The 3′ end of histone mRNA is oligouridylated during its degradation, although it is not known whether this is an essential step. The authors introduced firefly luciferase reporter mRNAs containing the histone 3′ UTR SL (Luc-SL) and either a normal or hDcp2-resistant cap into S-phase HeLa cells. Both mRNAs were translated, and translation initially protected the mRNAs from degradation, but there was a lag of ∼40 min with the uncleavable cap compared to ∼8 min for the normal cap before rapid decay. Knockdown of terminal uridyltransferase (TUTase) 4 but not TUTase 3 slowed the decay process in control cells. Both Luc-SL and its 5′ decay intermediates were oligouridylated. Preventing oligouridylation by 3′-deoxyadenosine (cordycepin) addition to the mRNA slowed degradation, suggesting oligouridylation initiates degradation.
Keywords: hDcp2-resistant cap analog, hydroxyurea, oligouridylation, TUTase 4, cordycepin
Abstract
Metazoan replication-dependent histone mRNAs are only present in S-phase, due partly to changes in their stability. These mRNAs end in a unique stem–loop (SL) that is required for both translation and cell-cycle regulation. Previous studies showed that histone mRNA degradation occurs through both 5′→3′ and 3′→5′ processes, but the relative contributions are not known. The 3′ end of histone mRNA is oligouridylated during its degradation, although it is not known whether this is an essential step. We introduced firefly luciferase reporter mRNAs containing the histone 3′ UTR SL (Luc-SL) and either a normal or hDcp2-resistant cap into S-phase HeLa cells. Both mRNAs were translated, and translation initially protected the mRNAs from degradation, but there was a lag of ∼40 min with the uncleavable cap compared to ∼8 min for the normal cap before rapid decay. Knockdown of hDcp2 resulted in a similar longer lag for Luc-SL containing a normal cap, indicating that 5′→3′ decay is important in this system. Inhibition of DNA replication with hydroxyurea accelerated the degradation of Luc-SL. Knockdown of terminal uridyltransferase (TUTase) 4 but not TUTase 3 slowed the decay process, but TUTase 4 knockdown had no effect on destabilization of the mRNA by hydroxyurea. Both Luc-SL and its 5′ decay intermediates were oligouridylated. Preventing oligouridylation by 3′-deoxyadenosine (cordycepin) addition to the mRNA slowed degradation, in the presence or absence of hydroxyurea, suggesting oligouridylation initiates degradation. The spectrum of oligouridylated fragments suggests the 3′→5′ degradation machinery stalls during initial degradation, whereupon reuridylation occurs.
INTRODUCTION
Histones play a critical role in eukaryotes in both packaging chromosomal DNA and regulating gene expression at the transcriptional level (Henikoff and Ahmad 2005; Marzluff et al. 2008). Synthesis of the canonical replicative histones (H2A, H2B, H3, and H4) occurs only when DNA is being synthesized (Marzluff et al. 2008) and is regulated primarily by changes in mRNA levels, which increase 35-fold as cells enter S-phase. At the end of S-phase or when DNA synthesis is inhibited, histone mRNAs are rapidly degraded (Harris et al. 1991; Whitfield et al. 2000). These mRNAs are unusual in that they contain a conserved 25- to 26-nt stem–loop (SL) at the 3′ end instead of poly(A) (Marzluff et al. 2008). The SL is recognized by a stem–loop binding protein (SLBP) that is essential for histone pre-mRNA processing as well as for both translation (Gallie et al. 1996; Sanchez and Marzluff 2002) and regulated stability (Harris et al. 1991). Recent studies provide new information for a mechanistic model for histone mRNA degradation (Kaygun and Marzluff 2005; Mullen and Marzluff 2008). Upon cessation of DNA synthesis, Upf1 is recruited to the SLBP, and this, in turn, recruits a terminal uridyltransferase (TUTase) that catalyzes 3′ oligouridylation. The oligo(U) tract forms a binding site for the Lsm1–7 heptamer, which recruits the machinery for decapping and bidirectional degradation of histone mRNA by exoribonucleases.
Several questions remain concerning this model. First, the fact that oligouridylated fragments of histone mRNA are observed (Mullen and Marzluff 2008) does not indicate whether 3′ oligouridylation is an initiating and obligatory step in histone mRNA degradation or a side reaction occurring for preexisting degradation intermediates. Second, the relative contributions of 5′→3′ and 3′→5′ degradation are not known, nor whether decapping is a rate-limiting step. Third, an oligo(U) tract promotes decapping of an RNA substrate in vitro (Song and Kiledjian 2007), but it is not known whether oligouridylation can also affect mRNA stability in cultured cells if decapping is prevented. Finally, translation of histone mRNA is required for its degradation (Graves et al. 1987; Kaygun and Marzluff 2005), but it is unclear whether it principally affects 5′→3′ or 3′→5′ pathways.
We have recently applied two technologies to study degradation of polyadenylated mRNAs that may also provide insight into SL-containing mRNAs. The first is the synthesis of mRNAs in vitro with different structural features and their introduction into mammalian cells by nucleoporation (Grudzien-Nogalska et al. 2007a). The mRNAs are incorporated into polysomes, efficiently translated within 15 min, and both their translation and degradation rates are sensitive to the nature of the cap and the poly(A) tract. The second is the development of cap analogs that cannot be cleaved by the catalytic subunit of the human Dcp1-Dcp2 decapping complex, hDcp2 (Grudzien-Nogalska et al. 2007b; Kowalska et al. 2008; Su et al. 2011). By blocking 5′→3′ degradation with a cleavage-resistant cap, one can observe 3′→5′ degradation in isolation. We applied these two technologies to achieve a better understanding of the mechanisms governing the rapid destruction of histone mRNAs when DNA synthesis ceases.
RESULTS
Decay of a polyadenylated reporter mRNA is modulated by 5′ and 3′ structural elements
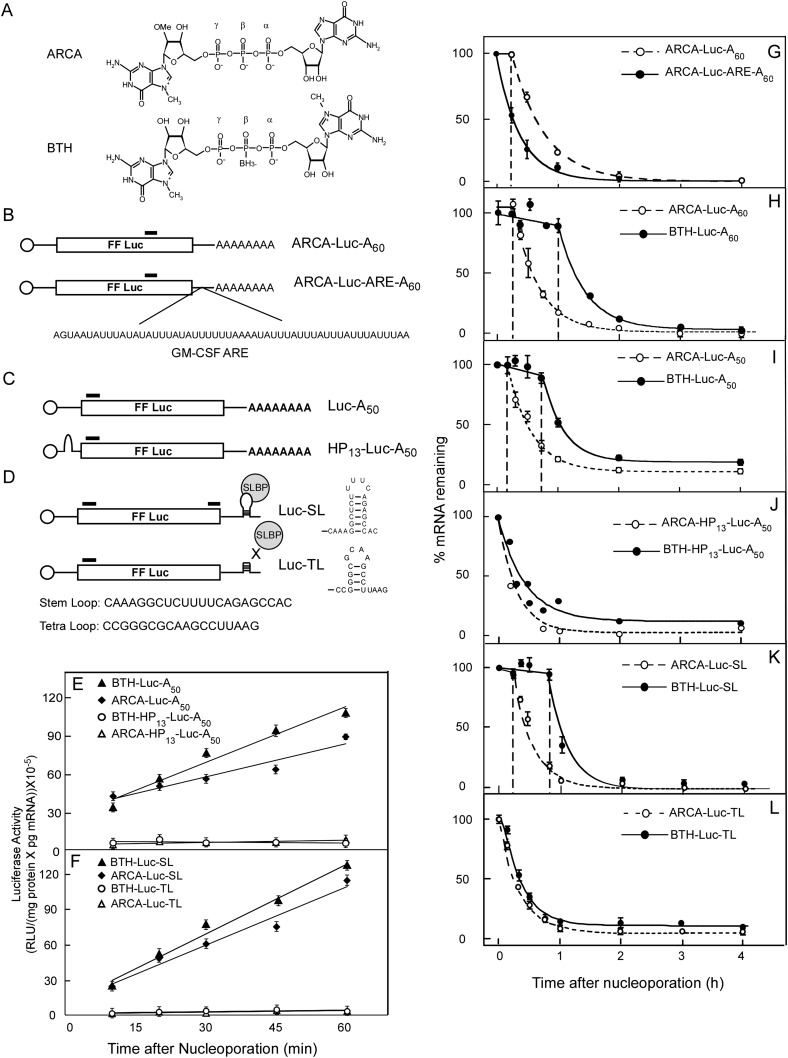
We synthesized mRNAs in vitro containing normal and cleavage-resistant caps, different 5′ UTRs, the coding region of firefly luciferase, and different 3′ UTRs. These were introduced into HeLa cells by nucleoporation, and the loss of sequences over time was followed by qRT-PCR. We have used this approach to introduce mRNAs into cultured mammalian cells previously and have validated the qRT-PCR method of quantifying mRNA by two alternative methods: Northern blotting and autoradiography of 32P-labeled mRNA (Grudzien et al. 2006; Grudzien-Nogalska et al. 2007a; Su et al. 2011). The ARCA (anti-reverse cap analog) (Fig. 1A) has the same characteristics as the natural cap with respect to translation and hydrolysis by hDcp2, but it has a modification at the C2′ position of the m7Guo moiety to prevent it from being incorporated into mRNA in the reverse orientation by T7 polymerase (Stepinski et al. 2001; Jemielity et al. 2003). mRNAs capped with the BTH analog (borano two-headed) are more efficiently translated than those capped with the ARCA and are completely resistant to hDcp2 hydrolysis in vitro (Su et al. 2011).
FIGURE 1.
Translation and decay of firefly luciferase (Luc) mRNA in HeLa cells. Luc mRNAs containing either a poly(A) tract (B,C) or the 3′ UTR of a human replicative histone (D) and either a normal cap (ARCA) or one that cannot be cleaved by hDcp2 (BTH) (A). (B) The AU-rich element (ARE) of GM-CSF mRNA was inserted in the 3′ UTR of ARCA-Luc-A60. Bars indicate a 90-nt sequence (nt 1082–1171) that is amplified during qRT-PCR (total length of mRNA, 1773 nt; initiation codon at nt 37; termination codon at nt 1627). (C) Luc mRNAs with (lower diagram) and without (upper diagram) a hairpin structure (HP13) in the 5′ UTR. Bars in the upper diagram indicate a 190-nt sequence (nt 90–279) that is amplified during qRT-PCR (total length of mRNA, 1763 nt; initiation codon at nt 37; termination codon at nt 1627). Bars in the lower diagram indicate a 190-nt sequence (nt 120–309) that is amplified during qRT-PCR (total length of mRNA, 1793 nt; initiation codon at nt 67; termination codon at nt 1657). (D) Luc mRNAs with a wild-type SL or mutated TL. Bars indicate a 190-nt sequence (nt 69–258) and 149-nt sequence (nt 1447–1595) that are amplified during qRT-PCR with 5′ and 3′ primer sets, respectively (total length of mRNA, 1653 nt; initiation codon at nt 16; termination codon at nt 1606). (E,F) Luc activity produced by introduction of the indicated mRNAs into HeLa cells by nucleoporation. Data are normalized for the amount of Luc mRNA present in the cells as determined by qRT-PCR and averaged from at least two separate experiments for each transcript. (G–L) Decay pattern of in vitro-synthesized Luc mRNAs in HeLa cells. Cells were lysed at the indicated times and 5′ Luc sequences measured by qRT-PCR, except in G, where 3′ sequences were followed. In G, H, I, and K, the decay is biphasic and consists of a lag phase and rapid-decay phase. Vertical dashed lines mark the boundary between lag and rapid-decay phases. The data for each transcript represent a single experiment, and error bars are for duplicate determinants at each time points. When open circles and open triangles coincide, the former is shown on top. Average values from multiple experiments are given in Tables 1 and 2.
We recognize that there may be stability features in endogenous mRNAs not present in exogenous reporter mRNAs, which are never as stable as long-lived endogenous mRNAs such as globin (Russell and Liebhaber 1996), ovalbumin (Palmiter and Carey 1974), and casein (Guyette et al. 1979). Furthermore, exogenous mRNAs do not go through classical nuclear processing events, although it has been demonstrated that nucleoporation delivers the majority of nucleic acids into the nucleus (Martinet et al. 2003; Kurosawa et al. 2012). These differences may well result in differences in the pathways utilized for degradation of endogenous versus exogenous mRNAs. We, therefore, tested several characteristics of exogenous mRNA decay in an attempt to validate its physiological relevance.
We inserted the AU-rich element (ARE) of granulocyte-macrophage colony-stimulating factor (GM-CSF) mRNA (Chen et al. 1995) into the 3′ UTR of Luc mRNA containing a 60-nt poly(A) tract (ARCA-Luc-A60) to generate ARCA-Luc-ARE-A60 (Fig. 1B). AREs are known to accelerate the degradation of endogenous c-Fos and GM-CSF mRNAs by causing poly(A) shortening (Shaw and Kamen 1986; Chen and Shyu 1995). mRNA loss was measured with a primer set that amplifies nt 1082–1171 (Fig. 1B, bars). The decay pattern for ARCA-Luc-A60 was biphasic, consisting of a lag phase of 9 ± 3 min, followed by a rapid-decay phase with t½ = 33.6 ± 0.5 min (Fig. 1G, open symbols; Table 1, line 1). The introduction of the ARE destabilized the mRNA (Fig. 1G, filled symbols); the lag phase disappeared, the t½ was reduced to 15.4 ± 2.0 min, and the relative translational efficiency was decreased to 0.7 ± 0.1 (Table 1, lines 1,2). This indicates that the mRNA degradation machinery triggered by an ARE is operational for the exogenous mRNA. The fact that ARCA-Luc-ARE-A60 is translated (Table 1) agrees with the known requirement for ARE-accelerated degradation (Curatola et al. 1995).
TABLE 1.
Translation and decay of polyadenylated mRNAs
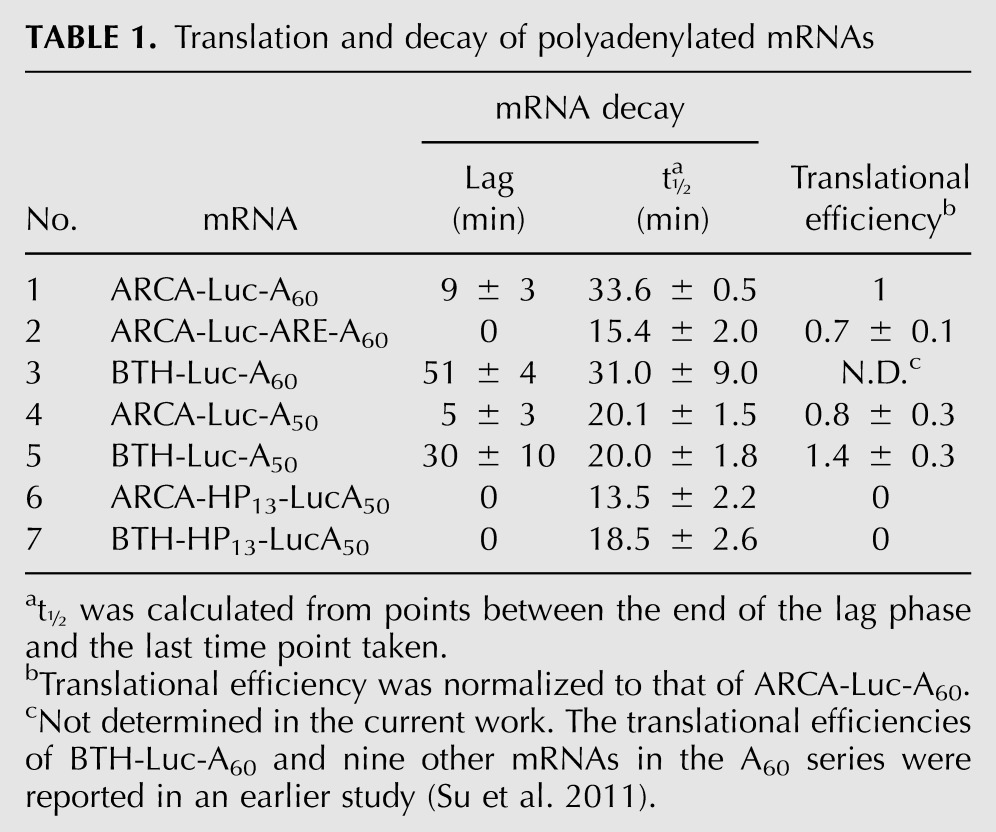
When we introduced ARCA-Luc-A60 and BTH-Luc-A60 into HeLa cells, both underwent biphasic decay (Fig. 1H), but the lag phase was greatly extended for BTH-Luc-A60 (51 ± 4 min) compared to ARCA-Luc-A60 (9 ± 3 min) (Table 1, lines 1,3). The t½ values for the rapid-decay phase, however, were statistically the same (31.0 ± 9.0 versus 33.6 ± 0.5 min). Since the BTH cap substantially stabilized the mRNA, we conclude that decapping and 5′→3′ hydrolysis are important determinants of the decay rate in our system. The eventual loss of BTH-Luc-A60 mRNA (after the lag) could be due to 3′→5′ hydrolysis by the exosome, loss of the cap by another decapping mechanism that is not blocked by the BTH modification (Xiang et al. 2009; Jiao et al. 2010; Lu et al. 2011), or endonuclease cleavage (Schoenberg 2011). These results indicate that decapping is an early and important step in the decay of polyadenylated mRNA in our system, as has been shown for endogenous mRNAs (Coller and Parker 2004; Chen and Shyu 2011).
A correlation between translation and stability of an mRNA has been observed in numerous systems (Parker and Sheth 2007). We tested the relationship between translation and stability in this system by inserting a 13-bp hairpin (HP13) into the 5′ UTR of Luc-A50 mRNA (Fig. 1C, lower diagram). This completely blocked translation of both ARCA-HP13-LucA50 and BTH-HP13-LucA50 (Fig. 1E, open symbols). The t½ of rapid-phase degradation was shorter for ARCA-HP13-Luc-A50 (13.5 ± 2.2 min) than for ARCA-Luc-A50 (20.1 ± 1.5 min) (cf. Fig. 1I and Fig. 1J, open symbols; Table 1, lines 4,6). More strikingly, the lag phases for both ARCA-Luc-A50 and BTH-Luc-A50 were abolished by introduction of HP13 (Fig. 1J; Table 1, lines 4–7). Despite the absence of a lag, BTH-HP13-Luc-A50 was degraded more slowly than ARCA-HP13-Luc-A50 (Fig. 1J, cf. closed and open symbols; Table 1, lines 6,7).
This experiment suggests that the lag phase represents the time that the mRNA is being translated and, hence, protected from degradation. There is little effect of BTH on the degradation of mRNAs that cannot be translated, suggesting that decapping is no longer rate-limiting for such mRNAs and that other processes which are normally retarded by translating ribosomes (exosome, endonucleases) become predominant. Overall, the dependence of the degradation rate on translation is another indicator that our reporter mRNAs mimic endogenous mRNAs.
The 3′ SL is required for translation and stabilization of a reporter mRNA
We utilized this experimental system but with a reporter mRNA containing the 3′ UTR of a human histone mRNA (Luc-SL). We followed the loss of both 5′ and 3′ sequences with different primer sets (Fig. 1D, bars) after introducing mRNAs into synchronous S-phase HeLa cells. Both ARCA-Luc-SL and BTH-Luc-SL decayed with biphasic kinetics (Fig. 1K), but the lags for both 5′ and 3′ sequences of BTH-Luc-SL (38.4 ± 7.0 and 30.0 ± 0.5 min, respectively) were considerably longer than those of ARCA-Luc-SL (7.8 ± 4.1 and 7.5 ± 4.8 min, respectively) (Table 2, lines 1,11). The loss of sequences during the rapid-decay phase was also somewhat slower for BTH-Luc-SL (t½ = 30.2 ± 2.4 and 21.5 ± 1.5 min for 5′ and 3′ sequences, respectively) than for ARCA-Luc-SL (t½ = 20.0 ± 3.1 and 18.5 ± 1.0 min, respectively). Interestingly, the t½ for loss of 5′ sequences of BTH-Luc-SL was longer than for 5′ sequences of ARCA-Luc-SL (Table 2, lines 1,11), but it was the same for 5′ sequences of ARCA-Luc-A50 and BTH-Luc-A50 (Table 1, lines 4,5). This may result from different modes of decay for SL-containing versus polyadenylated mRNAs.
TABLE 2.
Translation and decay of Luc-SL mRNAs with modified 3′ termini as a function of cell treatment
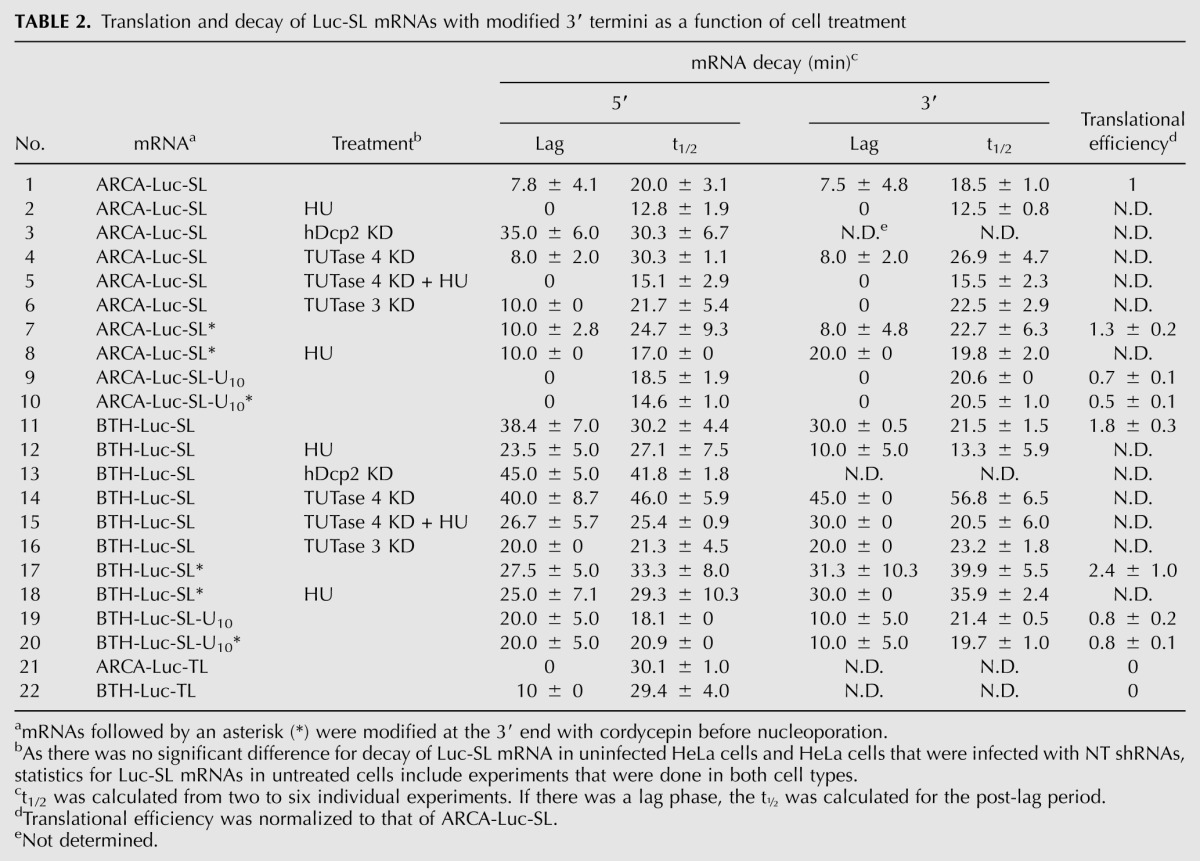
Replacing the SL with a different tetra-loop (TL) structure prevents binding of histone mRNA to the SLBP and inhibits translation (Gallie et al. 1996). A reporter mRNA containing the TL (Fig. 1D, lower diagram) was similarly inactive in translation (Fig. 1F, open symbols). The lag phase was reduced from 7.8 ± 4.1 min for ARCA-Luc-SL to 0 min for ARCA-Luc-TL (cf. Fig. 1K and Fig. 1L, open symbols; Table 2, lines 1,21), whereas the lag phase was reduced from 38.4 ± 7.0 min for BTH-Luc-SL to 10 ± 0 min for BTH-Luc-TL (cf. Fig. 1K and Fig. 1L, filled symbols; Table 2, lines 11,22). Thus, as in the case of the HP13-Luc-A50 mRNAs, decapping is a strong determinant in the degradation of ARCA-Luc-SL (since failure to decap delays degradation) but only for actively translated mRNAs. In the absence of translation, other processes that are not retarded by translation become dominant.
The SL is necessary and sufficient for mRNA destabilization after inhibition of DNA synthesis by hydroxyurea
Replication stress can be induced by treatment with the ribonucleotide reductase inhibitor hydroxyurea (HU), which halts S-phase progression (Ye et al. 2003). Endogenous histone mRNAs are stable during S-phase (Harris et al. 1991), but HU treatment results in their rapid decay with a t1/2 of ∼20 min (Sittman et al. 1983; Mullen and Marzluff 2008). We asked whether degradation of an SL-containing mRNA is also accelerated by HU. In a preliminary experiment, we demonstrated that synchronous HeLa cells continued to progress through the cell cycle after nucleoporation, albeit at a slower rate (Supplemental Fig. S1), and that DNA synthesis dropped ∼50%, after which it slowly recovered (Supplemental Fig. S2).
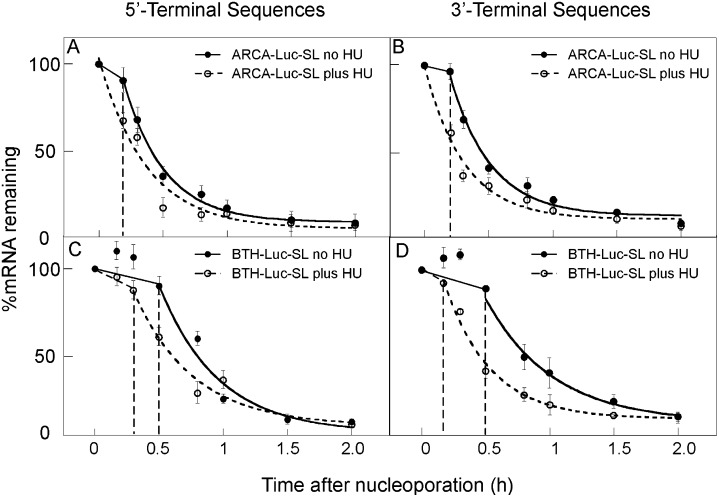
We introduced ARCA-Luc-SL and BTH-Luc-SL into mid-S-phase HeLa cells (Fig. 2). HU added immediately after nucleoporation accelerated the decay of both types of mRNA (Fig. 2, open symbols), but the effect of HU was greater on the loss of 3′ sequences (Fig. 2B,D) than on 5′ sequences (Fig. 2A,C). Furthermore, the lag phase for BTH-Luc-SL was shortened by HU (Table 2, lines 11,12), and this was more pronounced for 3′ sequences (30.0 ± 0.5 versus 10.0 ± 5.0 min) than for 5′ sequences (38.4 ± 7.0 versus 23.5 ± 5.0 min). For ARCA-Luc-SL, the lag phase was completely eliminated by HU, both for 5′ sequences (Fig. 2A) and 3′ sequences (Fig. 2B; Table 2, lines 1,2). The predominant effect of HU on loss of 3′ sequences is also suggested by an analysis of t½ values for the rapid-degradation phase. For ARCA-Luc-SL, where decapping can occur and degradation is bidirectional (Mullen and Marzluff 2008), HU shortens the t½ of both 5′ (12.8 ± 1.9 versus 20.0 ± 3.1 min) and 3′ sequences (12.5 ± 0.8 versus 18.5 ± 1.0 min) (Table 2, lines 1,2). However, for BTH-Luc-SL, where decapping is prevented and degradation is unidirectional, HU has little effect on the t½ of 5′ sequences (27.1 ± 7.5 versus 30.2 ± 2.4) but still shortens the t½ of 3′ sequences (13.3 ± 5.9 versus 21.5 ± 1.5 min) (Table 2, lines 11,12).
FIGURE 2.
Decay of Luc-SL mRNA is accelerated by HU treatment. ARCA-Luc-SL (A,B) or BTH-Luc-SL (C,D) were introduced into synchronous S-phase HeLa cells by nucleoporation, and the loss of 5′ (A,C) or 3′ (B,D) sequences was followed by qRT-PCR. Some cells were treated with HU immediately after nucleoporation (open symbols).
These results show that a reporter mRNA containing only the 3′ UTR of histone mRNA is destabilized by HU similar to endogenous histone mRNA and that HU acts mainly on the loss of 3′ sequences. The findings are also consistent with the previous report that knockdown of exosome components stabilizes histone mRNA after HU treatment more than knockdown of components involved in the 5′→3′ pathway (Mullen and Marzluff 2008).
Degradation of Luc-SL mRNA involves decapping by hDcp2
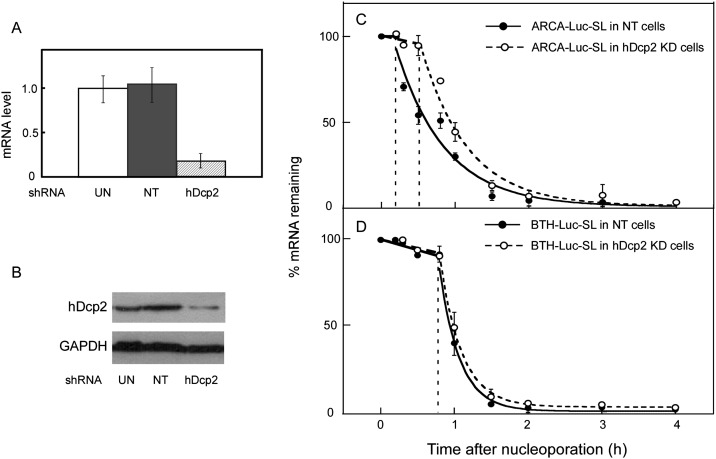
We interpret the observation that BTH-Luc-SL was stable for ∼38 min (Table 2, line 11) whereas ARCA-Luc-SL was stable for ∼8 min (Table 2, line 1) to mean that the lag phase is related to the time required to decap the mRNA. When decapping can occur (with ARCA-Luc-SL), it triggers 5′→3′ exonucleolytic hydrolysis (the rapid-phase decay). When decapping cannot occur (with BTH-Luc-SL), the rapid-decay phase is delayed, but the mRNA is eventually degraded by other processes. However, just because BTH is resistant to hDcp2 (Su et al. 2011), it does not mean that hDcp2 is responsible for decapping of ARCA-Luc-SL, since there are other mammalian decapping enzymes (Xiang et al. 2009; Jiao et al. 2010; Lu et al. 2011). We, therefore, knocked down hDcp2 in order to determine whether it was involved in the decay of ARCA-Luc-SL.
Infection of HeLa cells with a lentivirus expressing a shRNA directed against the coding sequence of hDcp2 mRNA reduced hDcp2 mRNA levels to ∼20% of untreated cells (Fig. 3A) and hDcp2 protein levels to ∼10% (Fig. 3B), whereas a control nontarget (NT) shRNA had no effect. ARCA-Luc-SL was more stable in hDcp2 knockdown cells compared to NT cells (Fig. 3C). The lag phase was elongated (35.0 ± 6.0 versus 7.8 ± 4.1 min) (Table 2, lines 1,3) as was the t½ of the rapid-decay phase (30.3 ± 6.7 versus 20.0 ± 3.1 min). In contrast, the lag phases in knockdown and NT cells for BTH-Luc-SL were statistically the same (Fig. 3D), whereas the t½ of the rapid-decay phase was longer in knockdown than NT cells (41.8 ± 1.8 versus 30.2 ± 2.4) (Table 2, lines 11,13). These results show that hDcp2 is involved in decapping of ARCA-Luc-SL but has no effect on BTH-Luc-SL. They also add further support to the idea that the lag phase reflects the time required for initiation of decapping, since the lag can be elongated by either the presence of a BTH cap or by depletion of hDcp2.
FIGURE 3.
Decay of Luc-SL mRNA involves decapping by hDcp2. HeLa cells were infected with lentiviruses expressing shRNA against hDcp2 or a nontarget shRNA (NT). (A) hDcp2 mRNA levels in uninfected (UN), NT, and hDcp2 KD cells. (B) Western blot of hDcp2 protein levels in UN, NT, and hDcp2 KD cells. ARCA-Luc-SL (C) and BTH-Luc-SL (D) were introduced into NT (filled symbols) or hDcp2 KD (open symbols) cells and the decay of 5′ sequences followed over time. Error bars represent the results of duplicate experiments. When error bars are not visible, they are smaller than the symbols.
Luc-SL mRNA is oligouridylated during degradation
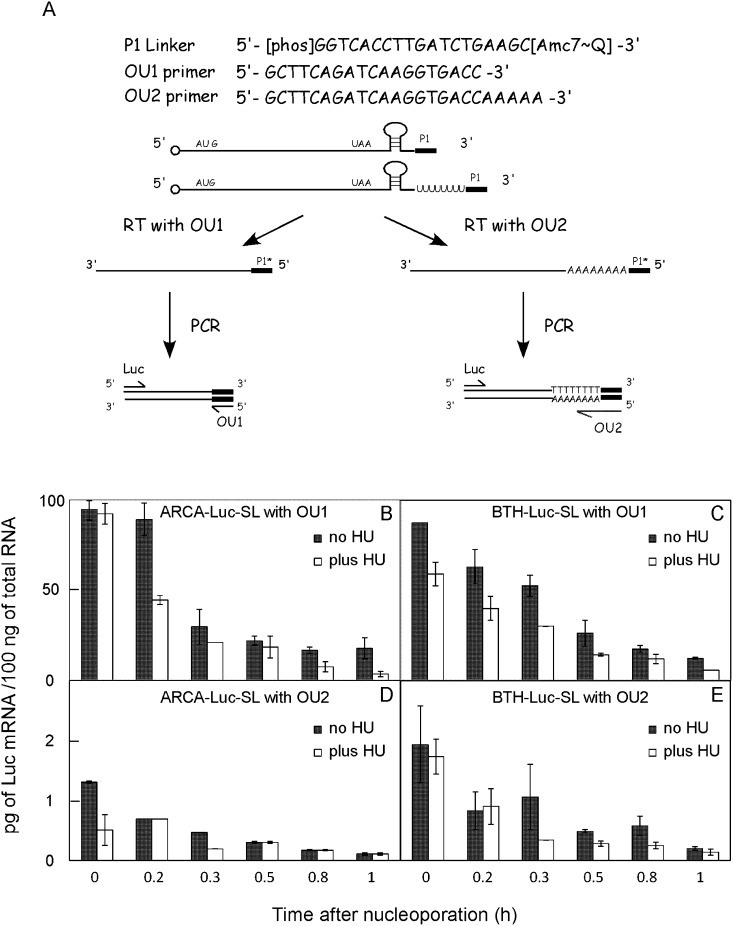
Fragments of histone mRNA with 3′ oligo(U) tracts downstream from the SL were found after treatment of cells with HU (Mullen and Marzluff 2008), but because this study examined steady-state populations, it was not possible to distinguish between oligouridylation being a rate-determining step in histone mRNA degradation versus an HU-induced side reaction, nor was it known whether oligouridylation occurred prior to decapping. We, therefore, asked whether Luc-SL was oligouridylated in our experimental system and whether this was affected by HU or decapping. We modified a previously published protocol for RNA ligation-coupled RT-PCR (LC-RT-PCR) (Charlesworth et al. 2004). Total RNAs extracted from cells after nucleoporation were ligated to a DNA linker termed P1 using T4 RNA ligase (Fig. 4A). Two alternative primers were used for reverse transcription. The OU1 primer is complementary to P1, so all RNAs ligated to P1 are copied into cDNA. The OU2 primer contains five A residues 3′ to the sequence complementary to P1, so only mRNAs that become ligated to P1 and end in a 3′ oligo(U) tract are copied. Our use of five A residues avoided excessive false positives resulting from the A tract being too short and a paucity of intermediates resulting from the A tract being too long, since Lsm1–7 binds to between five and 10 U residues (SM Lyons and WF Marzluff, unpubl.). Quantitative PCR (qPCR) was then performed using an upstream primer in the luciferase coding region (Luc in Fig. 4A) and either OU1 or OU2 as the downstream primer.
FIGURE 4.
Detection of oligouridylation of Luc-SL mRNA by ligation-coupled RT-PCR (LC-RT-PCR). (A) The LC-RT-PCR technique (see Materials and Methods). ARCA-Luc-SL (B,D) or BTH-Luc-SL (C,E) were introduced into synchronous S-phase HeLa cells by nucleoporation. RNAs were extracted, ligated to the P1 linker, amplified with either the OU1 (B,C) or OU2 (D,E) primer, and quantified by qPCR. Some cultures were treated with HU immediately after nucleoporation (open bars). Error bars represent duplicate experiments.
Sequences in ARCA-Luc-SL and BTH-Luc-SL amplified with the OU1 primer decreased with time (Fig. 4B,C, filled bars), and the decay of both mRNAs was accelerated by HU (Fig. 4B,C, open bars). Sequences from both mRNAs were also amplified with the OU2 primer, indicating some molecules are oligouridylated, but they were much less abundant, only 1%–2% of the OU1 products (Fig. 4D,E; note difference in y-axes). Unlike the OU1-amplified products, there was no significant effect of HU on the amount of OU2-amplified products of ARCA-Luc-SL (Fig. 4D) and only a marginal effect on those of BTH-Luc-SL (Fig. 4E).
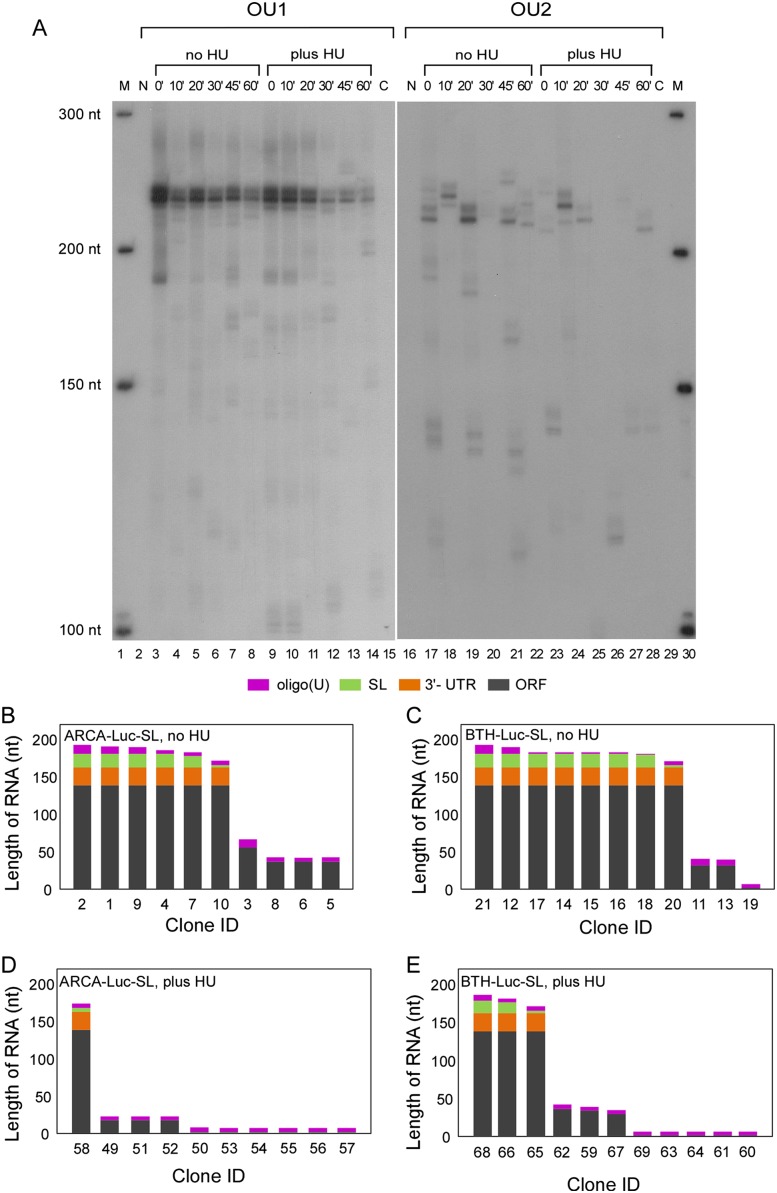
To examine the spectrum of products obtained by amplification of BTH-Luc-SL with OU1 and OU2 primers, we included [α-32P]dATP in the PCR reaction and analyzed the amplification products by polyacrylamide gel electrophoresis (PAGE) (Fig. 5A). The major product amplified with the OU1 primer (Fig. 5A, lanes 3–14) consisted of a cluster at 224, 229, and 233 nt, which is near the size expected from full-length mRNA. The reason for the heterogeneity is not known; it could represent addition of nontemplated nucleotide residues during synthesis of RNA by T7 RNA polymerase (Milligan et al. 1987) or loss of nucleotides downstream from the SL (Ross et al. 1986). Similar results were obtained with ARCA-Luc-SL (Supplemental Fig. S3).
FIGURE 5.
Analysis of oligouridylated RNAs. (A) BTH-Luc-SL was introduced into synchronous S-phase HeLa cells by nucleoporation and amplified by LC-RT-PCR with either the OU1 (lanes 3–14) or OU2 (lanes 17–28) primer except that [α-32P]dATP was included during the PCR reaction. Cells were treated without (lanes 3–8, 17–22) or with HU (lanes 9–14, 23–28). DNA markers are indicated as M (lanes 1,30). LC-RT-PCR products from RNA isolated from HeLa cells into which no Luc-SL mRNAs had been introduced are indicated as N (lanes 2,16). PCR reactions that did not receive any cDNA (nontemplate control) are indicated as C (lanes 15,29). The image represents a single gel, but the left half was exposed to film for 16 h, whereas the right half was exposed for 32 h. (B–E) LC-RT-PCR products from RNAs isolated at the 30-min time point in A and amplified with the OU2 primer were cloned and sequenced. The actual sequences are shown in Supplemental Figure S4, and summaries are shown for ARCA-Luc-SL (B,D) and BTH-Luc-SL (C,E). All sequences contained an oligo(U) tract (red) and various combinations of the SL (green), 3′ UTR upstream of the SL (orange), and ORF (dark gray).
In contrast to the LC-RT-PCR products obtained with the OU1 primer, those obtained with the OU2 primer were much less abundant and more heterogeneous (Fig. 5A, lanes 17–28; note difference in time of exposure in the figure legend). Some of the bands migrated faster than the major OU1 products, indicating that they were shortened at the 3′ end (since all products result from amplification with the common Luc upstream primer). However, some bands migrated slower than the major OU1 products, suggesting that they were derived from the addition of relatively long oligo(U) tracts. Similar OU2-amplified products were observed with and without HU treatment (Fig. 5A, cf. lanes 17–22 and lanes 23–28), which is consistent with the qRT-PCR results (Fig. 4D,E) and indicates that oligouridylation occurs in both HU-treated and untreated cells.
We cloned a selection of LC-RT-PCR products to determine their sequences. The complete sequences of the 3′ ends of the RNAs are shown in Supplemental Figure S4 and a schematic of the sequences, in Figure 5B–E. They represent a mixture of full-length RNAs and those lacking combinations of the SL (green), the 3′ UTR (orange), and the coding region up to the location of the upstream Luc primer (dark gray). (Shorter RNAs would not have been amplified because they lacked the sequence complementary to the upstream primer.) U-tracks were observed in all products (red). For ARCA-Luc-SL, roughly half of the clones sequenced containing the intact SL (Fig. 5B), in agreement with the nearly full-length major band in Supplemental Figure S3, lanes 3–8. Treatment with HU drastically reduced the number of SL-containing sequences (Fig. 5D). For BTH-Luc-SL, there were more long products containing the SL than for ARCA-Luc-SL (cf. Fig. 5C and Fig. 5B), in agreement with the quantitative data (cf. Fig. 4E and Fig. 4D). However, fewer long sequences were detected after HU treatment (Fig. 5E). There were relatively few OU2 products intermediate in size between the nearly full-length SL-containing RNAs and those that terminated near the upstream Luc primer (Figs. 5B–E).
Degradation of Luc-SL mRNA is retarded by knockdown of TUTase 4, but not TUTase 3, in the absence of HU
TUTase 1 and 3 were implicated in the oligouridylation of histone mRNA in one study (Mullen and Marzluff 2008), but TUTase 4 was implicated in another (Schmidt et al. 2011). (See Supplemental Table S1 for alternative TUTase names.) We, therefore, tested the effect of knocking down TUTases 3 and 4 in our system. After selection of stable cell lines with puromycin, we obtained 65% knockdown of TUTase 3 mRNA (Supplemental Fig. S5A) and 70% knockdown of TUTase 4 mRNA (Fig. 6A), whereas NT shRNA had no effect.
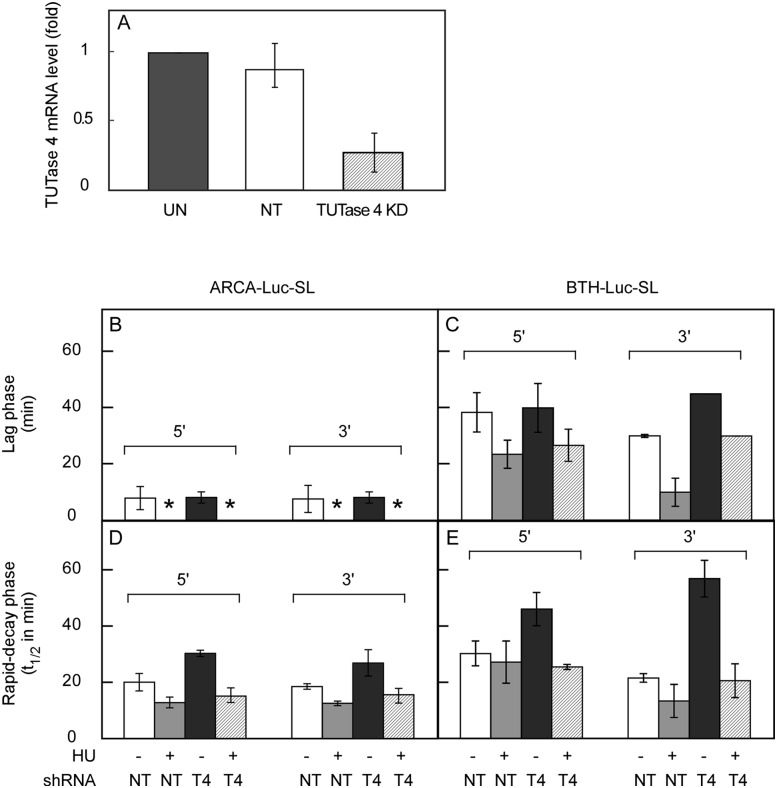
FIGURE 6.
Knocking down TUTase 4 stabilizes Luc-SL mRNA. HeLa cells were infected with lentiviruses expressing an shRNA against TUTase 4 or a NT shRNA. (A) TUTase 4 mRNA level in uninfected (UN), NT, and TUTase 4 KD cells. The length of the lag phase (B,C) and t½ (D,E) for decay of 5′ and 3′ sequence of ARCA-Luc-SL (B,D) and BTH-Luc-SL (C,E) was measured by qRT-PCR in NT and TUTase 4 KD (T4) cells for cells treated (+) or not treated (−) with HU. Asterisks indicate no lag was detected.
Knockdown of TUTase 3 did not retard the loss of sequences at either the 5′ or 3′ end of either ARCA-Luc-SL or BTH-Luc-SL (Supplemental Fig. S5B–E; Table 2, lines 1,6,11,16). In contrast, TUTase 4 knockdown significantly retarded the loss of sequences for both mRNAs in cells not treated with HU. For ARCA-Luc-SL mRNA, the lag phases for 5′ and 3′ sequences were not altered (8.0 ± 2.0 versus 7.8 ± 4.1 min and 8.0 ± 2.0 versus 7.5 ± 4.8 min, respectively) (Fig. 6B; Table 2, lines 1,4), but the t½ of the rapid-decay phase was lengthened (30.3 ± 1.1 versus 20.0 ± 3.1 min and 26.9 ± 4.7 versus 18.5 ± 1.0 min, respectively). For BTH-Luc-SL mRNA, the lag phase for 5′ sequences was not altered (40.0 ± 8.7 versus 38.4 ± 7.0 min) (Fig. 6C; Table 2, lines 11,14), but the t½ for the rapid-decay phase was lengthened from 30.2 ± 4.4 min to 46.0 ± 5.9 min (Fig. 6E; Table 2, lines 11,14). For the 3′ sequences of BTH-Luc-SL, TUTase 4 knockdown increased the lag phase from 30.0 ± 0.5 to 45.0 ± 0 min (Fig. 6C) and the t½ of the rapid-decay phase from 21.5 ± 1.5 to 56.8 ± 6.5 min, an increase of almost threefold (Fig. 6E; Table 2, lines 11,14).
Interestingly, the gains in mRNA stability resulting from TUTase 4 knockdown were prevented by addition of HU—the lag phase for both 5′ and 3′ sequences of ARCA-Luc-SL was abolished (Fig. 6B, asterisks) and t½ was reduced (Fig. 6D; Table 2, lines 4,5). Similarly, for BTH-Luc-SL, the lag phase for the loss of 5′ sequences was shortened by HU in both control cells (23.5 ± 5.0 versus 38.4 ± 7.0 min) and TUTase 4-knockdown cells (26.7 ± 5.7 versus 40.0 ± 8.7 min) (Fig. 6C), and the lengthening of the t½ (46.0 ± 5.9 versus 30.2 ± 4.4 min) was reversed by HU treatment (25.4 ± 0.9 min) (Fig. 6E; Table 2, lines 11,14,15). The dominant effect of HU over TUTase 4 knockdown is perhaps best seen in the loss of 3′ sequences of BTH-Luc-SL mRNA, where TUTase 4 knockdown lengthens the rapid-phase t½ from 21.5 ± 1.5 to 56.8 ± 6.5 min, but the t½ is still decreased by HU (20.5 ± 6.0 min) (Fig. 6E; Table 2, lines 11,14,15).
These results suggest that TUTase 4 plays an important role in Luc-SL mRNA turnover in S-phase cells. The fact that TUTase 4 knockdown stabilizes both ARCA-Luc-SL and BTH-Luc-SL indicates that the TUTase 4-mediated acceleration of 3′→5′ degradation does not require decapping. The observation that HU treatment abrogates the effect of TUTase 4 knockdown suggests that accelerated degradation of SL-containing mRNAs upon cessation of DNA synthesis proceeds through a pathway that does not involve TUTase 4.
TUTase 4 knockdown causes accumulation of stalled degradation products
To better understand the role of TUTase 4, we compared the spectrum of LC-RT-PCR products after nucleoporation of ARCA-Luc-SL or BTH-Luc-SL into cells expressing either NT or TUTase 4 shRNAs. For total products amplified with the OU1 primer (Fig. 7A, lanes 3–18), the major band in both sets of cells corresponded to an RNA of 231 nt, near the size expected from full-length RNA (Fig. 7A, open arrowhead). Oligouridylated products amplified with the OU2 primer were still detected, despite the TUTase 4 knockdown (Fig. 7A, lanes 27–34), the major bands consisting of a doublet at 223 and 219 nt (filled arrow). Both bands were more prominent and persisted longer in TUTase 4 knockdown cells than in NT cells.
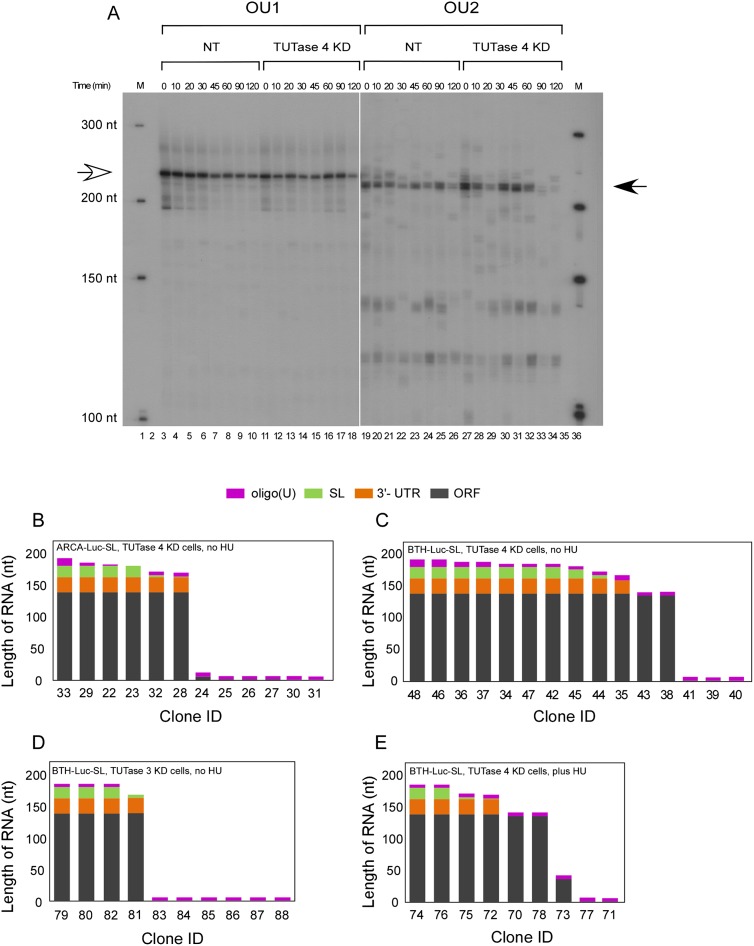
FIGURE 7.
Analysis of LC-RT-PCR products of Luc-SL mRNAs in TUTase knockdown (KD) cells. (A) LC-RT-PCR products derived from the degradation of BTH-Luc-SL were analyzed as in Figure 5 except that TUTase 4 KD cells (lanes 11–18, 27–34) or NT cells (lanes 3–10, 19–26) were used. The open arrow shows the position of the major OU1 product, while the filled arrow shows the position of the major OU2 product. DNA markers are indicated as M (lanes 1,36). The image represents a single gel, but the left half was exposed to film for 7 h, whereas the right half was exposed for 70 h. (B–E) Summary of LC-RT-PCR sequences as in Figure 5.
Cloning and sequencing of the LC-RT-PCR products revealed that more RNAs contained the SL for BTH-Luc-SL than ARCA-Luc-SL in TUTase 4 knockdown cells (cf. Fig. 7C and Fig. 7B). Also, more contained the SL for BTH-Luc-SL in TUTase 4 knockdown cells (Fig. 7C) than in untreated cells (Fig. 5C), in agreement with the stronger signals observed on the sequencing gel (Fig. 7A, cf. lanes 27–32 and lanes 19–26). This suggests that stalling of 3′→5′ degradation occurs more often at the SL/SLBP complex when levels of TUTase 4 are reduced. Products that contain full coding sequence but no 3′ UTR were observed only in TUTase 4 knockdown cells (Fig. 7; clones #38, 43, 70, and 78) but not in uninfected cells (Fig. 5), consistent with a model wherein 3′→5′ degradation stalls more often at the termination codon when levels of TUTase 4 are reduced. We also observed oligouridylated products in TUTase 3 knockdown cells (Fig. 7D) that were similar to those in untreated cells (Fig. 5C).
Blocking the 3′ end stabilizes histone mRNA
It is possible that 3′ oligouridylation is one of a series of sequential and obligatory steps in the degradation pathway, but it is also possible that histone mRNA fragments are produced by a pathway that does not involve oligouridylation and are only subsequently oligouridylated. In the first case, preventing oligouridylation should retard the rate of Luc-SL degradation, whereas, in the second, it should have no effect. To distinguish between these possibilities, we synthesized SL-containing mRNAs with a chain terminating nucleoside at the 3′ end, namely, 3′-deoxyadenosine (cordycepin), to block addition of the 3′ oligo(U) tract.
ARCA-Luc-SL was synthesized in vitro as in previous experiments, but the 3′ end was then blocked by adding cordycepin with yeast poly(A) polymerase (PAP) (ARCA-Luc-SL*). Performing a second PAP reaction on an aliquot in the presence of [α-32P]ATP indicated that >95% of the mRNAs were blocked (Supplemental Fig. S6A). When introduced into HeLa cells by nucleoporation (Fig. 8A), ARCA-Luc-SL* (filled symbols) decayed more slowly than ARCA-Luc-SL (open symbols). The stabilization was more apparent at later time points and for 3′ sequences (Fig. 8A, lower panel) than for 5′ sequences (upper panel), suggesting that oligouridylation affects the 3′→5′ pathway more than the 5′→3′ pathway. BTH-Luc-SL* was also more stable than BTH-Luc-SL (Fig. 8B), and again, this was more pronounced at later time points and affected 3′ more than 5′ sequences. For instance, the t½ for the rapid-decay phase was lengthened from 21.5 ± 1.5 to 39.9 ± 5.5 min for 3′ sequences but only from 30.2 ± 4.4 to 33.3 ± 8.0 for 5′ sequences (Table 2, lines 11,17). The fact that cordycepin modification increased the stability of BTH-Luc-SL is further evidence that oligouridylation primarily affects the 3′→5′ pathway; if it primarily affected the 5′→3′ pathway, the cordycepin modification would not have changed the decay kinetics, since BTH-Luc-SL cannot be decapped.
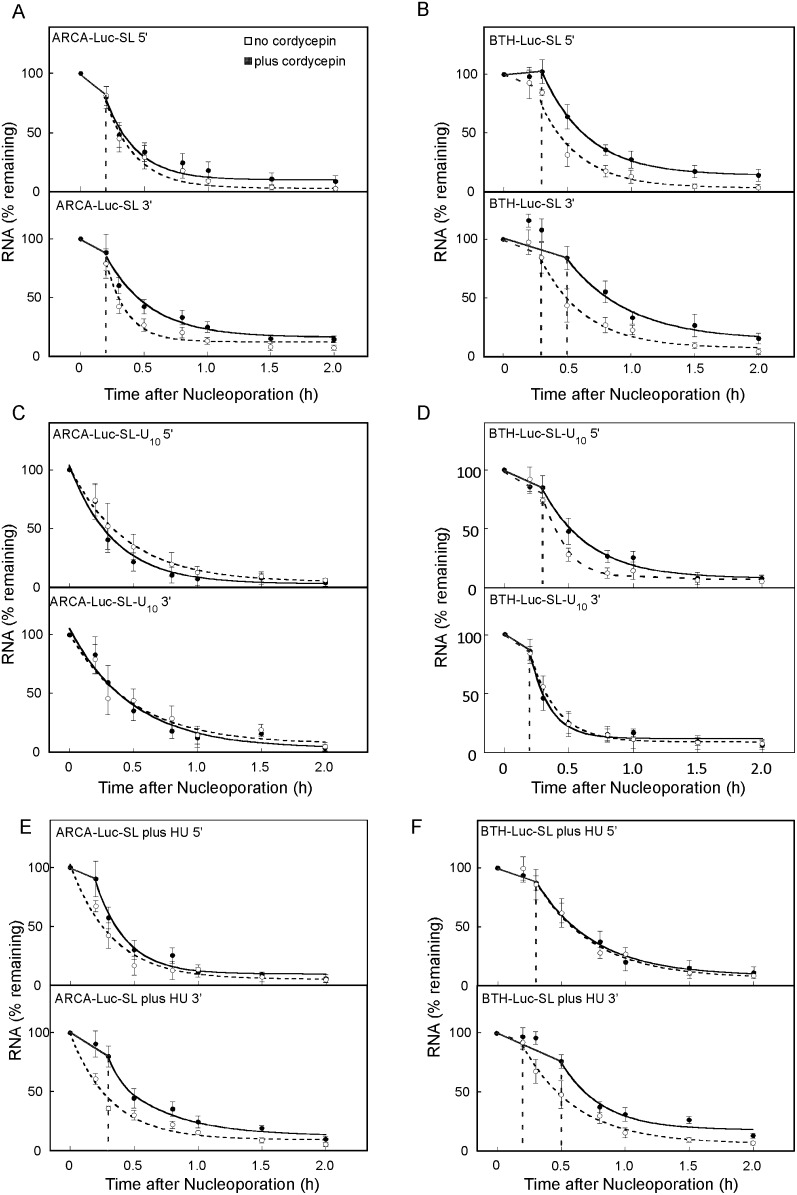
FIGURE 8.
Blocking the 3′ end of ARCA-Luc-SL and BTH-Luc-SL with the chain-terminator cordycepin slows their degradation. (A) Loss of 5′ and 3′ sequences for ARCA-Luc-SL with (filled symbols) or without (open symbols) cordycepin modification. (B) Same as A except that BTH-Luc-SL was introduced. (C) Same as A except the ARCA-Luc-SL was synthesized in vitro to contain a 3′ (U)10 tract. (D) Same as C except BTH-Luc-SL-U10 was introduced. Addition of HU immediately after nucleoporation destabilized ARCA-Luc-SL (E) and BTH-Luc-SL (F) (open symbols) but did not destabilize these RNAs if they were first modified with cordycepin (filled symbols).
We interpret the increased stability that is achieved by cordycepin modification as resulting from prevention of oligouridylation, but it is possible that cordycepin modification interferes with some other step that does not involve oligouridylation, for instance, degradation by a 3′→5′ exonuclease that requires a free 3′ OH. To rule this out, we synthesized a preuridylated reporter mRNA by inserting 10 T residues in the DNA template after the sequence for Luc-SL, resulting in an mRNA that contains 10 U residues located 3′ from the SL. Both ARCA-Luc-SL-U10 and BTH-Luc-SL-U10 were synthesized, and each was modified with cordycepin to produce ARCA-Luc-SL-U10* and BTH-Luc-SL-U10*, respectively. If 3′ oligouridylation increases the rate of mRNA degradation, the preuridylated mRNAs will not be affected when additional oligouridylation is prevented by cordycepin modification. ARCA-Luc-SL-U10 had no lag phase and similar t1/2 for rapid decay as ARCA-Luc-SL (cf. Fig. 8C and Fig. 8A, open symbols; Table 2, lines 1,9), but cordycepin did not diminish the degradation rate of ARCA-Luc-SL-U10 (Fig. 8C, cf. filled and open symbols). For BTH-Luc-SL-U10, a shorter lag phase was observed compared to BTH-Luc-SL (cf. Fig. 8D and Fig. 8B, open symbols). Since 5′→3′ degradation is blocked for this mRNA, we interpret this to mean that preuridylation accelerates the rate of 3′→5′ degradation, something we cannot observe with ARCA-Luc-SL-U10 because 5′→3′ degradation is so fast. Importantly, however, cordycepin incorporation did not affect the rate at which either 5′ or 3′ sequence was lost from BTH-Luc-SL-U10 mRNAs (Fig. 8D; Table 2, lines 19,20). These results indicate that oligouridylation per se accelerates the rate of decay and not some other process that is blocked by cordycepin modification.
Because HU does not appear to destabilize SL-containing mRNAs through a pathway involving TUTase 4 (Fig. 6), we expected that cordycepin-modified mRNA would still be destabilized by HU. Surprisingly, we found just the opposite. For BTH-Luc-SL*, HU treatment had no effect on the rate of loss of either 5′ or 3′ sequences or either the lag phase or the rapid-decay phase (cf. Fig. 8F and Fig. 8B, filled symbols; Table 2, lines 17,18). This was also true for ARCA-Luc-SL* (cf. Fig. 8E and Fig. 8A, filled symbols). In fact, the lag phase for decay of 3′ sequences was actually lengthened by HU (20.0 ± 0 versus 8.0 ± 4.8 min) (Table 2, lines 7,8). These results indicate that destabilization of Luc-SL mRNAs by HU treatment occurs through a pathway that involves oligouridylation, since inhibition of oligouridylation by cordycepin prevents the action of HU. The implication is that there are enzymes in addition to TUTase 4 that oligouridylate Luc-SL mRNAs.
To ensure that 3′ addition of a cordycepin to mRNA did not impair its ability to program protein synthesis, we compared the translational efficiency in HeLa cells of ARCA-Luc-SL to ARCA-Luc-SL* as well as BTH-Luc-SL to BTH-Luc-SL* and found cordycepin modification had no effect (Supplemental Fig. S6B; Table 2, lines 1,7,11,17). Interestingly, we observed that translational efficiencies for ARCA-Luc-SL-U10 and BTH-Luc-SL-U10 were approximately half those of their unmodified counterparts (Supplemental Fig. S6C; Table 2, lines 1,9,11,19). However, incorporation of cordycepin did not alter their translational efficiencies (Supplemental Fig. S6C; Table 2, lines 9,10,19,20). The diminished translation of preuridylated mRNAs may be due to interference in the SL-SLBP interaction by proteins that bind to oligo(U), such as Lsm1–7, which are also known to recruit inhibitors of translation (Coller and Parker 2005).
DISCUSSION
Previous studies have suggested that histone mRNA degradation is initiated by oligouridylation at the 3′ end followed by decapping and 5′→3′ exonuclease degradation and, at least on some molecules, by 3′→5′ degradation by the exosome (Mullen and Marzluff 2008). Early reports suggested that histone mRNA degradation occurs by a 3′→5′ pathway after inhibition of DNA synthesis (Ross et al. 1986, 1987). In mammalian cell extracts, degradation of polyadenylated mRNAs occurs primarily 3′→5′ (Mukherjee et al. 2002), although the contribution of the two pathways in vivo is not well understood. For degradation of histone mRNA, it is not known whether oligouridylation is essential for degradation or to what extent decapping plays a role in histone mRNA degradation. It is also not known whether degradation initiated by oligouridylation is important only for the rapid degradation that occurs when DNA replication is inhibited and at the end of S-phase, or whether oligouridylation is also involved in degradation during S-phase when histone mRNA is relatively stable. Our studies here, by introducing into cells RNAs that have been chemically modified to either prevent decapping, prevent oligouridylation, or bypass the need for oligouridylation by preuridylation, provide new insights into the histone mRNA degradation process in intact cells. Both decapping and 3′→5′ degradation play important quantitative roles in histone SL-containing mRNA turnover, not only in S-phase but also when DNA synthesis is inhibited. When modification of the 3′ end of histone SL-containing mRNA is prevented by addition of cordycepin, degradation is slowed, indicating that modification of the 3′ end is a critical feature of histone SL-containing mRNA degradation. Introduction of a preuridylated histone SL-containing mRNA (with or without cordycepin blocking the 3′ end) destabilized the mRNA, indicating the importance of the oligouridylation reaction for both pathways. Finally, histone SL-containing mRNAs are oligouridylated in S-phase both before and after treatment of cells with HU.
In earlier studies, we introduced polyadenylated mRNAs into mammalian cells to evaluate the ability of different cap analogs to support translation (Grudzien et al. 2006; Grudzien-Nogalska et al. 2007b; Su et al. 2011). Here, we have similarly followed the fate of a reporter mRNA ending in a histone SL. The reporter mRNA has a cap and the SL that is essential for translation of histone mRNAs but differs from endogenous histone mRNAs in two significant ways: the ORF is Luc rather than histone, and the reporter mRNA does not have a “nuclear history” (Kitamura et al. 1998), which can affect the cytoplasmic function of an mRNA. However, several observations support the idea that the metabolism of SL-containing reporter mRNAs occurs by pathways shared with endogenous histone mRNAs. First, the SL confers efficient translation to Luc-SL mRNA (Fig. 1F), which indicates it binds SLBP (Williams et al. 1994; Gallie et al. 1996; Sanchez and Marzluff 2002). Second, Luc-SL mRNA is destabilized by HU (Fig. 2A–D), as are endogenous histone mRNAs (Pandey and Marzluff 1987). Third, degradation intermediates of Luc-SL mRNAs are found with 3′ oligo(U) tracts (Figs. 5, 7), as are those of endogenous histone mRNAs (Mullen and Marzluff 2008).
The use of an exogenous reporter mRNA affords several experimental advantages that cannot be realized with plasmids expressing histone mRNAs. One can study the kinetics of mRNA decay without ongoing synthesis as an interfering factor. One can modify mRNAs at the 5′ or 3′ termini with cap analogs or chain-terminating 3′ nucleoside residues. One can alter translational efficiency of the mRNA and determine how it affects decay. Finally, one can follow the degradation of a homogenous cohort of initial intact mRNA molecules rather than a mixture of intermediates.
Translation and degradation of the Luc-SL mRNA
These experiments provide insight into how the translational state of Luc-SL affects the rate and modes of degradation. Initial studies suggested that the first detectable step in human H4 histone mRNA decay occurs at the 3′ terminus and that degradation proceeds mainly in the 3′→5′ direction (Ross et al. 1986). More recently, histone mRNAs were shown to be degraded by both 5′→3′ and 3′→5′ pathways (Mullen and Marzluff 2008), and detection of histone mRNA fragments missing both 5′ and 3′ sequences indicated that degradation could occur bidirectionally on the same molecule (Mullen and Marzluff 2008). However, it was not clear which pathway is predominant. The requirement for deadenylation of polyadenylated mRNAs as a prerequisite for decapping provides a precedent for events at the 3′ end affecting 5′→3′ degradation (Muhlrad et al. 1994).
Histone and poly(A)-containing mRNAs introduced by nucleoporation are translated and are initially relatively stable, but then they enter a rapid phase of degradation (Fig. 1). We interpret the lag phase as the time prior to initiation of decapping, whereas the rapid-decay phase reflects cessation of translation as a result of initial events in degradation. We found that the lag preceding the initiation of rapid-phase degradation increases from ∼10 to ∼40 min when a normal cap (ARCA-Luc-SL) is replaced with an uncleavable one (BTH-Luc-SL), consistent with decapping being important for governing the overall rate of Luc-SL mRNA degradation. BTH-Luc-SL is translated longer than ARCA-Luc-SL but still eventually enters a rapid-decay pathway, likely the 3′→5′ pathway.
When translation is prevented by mutation of the SL to TL, there is rapid degradation, suggesting that when exogenous mRNAs are introduced into cells but not translated, they are rapidly degraded. The lag either disappears completely (in the case of ARCA-Luc-TL) or is shortened (in the case of BTH-Luc-TL). We interpret this to mean that the mRNA is protected from degradation when it is translated, possibly because circularization blocks access of the exosome, thereby preventing the activation of decapping (Coller and Parker 2005).
Oligouridylation of histone mRNA
Oligouridylation of histone mRNAs occurs during S-phase upon HU treatment (Mullen and Marzluff 2008), but the mechanism and role of oligouridylation in histone mRNA degradation is not well understood. Our data indicate that the SL alone is necessary and sufficient to trigger oligouridylation, independent of the nature of the ORF, and that oligouridylation accelerates degradation. Preventing oligouridylation by blocking the 3′ end of the mRNA with cordycepin stabilized Luc-SL. Coupling that modification with the BTH cap analog resulted in even greater stabilization of the mRNA under normal conditions and also prevented the acceleration of degradation resulting from HU treatment. This result is consistent with oligouridylation being an important step for rapid degradation after HU treatment. Oligouridylation is required for both 5′→3′ and 3′→5′ degradation. After HU treatment, there is a substantial decrease (30 to 10 min) in the lag phase for the loss of 3′ sequences in BTH-Luc-SL, suggesting that the 3′→5′ pathway becomes particularly important after HU treatment.
The TUTase involved in oligouridylation of histone mRNA has not been definitively identified. One group reported that knocking down TUTase1 or TUTase 3 slowed histone mRNA degradation (Mullen and Marzluff 2008), while another found this for TUTase 4 (Schmidt et al. 2011). The effect of TUTase 1 knockdown has since been shown to result from an effect on cell growth (PE Lackey, MK Slevin, and WF Marzluff, unpubl.). Here, we found that knockdown of TUTase 4 slowed the degradation of Luc-SL mRNA during normal S-phase progression, suggesting that it plays a role in this pathway. However, knockdown of TUTase 4 did not affect the increased degradation of the Luc-SL mRNA that results from HU treatment, suggesting that cell-cycle regulated degradation may occur by a different pathway, possibly involving a different TUTase.
We detected oligouridylated intermediates from Luc-SL mRNAs in both control and TUTase 4 knockdown cells. Cloning of LC-RT-PCR products indicated that the RNAs had oligo(U) tracts added at multiple sites, with the majority at or near the SL. Many of the RNAs had undergone partial degradation into the SL (likely as a result of 3′→5′ exonuclease activity) and had oligo(U) added at various places in the SL. We interpret these findings to indicate exonuclease stalling and oligouridylation “repriming” for the continued degradation of the mRNA, as previously suggested (Mullen and Marzluff 2008). The occurrence of reuridylation is plausible since an initial round of 3′→5′ degradation may be stalled by RNA secondary structure or proteins bound to the RNA, leading to dissociation of degradation machinery; another round of oligouridylation would be needed to recruit the machinery again and reinitiate the degradation process. Different TUTases may be needed for initial and reoligouridylation, and knocking them down may have different effects on the rate of decay. Finally, the fact that the pattern of oligouridylated products was the same for BTH-Luc-SL and ARCA-Luc-SL indicates that oligouridylation is not dependent on prior decapping.
Blocking the 3′ end stabilizes Luc-SL
It is well established that cordycepin terminates RNA synthesis when incorporated by RNA polymerase and polyadenylation when incorporated by poly(A) polymerase (Horowitz et al. 1976; Muller et al. 1977). It will also prevent oligouridylation, regardless of which TUTases are responsible, and here, we find that it slows Luc-SL degradation. This provides evidence that oligouridylation is an obligatory step in Luc-SL degradation. The fact that we observe oligouridylated products in cells that are not treated with HU indicates that cessation of DNA synthesis is not a requirement for oligouridylation. Yet, even when decapping is blocked by an uncleavable cap and 3′ oligouridylation is blocked by cordycepin, BTH-Luc-SL* is still ultimately degraded, albeit at a slower rate. This may be due to endonucleolytic cleavage or enzymes capable of removing the BTH cap. Consistent with the observation that blocking oligouridylation stabilizes Luc-SL, we find that providing a preuridylated mRNA accelerates Luc-SL degradation. Thus oligouridylation is likely the rate-limiting step for histone mRNA degradation.
Besides the insights into the mechanism of mRNA degradation provided by the cordycepin modification, there are potential practical applications. In vitro-synthesized mRNAs encoding tumor antigens can be used to elicit an immune response in dendritic cells, and vaccination with such mRNAs has already entered clinical testing for the treatment of cancer (Weide et al. 2008; Diken et al. 2011). The use of a cleavage-resistant cap increases mRNA stability and translational efficiency of exogenous mRNA in mice and enhances priming and expansion of naive antigen-specific T-cells (Kuhn et al. 2010, 2011). In Saccharomyces pombe, oligouridylation of poly(A)-containing mRNA by the noncanonical poly(A) polymerase Cid1 has been demonstrated (Rissland and Norbury 2008, 2009). In preliminary experiments, we have observed that polyadenylated reporter mRNAs are also stabilized by cordycepin modification when introduced into HeLa cells (W Su and RE Rhoads, unpubl.). We, therefore, speculate that such an mRNA would have improved stability and translational efficiency in mice, thus enhancing the immune response.
MATERIALS AND METHODS
Materials
All common reagents were of analytical grade. ARCA and BTH cap analogs were synthesized as previously described (Jemielity et al. 2003; Su et al. 2011).
In vitro synthesis of mRNA
pLuc-A60 was constructed as previously described (Grudzien et al. 2006). pT7-Luc-ARE-A60 was constructed by inserting an ARE (Fig. 1B; Chen et al. 1995) into the 3′ UTR of Luc-A60 at the BamHI site immediately upstream of the poly(A) tract. pT7-Luc-SL and pT7-Luc-TL were constructed and linearized as previously described (Gallie et al. 1996; Ling et al. 2002). pT7-Luc-A50 and pT7-HP13-Luc-A50 (Niepel et al. 1999) were generously provided by Daniel Gallie (University of California, Riverside). The linearized plasmids served as templates for in vitro synthesis of mRNAs as previously described (Su et al. 2011).
Lentiviral shRNA transduction
Plasmids for construction of lentiviruses containing various shRNAs were introduced into cells by either calcium-phosphate (Kingston et al. 2003) or Lipofectamine 2000 (Invitrogen). Plasmids encoding shRNA against hDcp2 were purchased from Sigma (MISSION TRC shRNA, Sigma-Aldrich). pLKO1 plasmids containing shRNA to TUTase 3 and 4 and lentiviral packaging plasmids were obtained from the University of North Carolina lenti-shRNA core. HeLa cells were infected with lentiviruses expressing shRNAs by using the shRNA targeting protocol (Sigma-Aldrich). Infected cells were selected with 2 μg/mL puromycin 24 h post-infection and kept in puromycin for at least 4 d prior to analysis. Infecting with lentiviruses expressing shRNA against TUTase 3 and 4 resulted in 10% and 30% survival of HeLa cells, respectively.
Cell culture and nucleoporation
HeLa cells were cultured as previously described (Su et al. 2011). Asynchronous cells were seeded 1 d prior to nucleoporation onto 150-mm dishes at a density such that cells would reach 70% confluency the next day. Cells for synchronization (1 × 106) were seeded onto 150-mm dishes and synchronized by double-thymidine block (Jackman and O'Connor 2001). Cells were released from double-thymidine block on the day of nucleoporation, detached from plates 3 h after release (middle of S-phase), and subjected to nucleoporation as described previously (Su et al. 2011).
Measurement of translational efficiency and mRNA decay in HeLa cells
For assay of translational efficiency in cultured cells, aliquots of 0.5 × 105 cells were shaken in 1.5-mL Eppendorf tubes at 37°C for various times after nucleoporation. Total protein was extracted and luciferase activity measured and normalized to the amount of luciferase mRNA delivered into the cell at 0-time, as described previously (Grudzien-Nogalska et al. 2007a). For measurement of mRNA stability over periods of <1 h, cells were shaken in Eppendorf tubes as described above. For periods >1 h, cells were plated onto 35-mm cell culture dishes and incubated at 37°C in 5% CO2. The extraction of total RNA was performed as described previously (Grudzien-Nogalska et al. 2007a). Luciferase mRNA was quantified by qRT-PCR by using primers designed with the Beacon Designer tool (Bio-Rad). Sequences from the 5′ end of Luc mRNA were amplified with 5′-GGATGGAACCGCTGGAGAG-3′ and 5′-GCATACGACGATTCTGTGATTTG-3′. Sequences from the 3′ end of Luc mRNA were amplified with 5′-ATCGTGGATTACGTCGCCAGTCAA-3′ and 5′-TTTCCGCCCTTCTTGGCCTTTATG-3′. Human 18S rRNA levels were measured by the same method and in the same RNA samples with primers 5′-CGAGCCGCCTGGATACC-3′ and 5′-CAGTTCCGAAAACCAACAAAATAGA-3′. Amplification and detection were performed with the iCycler IQ real time PCR detection system in 25-μL reaction mixtures containing 5 μL of the transcription reaction mixture (50 ng of cDNA), 12.5 μL of IQ SYBRgreen Supermix, and 0.3 mM primers (Bio-Rad). The incubation conditions consisted of 3 min at 95°C for polymerase activation and 40 cycles, each of 15 sec at 95°C and 1 min at 60°C. Luciferase mRNA levels were calculated using the absolute standard curve method as described in User Bulletin No. 2 for the ABI Prism 7700 Sequence Detection System. Luciferase mRNA was then normalized for the amount of 18S rRNA in each sample, an indicator of total cellular RNA purified from each cell extract. KaleidaGraph (version 3.06, Synergy Software) was used for nonlinear least-squares fitting of decay data.
LC-RT-PCR and PAGE
LC-RT-PCR was performed as described (Charlesworth et al. 2004) but with modifications. The reaction mixture (10 μL) contained 1 μg of total RNA, 50 μM P1 linker [5′-(Phos)GGTCACCTTGATCTGAAGC(AmC7-Q)-3′] (MWG-Operon), 1 μL of T4 RNA ligase (New England Biolabs), and 1× ligation buffer. RNA and the P1 linker were heated to 65°C for 10 min, cooled on ice for 2 min prior to addition of T4 RNA ligase, incubated at 37°C for 30 min, and treated at 65°C for 15 min to inactivate the enzyme. The reverse transcription reaction mixture (5 μL), containing 2 μL of ligated RNA, 1 μL of specific primer (10 mM), and 0.2 μL of 10 mM dNTPs, was heated at 65°C for 5 min and cooled on ice for 2 min, after which 0.5 μL of 0.1 M DTT, 2 μL of 5× first strand buffer (Invitrogen), and Superscript III reverse transcriptase (Invitrogen) were added in a final volume of 10 μL. Two specific primers, OU1 and OU2, were utilized for reverse transcription separately (Fig. 4A). For some reactions, PCR products were labeled in a 25-μL reaction containing 1 μL of cDNA, 2.5 μL of Platinum PCR reaction buffer, 0.75 μL of 50 mM MgCl2, 0.5 μL of 10 mM dNTPs, 0.5 μL of 10 mM upstream Luc primer (5′-ATCGTGGATTACGTCGCCAGTCAA-3′), 0.5 μL of either 10 mM OU1 or OU2 primer, 0.5 μL of [α-32P]dATP, and 0.1 μL Platinum Taq Polymerase (Invitrogen). The PCR reaction was performed by 35 cycles at 94°C for 30 sec, 55.1°C for 1 min, and 72°C for 1 min. 32P-labeled PCR products were analyzed by PAGE as described previously (Su et al. 2011), except that the gel contained 8% polyacrylamide and 8 M urea. DNA markers were prepared by labeling an ultra low range DNA ladder (10–300 bp) (Fisher Scientific) with T4 polynucleotide kinase and [γ-32P]ATP by the exchange protocol as described by the manufacturer.
Cloning and sequencing of LC-RT-PCR products
LC-RT-PCR products (1 μL) were inserted into the TOPO vector according to the manufacturer's protocol (Invitrogen). Colonies were screened by colony hybridization using radiolabeled probes (Sambrook et al. 1989). Plasmids with Luc-SL inserts were sequenced at Iowa State University.
Cordycepin incorporation
Incorporation of cordycepin was performed in a 100-μL reaction mixture that contained 0.2 μM ARCA-Luc-SL or BTH-Luc-SL, 1× PAP reaction buffer (Affymetrix), 100 μM cordycepin 5′-triphosphate (Sigma), 1 unit/μL of RNase Inhibitor (Applied Biosystems), and 2400 units of yeast PAP (Affymetrix). The reaction mixture was incubated at 37°C for 1 h. RNAs were precipitated by adding two volumes of Precipitation/Inactivation buffer (Ambion), kept at −20°C for at least 1 h, collected by centrifugation at 13,000g at 4°C for 20 min, and dissolved in 30 μL of DEPC-treated water. The RNA concentration was determined by UV absorbance at 260 nm, and RNA integrity was verified by electrophoresis on 1.2% agarose gels containing formaldehyde.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Ewa Grudzien-Nogalska and Ann-Bin Shyu for helpful discussions, Eric First for assistance with RNA sequencing gels, and Daniel Gallie for providing plasmids pT7-Luc-A50 and pT7-HP13-Luc-A50. This work was supported by grants R01GM20818 (R.E.R.), R01GM29832 (W.F.M), F32GM87059 (W.F.M), and a fellowship (M.K.S.) from the National Institutes of Health, grants N N204 089438 and N N301 096339 from the Polish Ministry of Science and Higher Education (E.D. and J.J.), and a scholarship from the Foundation for Polish Science International Ph.D. Projects Program and EU European Regional Development Fund (M.Z.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.034470.112.
REFERENCES
- Charlesworth A, Cox LL, MacNicol AM 2004. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J Biol Chem 279: 17650–17659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB 1995. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem Sci 20: 465–470 [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB 2011. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA 2: 167–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Xu N, Shyu AB 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: Different deadenylation kinetics and uncoupling from translation. Mol Cell Biol 15: 5777–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R 2004. Eukaryotic mRNA decapping. Annu Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatola AM, Nadal MS, Schneider RJ 1995. Rapid degradation of AU-rich element (ARE) mRNAs is activated by ribosome transit and blocked by secondary structure at any position 5′ to the ARE. Mol Cell Biol 15: 6331–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diken M, Kreiter S, Selmi A, Britten C, Huber C, Tureci Ö, Sahin U 2011. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther 18: 702–708 [DOI] [PubMed] [Google Scholar]
- Gallie DR, Lewis NJ, Marzluff WF 1996. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res 24: 1954–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves RA, Pandey NB, Chodchoy N, Marzluff WF 1987. Translation is required for regulation of histone mRNA degradation. Cell 48: 615–626 [DOI] [PubMed] [Google Scholar]
- Grudzien E, Kalek M, Jemielity J, Darzynkiewicz E, Rhoads RE 2006. Differential inhibition of mRNA degradation pathways by novel cap analogs. J Biol Chem 281: 1857–1867 [DOI] [PubMed] [Google Scholar]
- Grudzien-Nogalska E, Jemielity J, Kowalska J, Darzynkiewicz E, Rhoads RE 2007a. Phosphorothioate cap analogs stabilize mRNA and increase translational efficiency in mammalian cells. RNA 13: 1745–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudzien-Nogalska E, Stepinski J, Jemielity J, Zuberek J, Stolarski R, Rhoads RE, Darzynkiewicz E 2007b. Synthesis of anti-reverse cap analogs (ARCAs) and their applications in mRNA translation and stability. Methods Enzymol 431: 203–227 [DOI] [PubMed] [Google Scholar]
- Guyette WA, Matusik RJ, Rosen JM 1979. Prolactin-mediated transcriptional and post-transcriptional control of casein gene expression. Cell 17: 1013–1023 [DOI] [PubMed] [Google Scholar]
- Harris ME, Bohni R, Schneiderman MH, Ramamurthy L, Schumperli D, Marzluff WF 1991. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol Cell Biol 11: 2416–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K 2005. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol 21: 133–153 [DOI] [PubMed] [Google Scholar]
- Horowitz B, Goldfinger B, Marmur J 1976. Effect of cordycepin triphosphate on the nuclear DNA-dependent RNA polymerases and poly(A) polymerase from the yeast Saccharomyces cerevisiae. Arch Biochem Biophys 172: 143–148 [DOI] [PubMed] [Google Scholar]
- Jackman J, O'Connor PM 2001. Methods for synchronizing cells at specific stages of the cell cycle. In Current protocols in cell biology. J. Wiley, Hoboken, NJ. doi: 10.1002/0471143030.cb0803s00 [Google Scholar]
- Jemielity J, Fowler T, Zuberek J, Stepinski J, Lewdorowicz M, Niedzwiecka A, Stolarski R, Darzynkiewicz E, Rhoads RE 2003. Novel “anti-reverse” cap analogues with superior translational properties. RNA 9: 1108–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Xiang S, Oh C, Martin C, Tong L, Kiledjian M 2010. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF 2005. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 12: 794–800 [DOI] [PubMed] [Google Scholar]
- Kingston RE, Chen CA, Rose JK 2003. Calcium phosphate transfection. In Current protocols in molecular biology, pp 9.1.1–9.1.11. J. Wiley, Hoboken, NJ [Google Scholar]
- Kitamura T, Ogawa W, Sakaue H, Hino Y, Kuroda S, Takata M, Matsumoto M, Maeda T, Konishi H, Kikkawa U, et al. 1998. Requirement for activation of the serine-threonine kinase Akt (protein kinase B) in insulin stimulation of protein synthesis but not of glucose transport. Mol Cell Biol 18: 3708–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalska J, Lewdorowicz M, Zuberek J, Grudzien-Nogalska E, Bojarska E, Stepinski J, Rhoads RE, Darzynkiewicz E, Davis RE, Jemielity J 2008. Synthesis and characterization of mRNA cap analogs containing phosphorothioate substitutions that bind tightly to eIF4E and are resistant to the decapping pyrophosphatase DcpS. RNA 14: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn AN, Diken M, Kreiter S, Selmi A, Kowalska J, Jemielity J, Darzynkiewicz E, Huber C, Tureci O, Sahin U 2010. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther 17: 961–971 [DOI] [PubMed] [Google Scholar]
- Kuhn AN, Diken M, Kreiter S, Vallazza B, Tureci Ö, Sahin U 2011. Determinants of intracellular RNA pharmacokinetics: Implications for RNA-based immunotherapeutics. RNA Biol 8: 35–43 [DOI] [PubMed] [Google Scholar]
- Kurosawa A, Saito S, Mori M, Adachi N 2012. Nucleofection-based gene targeting in human pre-B cells. Gene 492: 305–308 [DOI] [PubMed] [Google Scholar]
- Ling J, Morley SJ, Pain VM, Marzluff WF, Gallie DR 2002. The Histone 3′-terminal stem-loop-binding protein enhances translation through a functional and physical interaction with eukaryotic initiation factor 4G (eIF4G) and eIF3. Mol Cell Biol 22: 7853–7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Zhang J, Li Y, Li Z, Zhang N, Xu X, Wang T, Guan Z, Gao GF, Yan J 2011. hNUDT16: A universal decapping enzyme for small nucleolar RNA and cytoplasmic mRNA. Protein Cell 2: 64–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet W, Schrijvers DM, Kockx MM 2003. Nucleofection as an efficient nonviral transfection method for human monocytic cells. Biotechnol Lett 25: 1025–1029 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ 2008. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J, Groebe D, Witherell G, Uhlenbeck O 1987. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15: 8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Decker C, Parker R 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev 8: 855–866 [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J 21: 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller WEG, Seibert G, Beyer R, Breter HJ, Maidhof A, Zahn RK 1977. Effect of cordycepin on nucleic acid metabolism in L5178Y cells and on nucleic acid-synthesizing enzyme systems. Cancer Res 37: 3824–3833 [PubMed] [Google Scholar]
- Niepel M, Ling J, Gallie D 1999. Secondary structure in the 5′-leader or 3′-untranslated region reduces protein yield but does not affect the functional interaction between the 5′-cap and the poly(A) tail. FEBS Lett 462: 79–84 [DOI] [PubMed] [Google Scholar]
- Palmiter RD, Carey NH 1974. Rapid inactivation of ovalbumin messenger ribonucleic acid after acute withdrawal of estrogen. Proc Natl Acad Sci 71: 2357–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey NB, Marzluff WF 1987. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol 7: 4557–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U 2007. P bodies and the control of mRNA translation and degradation. Mol Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ 2008. The Cid1 poly(U) polymerase. Biochim Biophys Acta 1779: 286–294 [DOI] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16: 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Peltz SW, Kobs G, Brewer G 1986. Histone mRNA degradation in vivo: The first detectable step occurs at or near the 3′ terminus. Mol Cell Biol 6: 4362–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Kobs G, Brewer G, Peltz SW 1987. Properties of the exonuclease activity that degrades H4 histone mRNA. J Biol Chem 262: 9374–9381 [PubMed] [Google Scholar]
- Russell JE, Liebhaber SA 1996. The stability of human β-globin mRNA is dependent on structural determinants positioned within its 3′ untranslated region. Blood 87: 5314–5323 [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sanchez R, Marzluff WF 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol Cell Biol 22: 7093–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M-J, West S, Norbury CJ 2011. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 17: 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR 2011. Mechanisms of endonuclease-mediated mRNA decay. Wiley Interdiscip Rev RNA 2: 582–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G, Kamen R 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46: 659–667 [DOI] [PubMed] [Google Scholar]
- Sittman DB, Graves RA, Marzluff WF 1983. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci 80: 1849–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M-G, Kiledjian M 2007. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA 13: 2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepinski J, Waddell C, Stolarski R, Darzynkiewicz E, Rhoads RE 2001. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogues 7-methyl(3′-O-methyl)GpppG and 7-methyl(3′-deoxy)GpppG. RNA 7: 1486–1495 [PMC free article] [PubMed] [Google Scholar]
- Su W, Slepenkov S, Grudzien-Nogalska E, Kowalska J, Kulis M, Zuberek J, Lukaszewicz M, Darzynkiewicz E, Jemielity J, Rhoads RE 2011. Translation, stability, and resistance to decapping of mRNAs containing caps substituted in the triphosphate chain with BH3, Se, and NH. RNA 17: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide B, Garbe C, Rammensee HG, Pascolo S 2008. Plasmid DNA- and messenger RNA-based anti-cancer vaccination. Immunol Lett 115: 33–42 [DOI] [PubMed] [Google Scholar]
- Whitfield ML, Zheng L-X, Baldwin A, Ohta T, Hurt MM, Marzluff WF 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol 20: 4188–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AS, Ingledue TC, Kay BK, Marzluff WF 1994. Changes in the stem-loop at the 3′ terminus of histone mRNA affects its nucleocytoplasmic transport and cytoplasmic regulation. Nucleic Acids Res 22: 4660–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L 2009. Structure and function of the 5′-3′ exoribonuclease Rat1 and its activating partner Rai1. Nature 458: 784–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD 2003. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell 11: 341–351 [DOI] [PubMed] [Google Scholar]