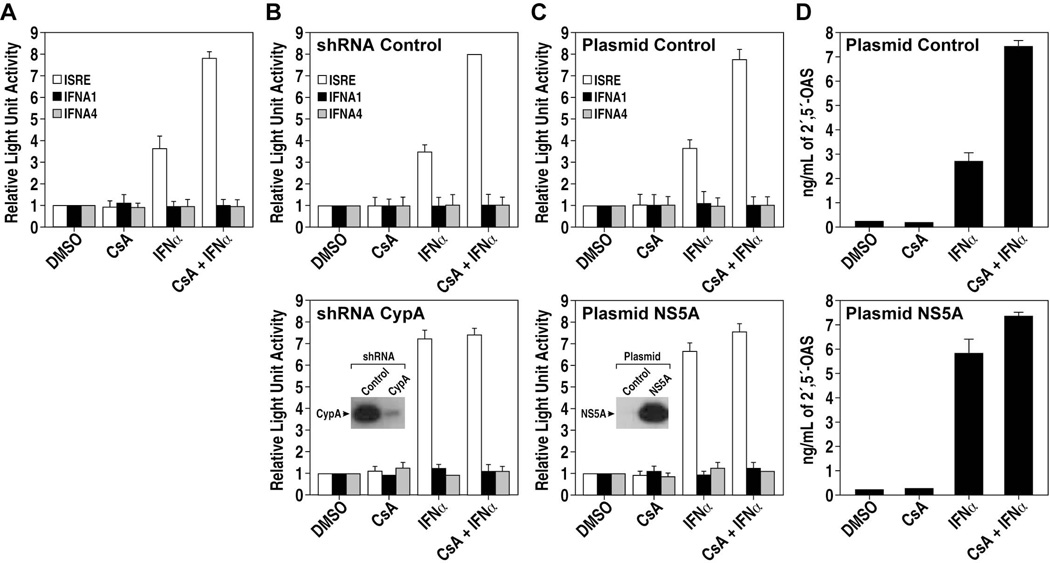
IRF9 mediates the ISGF3 complex to bind to IFN-stimulated response elements (ISREs) [46]. We thus asked whether CypA, by interacting with IRF9, influences IFN-induced transcriptional activities. To address this issue, HepG2 cells were pretreated with DMSO, IFN, CsA or a combination of IFN and CsA. We used HepG2 cells rather than Huh7 cells simply because we found that the reporter gene assay was constantly more robust in HepG2 than Huh7 cells (data not shown). Cells were then transfected with plasmids encoding luciferase driven either by an ISRE, IFNA1 or IFNA4 promoter. The ISRE promoter is activated by IRF9, whereas IFNA1 or IFNA4 promoters by IRF3 and 7 [47]. Neither IFN nor CsA have any effect on IFNA1 and IFNA4 promoter activities (Figure 4A). IFN triggers ISRE-mediated transcriptional induction, whereas CsA alone did not (Figure 4A). In contrast, CsA enhances the IFN-induced transcriptional activity driven by ISRE. We obtained a similar effect upon CypA siRNA knockdown (Figure 4B), suggesting that CypA somehow restrains the IFN-induced ISRE transcriptional activity. Overexpression of NS5A also enhances the IFN-induced ISRE transcriptional activity (Figure 4C). To further suggest that competition of NS5A with IRF9 for CypA binding modulates signaling through ISRE, we repeated a similar experiment, but this time, we examined the expression of an IFN-stimulated gene such as the 2',5'-oligoadenylate synthetase (2’,5’-OAS). Importantly, we obtained similar results (Figure 4D), further suggesting that NS5A and IRF9 competition for CypA can influence the IFN response, at least in vitro. Importantly, both CypA knockdown and NS5A overexpression induce IFN-induced ISRE transcriptional levels similar to those of CsA. In contrast to CypA shRNA and NS5A plasmid, control shRNA and control plasmid do not influence the IFN-induced ISRE transcriptional activities (Figures 4B and C). Altogether these data suggest that inhibition of CypA-IRF9 interactions by i) the Cyp inhibitor CsA; ii) the CypA knockdown; or iii) NS5A expression (Figure 4), enhances IFN-induced ISRE transcriptional activities.
Fig. 4. CypA influences IFN-induced transcriptional activities.
A. HepG2 cells were pretreated with DMSO, IFN, CsA or a combination of IFN and CsA. Cells were transfected with luciferase plasmid driven by ISRE, IFNA1 or IFNA4 promoter. Luciferase activity was analyzed in lysates after 24 h. B. Same as A, except that cells were transfected with control shRNA or shRNA CypA [35]. C. Same as A, except that cells were transfected with control (pcDNA3-betagalactosidase) or NS5A plasmid (pcDNA3-NS5A). D. Same as C, except that 2’,5’-OAS levels in lysates were quantified by ELISA. Errors bars of panels represent standard errors of triplicates.
CONCLUSIONS
Based on recent evidence suggesting that CypA neutralization in HCV-infected patients modulates the IFN response [34], we investigated here the possibility that CypA associates with components of the IFN response. We demonstrated that CypA associates with IRF9. We showed that CypA directly binds IRF9 via its enzymatic hydrophobic pocket. We identified the IAD of IRF9 as a major CypA-binding site. Cyp inhibitors block IRF9-CypA interactions. We also obtained evidence that CypA modulates IFN-induced transcription. This study thus provides the first demonstration that CypA binds to a component of the JAK/STAT pathway.
It is important to note that we were unable to detect an interaction between endogenous IRF9, STAT1, STAT2 and CypA in the absence of IFN (data not shown). This is likely due to the fact that in the absence of IFN, IRF9 is poorly or not expressed, at least not detectable by Western blotting. Moreover, it is worthy to emphasize that Huh7 cells are not “physiological” cells to examine the interplay between IRF9, STAT1, STAT2 and CypA during viral replication. The ideal situation would be to examine this interaction in primary hepatocytes of HCV-infected patients. The CypA/IRF9/STAT1/STAT2 interactions likely occur at very specific stages of HCV infection. Indeed, it is likely that primary hepatocytes develop an innate response (IFN response) during the early steps of HCV infection, allowing IRF9 expression. At that time, CypA and IRF9 should interact. If the virus is able to “survive” or counteract the innate response, NS5A will be expressed, and therefore, be in a position to either bind free CypA (which is very abundant in a cell) or to disrupt CypA-IRF9 complexes. Based on our in vitro competition experiments, large amounts of NS5A are required to disrupt CypA-IRF9 complexes. Thus, it is unlikely that NS5A will disrupt all existing CypA-IRF9 complexes, except eventually during the peak of viral replication in hepatocytes.
A previous study suggested an indirect role for CypA in the IFN response. Specifically, HIV-1 induces an antiviral type-I IFN response that renders dendritic cells permissive to infection [48]. This response was dependent on the interaction between HIV-1 capsid and CypA as well as on the activation of IRF3. It is unlikely that CypA regulates IRF3 transcription activity directly since it does not bind IRF3 [49]. Here, we provide a direct link between CypA and the innate response. Indeed, we obtained evidence that suggest that CypA can modulate the IFN-induced transcription activity. However, we do not yet know the specific biochemical consequences of CypA-IRF9 interactions. One possibility is that CypA catalyzes conformational changes in IRF9 that lead to modifications such as acetylation or phosphorylation. Conformational changes also could influence the association of IRF9 with other cellular partners. Additionally, CypA-mediated conformational changes of IRF9 could affect either its affinity to ISREs, subcellular localization, or half-life. Therefore, further work is required to determine the true action of CypA on IRF9 either in a cellular and/or viral infection context.
We found that recombinant NS5A from genotypes 1a and 1b compete similarly with IRF9 for CypA binding (data not shown). This is in accordance with our previous work that showed that NS5A from various genotypes bind similarly to CypA and that all NS5A-CypA interactions are prevented by Cyp inhibitors [23]. It is thus unlikely that the interplay between NS5A, IRF9 and CypA could explain why different genotypes are more or less sensitive to IFN.
The consequence of recruitment of CypA by NS5A in HCV-infected cells, and action of CypA on NS5A, also remain to be fully elucidated. The impact of NS5A-CypA binding on IRF9 function will depend in part on the relative levels of NS5A, IRF9, and CypA in a cell. The experiments reported here, in which NS5A is expressed at high levels within the cell, resulted in increased levels of ISRE-driven gene expression, may most resemble the situation in acute HCV infection when the cell mounts an initial innate immune response to the virus. Note that the initial pulldown of IRF9 with CypA-Strep was conducted in JFH-1-infected cells. The impact of competition for CypA binding between NS5A and IRF9 in chronic infections, when others reported NS5A interactions have been established [20, 21], remains to be investigated.
We showed that overexpression of NS5A, slightly, but significantly enhances IFN-induced ISRE transcriptional activities, suggesting that NS5A promotes IFN-induced signaling through the JAK-STAT pathway. This apparently contrasts with previous reports [50, 51]. How could we reconcile these apparent conflicting results? First, it is important to note that the experimental design greatly differ between studies. For example, we used HepG2, whereas the other studies used Hep3B, COS7 or HeLa [50, 51]. This clearly could make a difference given that Lan et al. found that NS5A influences the ISRE response in Hep3B, but not COS7 cells [51]. Moreover, it is likely that the levels of NS5A, CypA, IRF9 and ISRE copy numbers differ between studies. The order of expression or addition of the various components also greatly differs between studies. In our study, cells were initially transfected for 3 days with NS5A to allow its expression and establishment of CypA-NS5A interactions, then transfected with the ISRE reporter for 2 days and treated with IFN for 24h. In sharp contrast, in the other studies, cells were co-transfected with NS5A and the ISRE reporter at the same time, and after an unspecified period of time post-transfection, IFN was added for either 0.5 [51] or 24 h [50]. Altogether, these diverse experimental methodologies may explain the apparent conflicting models. It is likely that NS5A could modulate multiple host-viral interactions and thus may lead to different effect depending on the conditions of the cells and status of the IFN pathway. The specific function of NS5A described in this study is regarding its interaction with CypA, which appeared to have a positive impact on the activation of the IFN pathway due to its competitive binding to IRF-9.
In conclusion, this study provides the first demonstration that CypA associates directly with IRF9, a major component of the JAK-STAT pathway, and may regulate the IFN-induced transcription. This study also reveals a novel potential of HCV to modulate the IFN response via NS5A.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following: C. Cameron for pET26Ub-NS5A-His Con1 plasmid, A. García-Sastre for Flag-STAT1 and -STAT2 plasmids, B. R. tenOever for IRF9 plasmid, P. Targett-Adams for NS5A plasmid, R. Bartenschlager for Huh7-Luc/Neo ET cells, G. Stark for 2fTGH and U2A cells, Novartis, SCYNEXIS and Biotica for alisporivir, SCY-635 and sanglifehrin B, respectively, F. Chisari for JFH-1-Huh7.5.1 cells and for careful reading of the manuscript. This is publication no. 21644 from the Department of Immunology & Microbial Science, The Scripps Research Institute, La Jolla, CA.
We acknowledge financial support from the U.S. Public Health Service grant no. AI087746 (P.A.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No conflict of interest
REFERENCES
- 1.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 2.Loo YM, Gale M., Jr. Viral regulation and evasion of the host response. Curr Top Microbiol Immunol. 2007;316:295–313. doi: 10.1007/978-3-540-71329-6_14. [DOI] [PubMed] [Google Scholar]
- 3.Bonjardim CA, Ferreira PC, Kroon EG. Interferons: signaling, antiviral and viral evasion. Immunol Lett. 2009;122:1–11. doi: 10.1016/j.imlet.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baum A, Garcia-Sastre A. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids. 2010;38:1283–1299. doi: 10.1007/s00726-009-0374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy DE, Marie IJ, Durbin JE. Induction and Function of Type I and III Interferon in Response to Viral Infection. Curr Opin Virol. 2011;1:476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pflugheber J, Fredericksen B, Sumpter R, Jr., Wang C, Ware F, Sodora DL, et al. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc Natl Acad Sci U S A. 2002;99:4650–4655. doi: 10.1073/pnas.062055699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fredericksen B, Akkaraju GR, Foy E, Wang C, Pflugheber J, Chen ZJ, et al. Activation of the interferon-beta promoter during hepatitis C virus RNA replication. Viral Immunol. 2002;15:29–40. doi: 10.1089/088282402317340215. [DOI] [PubMed] [Google Scholar]
- 9.Sumpter R, Jr., Loo YM, Foy E, Li K, Yoneyama M, Fujita T, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. Journal of virology. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 11.Biron CA. Initial and innate responses to viral infections--pattern setting in immunity or disease. Curr Opin Microbiol. 1999;2:374–381. doi: 10.1016/s1369-5274(99)80066-6. [DOI] [PubMed] [Google Scholar]
- 12.Bigger CB, Brasky KM, Lanford RE. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. Journal of virology. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, et al. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bigger CB, Guerra B, Brasky KM, Hubbard G, Beard MR, Luxon BA, et al. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. Journal of virology. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. Journal of virology. 2005;79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foy E, Li K, Sumpter R, Jr., Loo YM, Johnson CL, Wang C, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin W, Kim SS, Yeung E, Kamegaya Y, Blackard JT, Kim KA, et al. Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. Journal of virology. 2006;80:9226–9235. doi: 10.1128/JVI.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kriegs M, Burckstummer T, Himmelsbach K, Bruns M, Frelin L, Ahlen G, et al. The hepatitis C virus non-structural NS5A protein impairs both the innate and adaptive hepatic immune response in vivo. The Journal of biological chemistry. 2009;284:28343–28351. doi: 10.1074/jbc.M109.038877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polyak SJ, Khabar KS, Paschal DM, Ezelle HJ, Duverlie G, Barber GN, et al. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. Journal of virology. 2001;75:6095–6106. doi: 10.1128/JVI.75.13.6095-6106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gale MJ, Jr., Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, et al. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 22.Hanoulle X, Badillo A, Wieruszeski JM, Verdegem D, Landrieu I, Bartenschlager R, et al. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. The Journal of biological chemistry. 2009;284:13589–13601. doi: 10.1074/jbc.M809244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterji U, Lim P, Bobardt MD, Wieland S, Cordek DG, Vuagniaux G, et al. HCV resistance to cyclosporin A does not correlate with a resistance of the NS5A-cyclophilin A interaction to cyclophilin inhibitors. Journal of hepatology. 2010;53:50–56. doi: 10.1016/j.jhep.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandes F, Poole DS, Hoover S, Middleton R, Andrei AC, Gerstner J, et al. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology. 2007;46:1026–1033. doi: 10.1002/hep.21809. [DOI] [PubMed] [Google Scholar]
- 25.Yang F, Robotham JM, Grise H, Frausto S, Madan V, Zayas M, et al. A major determinant of cyclophilin dependence and cyclosporine susceptibility of hepatitis C virus identified by a genetic approach. PLoS Pathog. 2010;6:e1001118. doi: 10.1371/journal.ppat.1001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coelmont L, Hanoulle X, Chatterji U, Berger C, Snoeck J, Bobardt M, et al. DEB025 (Alisporivir) inhibits hepatitis C virus replication by preventing a cyclophilin A induced cis-trans isomerisation in domain II of NS5A. PLoS One. 2010;5:e13687. doi: 10.1371/journal.pone.0013687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdegem D, Badillo A, Wieruszeski JM, Landrieu I, Leroy A, Bartenschlager R, et al. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic alpha-helical propensity and is a substrate of cyclophilin A. The Journal of biological chemistry. 2011;286:20441–20454. doi: 10.1074/jbc.M110.182436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foster TL, Gallay P, Stonehouse NJ, Harris M. Cyclophilin A interacts with domain II of hepatitis C virus NS5A and stimulates RNA binding in an isomerase-dependent manner. Journal of virology. 2011;85:7460–7464. doi: 10.1128/JVI.00393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waller H, Chatterji U, Gallay P, Parkinson T, Targett-Adams P. The use of AlphaLISA technology to detect interaction between hepatitis C virus-encoded NS5A and cyclophilin A. J Virol Methods. 2010;165:202–210. doi: 10.1016/j.jviromet.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandes F, Ansari IU, Striker R. Cyclosporine inhibits a direct interaction between cyclophilins and hepatitis C NS5A. PLoS One. 2010;5:e9815. doi: 10.1371/journal.pone.0009815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallay PA. Cyclophilin inhibitors: a novel class of promising host-targeting anti-HCV agents. Immunol Res. 2011 doi: 10.1007/s12026-011-8263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawitz E, Godofsky E, Rouzier R, Marbury T, Nguyen T, Ke J, et al. Safety, pharmacokinetics, and antiviral activity of the cyclophilin inhibitor NIM811 alone or in combination with pegylated interferon in HCV-infected patients receiving 14 days of therapy. Antiviral Res. 2011;89:238–245. doi: 10.1016/j.antiviral.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Flisiak R, Jaroszewicz J, Flisiak I, Lapinski T. Update on alisporivir in treatment of viral hepatitis C. Expert Opin Investig Drugs. 2012;21:375–382. doi: 10.1517/13543784.2012.658641. [DOI] [PubMed] [Google Scholar]
- 34.Hopkins S, Scorneaux B, Huang Z, Murray MG, Wring S, Smitley C, et al. SCY-635, a novel nonimmunosuppressive analog of cyclosporine that exhibits potent inhibition of hepatitis C virus RNA replication in vitro. Antimicrob Agents Chemother. 2010;54:660–672. doi: 10.1128/AAC.00660-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatterji U, Bobardt M, Selvarajah S, Yang F, Tang H, Sakamoto N, et al. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. The Journal of biological chemistry. 2009;284:16998–17005. doi: 10.1074/jbc.M109.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saphire AC, Gallay PA, Bark SJ. Proteomic analysis of human immunodeficiency virus using liquid chromatography/tandem mass spectrometry effectively distinguishes specific incorporated host proteins. J Proteome Res. 2006;5:530–538. doi: 10.1021/pr050276b. [DOI] [PubMed] [Google Scholar]
- 37.Galigniana MD, Morishima Y, Gallay PA, Pratt WB. Cyclophilin-A is bound through its peptidylprolyl isomerase domain to the cytoplasmic dynein motor protein complex. The Journal of biological chemistry. 2004;279:55754–55759. doi: 10.1074/jbc.M406259200. [DOI] [PubMed] [Google Scholar]
- 38.Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci U S A. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kallen J, Walkinshaw MD. The X-ray structure of a tetrapeptide bound to the active site of human cyclophilin A. FEBS Lett. 1992;300:286–290. doi: 10.1016/0014-5793(92)80865-e. [DOI] [PubMed] [Google Scholar]
- 40.Zydowsky LD, Etzkorn FA, Chang HY, Ferguson SB, Stolz LA, Ho SI, et al. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1992;1:1092–1099. doi: 10.1002/pro.5560010903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paeshuyse J, Kaul A, De Clercq E, Rosenwirth B, Dumont JM, Scalfaro P, et al. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43:761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- 42.Sanglier JJ, Quesniaux V, Fehr T, Hofmann H, Mahnke M, Memmert K, et al. Sanglifehrins A, B, C and D, novel cyclophilin-binding compounds isolated from Streptomyces sp. A92-308110. I. Taxonomy, fermentation, isolation and biological activity. J Antibiot (Tokyo) 1999;52:466–473. doi: 10.7164/antibiotics.52.466. [DOI] [PubMed] [Google Scholar]
- 43.Ochiai T, Nakajima K, Nagata M, Suzuki T, Asano T, Uematsu T, et al. Effect of a new immunosuppressive agent, FK 506, on heterotopic cardiac allotransplantation in the rat. Transplant Proc. 1987;19:1284–1286. [PubMed] [Google Scholar]
- 44.Hennig L, Christner C, Kipping M, Schelbert B, Rucknagel KP, Grabley S, et al. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry. 1998;37:5953–5960. doi: 10.1021/bi973162p. [DOI] [PubMed] [Google Scholar]
- 45.Qin BY, Liu C, Lam SS, Srinath H, Delston R, Correia JJ, et al. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat Struct Biol. 2003;10:913–921. doi: 10.1038/nsb1002. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen H, Hiscott J, Pitha PM. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 47.Lin R, Genin P, Mamane Y, Hiscott J. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol Cell Biol. 2000;20:6342–6353. doi: 10.1128/mcb.20.17.6342-6353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, Littman DR. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010;467:214–217. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Obata Y, Yamamoto K, Miyazaki M, Shimotohno K, Kohno S, Matsuyama T. Role of cyclophilin B in activation of interferon regulatory factor-3. The Journal of biological chemistry. 2005;280:18355–18360. doi: 10.1074/jbc.M501684200. [DOI] [PubMed] [Google Scholar]
- 50.Geiss GK, Carter VS, He Y, Kwieciszewski BK, Holzman T, Korth MJ, et al. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. Journal of virology. 2003;77:6367–6375. doi: 10.1128/JVI.77.11.6367-6375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lan KH, Lan KL, Lee WP, Sheu ML, Chen MY, Lee YL, et al. HCV NS5A inhibits interferon-alpha signaling through suppression of STAT1 phosphorylation in hepatocyte-derived cell lines. Journal of hepatology. 2007;46:759–767. doi: 10.1016/j.jhep.2006.11.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.