Abstract
The ability of the human body to naturally recover from coronary heart disease is limited because cardiac cells are terminally differentiated, have low proliferation rates, and low turnover rates. Cardiovascular tissue engineering offers the potential for production of cardiac tissue ex vivo but is currently limited by several challenges: (i) Tissue engineering constructs require pure populations of seed cells, (ii) Fabrication of 3-D geometrical structures with features of the same length scales that exist in native tissue is non-trivial, and (iii) Cells require stimulation from the appropriate biological, electrical and mechanical factors. In this review, we summarize the current state of microfluidic techniques for enrichment of subpopulations of cells required for cardiovascular tissue engineering, which offer unique advantages over traditional plating and FACS/MACS-based enrichment. We then summarize modern techniques for producing tissue engineering scaffolds that mimic native cardiac tissue.
Keywords: Cardiovascular Tissue Engineering, Cell Enrichment, Scaffolds, Immunocapture, Hydrogel, Cardiomyocyte, Endothelial Progenitor Cell, Fibroblast, Microfluidic
1. Introduction
Nearly 83 million American adults have one ore more types of cardiovascular disease, with over 16 million suffering from coronary heart disease (Roger et al., 2012). Coronary heart diseases can lead to the loss of heart tissue, including cells like cardiomyocytes, which do not regenerate rapidly in mammals. Cardiomyocytes are considered to be terminally differentiated, have low proliferation rates (Soonpaa and Field, 1998), and low turn-over rates (Bergmann et al., 2009). Because the human body cannot natively regenerate cardiac tissue, there is a critical need for the ability to engineer cardiac tissue constructs ex vivo. Engineered 3-D tissue constructs have the potential to serve as models for cardiac tissue to study the function of diseased and normal tissue and the effects of drugs. The ultimate goal is for such constructs to be used to generate transplantable tissue that can replace diseased tissue in patients.
Cardiac tissue engineering approaches require cells that are truly doing the work of regeneration, and suitable matrices that will allow efficient application of these cells to the body. The cells come from either primary sources or through stem cell differentiation. Once a suitable cell population is derived, separation methods are required to allow for enrichment and isolation of a desired cell sub-type so that a defined tissue culture can be achieved. If a cell type is present in large quantities in a given sample and a surface marker is known, we can rely on established methods such as FACS or MACS to achieve isolation of large quantities of cells. However, for smaller tissue samples and for cells where surface markers are not known, new microfluidic methods are required for separation. Matrices for cell delivery and culture can be made from synthetic or natural materials and their mechanical, electrical and adhesive properties can be tuned for a specific application.
For the ultimate envisioned clinical application, the cells can either be isolated based on the expression of surface markers tagged with magnetic particles or in a label-free fashion. The cells whose components are labeled with fluorescent probes cannot be used in clinical applications due to the unknown long-term effects of these organic probe molecules in humans. These restrictions pose significant challenges for sorting of cells based on intracellular marker expression. Antibody staining of intracellular markers such as contractile proteins requires cell permeabilization which unfortunately renders the cells non-viable and unusable for cell therapy. On the other hand, genetic labeling of cells for clinical applications cannot be performed in humans due to ethical concerns. It is in these areas that microfluidic cell separation can truly yield methods required for robust cell separation consistent with clinical application requirements.
1.1. Cellular Composition and Cell Markers in Native Cardiac Tissue
The native myocardium (cardiac muscle) is a highly differentiated tissue composed of CM, FB and SMC with a dense supporting vasculature and collagen-based extracellular matrix, with an average cell density of 1–10 × 108 cells/cm3. The CM form a three-dimensional syncytium that enables propagation of electrical signals across specialized intracellular junctions to produce coordinated mechanical contractions that pump blood forward. Only 20–40% of the cells in the heart are CM, but they occupy 80–90% of the heart volume (Nag, 1980). Cardiac FBs constitute most of the non-myocytes in the myocardium. The main roles of FB are to secrete the components of the (ECM) and transmit mechanical force by the receptor mediated connections to the ECM (Sussman et al., 2002). ECs line the blood vessels of the dense myocardial vasculature and engage in cross-talk with CM via numerous secreted factors (Parratt et al., 1997; Shah et al., 1997). Upon harvesting the heart tissue, cells are isolated by treatment with trypsin followed by a series of collagenase digestions resulting in a suspension of rounded cells. As reported by Naito et al. (2006), and in agreement with our findings (Radisic et al., 2008), after digestion the native heart isolate has 47% CM, 48% FB, 3%SMC and 2% EC. CM are the largest cells in this suspension with diameters ranging from 13–21 µm. The non-myocytes collectively range in size from 3–15 µm (this range includes FB, SMC, EC and progenitor cells). EC and FB have established surface markers (e.g. CD31 and DDR2 respectively), thus they conducive to FACS and MACS provided the abundant source of cells. The surface markers for muscle-cell types (e.g. CM and SMC) are still under active investigation. For example, Signal Regulatory Protein α (SIRPA) was recently described as a novel surface marker for human and mouse cardiomyocytes (Dubois et al., 2011). Until surface markers for these proteins are more clearly established, novel methods for separation of muscle cells in a label-free fashion must be developed and used.
This native heart isolate can be further enriched for CM in a technique commonly referred to as pre-plating. In pre-plating, the cell suspension is kept in a tissue culture plate for a period of 15–75 min to allow removal of FB and SMC by fast and preferential attachment to the tissue culture plastic surface while CM and EC remain in suspension (Laugwitz et al., 2005; Wang, 2004). It was also reported that the heart contains small numbers of resident cardiac progenitor cells (~ 1 in 107 cells) (Beltrami et al., 2003; Laugwitz et al., 2005; Linke et al., 2005; Oh et al., 2003; Orlic et al., 2001). Beltrami et al. (2003) reported that small cells positive for a stem-cell surface marker c-kit isolated from adult rat hearts and expanded under limited dilution gave rise to CM, SMC and EC when injected into ischemic myocardium. C-kit+ cells can be isolated by magnetic separation using beads coated with anti-c-kit antibody. Oh et al. (2003) reported that adult mouse heart cells expressing stem cell antigen-1 (Sca-1) on their surface can selectively give rise to CM upon treatment with 5-azacytidine. Yet, Sca-1 is not expressed in humans. Of particular interest is a resident cardiac progenitor marker isl1 (Laugwitz et al., 2005). During embryonic development, isl1+ cells contribute substantially to the formation of the right ventricle, atria, outflow tracts and part of the left ventricle in the second wave of CM formation (Evans D Fau Miller et al., 1988; C.L.Cai et al., 2003). Laugwitz et al. (2005) demonstrated that several hundreds of isl1+ cells remain in neonatal and adult mouse hearts as well as the hearts of newborn human patients. Since isl1 is a transcription factor, isl1+ cells were isolated from mouse hearts using genetic labeling in conjunction with fluorescence activated cell sorting (FACS). They were smaller than CM and plated together with FB in the pre-plating procedure. Several days (4–6) after pre-plating, isl1+ cell fraction increased to 0.5% and then reached 8% at 15–18 days in culture (Laugwitz et al., 2005), which is the expected percentage of isl1+ cells in the input cell population for microfluidic enrichment. Isl1+ cells differentiated at high frequency (~ 25%) when in direct contact with either alive or fixed CMs (Laugwitz et al., 2005).
While EC and FB can be isolated from primary sources, cardiomyocytes are considered to be terminally differentiated and they cannot be expanded if primary post-natal sources are used. Studies from a number of groups have shown that it is possible to generate CMs from mouse (Kattman et al., 2006), human ESCs (Yang et al., 2008) and human iPSCs (Zhang et al., 2009). The most efficient and reproducible protocols to date are those that have replicated the signaling pathways that regulate lineage commitment in the early embryo. With this approach, the earliest stages of cardiovascular development in ESC differentiation cultures were mapped identifying a multipotent cardiovascular progenitor that displays the capacity to generate cardiac and vascular progeny (Kattman et al., 2006; Yang et al., 2008). In mouse Flk1 and in humans KDR expression can be used to enrich for cardiac specified mesoderm (Kouskoff et al., 2005). When isolated from the differentiated embryoid bodies (EBs) and cultured as a monolayer, these progenitors generate contracting CMs (Kattman et al., 2006; Yang et al., 2008). As these progenitors differentiate, they progress through the developmental stages thought to be involved in the establishment of the cardiovascular lineages in vivo, for which specific cytokines are required. The combination of activin A and BMP4 on Days 1–4 of EB differentiation induces a primitive-streak-like population and mesoderm development. Subsequent application of WNT inhibitor DKK1 and KDR ligand VEGF165 significantly enhances the differentiation of KDR+ progenitors into CMs (Kattman et al., 2006; Yang et al., 2008) while bFGF is added to support continued expansion of cardiovascular lineages.
1.2. Importance of Cell Isolation in Vascular Tissue Engineering
Cell-based therapeutic approaches aim to identify and utilize cells that are capable of promoting regeneration of important cell types, such as cardiomyocytes, or structural elements of cardiac tissue. Cell-based regenerative approaches, which can be employed independently in vivo by cell implantation, or as a precursor to in-vitro cultivation, have seen a major boost in the last ten years in the cardiac (Beltrami et al., 2003; Laugwitz et al., 2005; Linke et al., 2005; Oh et al., 2003; Orlic et al., 2001) and skin areas (Morris et al., 2004; Oshima et al., 2001). In these studies, fluorescence- or magnet-activated cell sorting (FACS or MACS) are used to isolate the stem/progenitor cells. Both of these techniques are highly developed and can be utilized in positive or negative selection modes of separation. Positive selection is desirable when markers for the target cells are known, such as c-kit (Beltrami et al., 2003; Linke et al., 2005; Oh et al., 2003; Orlic et al., 2001), sca-1 (Linke et al., 2005; Oh et al., 2003), and MDR1 (Linke et al., 2005) for cardiac progenitor cells, or P-cadherin, CD34, or the α6-integrin in the case of skin stem cells (Rhee et al., 2006). The negative selection approach can be utilized when target cell surface markers are not known (for example, islet-1+ cardiac progenitor cells are can be identified only on the basis of this intracellular marker (Laugwitz et al., 2005)). A common aspect of either selection approach is the requirement for pre-processing incubation to attach the fluorescent or magnetic antibody tags which facilitate separation. FACS-based systems are expensive and typically require core facility infrastructure. MACS systems are less expensive but are not portable. Furthermore, when the number of cells to be isolated is very small (order of 10–100 cells in a given sample), these techniques cannot be utilized reliably.
The potential ability of endothelial progenitor cells (EPCs) to help in repairing damaged endothelium and promoting angiogenesis has led to exploration of their use in cardiac therapeutics (Rehman, 2011). In vascular tissue engineering using EPCs, these cells are removed from whole blood in two steps, centrifugation followed by pre-plating (Kaushal et al., 2001). In the centrifugation step, the EPCs are separated along with leukocytes from other blood components, and in the pre-plating step, the EPCs adhere to fibronectin-coated culture dishes and are thus separated from leukocytes. The pre-plating process is inherently prone to several shortcomings. (i) The process is inefficient in that there is no way to prevent undesired cell types from being present in the enriched supernatant (for cardiac cells) or in the pre-plate (for EPCs). (ii) This technique is not systematic in that there are few parameters that can be manipulated to control the enrichment process and make it reproducible. (iii) Lastly, the process does not allow distinction between multiple cell types of interest that may be present in the supernatant or the pre-plate. For example, cardiac progenitor cells have been observed in the plated fraction of cardiac cells and are generally studied in this type of environment since further separation is difficult (Laugwitz et al., 2005). Microfluidic cell isolation techniques have the potential to eliminate these shortcomings, and in addition can be used to isolated rare cells that are inaccessible to FACS and MACS systems. Such techniques are discussed in the context of cardiovascular regenerative medicine in the next section.
2. Microfluidic Platforms for Cell Enrichment in Cardiovascular Regenerative Medicine Applications
One of the key challenges associated with cardiovascular regenerative medicine is isolation and enrichment of rare subtypes of cells, which is necessary for construction of scaffolds for potential in vivo experiments and treatments. There are several reasons why pure populations of cells are desirable for regenerative medicine in both clinical and research applications (Kaushal et al., 2001; Kolvenbach et al., 2007; O et al., 2011). For example: (i) minimization of foreign or unwanted tissue is critical for tissue cultures that are intended for implantation, (ii) stem cell differentiation is influenced by surrounding cell types; pure populations are necessary for controlled differentiation, and (iii) some important cell types are rare (e.g. EPCs, CTCs), and cannot be analyzed in bulk samples owing to noise/interference from the dominant cell types. Major challenges in isolating cell types include: (i) conventional cell isolation methods are time consuming and/or labor intensive, (ii) different cell types can be difficult to distinguish from each other, and (iii) the number of desired cells in a clinical/biological sample may be extremely limited.
Microfluidic cell isolation methods offer unique advantages over conventional methods, as they are relatively low cost, high throughput, can be used with small (1 µL or less) sample volumes, and in many cases are capable of isolating extremely rare subpopulations of cells. They also have the potential to reduce the time and labor required for cell isolation, and to distinguish between cell types that are difficult to isolate using conventional FACS or plating isolation methods. This section of the review describes microfluidics-based methods for cell enrichment in cardiovascular regenerative medicine applications. A comprehensive review of microfluidic cell enrichment methods is beyond the scope of this paper; here we focus on methods that are adapted for or can be potentially adapted for enrichment of cells relevant to regenerative tissue engineering.
2.1. Methodology
Several metrics are used to characterize the efficacy of cell isolation methods. Here we summarize and define the most common quantifiers used to characterize cell isolation. In a heterogeneous population of cells, target cells are the desired subpopulation of cells to be isolated. The purity of a cell suspension is defined as:
| (1) |
Enrichment refers to an increase in purity, often denoted as an enrichment factor:
| (2) |
The yield is the total number of target cells obtained after isolation. The overall efficiency of the isolation is:
| (3) |
Note that some define yield as the quantity given by Equation 3. For microfluidic devices in which cells can be captured and released, capture and release efficiencies can be defined separately.
| (4) |
| (5) |
The overall efficiency can be written in terms of the capture and release efficiencies:
| (6) |
The throughput of a cell isolation system is a measure of the speed of the technique, and is usually given as a number of sample cells processed per unit time.
2.2. Static Cell Capture and Adhesion Properties
Isolation of subtypes of cells in microfluidic devices is commonly accomplished by immobilizing capture molecules, moieties that have an affinity for a surface protein that is expressed in high numbers in the chosen target cells, onto the walls of a microfluidic channel (Pratt et al., 2011). When a cell suspension (which may come from tissue digest, whole blood, cell culture, etc.) is flowed into such a channel, target cells adhere to the functionalized surface preferentially over non-target cells. Some non-target cells may non-specifically bind to the channel surfaces, but they may be removed via application of shear stress, i.e. by flowing media or buffer through the system. Trapped target cells can be stained, enumerated, and/or lysed for further analysis. Hansmann et al. (2011), for example, used anti-CD34 coated microfluidic channels to capture and enumerate endothelial progenitor cells (EPCs) from whole blood, demonstrating their potential use as a diagnostic or prognostic indicator for cardiovascular disease. Ng et al. (2010) also used anti-CD34 in devices to capture EPCs, and in addition developed an on-chip impedance-based detection method. Application of larger shear stresses will remove captured target cells from the surface, but may also damage cells, and/or alter phenotypic expression levels. In experiments where cells are sensitive to shear stress, alternative capture/release mechanisms are preferred (see next section).
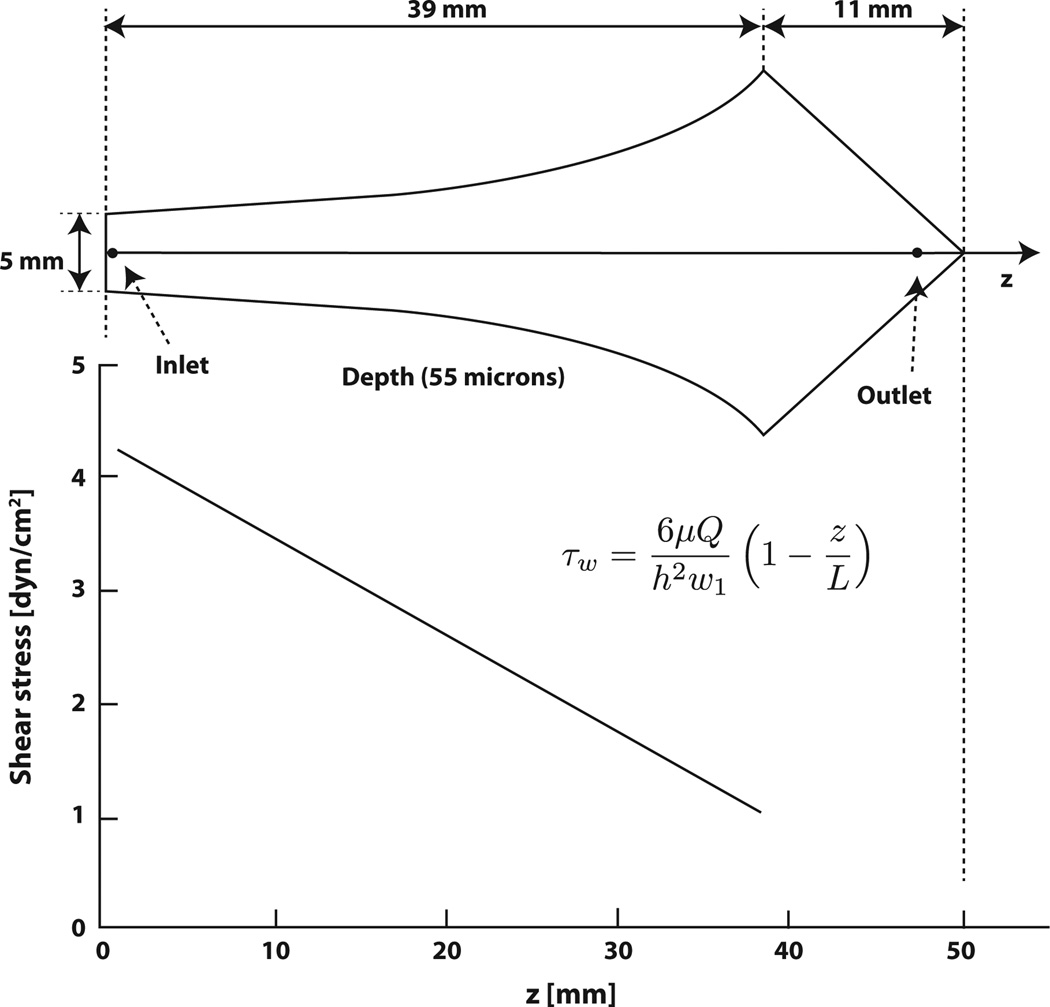
In order to optimize cell-capture devices for maximum throughput while maintaining high capture efficiency, it is critical that cell adhesion as a function of shear stress be characterized for each combination of cell type and capture molecule. Such an analysis also informs shear stress parameters for capturing and removing target cells. For capture, the shear stress must be large enough to remove non-specifically bound cells, but small enough so that target cells are not removed. Usami et al. (1993) designed a flow chamber based on Hele-Shaw flow with a linear drop in shear stress from inlet to outlet; Murthy et al. (2004) and others fabricated microfluidic devices based on the design of Usami et al., functionalized them with capture molecules, and used them to evaluate cell adhesion as a function of shear stress (Figure 1). The advantage of these Hele-Shaw cell devices is that they give the researcher access to a range of shear stresses in one device and in a single experiment.
Figure 1.
Scheme of Hele-Shaw flow chamber geometry and shear stress profile for the device geometry designed by Usami et al. (1993) and developed by Murthy et al. (2004). Figure adapted from Murthy et al. (2004).
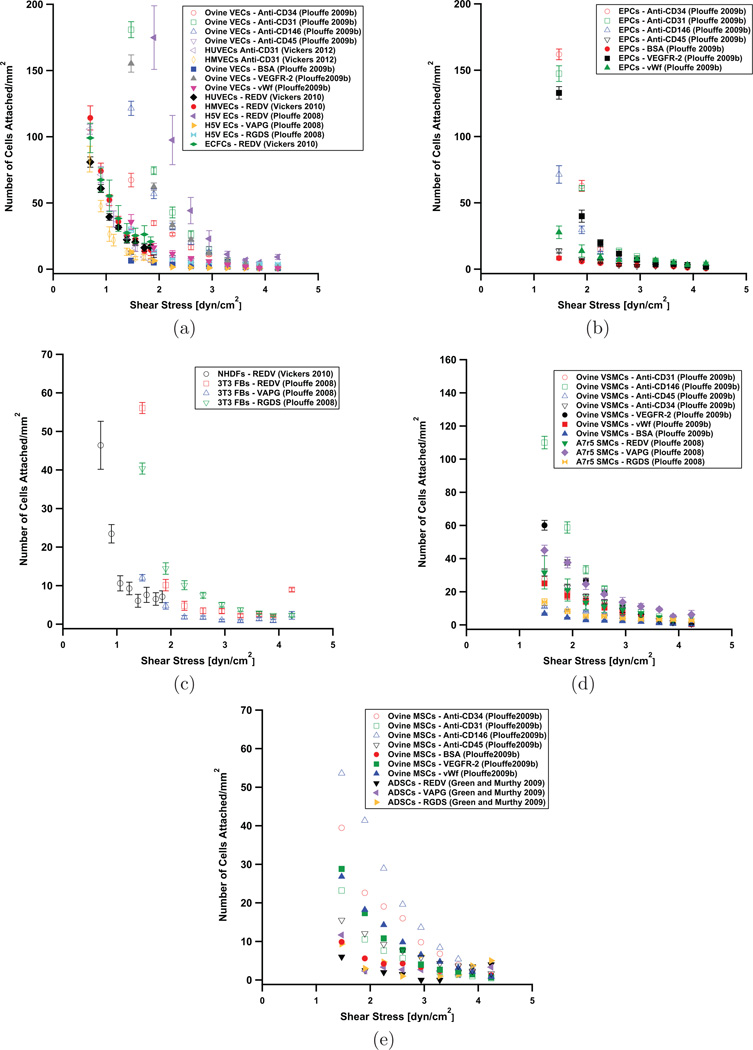
Figure 2 summarizes cell adhesion data for cell types relevant to cardiovascular regenerative medicine, with several capture molecules. Plouffe et al. (2007) and Green and Murthy (2009) characterized the adhesion of smooth muscle cells (SMCs), endothelial cells (ECs), fibroblasts (FBs), and adipose-derived stem cells (ADSCs) to surfaces coated with the peptide sequences, REDV, VAPG, and RGDS, as a function of shear stress. ECs preferentially bind to REDV surfaces, while SMCs preferentially bind to VAPG surfaces. Fibroblasts tend to pass through the system without binding. Vickers and Murthy (2010) demonstrated the ability to distinguish between three different types of endothelial cells based on differences in their surface receptor density, as measured by examining their adhesion to REDV peptide functionalized surfaces as a function of shear stress; the cell types examined were human umbilical vein endothelial cells (HUVECs), human microvascular endothelial cells (HMVECs), and endothelial colony forming cells (ECFCs). Vickers et al. (2012) used cell adhesion data for HUVECs and HMVECs on anti-CD31 coated surfaces to inform design of an enrichment platform for HMVECs and HUVECs (Figure 3). In developing a platform to capture endothelial progenitor cells (EPCs) for diagnostics/prognostics applications, Plouffe et al. (2009b) examined adhesion as a function of shear stress for EPCs, ECs, SMCs, and mesenchymal stem cells (MSCs) on surfaces coated with BSA, VEGFR-2, vWF, anti-CD34, anti-CD31, anti-CD146, and anti-CD45. In general, endothelial cells can remain adhered to surfaces at higher shear stress than other cells, such as stem cells. The data in Figure 2 can be used to determine an appropriate surface coating-shear stress combination for microfluidic cell capture devices; a shear stress must be chosen such that target cells adhere in large numbers, and non-target cells do not. Furthermore, these shear stress experiments can be used to give a measure of relative surface receptor expression levels for a given cell type (or for a comparison of two or more cell types) under different conditions; higher expression will lead to greater adhesion.
Figure 2.
Cell adhesion on a variety of functionalized surfaces as a function of shear stress for (a) endothelial cells, (b) endothelial progenitor cells, (c) fibroblasts, (d) smooth muscle cells, and (e) stem cells. Open symbols denote antibodies as the capture molecules, and close symbols represent peptides or other proteins.
Figure 3.
Two-stage microfluidic system for separation of cells with different densities of surface receptors. Each device stage has a different surface density of capture molecules, resulting in enrichment of a particular cell type. Figure from Vickers et al. (2012) (Permission Pending).
2.3. Enrichment by Cell Capture and Release
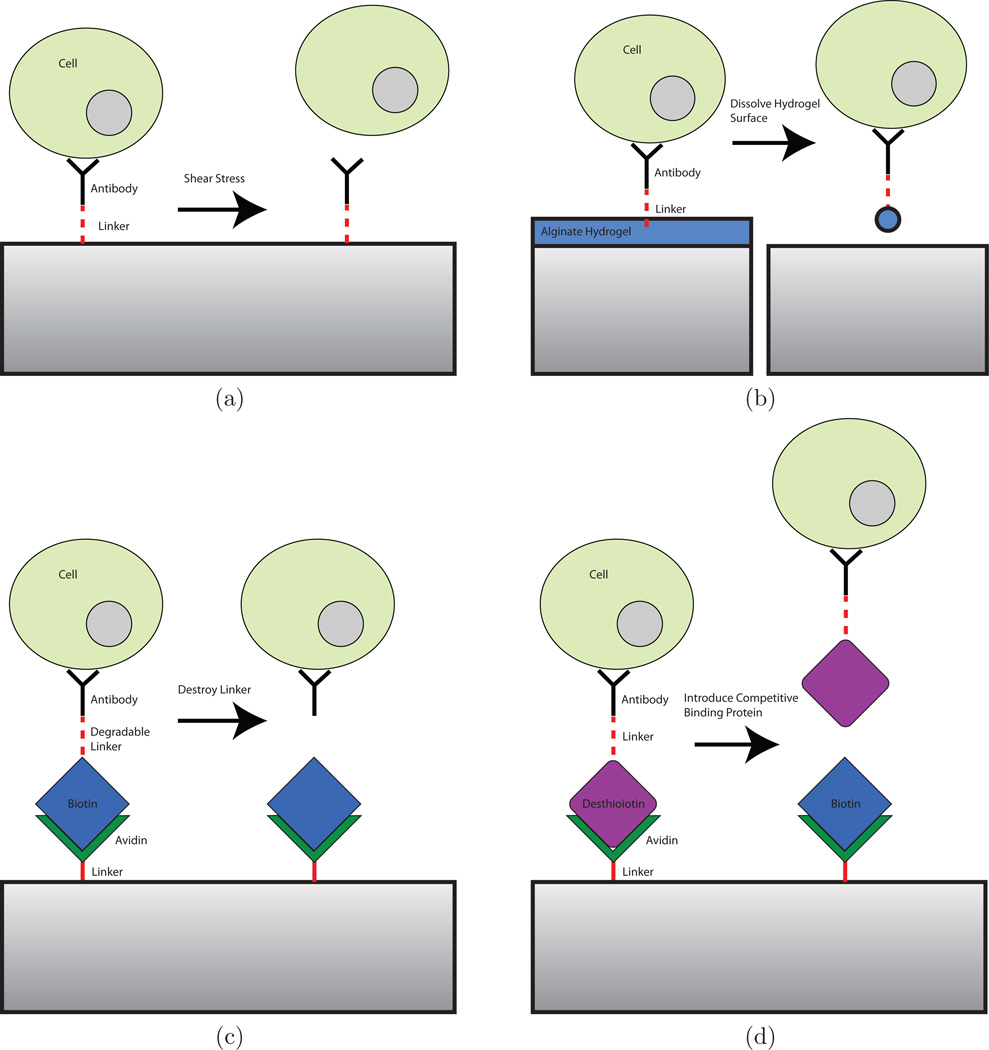
While conventional antibody-based capture mechanisms have been highly successful in trapping cells in microfluidic devices, releasing specifically captured cells for subsequent analysis and culture is a significant challenge. This limits the use of antibody-based positive selection mechanisms for cell enrichment. Covalently-bound surface antibodies cannot be removed without causing significant damage to cells. In some cases where the interaction between an antibody and the target cell surface receptor is sufficiently weak, captured cells can be removed by subjecting them to shear stress (Murthy et al., 2004; Plouffe et al., 2007, 2009b; Vickers et al., 2011; Santana et al., 2012). Optimization of the strength of antibody-antigen interaction (by varying surface antibody concentration, for example) and the magnitude of shear stress applied is non-trivial; weak antibody-cell interactions can lead to non-specific binding of cells and reduced purity, while high shear stresses can damage cells and remove target cells, leading to reduced efficiency. A number of alternatives to covalent surface attachment of antibodies for selective capture and subsequent release of target cells have emerged, including degradable surface coatings functionalized with capture molecules, cleavable linkages between capture antibodies and binding proteins (such as biotin), and reversible binding of capture molecules (Figure 5). Capture/release mechanisms that have been developed along with potential capture/release methods are summarized in Table 1. These techniques are less damaging to cells than application of shear stress, preserving viability, function, and phenotypic identity.
Figure 5.
Schemes of potential cell capture/release mechanisms. (a) Shear stress is applied to remove cells from capture antibodies. (b) Capture molecules are functionalized to a degradable surface. When the surface is removed, cells are released. (c) A photo- or electrodegradable linker between antibodies and other proteins is used to tag specific cells with biotin. Cells are released when the linker molecule is destroyed. (d) Avidin-desthiobiotin binding is disrupted by competitive binding from biotin, releasing captured cells.
Table 1.
Summary of potential cell capture and release mechanisms. Release efficiency refers to the number of cells released divided by the number of cells captured. Efficiency refers to the number or released cells divided by the total number of target cells in the initial sample (measured, for example, with a FACS-based flow cytometer). Efficiency is equivalent to the product of capture efficiency and release efficiency.
| Demonstrated Selective Cell Capture/Release Methods | ||||||
|---|---|---|---|---|---|---|
| Platform | Release Mechanism |
Capture Molecule |
Efficiency | Release Efficiency |
Sample | Target Cells |
| Ionically Crosslinked Alginate Hydrogels (Hatch et al., 2011) | Chelation with EDTA | Anti-CD34 Antibody | 33% | - | Whole Blood | EPCs |
| Ionically Crosslinked Alginate Hydrogels (2 Stages) (Hatch et al., 2012) | Chelation with EDTA | Anti-CD34 and Anti-FLk1 Antibodies | 45% | - | Whole Blood | EPCs |
| Ionically crosslinked alginate (Plouffe et al., 2009a) | Chelation with EDTA | RGDS peptide | - | 97% | Cell Suspension (10 × 104 cells/mL) | Cardiac Fibroblasts |
| Photo-crosslinked Alginate Hydrogels (Shah et al., 2012) | Dissolution with Alginate Lyase | Anti-EPCAM Antibody | - | 99% | Whole Blood | Spiked PC3 cells |
| Ligands Bound with an Electroactive Functional Group (Yeo et al., 2001) | Voltage Applied to Surface | RGD peptide | - | 70% Initially, ~100% with time | Cell Suspension - Cells were cultured on chip for 30 min | 3T3 Swiss Fibroblast Cells |
| PNIPAAm temperature responsive polymer (Gurkan et al., 2011) | Release of Adsorbed Neutravidin by Cooling to < 32° C | Biotinylated Human Anti-CD4/CD34 | - | 59% | Buffy Coat | CD4+ cells for HIV monitoring, EPCs |
| Potential Methods for Selective Cell Capture/Release | ||||||
| Platform | Release Mechanism | Target/Application | ||||
| Photolabile PEG Hydrogels (Kloxin et al., 2009, 2010) | UV Irradiation (405 nm) | Dynamically Tunable Cell Culture Platforms | ||||
| Desthiobiotin/Avidin (Hirsch et al., 2002) | Displacement via Competitive Binding of Biotin | Initial studies with proteins | ||||
| Photolabile Protein Crosslinking Reagents (Senter et al., 1985) | Photocleave protein (e.g. Biotin)-antibody linker | Antibody-Toxin Conjugates | ||||
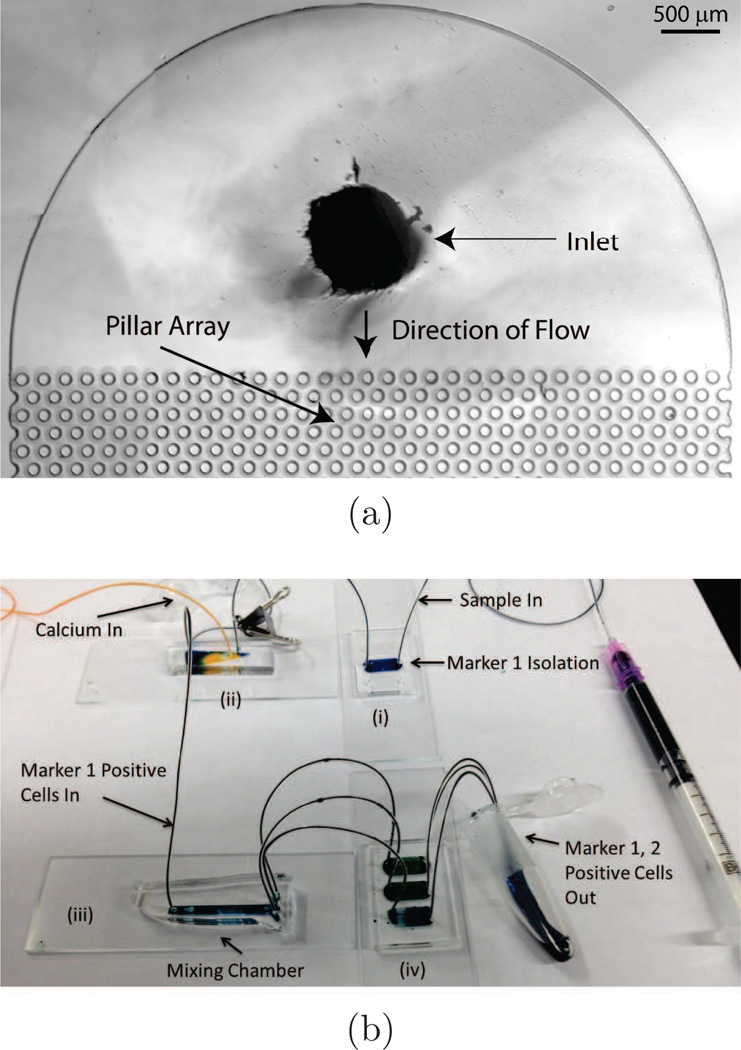
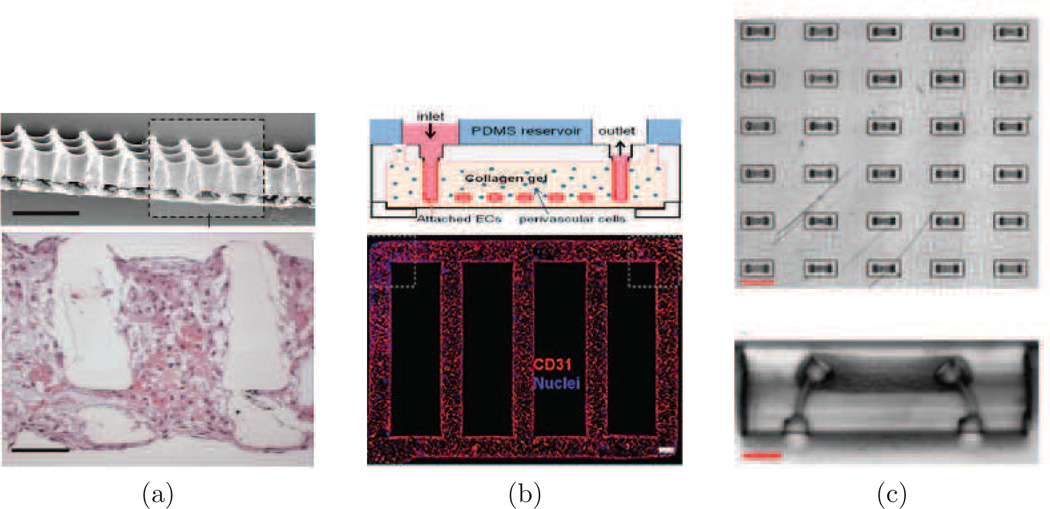
Aliginate hydrogels are commonly used in tissue engineering scaffolds, but have recently been utilized as degradable surface coatings for microfluidic channels in cell-enrichment platforms (Plouffe et al., 2009a; Hatch et al., 2011; Shah et al., 2012). Alginate-based hydrogels have been successfully conjugated with peptide (Plouffe et al., 2009a) and antibody (Hatch et al., 2011; Shah et al., 2012) capture molecules. Plouffe et al. (2009a) functionalized alginate hydrogels with a peptide sequence (RGDS) that binds to cardiac fibroblasts. The hydrogel was ionically crosslinked with Ca2+ ions on the walls of straight microfluidic channels. Cells in suspension that were exposed to the hydrogel surface adhered to the RGDS tetrapeptides, and afterward they were released by introduction of a chelator; in this case ethylenediaminetetraacetic acid (EDTA) was used. Hatch et al. (2011) designed a hydrogel surface coating using ionically-crosslinked alignate, and 4-armed poly(ethylene glycol) (PEG) molecules that increased the number of conjugation sites for capture antibodies (Figure 4a). The ionically-crosslinked hydrogel was also dissolved with EDTA to release bound cells. When conjugated with anti-CD34 and introduced into microfluidic devices with a pillar-array geometry, these hydrogels captured and released endothelial progenitor cells (EPCs) from whole human blood drawn from heparin tubes with 33% efficiency, 90% viability, and 74% purity. Hatch et al. (2012) refined this system and also explored processing blood through two of these pillar-array devices in series (where the first capture antibody was anti-CD34, and the second was anti-Flk1), improving purity and overall efficiency (Figure 4b). Shah et al. (2012) spin coated alginate hydrogels onto substrates and incubated them with a photoinitiator for photo-crosslinking. Such a hydrogel is stable against EDTA, and can be dissolved with alginate lyase. This procedure is therefore suitable for use with EDTA-treated blood, though it is more complicated and less cost-effective owing to the use of an enzyme for hydrogel degradation. Using anti-EpCAM as the capture antibody, Shah et. al. recovered spiked PC3 cells from whole blood with 99% release efficiency and 98.9% viability.
Figure 4.
(a) Micrograph of the inlet of a microfluidic device for selective capture and release of cells using degradable hydrogels. The pillar-array geometry increases the likelihood of collisions between the cells and the hydrogel coating. The device is symmetrical, where the outlet is similar to the inlet. (b) Sequence of devices for adhesion-based microfluidic separation of cells against multiple surface markers. Following capture and release from device (i), cells expressing marker 1 enter device (ii) where a calcium chloride solution is co-injected to neutralize the EDTA present in the cell suspension. Device (iii) mixes the calcium chloride solution and cell suspension. Device (iv) captures cells against marker 2, which can then be eluted out using an injection of EDTA solution. Figure from Hatch et al. (2012) (Permission Pending).
Temperature-repsonsive polymers have also been explored as a degradable surface coating for releasing adhered cells (Yamato et al., 2001, 2002; Ernst et al., 2007; Ma et al., 2010; Gurkan et al., 2011; Yang et al., 2012). Poly(N-isopropylacrylamide) copolymer (PNIPAAm) has been used as a coating in tissue-culture applications as part of an alternative method to trypsinization for removing adhered cells (Yamato et al., 2001, 2002; Ernst et al., 2007; Ma et al., 2010; Yang et al., 2012). It is hydrophobic at 37 °C and hydrophilic at 20 °C, so cells can be released by reducing the temperature of the system. This is useful in cases where digestion of extracellular matrix proteins and membrane proteins by proteolytic enzymes, such as trypsin, must be minimized. Yang et al. (2012), for example, demonstrated that mesenchymal stem cells from rat bone marrow (BM-MSCs) and human adipose tissue (ATMSCs) cultured on PNIPAAm copolymer films and recovered by reducing the temperature of the polymer were not significantly different in terms of morphology, immunophenotype, and osteogenesis/adipogenesis than cells released by trypsinization (Yang et al., 2012). These cells, however, had higher viability, stronger proliferation, and higher differentiation than those removed by trypsinization. Gurkan et al. (2011) extended the use of PNIPAAm copolymers to selective cell enrichment by incorporating cell-capture proteins into PNIPAAm films in microfluidic channels. Neutravidin and biotinylated antibody (humant anti-CD4 or anti-CD34) were immobilized on PNIPAAm surfaces in microfluidic channels at 37 °C. After buffy coat samples containing target cells were introduced into the microchannels, and a rinsing step, captured cells were released by reducing the temperature to less than 32 °C. CD4+ cells were released with 59% efficiency, and they were captured with 91% purity. Cell capture and release using PNIPAAm or other thermally-responsive polymers is useful when it is imperative that the cell sample be exposed to a minimal amount of shear stress and/or changing solvent conditions. These polymers are relatively expensive, however, and challenging to implement in non-trivial microfluidic channel geometries, limiting their utility for use with complex heterogeneous cell suspensions, including clinically relevant samples such as whole blood.
Several other methods for releasing captured targets exist, including photodegradable hydrogels (Kloxin et al., 2009, 2010), photodegradable linkers (Senter et al., 1985), electroactive linkers (Yeo et al., 2001), and desthiobiotin/avidin binding (Hirsch et al., 2002), though the use of these methods in selective cell enrichment has been minimal or non-existent to this point. Kloxin et al. (2009, 2010) created photodegradable hydrogels to create cell-culture platforms with tunable physical properties. By synthesizing a photodegradable, PEG-based cross-linking monomer and a photoreleasable peptide tether, a hydrogel with dynamically tunable modulus and peptide presentation was created. The hydrogel can be degraded by exposure to UV (405 nm) light. By combining such a photodegradable PEG hydrogel with capture molecules (antibodies or peptides), such a hydrogel can be used for cell enrichment, where release of captured cells is achieved by exposure to UV light. The effect of UV exposure on cell viability, morphology, and gene expression for such a procedure is not well-characterized. Methods for creating photocleavable linkages between antibodies and other proteins are available, and well-characterized (Senter et al., 1985). Avidin covalently bound to surfaces on microfluidic channels combined with capture antibodies linked to biotin via photocleavable linkers can be used as a capture/release mechanism. Electroactive linkers for attaching capture peptides to surfaces have also been demonstrated, and have been used to capture and release 3T3 fibroblasts (Yeo et al., 2001). Typically, avidin-biotin binding is irreversible under conditions amenable to maintaining cell viability. Binding between desthiobiotin and avidin, however, is much weaker (Hirsch et al., 2002), and is reversible. In a cell-capture mechanism where avidin is immobilized onto a surface and cells are tagged with an antibody-desthiobiotin conjugate, cells can be released by introducing biotin into solution for competitive binding. The challenge in this approach is that flow-induced shear stress may be sufficient to displace captured cells owing to the weaker avidin-desthiobiotin interaction, reducing yield and efficiency.
2.4. Continuous-Flow Cell Enrichment Techniques
There are numerous techniques for continuous-flow enrichment of subtypes of cell populations (Pratt et al., 2011). A complete review of all continuous-flow enrichment techniques, however, is beyond the scope of this study. Here we restrict ourselves to non-electrokinetic techniques, with emphasis on cell enrichment for applications in cardiovascular regenerative medicine. The cell-capture methods we have described in the previous sections rely on positive selection based on an immobilized antibody, i.e. cells that express surface markers with an affinity for the capture molecule are removed from the sample stream, and are either analyzed on-chip or released later for further analysis. Different methods have been employed to enrich cell populations from heterogeneous suspensions in a continuous flow fashion (Table 2), including size-based sorting, negative selection (unwanted cells are depleted from the sample stream), and tagging cells with antibodies conjugated to fluorescent or magnetic markers to activate sorting mechanisms (as is the case in traditional FACS). Continuous-flow methods have the potential to be higher throughput and better-suited to batch processing than capture/release methods.
Table 2.
Continuous-flow cell enrichment methods.
| Platform | Enrichment Mechanism |
Flow Rate |
Efficiency | Purity | Sample | Target Cells |
|---|---|---|---|---|---|---|
| Flared microchannel with sieves (Murthy et al., 2006) | Size-Based Sorting | 20 µL/min | - | 87% | Cell suspsension from neonatal rat ventricle tissue; non-myocytes were enriched in a first step by conventional pre-plating | Non-myocytes (primarily cardiac fibroblasts) |
| Deterministic Lateral Displacement (Davis et al., 2006) | Size-Based Sorting | 1 µL/min | - | 100% of lymphocytes and monocytes removed | Whole Blood | Blood Fractionation |
| Deterministic Lateral Displacement (Green et al., 2009) | Size-Based Sorting | 20 µL/min | 90% | 97% | 50:50 mixture of H1975 cells and 3T3 fibroblasts at 5 × 105 cells/mL, intended to model digested rat cardiac tissue | H1975 cells (Model for Cardiomyocytes) |
| Deterministic Lateral Displacement (Zhang et al., 2012) | Size-Based Sorting | 80 µL/min | 55% | 91% | Digested Neonatal Rat Cardiac Tissue | Cardiomyocytes |
| 3 Stages of Spiral Microfluidic Channels with Immobilized REDV, VAPG, and RGDS peptide sequences (Green and Murthy, 2009) | Negative Selection | 7.54 µL/min | 54.9% | 8.9% | 0.33 : 0.33 : 0.33 : 0.01 ECs : SMCs : FBs : ADSCs at 1 × 106 cells/mL in PBS | ADSCs |
| Magnetic Sorting (Plouffe et al., 2012) | Cells are Tagged with anti-CD133 functionalized Magnetic Beads | 120 µL/min | 96.5% (HSCs) and 95.5% (EPCs) | - | Whole Blood | HSCs and EPCs |
Separation or fractionation of cells based on size is attractive because size-based enrichment techniques usually do not require tagging or pre-processing of sample cells. Size-based sorting methods are particularly useful for enriching populations of cardiomyocytes, as they are larger on average as compared to non-myocytes (17 ± 4 µm as compared to 12 ± 3 µm) (Murthy et al., 2006; Green et al., 2009). Murthy et al. (2006) patterned microsieves in a microfluidic device that filtered out 13% of cardiomyocytes from a suspension derived from rat ventricle tissue. Deterministic lateral displacement (DLD) is another size-based enrichment technique, which takes advantage of asymmetric bifurcation of laminar flow around pillar obstacles in a microfluidic channel to effect size-based sorting of particles or cells (Huang et al., 2004; Inglis et al., 2006). DLD is proficient for batch fractionation of blood (Davis et al., 2006). In a model system for cardiomyocytes in heart tissue digest, Green et al. (2009) enriched larger H1975 cells from a heterogeneous suspension with 97% purity. Moving from the model system to a biological sample, Zhang et al. (2012) processed digested neonatal rat cardiac tissue with DLD and enriched cardiomyocytes to 91% purity. Size-based sorting techniques are useful when the target cell population is of a clearly different size than other cells in a heterogeneous sample. However, owing to the inherent variability in cell sizes, such techniques are less useful for separating two cell types of similar size.
Negative selection techniques have several advantages over positive selection techniques; they eliminate the need for a mechanism to release trapped cells, and they have the potential to operate at higher throughput in continuous flow. They are less selective, however, so they cannot be used to enrich very rare sub-populations of cells, and are therefore most suited to early-stage batch processing of samples. In negative selection techniques, unwanted cells are trapped in a microfluidic device by selective capture molecules (peptides or antibodies) and depleted from the sample as it flows through the device (Plouffe et al., 2008; Green and Murthy, 2009). Plouffe et al. (2008) designed a system for depletion of endothelial cells (ECs), smooth muscle cells (SMCs), and fibroblasts (FBs) by functionalizing microfluidic devices with the peptide sequences REDV, VAPG, and RGDS, respectively. Green and Murthy (2009) used a spiral device geometry to optimize this platform, and demonstrated greater than 5-fold enrichment of adipose-derived stem cells (ADSCs) from a heterogeneous suspension containing ECs, SMCs, FBs, and only 1.6% ADSCs. Despite the significant enrichment, the purity of ADSCs in the output was only 8.9% with 55% efficiency, demonstrating that negative selection is not suitable for rare cell isolation; all of the target cells would be lost due to non-specific binding before a pure population could be obtained.
Fluorescence-activated cell sorting (FACS) has, to this point, been the standard technique used for cell isolation and enumeration, and is ubiquitous owing to its versatility, robustness, and ability to produce highly pure cell populations based on factors such as size and biomarker affinity. In FACS and magnetically-activated cell sorting (MACS), cells are labeled or tagged with a fluorescent or magnetic marker linked to an antibody that binds to a biomarker of interest. When utilized in microfluidic devices, this technique is powerful, and is capable of enriching extremely rare cells with high throughput; the key disadvantage is that pre-processing is required (cells must be tagged). Plouffe et al. (2012) developed a microfluidic MACS system that utilizes optimized electromagnets. They demonstrated enrichment of as few as 50 MCF-7 cells/mL spiked into whole blood with 93% efficiency and 78% purity. The same system also isolated hematopoietic stem cells (HSCs) and endothelial progenitor cells (EPCs) from whole blood with 96.5% and 95.5% efficiency, respectively. Magnetophoretic separation, based on the Veridex CellSearch system, has also been used to isolate circulating endothelial cells (CECs) from blood (Damani et al., 2012), but isolation of viable CECs from a purely microfluidic system has yet to be demonstrated.
3. Development and Functionalization of Cardiovascular Tissue Scaffolds
When assembling individual cells into a functional tissue, the extra-cellular-matrix (ECM) plays a critical role in regulating the structural and mechanical properties of the entire tissue as well as providing structural and molecular signaling to guide the assembly of the surrounding cells. Native cardiac tissues are composed of well-aligned bundles of cardiomyocytes surrounded by cardiac fibroblasts and microvasculature as support. An ideal engineered matrix would simulate such structures as cells populate the matrix to form an engineered tissue.
3.1. Scaffolds for Anisotropic Tissue Assembly
Scaffolds have been shown to be critical in controlling the structural alignment of cardiomyocytes in engineered tissue. The control is often achieved by topographical features patterned on the scaffold. For instance, using replicate molding techniques, photocrosslinkable collagen-chitosan hydrogels with microgrooves of 10 µm, 20 µm, and 100 µm in width were fabricated from poly(dimethylsiloxane) (PDMS) molds (Chiu et al., 2012). Seeded cardiomyocytes aligned in the direction of the grooves, resulting significant reduction in their electrical excitation thresholds as compared to cells grown on smooth control gels. This is evidence for the importance of cell alignment in cardiac functions. In addition, smaller groove sizes (10 µm) provide a higher yield of functional tissue, which implies that larger grooves could interfere with cellular communication and gap junction formation (Chiu et al., 2012). However, hydrogel substrates may not be suitable for in vivo applications owing to their low mechanical strength.
Other studies have focused on the interactive effects of contact guidance and electrical stimulation on cardiac cell elongation and orientation on two dimensional (2-D) surfaces. Au et al. (2007), for example, utilized lapping paper polyvinyl surfaces that can be rapidly abraded resulting in unidirectional micro-grooves (3–13 µm), which help guide the elongation and orientation of cardiomyocytes and cardiac fibroblasts. By incorporating electrical stimulation, either parallel or perpendicular to the grooves, the study revealed that contact guidance has a much stronger effect on cardiac cell alignment than electrical stimulation, and that cells always orient along the grooves, even when electric fields are applied perpendicular to the grooves (Heidi Au et al., 2009). Furthermore, electrical stimulation has an additive effect on cell elongation if fields are applied in parallel with the grooves. Lastly, smaller groove sizes (1 µm) have a stronger effect on cell elongation than larger grooves (4 µm). Immunostaining of vinculin (a focal adhesion protein that links actin fibers to integrin receptors) revealed the localized positioning of the protein along the grooves (Heidi Au et al., 2009). The stronger effects seen from smaller grooves may be explained by a higher number of grooves underneath each cell, which could induce a greater effect on the localization of vinculin.
Elastomeric scaffolds have also been used to engineer aligned cardiac cell structures, and in addition provide near-physiological mechanical properties (Engelmayr et al., 2008; Guillemette et al., 2010; Wang et al., 2002). For instance, poly(glycerol sebacate) (PGS) was microfabricated into porous three dimensional (3-D) scaffolds with controlled structure, stiffness, and anisotropy (Engelmayr et al., 2008). By fabricating accordion-like honeycomb shaped pores, the mechanical properties (effective stiffness, anisotropy ratio, and strain-to-failure) of the scaffold were engineered to closely match that of native adult rat right ventricular myocardium (Figure 6a). Seeded cardiomyocytes followed the structural contour of the scaffold and formed aligned tissue, resulting in directionally-dependent electrical excitation thresholds. Although synchronized contraction of the cardiac cells was established, these contractions were not able to induce in-plane compression of the scaffold macroscopically (Engelmayr et al., 2008). This is due to the large compressive resistance of the scaffold, which must be reduced by further decreasing the strut width of the scaffold. Therefore, there is a need to create submicron-sized structures that can help guide cell alignment without compromising the overall scaffold compressibility.
Figure 6.
Cardiovascular tissue engineering. (a) Accordion-like honeycomb scaffolds for anisotropic cardiac tissue engineering (Engelmayr et al., 2008). (b) Microfluidic vessel network (Zheng et al., 2012). (c) Microfluidic tissue gauges used to measure temporal contractility response of micro-tissue (Legant et al., 2009).
Electrospun nanofibers were used to create submicron topographical features in 3-D (Li and Xia, 2004; Orlova et al., 2011; Xu et al., 2004). Spatially aligned nanofibers have been demonstrated to help guide the overall structural assembly of cardiomyocytes in 3-D. The best aligned tissue construct was created with a fiber diameter of ~ 300 nm with an interfiber distance of less than 30 µm (Orlova et al., 2011). This technique generates fibers on the same length scale as that of native extra cellular matrix. It has the advantage of providing topographical cues to cellular assembly without introducing large impedance to tissue contraction.
To further strengthen cardiomyocytes connectivity and conductivity in engineered 3-D tissue, gold nanowires have been incorporated into bioscaffolds made of synthetic materials, such as alginate and poly(lactic acid) (PLA), which both have poor conductivity (Dvir et al., 2011). These embedded gold nanowires have shown to help bridge the electrical communication between disconnected cardiomyocytes. Engineered tissues using this material have shown to contract synchronously and induce better cell alignment under electrical stimulation, while pristine alginate scaffolds tend to result in asynchronous local contractions. In addition, due to the increased electrical conductivity, engineered tissue with nanowires also results in elevated connexion43 and actinin expression. This method is one of the first to demonstrate that the inclusion of inorganic nanomaterials into polymer scaffolds can help the function of engineered tissues. However, more studies are required to examine the feasibility of this method for clinical applications.
3.2. Scaffolds for Vascularization
Microvasculature is another critical component in tissue engineering. Native tissue not only has well-aligned cardiomyocyte bundles, but also includes microvasculature surrounding the cardiomyocyte fibres. In addition to the role of providing oxygen and nutrients to the surrounding tissues, microvasculature, specifically endothelial cells, have been shown to enhance cardiomyocyte function by paracrine signaling (Narmoneva et al., 2004). Therefore, an ideal scaffold should also provide and induce rapid vascularization in engineered tissue to support cell survival and enhance cardiomyocyte functionality.
We have developed a method for inducing vascularization in engineered tissue by introducing angiogenic growth factors, such as vascular endothelial growth factor (VEGF) and Angiopoietin-1 (Ang1). Immobilizing these growth factors on collagen scaffolds (Figure 7) helps to establish sustained presence, induce endothelial cell proliferation and survival, and improve formation of tubular structures in 3-D scaffolds (Chiu and Radisic, 2010b). Covalently immobilizing both VEGF and Ang1 provides significantly better tube formation and endothelial proliferation as compared to immobilizing VEGF or Ang1 independently. In addition, immobilizing growth factors onto scaffolds results in enhanced cell proliferation compared to using soluble growth factors, indicating the lasting effect of signal activation due to these immobilized growth factors (Chiu and Radisic, 2010a; Chiu et al., 2011). Gradients of VEGF-165 were shown to direct endothelial cell migration toward higher concentrations of VEGF, but proliferation was the same as in scaffolds with uniformly distributed VEGF (Odedra et al., 2011). Through the use of VEGF gradients, cells cell density at the center of an engineered scaffold can be increased.
Figure 7.
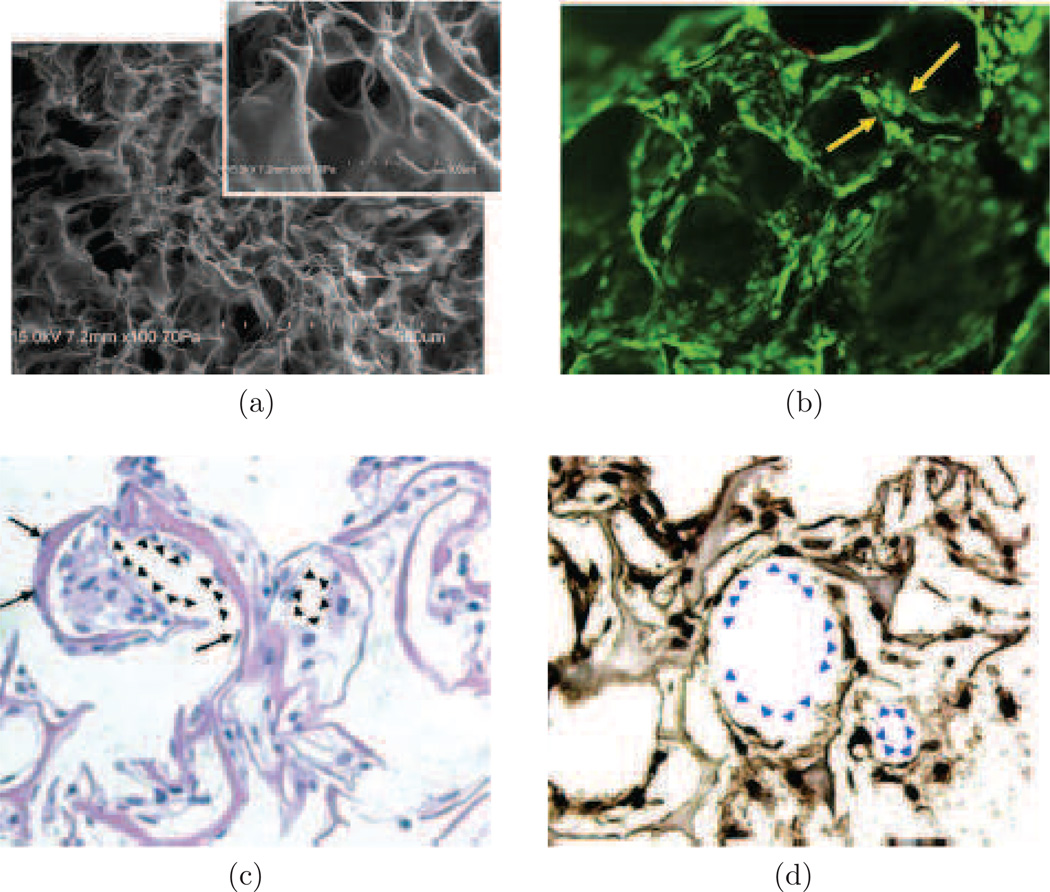
Characterization of functionalized collagen scaffolds with immobilized VEGF+Ang1 growth factors. (a) SEM image of collagen sponge. (b) Tube formation H5V cells on collagen scaffold immobilized with VEGF+Ang1. (c) and (d) show cell morphology and organization after 7 days of cultivation in vitro, using (c) hemotoxylin and eosin staining, and (d) Von Willebrand factor staining. Figure adapted from Chiu and Radisic (2010a).
The encapsulation and prolonged release of Thymosin β4 (Tβ4) (an angiogenic and cardioprotective 43 amino acid peptide) was achieved with a collagen-chitosan hydrogel to induce migration and outgrowth of CD-31 positive capillaries from mouse and rat epicardial explants in vitro (Chiu, 2011). In addition, implanted Tβ4 encapsulated collagen-chitosan hydrogels promoted enhanced angiogenesis in vivo as compared to collagen-only hydrogels. Surface charges and interconnected pore structures in the collagen-chitosan hydrogels were used to entrap Tβ4 molecules and resulting in a prolonged release profile with nearly zero-order kinetics. Although some success of vascularization was achieved by functionalizing scaffolds or hydrogels with growth factors, a perfusable and functional vasculature network that can be incorporated into functional cardiac tissue is yet to be demonstrated.
Other top down approaches that have been developed have involved direct design and fabrication of perfusable microvasculature structures. Microchannel networks have been fabricated with both biodegradable elastomers (Fidkowski et al., 2005; King et al., 2004) and hydrogels (Choi et al., 2007; Tang et al., 2004; Zheng et al., 2012), and then were subsequently seeded with endothelial cells to form functional vasculature. These approaches allow for rapid vasculature formation that can support surrounding cells. However, the size of the microvasculature is usually in the range of 100 µm, which is significantly larger than the typical size of capillaries, which have diameters on the order of 10 µm. Zheng et al. (2012) recently demonstrated interaction between pericytes and seeded endothelial cells in microfabricated microvasculature for the first time (Figure 6b). By adding this critical interaction, a more complete model of microvasculature can be generated to study vasculature stability and remodeling in vitro.
Many of the current top down methods for engineering vasculature are only a variation of layer-by-layer assembly methods, which involve laminating two layers together to form a closed channel network. This process is slow and often difficult to scale up to a 3-D network. In contrast, sacrificial molding methods provide an attractive alternative. Using sacrificial molds, a closed-channel network can be fabricated by casting a cell-encapsulated gel onto a sacrificial patterned network, which can then be subsequently removed under mild conditions to leave behind a closed-channel network. However, this approach has generally been limited to 2-D structures due to the lack of suitable materials; the material needs to be strong enough to form a rigid 3-D lattice, yet also must dissolvable under mild conditions. Miller et al. (2012) overcame this limitation by designing a carbohydrate composite that can satisfy all of these criteria. The carbohydrate composite includes mixtures of glucose and sucrose to form a simple and inexpensive glass, with the addition of dextran to further reinforce the glass and improve its temperature stability (Miller et al., 2012). Using this material, a micro-channel lattice can be printed with a 3-D printer in one simple step, eliminating the tedious process of layer-by-layer assembly. Using a 3-D printed scaffold composed of this material, endothelial cells were cultured to line the channel network, which sustained the metabolic function of embedded primary rat hepatocytes in the surrounding matrices. However one limitation with this approach, as well as other hydrogel moulding based approaches in vascularisation, is the inability to achieve the cellular density of native tissues (500 million cells/mL) (Zheng et al., 2012). To achieve physiological cell densities, extensive gel compaction and remodeling must be carried out to allow cells to form a functional tissue. Such remodeling, unfortunately, would likely interfere with the fabricated vasculature network which is weak without pericyte support. The addition of pericytes supporting the endothelialized network could be a potential solution. However, the time it takes for pericytes to stabilize the vasculature might exceed the time of gel compaction. Therefore, its feasibility in practice is yet to be demonstrated.
3.3. Functionalization of Scaffolds for Improved Tissue Performance
In addition to tissue structure and vascularization, cell survival is of critical importance in tissue engineering. QHREDGS peptides have been shown to provide cardioprotective properties (Rask et al., 2010a,b). The QHREDGS peptide sequence is a part of the putative integrin binding site of Ang1, a growth factor that is angiogenic and also cardioprotective. Immobilizing QHREDGS peptide to hydrogles has shown significant improvements in cell viability during culture in vitro. In addition, QHREDGS seems to promote cardiomyocyte elongation and enhance the contractile functionality of cardiomyocytes in vitro. Therefore, QHREDGS peptide is expected to improve tissue cultures when incorporated into cardiac tissue scaffolds. In our work with mouse models, QHREDGS-modified chitosan-collagen hydrogels were able to localize at the site of subcutaneous injection and localize cells (Reis et al., 2012).
In addition to functionlization of tissue scaffolds with growth factors or peptides, the functionality of engineered tissue can also be enhanced by pre-seeding scaffolds with non-myocytes to provide a more suitable initial environment for the subsequent seeding of cardiomyocytes (Radisic et al., 2007). The exact cellular interactions are yet to be elucidated, however signaling by VEGF-A released by fibroblasts and endothelial cells during pre-culture has been found to be of significance (Iyer et al., 2012). Additionally, the generation of extracellular matrix by the pre-seeded fibroblasts contributes to the better attachment and elongation of subsequently seeded cardiomyocytes. This implies an underling cellular interaction between myocytes and non-myocytes where non-myocytes play a supportive role.
3.4. Electrical and Mechanical Stimulation
Electrical signals play a critical role in orchestrating the synchronized contraction of cardiac muscle tissue. Cardiomyocytes respond to applied electrical stimulation by synchronous contractions. Over time, this leads to preferred structural alignment of tissue in the direction of the applied electric field; this reduces the excitation threshold voltage and increases the amplitude of contraction (Radisic et al., 2004; Tandon et al., 2009). However, electrical stimulation does not only simply induce cell alignment. On a molecular level, it also enhances cardiomyocyte performance by increasing the level of connexin-43(Cx-43), troponin I (Tn-I), myosin heavy chain (MHC), and sarcomeric α-actin, which are the components directly related to muscle contraction. Electrical stimulation also significantly enhances electrical impulse propagation of the engineered cardiac tissue. The average conduction velocity of stimulated tissue is significantly higher than in non-stimulated tissue (Radisic et al., 2009). This result is directly correlated to the better structural organization and increased gap junction density of the engineered tissue cultivated in the presence of electrical stimulation. Symmetric biphasic electrical stimulation was also shown to induce better cell function than using monophasic stimulation (Chiu et al., 2008). It is thought that biphasic pulses are more biocompatible, owing to their lower cumulative energy requirements and the ability to produce charge-balanced pulses.
Tension and mechanical stimulation also play critical roles in engineered cardiac tissue. Like all muscle cells, cardiomyocytes elongate with surface tension. Well aligned cardiac fibres have been engineered by utilizing tension created by gel compaction during hydrogel matrix remodeling (Boudou et al., 2012; Legant et al., 2009; Zimmermann et al., 2000). Initially, cardiac sheets with aligned cardiomyocytes were formed by embedding cardiomyocytes into a collagen gel and compacting them around two glass tubes (Eschenhagen et al., 1997). Later, this technique was altered to form tissue rings that were installed into a mechanical stretcher to mechanically stimulate the engineered tissue (Zimmermann et al., 2000). Thick cardiac tissue (1–4 mm) grown using this method was tested in vivo on myocardial infarcts of immunosuppressed rats (Zimmermann et al., 2006). The engineered heart tissue integrated successfully with the native myocardium with electrical coupling, without signs of arrhythmia induction, and induced systolic wall thickening of the infracted myocardial segment. This study is one of the first indicating that artificially engineered cardiac tissue can survive and support contractile function of infracted hearts in vivo upon implantation (Zimmermann et al., 2006).
Similar compaction methods were also utilized with fibrin gel to construct both aligned and isotropic configurations around a tubular mold (Black et al., 2009). The goal of this study was to examine the functional benefits of having an aligned myocardium. Aligned tissue constructs improved contraction in the aligned direction by 181%, which was even higher than what would be expected from simply aligning all the cells in the force measurement direction in an isotropic model. This implies that biological factors were also involved in the greater contraction forces experienced in the aligned model. Western blotting analysis on connexin 43(Cx43) and phosphorylated Cx43 (a less functional derivative of Cx43) revealed a decrease in phosphorylated Cx43 in the aligned model, which implies improved gap junction formation and function in the aligned model.
Using this gel compaction method, many studies have focused on generating microtissue in vitro as a 3-D heart model to study mechanotransduction, cellular forces, and drug screening. For instance, micro-cantilevers were used to shape the remodeling of a collagen gel embedded with cardiomyocytes (Figure 6c) (Legant et al., 2009). Forces can be correlated to the degree of bending from the micro-cantilevers. Forces from both gel/tissue remodeling and cellular contraction can be measured using this method in real time and with high throughput. These measurements will help in elucidating fundamental features of myocardial biology and in screening for drug-induced changes (Boudou et al., 2012).
3.5. Current Challenges
Numerous studies have been done on various aspects of scaffold design. Many of these studies often only focus on one or two aspects at a time. However, the complexity of cardiac tissue requires careful attention to physical structure, biofunctionality, vascularization, and electical/mechanical stimulation simultaneously. Some of major challenges in simultaneous incorporation of necessary features are summarized in Table 3.
Table 3.
Challenges in Cardiovascular Tissue Engineering.
| Key Challenges | Expectations |
|---|---|
| Cardiac Tissue Vascularization | A rapid vascularization method that can support an engineered cardiac tissue assembled with cardiomyocytes at physiological density and anisotropy |
| Mechano-electrical Stimulation on Engineered Cardiac Tissue | A platform that can provide both electrical and mechanical stimulation to engineered tissue and a potential to scale up to thick cardiac tissue by incorporating perfusable microvasculature. |
| Cardiac Scaffold | A scaffold that has robust mechanical properties matching that of native heart tissue but also allows cells to contract freely without impedance. |
From a design standpoint, addressing all of the needs for functional cardiac tissue is challenging, because different aspects of design often conflict with each other. For example, incorporating a topographical signal in a 3-D scaffold to induce cell alignment, while at the same time providing vascularization, might be challenging in practice. In this case, electrical stimulation can be utilized to achieve cell alignment, while scaffolds only need to be engineered to incorporate vasculature. Simultaneous use of several of the techniques described in this review is challenging, but has the potential to help in producing scaffolds that more closely mimic live tissue.
4. Conclusions
Microfluidic methods for cell enrichment offer unique advantages over traditional plating techniques and conventional FACS/MACS-based sorting, including simplicity, higher throughput, and the potential for very rare cell enrichment. Antibody- and peptide-functionalized devices have been shown to provide a means for measuring adhesion properties that are related to surface receptor density and binding interaction strength, as well as a means for capturing specific cells for diagnostics and analysis. When surface functionalization is combined with degradable surface coatings in microfluidic devices, captured cells can be released, cultured, and introduced into tissue engineering scaffolds for growth of new tissue. Microfluidic devices can also enrich subpopulations of cells in a continuous fashion, for even higher throughput analysis.
When enriched cardiac cell populations are introduced into advanced scaffolds, there is potential for production of functional, transplantable tissue. Modern microfabrication methods, advanced materials such as sacrificial molds, and electromechanical stimulation have all been used to create scaffolds that more closely mimic natural tissue environments. Scaffold geometry, biofunctionalization, and microvasculature are key components to creating successful cardiovascular tissue engineering scaffolds. Engineered tissue can be used to replace cells damaged as a result of cardiac disease; such tissue replacements will be critical in cardiovascular regenerative medicine in the future.
Acknowledgements
We would like to acknowledge our funding source, the NIH under R01 EB009327. We would also like to thank: Adam Hatch, Sean Kevlahan, and Dave Walsh for useful discussions; Jimmy Green, Brian Plouffe, and Dwayne Vickers for help in gathering and organizing data presented in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Vishal Tandon, Email: v.tandon@neu.edu.
Boyang Zhang, Email: bzhang7@gmail.com.
Milica Radisic, Email: m.radisic@utoronto.ca.
Shashi K. Murthy, Email: s.murthy@neu.edu.
References
- Au HT, Cheng I, Chowdhury MF, Radisic M. Interactive effects of surface topography and pulsatile electrical field stimulation on orientation and elongation of fibroblasts and cardiomyocytes. Biomaterials. 2007;28(29):4277–4293. doi: 10.1016/j.biomaterials.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114(6):763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Mhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LDr, Meyers JD, Weinbaum JS, Shvelidze YA, Tranquillo RT. Cell-induced alignment augments twitch force in fibrin gel-based engineered myocardium via gap junction modification. Tissue Eng Part A. 2009;15(10):3099–3108. doi: 10.1089/ten.tea.2008.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudou T, Legant WR, Mu A, Borochin MA, Thavandiran N, Radisic M, Zandstra PW, Epstein JA, Margulies KB, Chen CS. A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue engineering. Part A. 2012 doi: 10.1089/ten.tea.2011.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu LL, Iyer RK, King JP, Radisic M. Biphasic electrical field stimulation aids in tissue engineering of multicell-type cardiac organoids. Tissue Eng Part A. 2008 doi: 10.1089/ten.tea.2007.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu LL, Radisic M. Scaffolds with covalently immobilized vegf and angiopoietin-1 for vascularization of engineered tissues. Biomaterials. 2010a;31(2):226–241. doi: 10.1016/j.biomaterials.2009.09.039. [DOI] [PubMed] [Google Scholar]
- Chiu LL, Weisel RD, Li RK, Radisic M. Defining conditions for covalent immobilization of angiogenic growth factors onto scaffolds for tissue engineering. J Tissue Eng Regen Med. 2011;5(1):69–84. doi: 10.1002/term.292. [DOI] [PubMed] [Google Scholar]
- Chiu LLY. Controlled release of thymosin β4 using collagenchitosan composite hydrogels promotes epicardial cell migration and angiogenesis. Journal of Controlled Release. 2011;155:376–385. doi: 10.1016/j.jconrel.2011.05.026. [DOI] [PubMed] [Google Scholar]
- Chiu LLY, Janic K, Radisic M. Engineering of oriented myocardium on three-dimensional micropatterned collagen-chitosan hydrogel. The International journal of artificial organs. 2012;35:237–250. doi: 10.5301/ijao.5000084. [DOI] [PubMed] [Google Scholar]
- Chiu LLY, Radisic M. Scaffolds with covalently immobilized vegf and angiopoietin-1 for vascularization of engineered tissues. Biomaterials. 2010b;31:226–241. doi: 10.1016/j.biomaterials.2009.09.039. [DOI] [PubMed] [Google Scholar]
- Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD. Microfluidic scaffolds for tissue engineering. Nature Materials. 2007;6:908–915. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- C.L.Cai XXL, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damani S, Bacconi A, Libiger O, Chourasia AH, Serry R, Gollapudi R, Goldberg R, Rapeport K, Haaser S, Topol S, Knowlton S, Bethel K, Kuhn P, Wood M, Carragher B, Schork NJ, Jiang J, Rao C, Connelly M, Fowler VM, Topol EJ. Characterization of circulating endothelial cells in acute myocardial infarction. Science Translational Medicine. 2012;4(126) doi: 10.1126/scitranslmed.3003451. 126ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JA, Inglis DW, Morton KJ, Lawrence DA, Huang LR, Chou SY, Sturm JC, Austin RH. Deterministic hydrodynamics: taking blood apart. PNAS. 2006;103(40):14779–14784. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G. Sirpa is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nature Biotechnology. 2011;29(11):1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir T, Timko BP, Brigham MD, Naik SR, Karajanagi SS, Levy O, Jin H, Parker KK, Langer R, Kohane DS. Nanowired three-dimensional cardiac patches. Nat Nanotechnol. 2011;6(11):720–725. doi: 10.1038/nnano.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmayr GC, Cheng MY, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Accordion-like honeycombs for tissue engineering of cardiac anisotropy. Nature Materials. 2008;7:1003–1010. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst O, Lieske A, Jager M, Kankenau A. Control of cell detachment in a microfluidic device using a thermo-responsive copolymer on a gold substrate. Lab on a Chip. 2007;7:1322–1329. doi: 10.1039/b708619a. [DOI] [PubMed] [Google Scholar]
- Eschenhagen T, Fink C, Remmers U, Scholz H, Wattchow J, Weil J, Zimmermann W, Dohmen HH, Schafer H, Bishopric N, Wakatsuki T, Elson EL. Three-dimensional reconstitution of embryonic cardiomyocytes in a collagen matrix: a new heart muscle model system. FASEB J. 1997;11(8):683–694. doi: 10.1096/fasebj.11.8.9240969. [DOI] [PubMed] [Google Scholar]
- Evans D Fau Miller JB, Miller Jb Fau Stockdale FE, Stockdale FE. Developmental patterns of expression and coexpression of myosin heavy chains in atria and ventricles of the avian heart. Developmental Biology. 1988;127(2):376–383. doi: 10.1016/0012-1606(88)90324-7. [DOI] [PubMed] [Google Scholar]
- Fidkowski C, Kaazempur-Mofrad MR, Borenstein J, Vacanti JP, Langer R, Wang YD. Endothelialized microvasculature based on a biodegradable elastomer. Tissue Engineering. 2005;11:302–309. doi: 10.1089/ten.2005.11.302. [DOI] [PubMed] [Google Scholar]
- Green JV, Murthy SK. Microfluidic enrichment of a target cell type from a heterogenous suspension by adhesion-based negative selection. Lab on a Chip. 2009;9:2245–2248. doi: 10.1039/b906768j. [DOI] [PubMed] [Google Scholar]
- Green JV, Radisic M, Murthy SK. Deterministic lateral displacement as a means to enrich large cells for tissue engineering. Analytical Chemistry. 2009;81:9178–9182. doi: 10.1021/ac9018395. [DOI] [PubMed] [Google Scholar]
- Guillemette MD, Park H, Hsiao JC, Jain SR, Larson BL, Langer R, Freed LE. Combined technologies for microfabricating elastomeric cardiac tissue engineering scaffolds. Macromolecular Bioscience. 2010;10:1330–1337. doi: 10.1002/mabi.201000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan U, Anand T, Tas H, Elkan D, Akay A, Keles H, Demirci U. Controlled viable release of selectively captured label-free cells in microchannels. Lab on a Chip. 2011;11(23):3979–3989. doi: 10.1039/c1lc20487d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansmann G, Plouffe BD, Hatch A, Gise Av, Sallmon H, Zamanian RT, Murthy AK. Design, validation, and clinical application of an endothelial progenitor cell microfluidic capture chip in patients with cardiovascular disease. Journal of Molecular Medicine. 2011;89:971–983. doi: 10.1007/s00109-011-0779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch A, Hansmann G, Murthy SK. Engineered alginate hydrogels for effective microfluidic capture and release of endothelial progenitor cells from whole blood. Langmuir. 2011;27:4257–4264. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch A, Pesko DM, Murthy SK. Tag-free microfluidic separation of cells against multiple markers. Analytical Chemistry. 2012;84(10):4618–4621. doi: 10.1021/ac300496q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidi Au HT, Cui B, Chu ZE, Veres T, Radisic M. Cell culture chips for simultaneous application of topographical and electrical cues enhance phenotype of cardiomyocytes. Lab Chip. 2009;9(4):564–575. doi: 10.1039/b810034a. [DOI] [PubMed] [Google Scholar]
- Hirsch JD, Eslamizar L, Filanoski BJ, Malekzadeh N, Haugland RP, Beechem JM, Haugland RP. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Analytical Biochemistry. 2002;308:343–357. doi: 10.1016/s0003-2697(02)00201-4. [DOI] [PubMed] [Google Scholar]
- Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304(5673):987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- Inglis DW, Davis JA, Austin RH, Sturm JC. Critical particle size for fractionation by deterministic lateral displacement. Lab on a Chip. 2006;6:655–658. doi: 10.1039/b515371a. [DOI] [PubMed] [Google Scholar]
- Iyer RK, Odedra D, Chiu LL, Vunjak-Novakovic G, Radisic M. Vegf secretion by non-myocytes modulates connexin-43 levels in cardiac organoids. Tissue Eng Part A. 2012 doi: 10.1089/ten.tea.2011.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11(5):723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kaushal S, Amiel GE, Guleserian KJ, Shapira OM, Perry T, Sutherland FW, Rabkin E, Moran AM, Schoen FJ, Atala A, Soker S, Bischoff J, Jr, JEM Functional small-diameter neovessels created using endothelial progenitor cells expanded ex vivo. Nature Medicine. 2001;7(9):1035–1040. doi: 10.1038/nm0901-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KR, Wang CCJ, Kaazempur-Mofrad MR, Vacanti JP, Borenstein JT. Biodegradable microfluidics. Advanced Materials. 2004;16 2007–+. [Google Scholar]
- Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324(3):59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloxin AM, Tibbitt MW, Anseth KS. Synthesis of photodegradable hydrogels as dynamically tunable cell culture platforms. Nature Protocols. 2010;5(12):1867–1887. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolvenbach R, Kreissig C, Ludwig E, Cagiannos C. Stem cell use in critical limb ischemia. J. Cardiovasc. Surg. (Torino) 2007;48(1):39–44. [PubMed] [Google Scholar]
- Kouskoff V, Lacaud G, Schwantz S, Fehling HJ, Keller G. Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc Natl Acad Sci U S A. 2005;102(37):13170–13175. doi: 10.1073/pnas.0501672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, Platoshyn O, Yuan JX, Evans S, Chien KR. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433(7026):647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legant WR, Pathak A, Yang MT, Deshpande VS, McMeeking RM, Chen CS. Microfabricated tissue gauges to measure and manipulate forces from 3d microtissues. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10097–10102. doi: 10.1073/pnas.0900174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Xia YN. Electrospinning of nanofibers: Reinventing the wheel? Advanced Materials. 2004;16:1151–1170. [Google Scholar]
- Linke A, Muller P, Nurzynska D, Casarsa C, Torella D, Nascimbene A, Castaldo C, Cascapera S, Bohm M, Quaini F, Urbanek K, Leri A, Hintze TH, Kajstura J, Anversa P. Stem cells in the dog heart are self-renewing, clonogenic, and multipotent and regenerate infarcted myocardium, improving cardiac function. Proc Natl Acad Sci U S A. 2005;102(25):8966–8971. doi: 10.1073/pnas.0502678102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Chen H, Li Z, He Q. Thermomodulated cell culture/harvest in polydimethylsiloxane microchannels with poly(n-isoporopylacrylamide)-grafted surface. Biomicrofluidics. 2010;4 doi: 10.1063/1.3516038. 044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DH, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater. 2012 doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Liu YP, Marles L, Yang ZX, Trempus C, Li SL, Lin JS, Sawicki JA, Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nature Biotechnology. 2004;22(4):411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Murthy SK, Sethu P, Vunjak-Novakovic G, Toner M, Radisic M. Size-based microfluidic enrichment of neonatal rat cardiac cell populations. Biomedical Microdevices. 2006;8:231–237. doi: 10.1007/s10544-006-8169-5. [DOI] [PubMed] [Google Scholar]
- Murthy SK, Sin A, Tompkins RG, Toner M. Effect of flow and surface conditions on human lymphocite isolation using microfluidic chambers. Langmuir. 2004;20:11649–11655. doi: 10.1021/la048047b. [DOI] [PubMed] [Google Scholar]
- Nag AC. Study of non-muscle cells of the adult mammalian heart - a fine-structural analysis and distribution. Cytobios. 1980;28(109):41–61. [PubMed] [Google Scholar]
- Naito H, Melnychenko I, Didie M, Schneiderbanger K, Schubert P, Rosenkranz S, Eschenhagen T, Zimmermann W-H. Optimizing engineered heart tissue for therapeutic applications as surrogate heart muscle. Circulation. 2006;114:172–178. doi: 10.1161/CIRCULATIONAHA.105.001560. [DOI] [PubMed] [Google Scholar]
- Narmoneva DA, Vukmirovic R, Davis ME, Kamm RD, Lee RT. Endothelial cells promote cardiac myocyte survival and spatial reorganization: implications for cardiac regeneration. Circulation. 2004;110(8):962–968. doi: 10.1161/01.CIR.0000140667.37070.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Reboud J, Wang KYP, Tang KC, Zhang L, Wong P, Moe KT, Shim W, Chen Y. Label-free impedance detection of low levels of circulating endothelial progenitor cells for point-of-care diagnosis. Biosensors and Bioelectronics. 2010;25:1095–1101. doi: 10.1016/j.bios.2009.09.031. [DOI] [PubMed] [Google Scholar]
- O E, Lee BH, Ahn HY, Shin JC, Kim HK, Kim M, Park IY, Park YG, Joe YA. Efficient nonadhesive ex vivo expansion of early endothelial progenitor cells derived from cd34+ human cord blood fraction for effective therapeutic vascularization. FASEB Journal. 2011;25(1):159–169. doi: 10.1096/fj.10-162040. [DOI] [PubMed] [Google Scholar]
- Odedra D, Chiu LL, Shoichet M, Radisic M. Endothelial cells guided by immobilized gradients of vascular endothelial growth factor on porous collagen scaffolds. Acta Biomater. 2011;7(8):3027–3035. doi: 10.1016/j.actbio.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, Garry DJ, Entman ML, Schneider MD. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci U S A. 2003;100(21):12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, Li B, Pickel J, McKay R, Nadal-Ginard B, Bodine DM, Leri A, Anversa P. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410(6829):701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- Orlova Y, Magome N, Liu L, Chen Y, Agladze K. Electrospun nanofibers as a tool for architecture control in engineered cardiac tissue. Biomaterials. 2011;32:5615–5624. doi: 10.1016/j.biomaterials.2011.04.042. [DOI] [PubMed] [Google Scholar]
- Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell. 2001;104(2):233–245. doi: 10.1016/s0092-8674(01)00208-2. [DOI] [PubMed] [Google Scholar]
- Parratt JR, Vegh A, Zeitlin IJ, Ahmad M, Oldroyd K, Kaszala K, Papp JG. Bradykinin and endothelial-cardiac myocyte interactions in ischemic preconditioning. The American Journal of Cardiology. 1997;80(3):124A–131A. doi: 10.1016/s0002-9149(97)00467-0. [DOI] [PubMed] [Google Scholar]
- Plouffe BD, Brown MA, Iyer RK, Radisic M, Murthy SK. Controlled capture and release of cardiac fibroblasts using peptide-functionalized alginate gels in microfluidic channels. Lab on a Chip. 2009a;9:1507–1510. doi: 10.1039/b823523f. [DOI] [PubMed] [Google Scholar]
- Plouffe BD, Kniaseva T, Mayer JE, Murthy SK, Sales VL. Development of microfluidics as endothelial progenitor cell capture technology for cardiovascular tissue engineering and diagnostic medicine. FASEB Journal. 2009b;23:3309–3314. doi: 10.1096/fj.09-130260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe BD, Mahalanabis M, Lewis LH, Klapperich CM, Murthy SK. Clinically relevant microfluidic magnetophoretic isolation of rare-cell populations for diagnostic and therapeutic monitoring applications. Analytical Chemistry. 2012;84:1336–1344. doi: 10.1021/ac2022844. [DOI] [PubMed] [Google Scholar]
- Plouffe BD, Njoka DN, Harris J, Liao J, Horick NK, Radisic M, Murthy SK. Peptide-mediated selective adhesion of smooth muscle and endothelial cells in microfluidic shear flow. Langmuir. 2007;23:5050–5055. doi: 10.1021/la0700220. [DOI] [PubMed] [Google Scholar]
- Plouffe BD, Radisic M, Murthy SK. Microfluidic depletion of endothelial cells, smooth muscle cells, and fibroblasts from heterogeneous suspensions. Lab on a Chip. 2008;8:462–472. doi: 10.1039/b715707j. [DOI] [PubMed] [Google Scholar]
- Pratt ED, Huang C, Hawkins BG, Gleghorn JP, Kirby BJ. Rare cell capture in microfluidic devices. Chemical Engineering Science. 2011;66:1508–1522. doi: 10.1016/j.ces.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisic M, Fast VG, Sharifov OF, Iyer RK, Park H, Vunjak-Novakovic G. Optical mapping of impulse propagation in engineered cardiac tissue. Tissue Engineering Part A. 2009;15:851–860. doi: 10.1089/ten.tea.2008.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisic M, Park H, Martens TP, Salazar-Lazaro JE, Geng W, Wang Y, Langer R, Freed LE, Vunjak-Novakovic G. Pre-treatment of synthetic elastomeric scaffolds by cardiac fibroblasts improves engineered heart tissue. J Biomed Mater Res A. 2007;86:713–724. doi: 10.1002/jbm.a.31578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisic M, Park H, Martens TP, Salazar-Lazaro JE, Geng W, Wang Y, Langer R, Freed LE, Vunjak-Novakovic G. Pre-treatment of synthetic elastomeric scaffolds by cardiac fibroblasts improves engineered heart tissue. J Biomed Mater Res A. 2008;86(3):713–724. doi: 10.1002/jbm.a.31578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisic M, Park H, Shing H, Consi T, Schoen FJ, Langer R, Freed LE, Vunjak-Novakovic G. Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds. PNAS. 2004;101(52):18129–18134. doi: 10.1073/pnas.0407817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask F, Dallabrida SM, Ismail NS, Amoozgar Z, Yeo Y, Rupnick MA, Radisic M. Photocrosslinkable chitosan modified with angiopoietin-1 peptide, qhredgs, promotes survival of neonatal rat heart cells. J Biomed Mater Res A. 2010a;95:105–117. doi: 10.1002/jbm.a.32808. [DOI] [PubMed] [Google Scholar]
- Rask F, Mihic A, Reis L, Dallabrida SM, Ismail NS, Sider K, Simmons CA, Rupnick MA, Weisel RD, Li R-K, Radisic M. Hydrogels modified with qhredgs peptide support cardiomyocyte survival in vitro and after sub-cutaneous implantation. Soft Matter. 2010b;6:5089. [Google Scholar]
- Rehman J. Chipping away at the surface of the endothelial progenitor cell (epc) mystery. J Mol Med (Berl) 2011;89(10):943–945. doi: 10.1007/s00109-011-0799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis LA, Chiu LL, Liang Y, Hyunh K, Momen A, Radisic M. A peptide-modified chitosan-collagen hydrogel for cardiac cell culture and delivery. Acta Biomater. 2012;8(3):1022–1036. doi: 10.1016/j.actbio.2011.11.030. [DOI] [PubMed] [Google Scholar]
- Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312(5782):1946–1949. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart disease and stroke statistics2012 update. Circulation. 2012;125(1):e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana SM, Liu H, Bander NH, Gleghorn JP, Kirby BJ. Immunocapture of prostate cancer cells by use of anti-psma antibodies in microdevices. Biomedical Microdevices. 2012;14:401–407. doi: 10.1007/s10544-011-9616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senter PD, Tansey MJ, Lambert JM, Blattler WA. Novel photocleavable protein crosslinking reagents and their use in the preparation of antibody-toxin conjugates. Photochemistry and Photobiology. 1985;42(3):231–237. [Google Scholar]
- Shah AM, Mebazaa A, Yang Z-K, Cuda G, Lankford EB, Pepper CB, Sollott SJ, Sellers JR, Robotham JL, Lakatta EG. Inhibition of myocardial crossbridge cycling by hypoxic endothelial cells: A potential mechanism for matching oxygen supply and demand? 1997;80:688–698. doi: 10.1161/01.res.80.5.688. [DOI] [PubMed] [Google Scholar]
- Shah AM, Yu M, Nakamura Z, Ciciliano J, Ulman M, Kotz K, Stott SL, Maheswaran S, Haber DA, Toner M. Biopolymer system for cell recovery from microfluidic cell capture devices. Analytical Chemistry. 2012;84:3682–3688. doi: 10.1021/ac300190j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonpaa MH, Field LJ. Survey of studies examining mammalian cardiomyocyte dna synthesis. Circulation Research. 1998;83:15–26. doi: 10.1161/01.res.83.1.15. [DOI] [PubMed] [Google Scholar]
- Sussman MA, McCulloch A, Borg TK. Dance band on the titanic - biomechanical signaling in cardiac hypertrophy. Circulation Research. 2002;91(10):888–898. doi: 10.1161/01.res.0000041680.43270.f8. [DOI] [PubMed] [Google Scholar]
- Tandon N, Cannizzaro C, Chao P-H, G H, Maidhof R, Marsano A, Au HTH, Radisic M, Vunjak-Novakovic G. Electrical stimulation systems for cardiac tissue engineering. Nat. Protocols. 2009;4:155–173. doi: 10.1038/nprot.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang MD, Golden AP, Tien J. Fabrication of collagen gels that contain patterned, micrometer-scale cavities. Advanced Materials. 2004;16 1345–+. [Google Scholar]
- Usami S, Chen H, Zhao Y, Chien S, Skalak R. Design and construction of a linear shear stress flow chamber. Annals of Biomedical Engineering. 1993;21(1):77–89. doi: 10.1007/BF02368167. [DOI] [PubMed] [Google Scholar]
- Vickers DAL, Chory E, Murthy SK. Separation of two phenotypically similar cell types via a single common marker in microfluidic channels. Lab on a Chip. 2012 doi: 10.1039/c2lc40290d. In Press. [DOI] [PubMed] [Google Scholar]
- Vickers DAL, Hincapie M, Hancock WS, Murthy SK. Lectin-mediated microfluidic capture and release of leukemic lymphocytes from whole blood. Biomedical Microdevices. 2011;2011(13):565–571. doi: 10.1007/s10544-011-9527-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers DAL, Murthy SK. Receptor expression changes as a basis for endothelial cell identification using microfluidic channels. Lab on a Chip. 2010;10:2380–2386. doi: 10.1039/c004870d. [DOI] [PubMed] [Google Scholar]
- Wang JX. Alpha-smooth muscle actin (sma) is required for mechanical force-induced p38 activation. FASEB Journal. 2004;18(8):C–39–C–39. doi: 10.1074/jbc.M410819200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ameer GA, Sheppard BJ, Langer R. A tough biodegradable elastomer. Nat Biotech. 2002;20:602–606. doi: 10.1038/nbt0602-602. [DOI] [PubMed] [Google Scholar]
- Xu C, Inai R, Kotaki M, Ramakrishna S. Electrospun nanofiber fabrication as synthetic extracellular matrix and its potential for vascular tissue engineering. Tissue Engineering. 2004;10:1160–1168. doi: 10.1089/ten.2004.10.1160. [DOI] [PubMed] [Google Scholar]
- Yamato M, Konno C, Utsumi M, Kikuchi A, Okano T. Thermally responsive polymer-grafted surfaces facilitate patterned cell seeding and co-culture. Biomaterials. 2002;23(2):561–567. doi: 10.1016/s0142-9612(01)00138-7. [DOI] [PubMed] [Google Scholar]
- Yamato M, Utsumi M, Kushida A, Konno C, Kikuchi A, Okano T. Thermo-responsive culutre dishes allow the intact harvest of multilayered keratinocyte sheets without dispase by reducing temperature. Tissue Engineering. 2001;7(4):473–480. doi: 10.1089/10763270152436517. [DOI] [PubMed] [Google Scholar]
- Yang L, Cheng F, Liu T, Lu JR, Song K, Jiang L, Wu S, Guo W. Comparison of mesenchmal stem cells released from poly(n-isopropylacrylamide) copolymer film and trypsinization. Biomedical Materials. 2012;7 doi: 10.1088/1748-6041/7/3/035003. 035003. [DOI] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a kdr+ embryonic-stem-cell-derived population. Nature. 2008;453(7194):524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yeo W-S, Hodneland C, Mrksich M. Electroactive monolayer substrates that selectively release adherent cells. ChemBioChem. 2001;2(7–8):590–593. doi: 10.1002/1439-7633(20010803)2:7/8<590::AID-CBIC590>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Zhang B, Green JV, Murthy SK, Radisic M. Label-free enrichment of functional cardiomyocytes using microfluidic deterministic lateral flow displacement. PLoS One. 2012;7(5):e37619. doi: 10.1371/journal.pone.0037619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104(4):e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, Lpez JA, Stroock AD. In vitro microvessels for the study of angiogenesis and thrombosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann WH, Fink C, Kralisch D, Remmers U, Weil J, Eschenhagen T. Three-dimensional engineered heart tissue from neonatal rat cardiac myocytes. Biotechnology and bioengineering. 2000;68:106–114. [PubMed] [Google Scholar]
- Zimmermann WH, Melnychenko I, Wasmeier G, Didie M, Naito H, Nixdorff U, Hess A, Budinsky L, Brune K, Michaelis B, Dhein S, Schwoerer A, Ehmke H, Eschenhagen T. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med. 2006;12(4):452–458. doi: 10.1038/nm1394. [DOI] [PubMed] [Google Scholar]