This paper investigates the regulation of alternative splicing of exon 6 of the Interleukin 7 receptor (IL7R) pre-mRNA, which gives rise to a membrane-bound (inclusion) or a soluble IL7R isoform (skipping). The authors identify several exonic and intronic cis-acting sequences and use a RNA chromatography–mass spec approach to identify trans-acting factors. This resulted in the identification of the cleavage and polyadenylation specificity factor (CPSF1), which binds a canonical poly(A) signal, AAUAAA, present immediately downstream from exon 6, and negatively regulates IL7R exon 6 inclusion. Thus, binding of a polyadenylation factor to an intronic region downstream from a regulated exon interferes with assembly of spliceosome at the 5′ splice site of the regulated exon.
Keywords: alternative splicing, CPSF1, IL7R, multiple sclerosis, polyadenylation
Abstract
Interleukin 7 receptor, IL7R, is expressed exclusively on cells of the lymphoid lineage, and its expression is crucial for the development and maintenance of T cells. Alternative splicing of IL7R exon 6 results in membrane-bound (exon 6 included) and soluble (exon 6 skipped) IL7R isoforms. Interestingly, the inclusion of exon 6 is affected by a single-nucleotide polymorphism associated with the risk of developing multiple sclerosis. Given the potential association of exon 6 inclusion with multiple sclerosis, we investigated the cis-acting elements and trans-acting factors that regulate exon 6 splicing. We identified multiple exonic and intronic cis-acting elements that impact inclusion of exon 6. Moreover, we utilized RNA affinity chromatography followed by mass spectrometry to identify trans-acting protein factors that bind exon 6 and regulate its splicing. These experiments identified cleavage and polyadenylation specificity factor 1 (CPSF1) among protein-binding candidates. A consensus polyadenylation signal AAUAAA is present in intron 6 of IL7R directly downstream from the 5′ splice site. Mutations to this site and CPSF1 knockdown both resulted in an increase in exon 6 inclusion. We found no evidence that this site is used to produce cleaved and polyadenylated mRNAs, suggesting that CPSF1 interaction with intronic IL7R pre-mRNA interferes with spliceosome binding to the exon 6 5′ splice site. Our results suggest that competing mRNA splicing and polyadenylation regulate exon 6 inclusion and consequently determine the ratios of soluble to membrane-bound IL7R. This may be relevant for both T cell ontogeny and function and development of multiple sclerosis.
INTRODUCTION
The interleukin 7 receptor (IL7R) α chain (IL7Rα), which, together with a common cytokine receptor γ-chain (γc), forms a transmembrane receptor for interleukin 7 (IL7), is expressed almost exclusively on cells of the lymphoid lineage and is crucial for development and maintenance of T cells (for review, see Mazzucchelli and Durum 2007). Binding of IL7 to its receptor triggers a signaling cascade which, among other functions, results in transcriptional induction of anti-apoptotic genes (e.g., Bcl-2) and redistribution of cell death proteins (e.g., Bax and Bad), thereby protecting T cells from apoptosis (Jiang et al. 2004, and references therein). Deficient expression of IL7R results in severe lymphopenia in both humans and mice (Peschon et al. 1994; Maki et al. 1996; Akashi et al. 1997; Puel et al. 1998), and variation in the IL7R gene predisposes to several autoimmune diseases (Gregory et al. 2007; Anderson et al. 2011; Mells et al. 2011), underscoring the crucial role for this receptor in T cell biology. A nonsynonymous single nucleotide polymorphism (SNP) (rs6897932; C→T, Thr244Ile) in IL7R has been previously reported to be a significant risk factor for multiple sclerosis (MS) (Gregory et al. 2007; The International Multiple Sclerosis Genetics Consortium 2007; Lundmark et al. 2007), a debilitating neurodegenerative disorder of the central nervous system (CNS) (Trapp and Nave 2008). SNP rs6897932 is located in exon 6 of IL7R and the disease-associated allele leads to decreased inclusion of exon 6 (Gregory et al. 2007). Given that exon 6 encodes the transmembrane domain of the receptor (Pleiman et al. 1991), its alternative splicing leads to a membrane-bound isoform (rIL7R) when exon 6 is included, and a soluble isoform (sIL7R) when exon 6 is skipped. sIL7R lacks both the transmembrane and cytoplasmic domains of the receptor (Goodwin et al. 1990; Korte et al. 2000; for review, see Evsyukova et al. 2010). Importantly, both isoforms are able to bind IL7 with high affinity (Goodwin et al. 1990; Rose et al. 2009). These findings suggest that reduced inclusion of exon 6 and the concomitant increase in sIL7R could be causally linked to immune dysfunction and MS. Interestingly, several reports also imply the importance of this isoform in infection with the human immunodeficiency virus (HIV) (Koesters et al. 2006; Rose et al. 2009; Crawley et al. 2010). Characterizing exon 6 splicing control would shed light not only on the regulation of IL7R expression and its role in T cell functioning, but may also be relevant to establishing a connection between IL7R splicing and MS.
To characterize splicing of the IL7R exon 6 we performed extensive mutagenesis to identify cis-acting regulatory elements, and used RNA affinity chromatography followed by quantitative mass spectrometry to identify trans-acting protein factors that bind exon 6 and surrounding intronic regions. In addition to several exonic elements, some of which abut on the previously studied SNP, we identified an intronic consensus polyadenylation signal AAUAAA in the direct vicinity of the 5′ splice site of exon 6. In mammalian cells, AAUAAA represents one of the core consensus sequences that is associated with mRNA 3′ end cleavage and polyadenylation, and is located 15–30 nucleotides (nt) upstream of the actual cleavage site (Lutz 2008; Lutz and Moreira 2011; Proudfoot 2011). This sequence is recognized by the multisubunit cleavage and polyadenylation specificity complex (CPSF), specifically by its largest subunit, CPSF1 (Keller et al. 1991; Murthy and Manley 1995). Mutations of the AAUAAA element have been shown to impair splicing of the terminal intron in vitro (Niwa and Berget 1991; Cooke et al. 1999), and subunits of CPSF have been shown to interact with components of the spliceosome (Lutz et al. 1996; Kyburz et al. 2006), suggesting that splicing and polyadenylation are interconnected and influence each other.
Here we show that mutation of the consensus polyadenylation sequence AAUAAA in intron 6 of IL7R enhanced the inclusion of exon 6. Consistent with these data, CPSF1 was identified in RNA affinity chromatography experiments with RNA templates that included the AAUAAA sequence, and RNAi-mediated depletion of CPSF1 resulted in an increase in exon 6 inclusion. Intronic polyadenylation (IPA) has been recently described as an important regulator of gene expression (Kaida et al. 2010; Vorlová et al. 2011; Berg et al. 2012) (for review, see Campigli Di Giammartino et al. 2011). However, we found no evidence that the consensus AAUAAA site in intron 6 of IL7R directs cleavage and polyadenylation within intron 6. We propose that assembly of polyadenylation complexes on this intronic polyadenylation site or CPSF1 binding per se interferes with spliceosome binding to the adjacent 5′ splice site, which would result in lower levels of the full-length IL7R mRNA and functional IL7R protein.
RESULTS
Multiple exonic cis-acting regulatory elements in IL7R exon 6 regulate its inclusion
We had previously shown that the SNP C→T (rs6897932) within IL7R exon 6 affects alternative splicing of this exon by increasing its skipping by approximately twofold in cell culture and by four- to fivefold when total RNA from MS patients is analyzed (Gregory et al. 2007; P Seth, SG Gregory, and MA Garcia-Blanco, unpubl.). Given that the SNP is significantly associated with MS, we were presented with the intriguing possibility that aberrant alternative splicing of IL7R exon 6 impacts IL7R function and contributes to the disease. We sought to characterize splicing regulation of the IL7R exon 6 in detail, since understanding of this splicing event may contribute to understanding of MS pathogenesis.
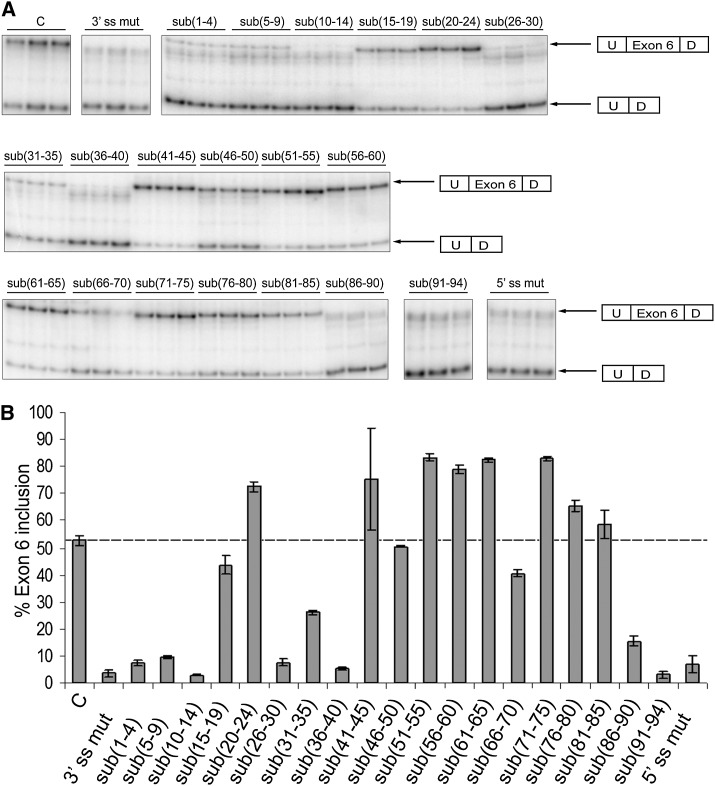
Several layers of regulation that govern splicing of pre-mRNAs exist in mammalian cells (for reviews, see Goldstrohm et al. 2001; Lynch 2004; Wang and Cooper 2007; Cooper et al. 2009). These are mediated by the combined action of 5′ and 3′ splice sites, branch point sequence, and additional regulatory sequences, which, based on their location and mode of action, are classified as either exonic/intronic splicing enhancers (ESEs or ISEs) or exonic/intronic splicing silencers (ESSs or ISSs) (Garcia-Blanco et al. 2004; Lynch 2004; Evsyukova et al. 2010). To dissect cis-acting exonic elements that are important for splicing of IL7R exon 6, we created substitution mutants spanning the whole length of exon 6 (sub(1–4) through sub(91–94)), in the context of the splicing reporter minigene pI-12 described previously (Carstens et al. 1998; Gregory et al. 2007). The reporter contains full-length exon 6 surrounded by 614 and 573 nt of IL7R introns 5 and 6, respectively, placed between two constitutively spliced exons U and D (Fig. 1). All of the substitutions were transversions and were introduced in the context of the MS-associated “C” allele at position 25 of the exon. The parental “C” minigene served as a positive control, while minigenes with disruptions of either the 3′ or 5′ splice sites (3′ ss mut and 5′ ss mut) served as negative controls. Control and mutant minigenes were transiently transfected into rat DT cells (Fig. 2) and human HeLa and Jurkat cells (data not shown), and exon 6 inclusion was ascertained by semiquantitative RT-PCR 48 h post-transfection. Consistent with our previous studies, 50%–55% of the transcripts containing the MS-associated “C” allele included exon 6 and, as expected, inclusion was minimal when either of the splice sites (3′ and 5′) was mutated (Fig. 2). Transfection with the substitution minigenes had multiple effects on exon 6 inclusion, with some substitutions (sub(5–9), sub(10–14), sub(26–30), sub(36–40)) resulting in a significant decrease in exon 6 inclusion (<10% inclusion), while others (sub(20–24), sub(41–45), sub(51–55) through sub(61–65), sub(71–75)) resulted in an increase to between 70% and 80% inclusion. While substitutions sub(1–4) and sub(91–94) also showed profound effects on inclusion (a decrease in inclusion to <10%), their effects likely disrupt integrity of the splice sites and were considered less informative. Additionally, several substitutions—sub(10–14), sub(15–19), sub(20–24), sub(26–30), sub(31–35), and sub(36–40)—were tested in the context of the “T” allele with effects on exon 6 inclusion being equivalent to those in the context of the “C” allele (data not shown). We thus performed all of the following studies with minigenes in the context of “C”. Importantly, three substitutions that impacted exon 6 inclusion were located in the vicinity of the MS-associated SNP. Two of these decreased inclusions revealing the presence of potential splicing enhancers, ESE1 and ESE2 (nt 5–14 and 26–40, respectively), and one increased inclusion, revealing the presence of a potential splicing silencer, ESS1 (nt 20–24). Given our interest in the MS-associated SNP, we first focused on these three exonic regulatory elements.
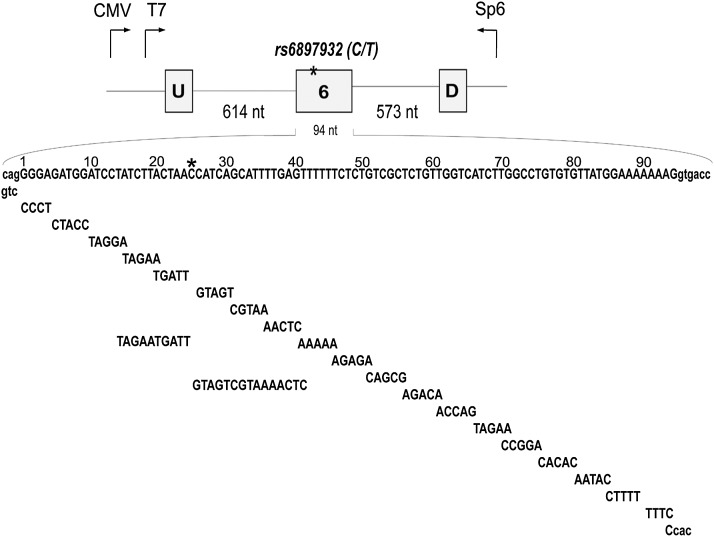
FIGURE 1.
IL7R minigene constructs. (Top) IL7R exon 6 (94 nt) and 614 and 573 nt of introns 5 and 6 were cloned into the pI-12 vector. The position of the MS-associated SNP rs6897932 is highlighted with an asterisk (*). U and D represent constitutively spliced upstream and downstream adenoviral exons, respectively. T7 and Sp6 represent sites for common PCR primers that were used to detect products that either included or skipped exon 6. (Bottom) Exon 6 was divided into short blocks that were either mutated or deleted. Larger sequence blocks that were mutated around the SNP are also shown. All substitutions/deletions were introduced in the context of the MS-associated “C” allele. Intronic sequences are shown in lowercase letters; exonic sequences are shown in uppercase letters. Sequences of the substitution mutations are shown below exon 6 sequence.
FIGURE 2.
Identification of the cis-acting regulatory elements within IL7R exon 6. Mutagenesis across exon 6 was performed to identify cis-acting regulatory elements that mediate exon 6 splicing. Control and mutant minigenes were transiently transfected into DT cells and total RNA was analyzed for exon 6 inclusion 48 h post-transfection. Percent exon 6 inclusion was determined from semiquantitative RT-PCRs as a ratio between the “included” and “included + skipped” products and plotted against each mutant. All transfections were performed in triplicate. Error bars represent standard deviation. (A) Gel images representing products of the semiquantitative RT-PCRs for different substitution mutants. (B) Quantification of the RT-PCR results shown in A.
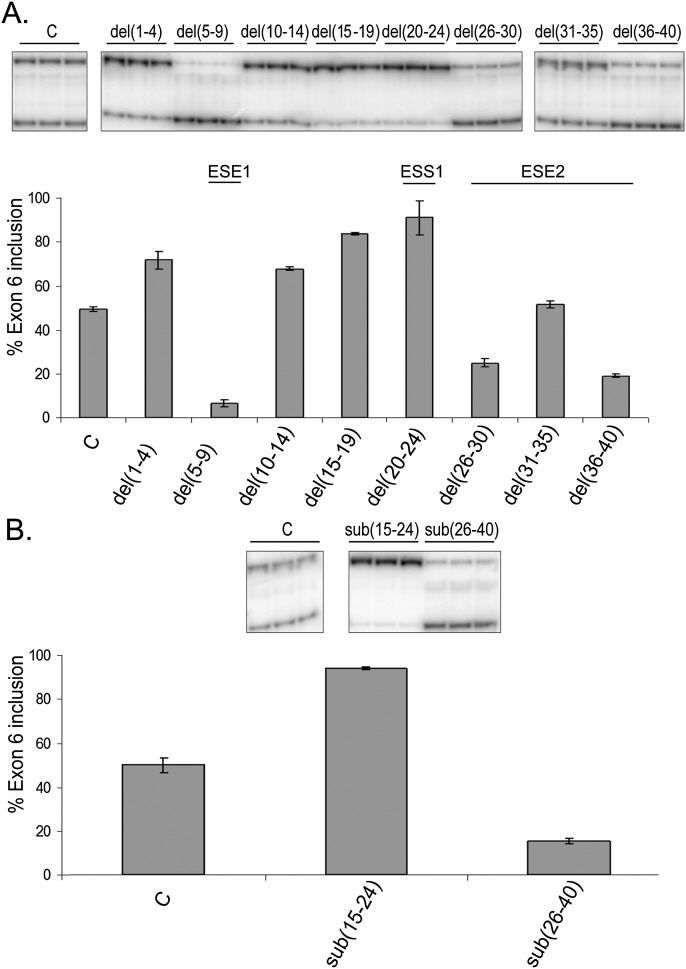
Since several of the substitutions had a significant effect on exon 6 inclusion, it was important to ensure that this effect was due to mutating existing regulatory elements rather than inadvertently introducing novel ones. To differentiate between these two possibilities, we introduced deletions of the same sequences that were substituted. Del(20–24) confirmed the presence of ESS1 (Fig. 3A). While del(10–14) failed to recapitulate the effects seen with sub(10–14), del(5–9) did show the same decreased inclusion seen with sub(5–9), supporting the existence of ESE1 (Fig. 3A). Compared with sub(1–4), deletion of the same nucleotides resulted in elevated inclusion of exon 6. This is most likely due to recreation of the 3′ splice site by the deletion (compare wild-type sequence, ccagGGGAGATGG, to the deletion sequence, ccagGATGG; intronic sequences are shown in lowercase). Deletion of nucleotides 31–35, within what we called ESE2, resulted in no change to exon 6 inclusion, but del(26–30) and del(36–40) recapitulated data from equivalent substitutions, which suggests that the regulatory sequence we previously labeled “ESE2” is composed of two different enhancer elements (Fig. 3A). Overall, deletion analyses confirmed the presence of three regulatory elements (ESS1 and ESE1 and ESE2) in the close vicinity of the MS-associated SNP.
FIGURE 3.
Deletion and substitution analysis confirms the presence of ESS1 and ESE2. Deletion and substitution mutants were transfected into DT cells, and exon 6 inclusion was analyzed by semiquantitative RT-PCR as described in Figure 2. (A) Inclusion of the IL7R exon 6 in the context of the deletion mutants. Only the area surrounding the MS-associated SNP (first 40 nt of exon 6) was analyzed. (B) Analysis of substitutions to the regions encompassing regulatory elements ESS1 (nt 15–24) and ESE2 (nt 26–40).
To further test for the presence of the two elements closest to the SNP, ESS1 and ESE2, we introduced larger substitutions. As expected, while substitution of the sequences corresponding to ESS1 (sub(15–24)) resulted in almost 100% exon 6 inclusion, substitution of the sequences corresponding to ESE2 (sub(26–40)) resulted in a profound decrease in inclusion (Fig. 3B). Together, deletion and substitution data suggest that at least two important cis-acting regulatory elements are present in the vicinity of the MS-associated SNP: ESS1, which is likely the silencer that we predicted was augmented by the MS-associated allele (Gregory et al. 2007) and the enhancer ESE2.
An intronic cis-acting element associated with 3′end formation and polyadenylation down-regulates IL7R exon 6 inclusion
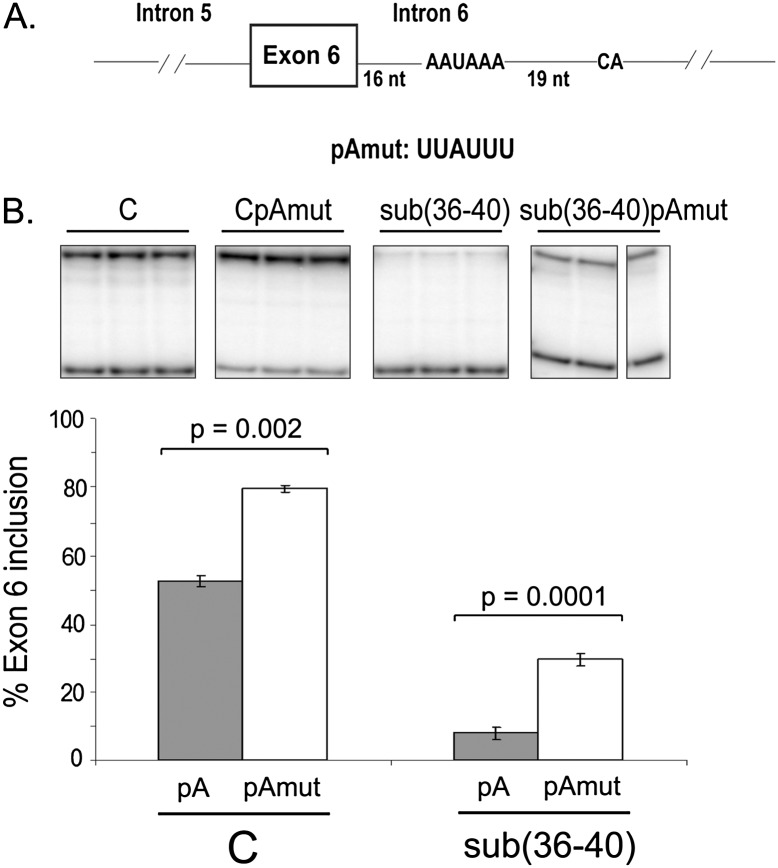
Examination of the sequences of exon 6 and introns 5 and 6 of IL7R revealed a consensus polyadenylation sequence, AAUAAA, 16 nt downstream from the 5′ splice site of exon 6 (Fig. 4A). In order to investigate whether or not intronic polyadenylation (IPA) contributed to the regulation of exon 6 splicing, we mutated the AAUAAA site (pAmut in Fig. 4A) within IL7R intron 6, and evaluated the effect of this mutation on exon 6 inclusion. Mutation of this consensus poly(A) signal resulted in a significant increase in exon 6 inclusion, from 50% to about 80% in the context of the “C” minigene and from <10%–30% in the context of the sub(36–40) minigene (Fig. 4B). Additionally, mutation of the consensus poly(A) signal in the context of the “T” allele at the position of the SNP resulted in an equivalent increase in exon 6 inclusion (up to 85%–90%) (data not shown).
FIGURE 4.
An intronic cis-acting element associated with 3′end cleavage and polyadenylation down-regulates IL7R exon 6 inclusion. (A) Schematics of the IL7R exon 6/intron 6 region showing the presence of the consensus polyadenylation signal AAUAAA in intron 6, 16 nt downstream from the exon 6 5′ splice site, followed by the cleavage signal (“CA”) 19 nt downstream from the poly(A) sequence. The substitution mutation used to assess effect of the intronic poly(A) signal on exon 6 inclusion is shown (pAmut). (B) Mutation to the intronic AAUAAA sequence results in increased inclusion of exon 6. Two minigenes, “C” and sub(36–40), were analyzed either with (right) or without (left) the poly(A) signal mutation in intron 6. Minigenes were transfected into HeLa cells, and exon 6 inclusion analyzed as in Figure 2.
To ensure that mutation of the intronic poly(A) site results in disruption of an existing regulatory element rather than introduction of a new one, we tested a deletion of the AAUAAA sequence as well as four additional substitutions in the context of the sub(36–40) minigene. While deleting this sequence resulted in a decrease in exon 6 inclusion (likely due to the introduction of a novel regulatory element), three of the new substitutions resulted in a significant increase in exon 6 inclusion (data not shown). We conclude that these data are most consistent with the intronic poly(A) site affecting splicing of exon 6 by suppressing its inclusion.
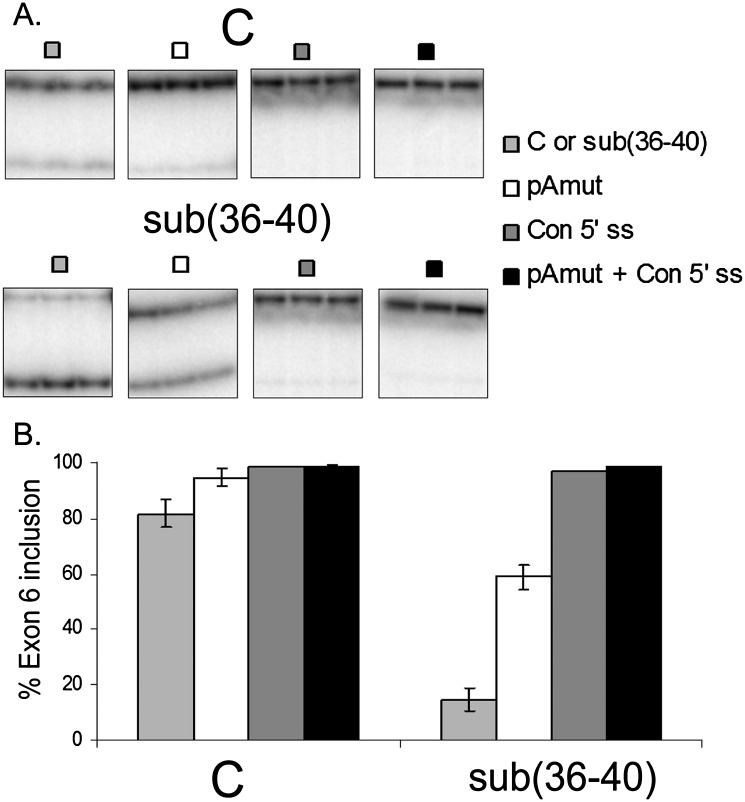
The 5′ splice site of IL7R exon 6 (AGgugacc, intronic sequences shown in lowercase) diverges from the 5′ splice consensus sequence (AGguPuagu, where Pu stands for purine) at the last two positions in the intron, and thus represents a “weak” 5′ splice site. Since the usage of intronic poly(A) signals has been shown to depend on the strength of the adjacent 5′ splice sites (Tian et al. 2007), we constructed minigenes in which the 5′ splice site was mutated to match the consensus sequence in “C” and sub(36–40) minigenes with or without intronic poly(A) site mutations (Fig. 5). The presence of the consensus 5′ splice site was sufficient to restore exon 6 inclusion to nearly 100%, independent of the minigene context (Fig. 5B), indicating that the inherent weakness of this 5′ splice site confers regulation by the adjacent intronic poly(A) signal.
FIGURE 5.
Effect of the consensus 5′ splice site on exon 6 inclusion. The exon 6 5′ splice site (AGgugacc, intronic sequences in lowercase) was mutated to match the consensus 5′ splice site sequence (AGgugagu) in the context of minigenes “C”, sub(36–40), CpAmut and sub(36–40)pAmut. The presence of the consensus 5′ splice site resulted in nearly 100% inclusion of exon 6 in all minigene contexts. (A) Representative gel images of the RT-PCR products. (B) Quantification of the RT-PCR results shown in A.
RNA affinity chromatography identifies cleavage and polyadenylation specificity factor subunit 1 (CPSF1) as a trans-acting protein factor that regulates exon 6 inclusion
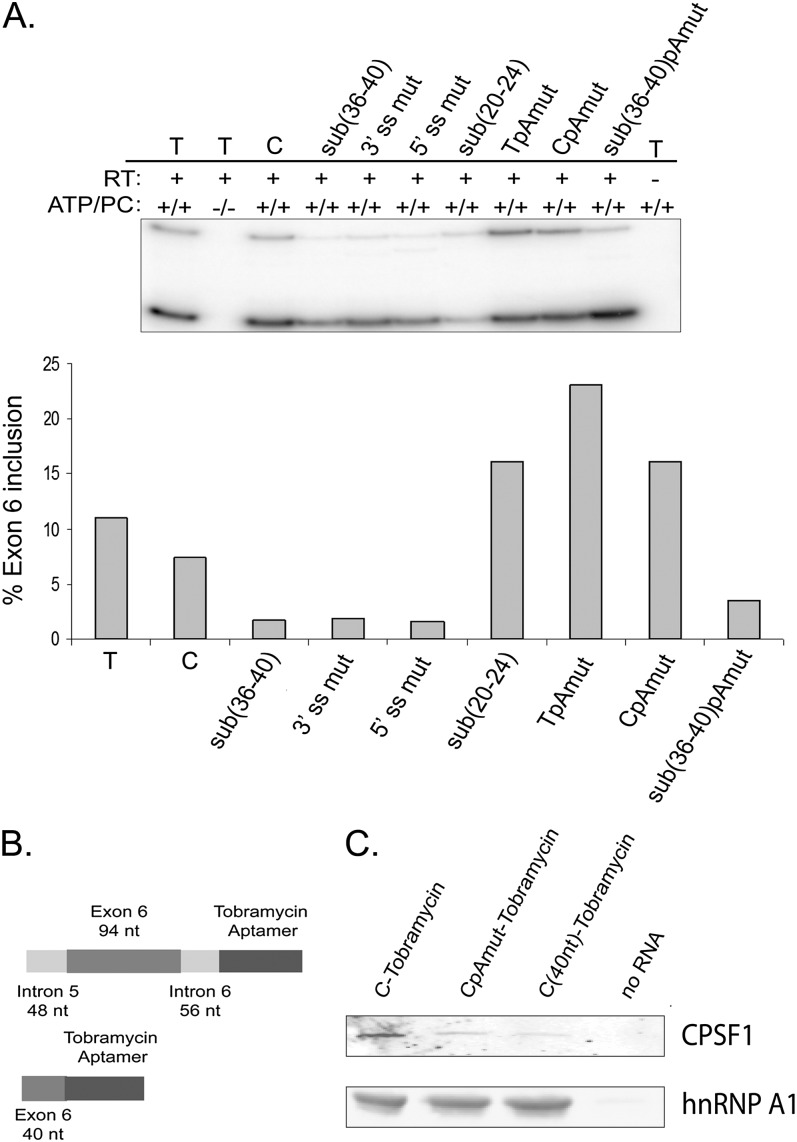
We decided to purify proteins that bound exon 6 and adjacent intronic regions from HeLa cell nuclear extracts. We first tested whether HeLa cell nuclear extracts contain the protein factors that are necessary to recapitulate exon 6 splicing regulation in vitro. To this end, we used RNAs that previously showed either significant increase in exon 6 inclusion (sub(20–24)) or a significant decrease in inclusion (sub(36–40)) in transfection experiments in HeLa cells. Minigene templates were transcribed in vitro, and transcripts were used in in vitro splicing reactions as outlined in the Materials and Methods. The “C” RNA and the 3′ or 5′ splice site mutants served as positive and negative controls, respectively. Although the overall efficiency of splicing was lower than in cultured cells, the pattern of exon 6 splicing regulation in vitro reproduced the pattern observed in transfected cell lines (Fig. 6A). Specifically, in vitro splicing of the sub(20–24) substrate, in which ESS1 is disrupted, resulted in an increase in exon 6 inclusion, while in vitro splicing of the sub(36–40) transcript, in which ESE2 is mutated, resulted in a significant decrease in inclusion (cf. Figs. 2B and 6A). The effect that the MS-associated “C” allele has on exon 6 inclusion as compared with the “T” allele, as well as the effect of the intronic poly(A) site mutation in the context of the “T,” “C,” and sub(36–40) minigenes, was reproduced in the in vitro system as well (Gregory et al. 2007) (cf. Figs. 4B and 6A). We thus concluded that HeLa cell nuclear extracts contain all of the necessary factors required to recapitulate exon 6 splicing in vitro, and used these extracts in all further chromatography experiments.
FIGURE 6.
In vitro splicing analysis and RNA affinity chromatography to identify trans-acting factors that regulate splicing of IL7R exon 6. (A) Splicing regulation of IL7R exon 6 is recapitulated in HeLa cell nuclear extracts in vitro. Substitution minigenes described in Figures 1, 2, and 4 were transcribed in vitro, and RNA transcripts used in in vitro splicing reactions with HeLa cell nuclear extracts. “C” and 3′ ss mut and 5′ ss mut minigenes were used as positive and negative controls, respectively. Each bar represents one in vitro splicing reaction. Exon 6 inclusion was analyzed by semiquantitative RT-PCR. This experiment was repeated several times with similar results. (B) RNA transcripts for tobramycin affinity chromatography. (Top) RNAs contained full-length IL7R exon 6 (94 nt), 48, and 56 nt of introns 5 and 6, respectively, followed by tobramycin/streptavidin aptamer sequences. (Bottom) A second set of RNAs contained the first 40 nt of IL7R exon 6, followed by aptamer sequences. Both RNAs were transcribed in vitro from the T7 promoter. (C) Western blot analysis of tobramycin RNA affinity chromatography elution fractions. “C”, CpAmut, and the first 40 nt of exon 6 were utilized for RNA affinity chromatography. Matrix without RNA (“No RNA”) was used as a negative control. The blots were probed with anti-CPSF1 antiserum and an anti-hnRNP A1 antibody.
To identify trans-acting protein factors that bind exon 6 and/or adjacent intronic regions we used tobramycin RNA affinity chromatography (Hartmuth et al. 2004; Ward et al. 2011; K Hartmuth, pers. comm.). Two RNAs were used for chromatography experiments. The first RNA contained full-length exon 6 (94 nt) flanked by 48 and 56 nt from introns 5 and 6, respectively. The second RNA contained only the first 40 nucleotides of exon 6 (Fig. 6B). IL7R sequences in both templates were followed by streptavidin/tobramycin aptamer sequences as described previously (Ward et al. 2011). A “no RNA” control was routinely used in all chromatography experiments to control for the specificity of protein binding. Chromatography experiments with each RNA were performed in duplicate. RNA–protein complexes were eluted with excess tobramycin and duplicate elutions submitted to Duke University Proteomics Core Facility for analysis.
Mass spectrometry analysis identified 38 proteins that bound the “full-length” exon 6 RNA and 66 that bound the first 40 nt of exon 6 but not the “no RNA” control matrices (Supplemental Table 1). We considered only proteins that were identified with 49%–95% probability with one or more unique peptides in each replicate, and zero peptides (or one to two peptides with 0%–49% probability of identification) in the “no RNA” control. The majority of proteins identified (31 in the experiment with RNAs that contain the “full-length” exon 6 and 34 in the experiment with RNAs that contain the first 40 nt of exon 6) were known RNA-binding proteins. Interestingly, cleavage and polyadenylation specificity factor 1 (CPSF1) was identified specifically with the “full-length” RNAs that contain the consensus polyadenylation sequence in intron 6 (Supplemental Table 1). Western blot analysis on chromatography elutions confirmed relatively efficient binding of CPSF1 to RNA that contained flanking intronic regions, but not to a corresponding RNA with mutation of the intronic poly(A) site. Moreover, the first 40 nt of exon 6 purified less CPSF1 than the larger RNA with intact poly(A) site, and each RNA bound similarly to hnRNP A1 (Fig. 6C). These observations confirm that the consensus intronic polyadenylation site in intron 6 binds CPSF1 in vitro.
CPSF1 regulates exon 6 splicing by promoting skipping of exon 6
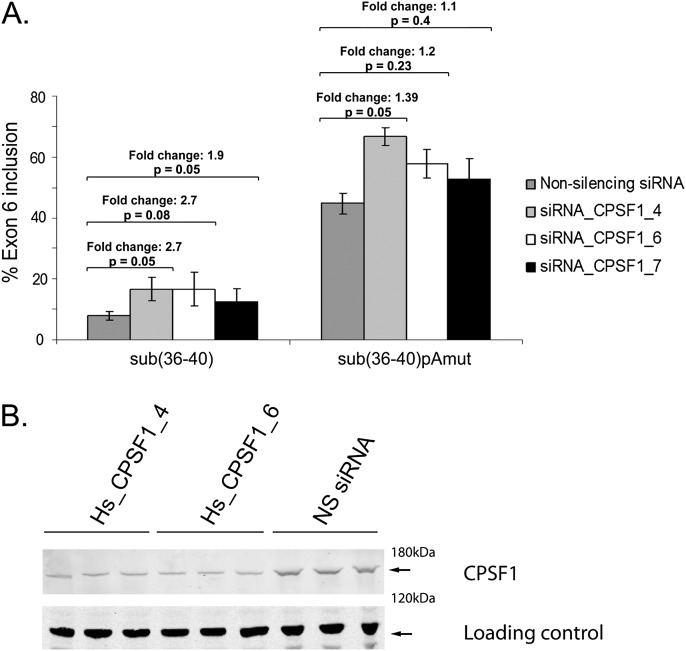
We next sought to test whether CPSF1 was involved in the splicing of IL7R exon 6. To this end we depleted CPSF1 in HeLa cells using siRNAs (Fig. 7B) and transfected sub(36–40) minigenes to determine the effect of CPSF1 depletion on inclusion of exon 6. Inclusion of exon 6 was increased over twofold upon depletion of CPSF1 with at least three different siRNAs (Fig. 7A). Importantly, this effect was significantly reduced for sub(36–40) RNA with a mutated AAUAAA signal (Fig. 7). We conclude that CPSF1 down-regulates IL7R exon 6 splicing and, in agreement with the RNA affinity chromatography data (Fig. 6C), is likely to do this via the AAUAAA 16 nt downstream from the exon.
FIGURE 7.
Effect of siRNA-mediated CPSF1 knockdown on exon 6 inclusion of minigenes sub(36–40) and sub(36–40)pAmut. (A) CPSF1 was depleted by three different siRNAs. A nonsilencing (Qiagen AllStars) siRNA served as control. siRNAs were transfected into HeLa cells, followed by minigene transfection 48 h later. Minigene sub(36–40)pAmut contained a substitution mutation to the polyadenylation signal AAUAAA in intron 6 (for mutation sequence, see Fig. 4A). Exon 6 inclusion was measured by semiquantitative RT-PCR, and fold change between control siRNA-treated and CPSF1 siRNA-treated cells was calculated for each minigene. Fold changes and corresponding P-values are shown. (B) Levels of CPSF1 knockdown by siRNAs Hs_CPSF1_4 and Hs_CPSF1_6 were determined by Western blotting. CPSF1 knockdown by siRNA Hs_CPSF1_7 resulted in similar levels of CPSF1 depletion (data not shown).
Intronic polyadenylation in intron 6 of IL7R is undetectable
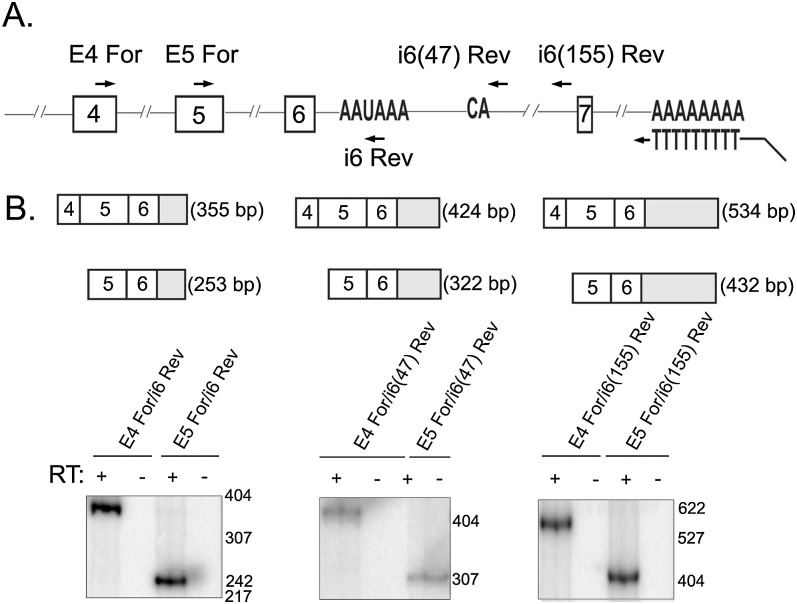
Two different but not mutually exclusive mechanisms could explain the effect of CPSF1 and the AAUAAA site on exon 6 inclusion: (1) Binding of the cleavage and polyadenylation machinery results in cleavage and polyadenylation of the intronic mRNA, in line with recent reports (Kaida et al. 2010; Vorlová et al. 2011; Berg et al. 2012), or (2) binding of CPSF1 and perhaps associated factors to the consensus poly(A) site occurs without cleavage and polyadenylation, but interferes with spliceosome binding to the adjacent 5′ splice site. We sought to determine whether the AAUAAA signal in intron 6 of IL7R is used to produce a previously unreported polyadenylated mRNA isoform. 3′ RACE experiments on total RNA from cell lines transfected with IL7R minigenes did not detect intronic polyadenylated isoforms (data not shown). We then performed 3′ RACE experiments on total RNA from Jurkat cells that naturally express IL7R (Fig. 8). Reverse transcription reactions were performed with an Oligo(dT)-Anchor primer and were followed by PCR with forward primer in exon 3 of IL7R and reverse primer that annealed to the anchor sequence incorporated by the RT primer (as described in the Materials and Methods). This initial PCR reaction was followed by nested PCR using several primer pairs (Fig. 8A). To distinguish between polyadenylated and intron-retaining IL7R transcripts, two different types of reverse primers were used for the nested PCRs: a reverse primer encompassing the AAUAAA site in intron 6 and two reverse primers downstream from the potential cleavage site (dinucleotide “CA” 19 nt downstream from the consensus polyadenylation signal). Nested PCR reactions with the reverse primer containing the AAUAAA site produced specific products that were confirmed by sequencing to correspond to transcripts containing spliced exons 4–6, with exon 6 extending into intron 6 (Fig. 8B). However, PCR with reverse primers downstream from the potential cleavage site also produced specific PCR products, suggesting that the RNA detected contains unspliced intron 6 and is not polyadenylated in intron 6. While we cannot exclude the possibility that a small amount of IL7R pre-mRNA is cleaved and polyadenylated at the poly(A) site in intron 6, the majority of the intronic PCR signal appears to derive from RNA transcripts that retain intron 6. We posit that inefficient splicing of this intron is due to both the weakness of the 5′ splice site and presence of the intronic polyadenylation signal in intron 6. These factors contribute to alternative splicing of exon 6.
FIGURE 8.
No evidence for intronic polyadenylation in intron 6 of IL7R. 3′ RACE was used to investigate intronic polyadenylated isoforms of IL7R in total RNA from Jurkat cells that naturally express IL7R. (A) Schematic of the IL7R pre-mRNA. RT reactions were performed with an Oligo(dT)-Anchor primer either in the presence or absence of RT enzyme. The first round of PCR was performed with a forward primer in exon 3 and a reverse primer that anneals to the anchor sequence incorporated by the Oligo(dT)-Anchor primer during the RT step. This was followed by nested PCR reactions using the primers indicated. (B, top) A diagram of the expected PCR products with primer pairs indicated in lower panels. (B, bottom) Nested PCR products using the indicated primer pairs were resolved on 5% native polyacrylamide gels and exposed to PhosphorImager screens for visualization.
DISCUSSION
Interleukin 7 receptor plays a critical role in T cell development and maintenance, with one of its major functions being protection of T cells from apoptosis (Mazzucchelli and Durum 2007). Underscoring its importance in T cell biology, aberrant expression of IL7R has been implicated in several human diseases such as severe combined immunodeficiency (SCID) (Puel et al. 1998; Kalman et al. 2004) and acute lymphoblastic leukemia (Korte et al. 2000; Zhang et al. 2012), while genetic variation in IL7R can predispose to such diseases as sarcoidosis (Heron et al. 2009), primary biliary cirrhosis (Mells et al. 2011), ulcerative colitis (Yamazaki et al. 2003; Anderson et al. 2011), MS (Gregory et al. 2007), and others, suggesting a larger role for this gene in the pathogenesis of autoimmunity.
IL7R is expressed exclusively on cells of the lymphoid lineage, and its expression is tightly regulated based on the developmental stage of the T cell and availability of the ligand (Mazzucchelli and Durum 2007). While transcriptional regulation of IL7R expression has been widely studied (Pleiman et al. 1991; Franchimont et al. 2002; Xue et al. 2002; Park et al. 2004), post-transcriptional mechanisms of IL7R regulation have only recently been uncovered. Several splicing isoforms have been reported for IL7R at the RNA level (Goodwin et al. 1990; Pleiman et al. 1991; Korte et al. 2000; Gregory et al. 2007; Rose et al. 2009), but efforts to characterize these splicing events in detail have not been made. Moreover, to the best of our knowledge, other mechanisms of post-transcriptional regulation of IL7R expression, such as regulation by alternative polyadenylation, have not been explored. Considering the proposed role of the soluble IL7R isoform in MS (Gregory et al. 2007) and infection with the HIV (Koesters et al. 2006; Rose et al. 2009; Crawley et al. 2010), it is important to characterize in detail potential IL7R isoforms and their contribution to T cell biology and disease. We uncovered several cis-acting regulatory elements within IL7R exon 6, which exclusively encodes the transmembrane domain of the receptor, and identified CPSF1 as a factor that influences exon 6 inclusion. Our data uncover a role for this polyadenylation factor in regulating exon 6 inclusion and suggest that competing splicing and polyadenylation may influence levels of soluble IL7R isoform.
Splicing and polyadenylation are intricately connected and influence one another. The CPSF complex interacts with components of the spliceosome (Lutz et al. 1996; Kyburz et al. 2006) and has been found to be required for splicing of single introns in vivo (Li et al. 2001). Correspondingly, several splicing factors have been found to influence polyadenylation (Lou et al. 1998; Vagner et al. 2000; Millevoi et al. 2002, 2006; Danckwardt et al. 2008). It was thus very plausible that, by binding to the consensus AAUAAA site in intron 6 of IL7R, CPSF1 influences splicing of exon 6.
Two potential mechanisms may explain the effect that CPSF1 has on exon 6 splicing. First, usage of the IL7R intron 6 poly(A) site might be an example of a sporadic intronic polyadenylation event that competes with splicing and is modulated by U1 snRNP abundance in a splicing-independent manner (Kaida et al. 2010; Vorlová et al. 2011; Berg et al. 2012). Alternatively, binding of CPSF1 to the intronic poly(A) site or assembly of the full CPSF complex on the poly(A) site in intron 6 may interfere with binding of U1 snRNP to the adjacent 5′ splice site. Interestingly, it has been established that the frequency of intronic polyadenylation events is elevated in the context of weak upstream 5′ splice sites (Tian et al. 2007). We have determined that the exon 6 5′ splice site is weak (Fig. 5), thus it is possible that reduced binding of U1 snRNP to this site allows for the assembly of the CPSF complex on the intronic AAUAAA site. Whatever the mechanism, assembly of the polyadenylation complex on the intronic poly(A) site in intron 6, together with the effect that the weak 5′ splice site has on exon 6 splicing, is expected to contribute to poor recognition of this exon by the splicing machinery. At the level of RNA, this would result in less full-length IL7R and, subsequently, less functional IL7R protein.
Bioinformatic studies have shown that up to 20% of human genes contain polyadenylation signals in their introns (Tian et al. 2007). Sequence analysis shows that poly(A) signals are present in several of the IL7R introns. In introns 3, 4, and 6, these signals are in the direct vicinity of the 5′ splice sites and are followed by a cleavage signal dinucleotide CA. If expressed, the resultant protein products from IPA in introns 3 and 4 will be soluble. In the context of the suggested role of the soluble IL7R in MS pathogenesis, it would be important to investigate these potential polyadenylation events and determine whether corresponding proteins are indeed produced. Alternative polyadenylation of IL7R may thus constitute yet another layer of regulation of this receptor that contributes to the repertoire of IL7R isoforms and acts together with splicing to regulate its expression.
Moreover, while, in general, cleavage and polyadenylation factors are expressed constitutively (Proudfoot 2011), some data suggest that several cleavage and polyadenylation factors may be expressed at different levels, depending on the developmental stage of the cell or its physiological state (Chuvpilo et al. 1999; Sandberg et al. 2008; Singh et al. 2009; Lutz and Moreira 2011). For example, cleavage stimulation factor CstF-64 is expressed at limiting levels in pre-B cells, which results in preferential splicing of a specific hydrophobic C terminus of the immunoglobulin antibody heavy-chain pre-mRNA and production of a membrane-bound isoform. In mature B cells, levels of CstF-64 are elevated, which allows for the recognition of an intronic polyadenylation signal and production of a secreted protein isoform (Takagaki et al. 1996; Takagaki and Manley 1998). It would be important to determine whether any of the components of the cleavage and polyadenylation machinery are differentially expressed during stages of T cell development and/or T cell proliferation and differentiation, and whether differential expression of these factors affects splicing of the IL7R exon 6. An intriguing possibility (currently being explored by our group) would be that changes in the expression of either specific splicing or polyadenylation factors that regulate IL7R exon 6 inclusion contribute to MS (or other pathologies) by stimulating exon 6 skipping (and, thus, soluble IL7R production).
In summary, we have identified an additional layer of post-transcriptional regulation of the clinically important interleukin 7 receptor. Further studies are needed to determine the exact mechanism of the effect that CPSF1 has on exon 6 splicing and characterize the full isoform repertoire for IL7R. These studies would not only expand our understanding of how expression of this important receptor is regulated, but would also shed light on its role in T cell biology and—potentially—its involvement in MS pathogenesis.
MATERIALS AND METHODS
Plasmids and cloning
All substitution and deletion mutants were made in the context of the cloning vector pI-12 described previously (Carstens et al. 1998; Gregory et al. 2007). Briefly, mutant constructs were made by chimeric PCRs and purified with QIAquick PCR purification kit (Qiagen). Forward primers corresponding to the T7 promoter (5′-TAATACGACTCACTATAGGGAGA-3′) or the upstream “U” sequence (5′-GCTCGGATCCACTAGTAACG-3′) and reverse primers corresponding to the Sp6 promoter (5′-ATTTAGGTGACACTATAGAA-3′) and the downstream “D” sequence (5′-GTTCGGAGGATGCATAGAGA-3′) were used as common primers. PCR chimeras and pI-12 vector were digested with XbaI and XhoI (NEB), purified with the QIAquick PCR purification kit, and ligated with T4 DNA Ligase (NEB). All PCR products were confirmed by sequencing by the Duke University Sequencing Facility.
Cell culture and transfections
Rat DT cells were cultured in low-glucose Dulbecco's Modified Eagle's Medium (DMEM, Invitrogen/Gibco) supplemented with 10% fetal bovine serum (Gemini) and antibiotics (penicillin/streptomycin, Invitrogen/Gibco). Human HeLa cells were cultured in high-glucose DMEM (Invitrogen/Gibco) with 10% fetal bovine serum and antibiotics. Human Jurkat cells were cultured in Roswell Park Memorial Institute Medium 1640 (RPMI1640) supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, 10 mM HEPES, glucose up to 4.5 g/L and antibiotics (all reagents were from Invitrogen/Gibco). Transient minigene transfections into DT and HeLa cell lines were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. A total of 25 ng of minigene DNA was transfected per well in 24-well format (24-well plates were from BD Biosciences). A total of 5 × 104–7.5 × 104 cells were seeded into each well 24 h prior to transfection. Minigene transfections into Jurkat cells were performed with FuGENE 6 (Roche) according to the manufacturer's instructions. Briefly, 2 × 106 cells were freshly passed and seeded into each well of a 6-well plate (BD Biosciences) immediately before transfection, and 1 μg of minigene DNA was used per well. Cells were harvested 48 h post-transfection. All transfections were performed in triplicate.
SiRNA duplexes and CPSF1 knockdown
SiRNA duplexes were synthesized by Qiagen in a FlexiTube format and resuspended in water to a final concentration of 10 μM upon arrival. CPSF1 siRNA sequences were as follows: Hs_CPSF1_4, 5′-CCAGATGATCAGCGTCAAGAA-3′; Hs_CPSF1_5, 5′-AGGGCGGATCTTGATCATGGA-3′; Hs_CPSF1_6, 5-CACGTGGAGTCTAAGGTGTAT-3′; and Hs_CPSF1_7, 5′-CCGAGCGTTCCACTTTGACAA-3′. AllStars Negative Control siRNA (Qiagen) was used as a nonsilencing siRNA control in all knockdown experiments. Transfections of siRNAs into HeLa cells were performed with Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's instructions. Briefly, 6 pmol of siRNA duplexes were diluted in 100 μL of Opti-MEM I Medium (Invitrogen/Gibco) without serum in each well of a 24-well plate. Next, 0.8 μL of Lipofectamine RNAiMax was added to each well. SiRNA/transfection reagent mixtures were mixed gently and incubated for 20 min at room temperature. HeLa cells were diluted to obtain 3 × 104 cells per 500 μL of medium and 500 μL was added to each well to obtain a final siRNA concentration of 10 nM. At 48-h post-transfection, cells were transfected with minigenes as described above. At 48 h post-minigene transfection, cells were harvested with TRIZOL Reagent (Invitrogen) for RNA analysis or with 1X RIPA buffer (see below) for protein analysis. Transfection with all four CPSF1 siRNAs resulted in a comparable decrease in CPSF1 levels as estimated by Western blotting. However, transfections of IL7R minigenes into HeLa cells in which CPSF1 was depleted with siRNA Hs_CPSF1_5, did not have any effect on exon 6 inclusion and were excluded from analysis.
RNA isolation and semiquantitative RT-PCR of transfected minigenes
Total RNA was extracted with TRIZOL Reagent (Invitrogen) according to the manufacturer's instructions and treated with TURBO DNase (Ambion/Life Technologies). A total of 2 μg of DNase-treated RNA was used to synthesize first-strand cDNA with 250 ng of random hexamer primers (Invitrogen) and Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT, Invitrogen) following the manufacturer's protocol. PCRs were performed as follows: 0.1–2 μg (1 μL) of the RT reaction was mixed with 200 nM forward (T7) and 200 nM reverse (Sp6) primers and 100 nM dNTPs in 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, and 0.25 μL of Taq polymerase was added. [α-32P]dCTP (3000 Ci/mmol, 10 mCi/mL, PerkinElmer) was added to a final concentration of 0.1 μCi/μL. PCR products were loaded onto 5% nondenaturing polyacrylamide/TBE gels, electrophoresed for 3 h at 120 V, dried using a gel dryer and exposed to Molecular Dynamics or Amersham Hyperfilm-MP PhosphorImager screens. Quantifications were performed using ImageQuant software (Molecular Dynamics). Percent exon 6 inclusion was calculated according to the following formula: % Inclusion: [(Inclusion product)/(Inclusion + Skipped product)] × 100. Each calculation was the result of an average of triplicate samples. Error bars represent standard deviations.
Protein extraction, antibodies, and Western blotting
Total protein was harvested by scraping cells in 1X RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl a pH 7.5, 1X Protease Inhibitors [Roche]) and subjecting cells to freeze/thaw lysis (freezing at –80°C followed by thawing at 37°C). Protein lysates were loaded directly onto NuPAGE 4%–12% Bis-Tris pre-cast gels (Life Technologies), transferred onto Protran nitrocellulose membranes (Whatman), and Western blot assays were performed according to standard techniques. Anti-hnRNP A1 mouse monoclonal antibody was from Abcam.
In vitro transcription and in vitro splicing
DNA templates for in vitro transcription reactions were prepared by restriction endonuclease digestion tobramycin aptamer were digested with DraIII. Digested templates were purified with the QIAquick PCR Purification Kit or the QIAquick Gel Extraction Kit (both from Qiagen). For small-scale in vitro transcriptions, 2 μg of digested DNA plasmid was mixed with 0.5 mM each of ATP, CTP, GTP, and 12.5 nM UTP, 1X Transcription Buffer (40 mM Tris-HCl at pH 7.5, 6 mM MgCl2, 2 mM spermidine-HCl, 5 mM NaCl), 20–40 units RNase Out (Invitrogen) and 40 units T7 RNA Polymerase Plus (Ambion/Life Technologies). [α-32P]UTP (3000 Ci/mmole, 10 mCi/mL, PerkinElmer) was added to the final concentration of 2 nCi/μL. Reactions were incubated at 37°C for 90 min, and RNA was extracted with Phenol:Chloroform:Isoamyl Alcohol (25:24:1, Invitrogen) and precipitated with 0.4 M NH4OAc and ethanol. One micoliter of each transcription reaction was taken out before and after phenol/chloroform extraction and used for scintillation counting on a Beckman LS-6500 liquid scintillation counter. Large-scale in vitro transcriptions (for RNAs used in affinity chromatography) were performed using Ambion's MEGAscript T7 High Yield Transcription Kit and high-concentration T7 RNA Polymerase Plus (200 u/μL, Ambion/Life Technologies) following the manufacturer's protocol. Reactions were incubated at 37°C for 6 h and RNA was extracted with RNeasy Mini Kit (Qiagen). In vitro transcripts that were uniformly labeled were run on denaturing 15% polyacrylamide/TBE gels (19:1) with 7 M Urea, to check for transcript integrity. Nonradioactively labeled transcripts (prepared by using the MEGAscript kit) were resolved on denaturing agarose gels to check for transcript integrity.
In vitro splicing reactions contained 100,000 counts per minute (cpm) of uniformly labeled RNA template, 1 mM ATP, 5 mM phosphocreatine, 64 mM KCl (total), 2 mM MgCl2, 40 units RNase Out (Invitrogen), and 32% HeLa nuclear extract (v/v). HeLa nuclear extract was prepared according to Dignam et al. (1983). Reactions were incubated at 30°C for 90 min and treated with Proteinase K (Ambion/Life Technologies) in 10 mM Tris-HCl at pH 7.5, 1% SDS, 0.15 mM NaCl, 10 mM EDTA, 25 ng/μL of tRNA (Sigma) and 50 μg of Proteinase K (Ambion/Life Technologies) for 15 min at 37°C, after which RNA was extracted with phenol:chloroform:isoamyl alcohol and precipitated with NH4OAc and ethanol as described above. RNA was air-dried and resuspended in 10 μL of water and treated with 2 units of TURBO DNase (Ambion/Life Technologies) according to the manufacturer's protocol. DNase reactions were incubated at 37°C for 30 min and extracted with phenol:chloroform:isoamyl alcohol and precipitated with NH4OAc and ethanol as described above. RNA was resuspended in 10 μL of water, and all 10 μL were used in an RT-PCR reaction performed in the same manner as the RT-PCR for total RNA extracted from the transfected cell lines (see above). Primers U and D, which annealed to the sequences of the adenovirus upstream (U) and downstream (D) exons present in the pI-12 vector, were used in PCR reactions.
Tobramycin RNA affinity chromatography and mass spectrometry
Tobramycin RNA affinity chromatography was performed according to Hartmuth et al. (2004), and a personal communication of K. Hartmuth. A 2-mL packed bead volume of NHS-activated Sepharose (GE Healthcare) was washed four times in ice-cold 1 mM HCl and mixed with 1 mL of 5 or 10 mM tobramycin in the coupling buffer (either 0.2 M NaHCO3, 0.5 M NaCl at pH 8.3, or 75 mM HEPES-KOH at pH 7.9, and 90% dimethylformamide) and incubated overnight with head-over-tail rotation at 4°C. The next day, beads were spun and blocked in either 0.2 M NaHCO3, 0.1 M NaCl, 1 M ethanolamine (pH 8.0), or 0.2 M Tris-HCl (pH 8.4) for 2 h at room temperature. The matrix was then washed two to three times with 9 mL of ice-cold PBS (pH 8.0) and then two times with ice-cold PBS/NaN3 and resuspended in 2 mL PBS/NaN3. Matrices were stored in PBS/NaN3 at 4°C for up to a month.
For chromatography experiments, 15–60 μL (packed bead volume) of tobramycin-containing matrix aliquots were blocked overnight in 20 mM Tris-HCl (pH 8.1), 1 mM CaCl2, 1 mM MgCl2, 0.2 mM DTT (together referred to as 1X binding buffer, or 1X BP), 300 mM KCl, 0.1 mg/mL tRNA, 0.5 mg/mL BSA, and 0.01% NP-40 at 4°C. The next day, in vitro synthesized RNA templates were mixed with 1X BP, 145 mM KCl, and 0.1 mg/mL tRNA, added to the tobramycin Sepharose matrix, and incubated for 1 h at 4°C with head-over-tail rotation. At the same time, appropriate amounts of HeLa nuclear extract were pre-cleared with an aliquot of the tobramycin-containing matrix. RNA immobilized on the matrix was washed three times with 1 mL of the wash buffer (1X BP, 145 mM KCl, 0.1% NP-40), and conventional in vitro splicing reactions were assembled in the presence of 1 mM ATP, 5 mM phosphocreatine, 64 mM KCl (total), 2 mM MgCl2, and 32% pre-cleared HeLa nuclear extract (v/v). Splicing mixes were added to the immobilized RNA, and reactions were incubated at 30°C for 5 min with head-over-tail rotation. Following incubation, reactions were immediately put on ice and then centrifuged at 250g at 4°C to collect supernatant fraction, followed by three washes with the protein wash buffer (1X BP, 150 mM KCl, 0.1% NP-40) and three washes with either PBS or elution buffer without tobramycin (see below). All spins were done at 4°C. RNA–protein complexes were eluted with elution buffer (1X BP, 5 or 10 mM tobramycin, 145 mM KCl, 2 mM MgCl2) for 20 min at 30°C. Supernatant, wash, and elution fractions were collected and used to precipitate protein as follows: an equal volume of phenol:chloroform:isoamyl alcohol was added and, upon phase separation, the lower organic phase was transferred to a fresh tube. Four volumes of ice-cold acetone were added, and precipitation was allowed to occur overnight at −20°C. The next day, samples were spun at maximum speed at 4°C, washed two times in 4:1 acetone:water, air-dried for 15 min, resuspended in the SDS-gel loading buffer (100 mM Tris-HCl at pH 6.8, 4% SDS, 0.02% [w/v] bromophenol blue, 20% [v/v] glycerol, and 200 mM DTT, added fresh before each use) and either loaded onto NuPAGE protein gels for silver staining or Western blotting analysis or stored at −20°C. For mass spectrometry experiments, 1X protease inhibitors (Roche) were added to the elution buffer, and the eluates were stored at 4°C overnight until they could be submitted for the mass spectrometry analysis the following day.
Mass spectrometry analysis of the chromatography eluates was performed by the Duke University Proteomics Core Facility. Quantification of the label-free mass spectrometry experiments was performed with Rosetta Elucidator software by Dr. Will Thompson of the Duke University Core Proteomics Facility.
3′RACE
3′RACE was performed according to conventional protocols. Briefly, reverse transcription was performed with total DNase-treated RNA from Jurkat cells, reverse Oligo(dT)-Anchor primer (5′-CAGCTGTAGCTATGCGCACAGTTTTTTTTTTTTTTTTV-3′, V=A,C,G), and M-MLV RT. A total of 1 μL of the RT reaction was used in the first PCR with the forward primer complementary to the IL7R exon 3 (5′-GTGGAGGTAAAGTGCCTGAATTTC-3′) and PCR-Anchor (5′-CAGCTGTAGCTATGCGCACAG-3′) as the reverse primer. Nested PCRs were performed with the following primer pairs: exon 4 forward (5′-TTAATGCACGATGTAGCTTACCG-3′) and intron 6 reverse (5′-TTTATTAGTTGAAGAAGGTCACCTTTTTTTCC-3′); exon 5 forward (5′-CCGGCAGCAATGTATGAGATTAAAG-3′) and intron 6 reverse; exon 4 forward and intron 6(47) reverse (5′-CGTGAAATGCCTTAATCCCC-3′); exon 5 forward and intron 6(47) reverse; exon 4 forward and intron 6(155) reverse (5′-CTTACTTTGGGGACAGCGTTTG-3′); and exon 5 forward and intron 6(155) reverse. PCR products were resolved on 5% nondenaturing polyacrylamide/TBE gels. Gels were run at 120 V for ∼3 h, dried using a gel dryer, and exposed to Molecular Dynamics PhosphorImager screens.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We are grateful to Dr. Jeffery Wilusz (Colorado State University) for the generous gift of CPSF1 antiserum. We thank Dr. Alex Ward (Duke-NUS Singapore) for providing sequence information on tobramycin/streptavidin aptamers, Dr. Will Thompson and members of Duke University Proteomics Core Facility for performing mass spectrometry and quantification of the mass spectrometry results, and Dr. Jason Somarelli for critical reading of this manuscript and many useful comments. We also thank Dr. Klaus Hartmuth (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany) for communicating unpublished results. S.G.G. and M.A.G.-B. acknowledge funding from NIH grant 5R01-NS060925.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.035410.112.
REFERENCES
- Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL 1997. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell 89: 1033–1041 [DOI] [PubMed] [Google Scholar]
- Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, Lee JC, Goyette P, Imelinski M, Latiano A, et al. 2011. Meta-analysis identifies 29 additinoal ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 43: 246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. 2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campigli Di Giammartino D, Nishida K, Manley JL 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens RP, McKeehan WL, Garcia-Blanco MA 1998. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol Cell Biol 18: 2205–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuvpilo S, Zimmer M, Kerstan A, Glöckner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F, et al. 1999. Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity 10: 261–269 [DOI] [PubMed] [Google Scholar]
- Cooke C, Hans H, Alwine JC 1999. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol Cell Biol 19: 4971–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G 2009. RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley AM, Faucher S, Angel JB 2010. Soluble IL-7Rα (sCD127) inhibits IL-7 activity and is increased in HIV infection. J Immunol 184: 4679–4687 [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE 2008. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J 27: 482–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsyukova I, Somarelli JA, Gregory SG, Garcia-Blanco MA 2010. Alternative splicing in multiple sclerosis and other autoimmune diseases. RNA Biol 7: 462–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchimont D, Galon J, Vacchio MS, Fan S, Visconti R, Frucht DM, Geenen V, Chrousos GP, Ashwell JD, O'Shea JJ 2002. Positive effects of glucocorticoids on T cell function by up-regulation of IL-7 receptor alpha. J Immunol 168: 2212–2218 [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco MA, Baraniak AP, Lasda EL 2004. Alternative splicing in disease and therapy. Nat Biotechnol 22: 535–546 [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Greenleaf AL, Garcia-Blanco MA 2001. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene 277: 31–47 [DOI] [PubMed] [Google Scholar]
- Goodwin RG, Friend D, Ziegler SF, Jerzy R, Falk BA, Gimpel S, Cosman D, Dower SK, March CJ, Namen AE, et al. 1990. Cloning of the human and murine interleukin-7 receptors: demonstration of a soluble form and homology to a new receptor superfamily. Cell 60: 941–951 [DOI] [PubMed] [Google Scholar]
- Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, Prokop A, Caillier SJ, Ban M, Goris A, Barcellos LF, et al. 2007. Interleukin 7 receptor α (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet 39: 1083–1091 [DOI] [PubMed] [Google Scholar]
- Hartmuth K, Vornlocher HP, Lührmann R 2004. Tobramycin affinity tag purification of spliceosomes. Methods Mol Biol 257: 47–64 [DOI] [PubMed] [Google Scholar]
- Heron M, Grutters JC, van Moorsel CHM, Ruven HJT, Huizinga TWJ, van der Helm-van Mil AHM, Claessen AME, van den Bosch JMM 2009. Variation in IL7R predisposes to sarcoid inflammation. Genes Immun 10: 647–653 [DOI] [PubMed] [Google Scholar]
- The International Multiple Sclerosis Genetics Consortium 2007. Risk alleles for multiple sclerosis identified by a genome-wide study. N Engl J Med 357: 851–862 [DOI] [PubMed] [Google Scholar]
- Jiang Q, Li WQ, Hofmeister RR, Young HA, Hodge DR, Keller JR, Khaled AR, Durum SK 2004. Distinct regions of the interleukin-7 receptor regulate different Bcl2 family members. Mol Cell Biol 24: 6501–6513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman L, Lindegren ML, Kobrynski L, Vogt R, Hannon H, Howard JT, Buckley R 2004. Mutations in genes required for T-cell development: IL7R, CD45, IL2RG, JAK3, RAG1, RAG2, ARTEMIS, and ADA and severe combined immunodeficiency: HuGE review. Genet Med 6: 16–26 [DOI] [PubMed] [Google Scholar]
- Keller W, Bienroth S, Lang KM, Christofori G 1991. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J 10: 4241–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koesters SA, Alimonti JB, Wachihi C, Matu L, Anzala O, Kimani J, Embree JE, Plummer FA, Fowke KR 2006. IL-7Rα expression on CD4+ T lymphocytes decreases with HIV disease progression and inversely correlates with immune activation. Eur J Immunol 36: 336–344 [DOI] [PubMed] [Google Scholar]
- Korte A, Köchling J, Badiali L, Eckert C, Andreae J, Geilen W, Kebelmann-Betzing C, Taube T, Wu S, Henze G, et al. 2000. Expression analysis and characterization of alternatively spliced transcripts of human IL-7Rα chain encoding two truncated receptor proteins in relapsed childhood ALL. Cytokine 12: 1597–1608 [DOI] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W 2006. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 23: 195–205 [DOI] [PubMed] [Google Scholar]
- Li Y, Chen Z-Y, Wang W, Baker CC, Krug RM 2001. The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA 7: 920–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Neugebauer KM, Gagel RF, Berget SM 1998. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol 18: 4977–4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark F, Duvefelt K, Iacobaeus E, Kockum I, Wallström E, Khademi M, Oturai A, Ryder LP, Saarela J, Harbo HF, et al. 2007. Variation in interleukin 7 receptor α chain (IL7R) influences risk of multiple sclerosis. Nat Genet 39: 1108–1113 [DOI] [PubMed] [Google Scholar]
- Lutz CS 2008. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol 3: 609–617 [DOI] [PubMed] [Google Scholar]
- Lutz CS, Moreira A 2011. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip Rev RNA 2: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Murthy KGK, Schek N, O'Connor JP, Manley JL, Alwine JC 1996. Interaction between the U1 snRNP-A and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev 10: 325–337 [DOI] [PubMed] [Google Scholar]
- Lynch KW 2004. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol 4: 931–940 [DOI] [PubMed] [Google Scholar]
- Maki K, Sunaga S, Ikuta K 1996. The V-J recombination of T cell receptor-γ genes is blocked in interleukin-7 receptor-deficient mice. J Exp Med 184: 2423–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucchelli R, Durum SK 2007. Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol 7: 144–154 [DOI] [PubMed] [Google Scholar]
- Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, Heneghan MA, Neuberger JM, Donaldson PT, Day DB, et al. 2011. Genome-wide association study identified 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet 43: 329–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Geraghty F, Idowu B, Tam JL, Antoniou M, Vagner S 2002. A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep 3: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S 2006. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ processing machineries. EMBO J 25: 4854–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KG, Manley JL 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev 9: 2672–2683 [DOI] [PubMed] [Google Scholar]
- Niwa M, Berget SM 1991. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev 5: 2086–2095 [DOI] [PubMed] [Google Scholar]
- Park JH, Yu Q, Erman B, Appelbaum JS, Montoya-Durango D, Grimes HL, Singer A 2004. Suppression of IL7Rα transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity 21: 289–302 [DOI] [PubMed] [Google Scholar]
- Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med 180: 1955–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiman CM, Gimpel SD, Park LS, Harada H, Taniguchi T, Ziegler SF 1991. Organization of the murine and human interleukin-7 receptor genes: two mRNAs generated by differential splicing and presence of a type I-interferon-inducible promoter. Mol Cell Biol 11: 3052–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ 2011. Ending the message: poly(A) signals then and now. Genes Dev 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A, Ziegler SF, Buckley RH, Leonard WJ 1998. Defective IL7R expression in T-B+NK+ severe combined immunodeficiency. Nat Genet 20: 394–397 [DOI] [PubMed] [Google Scholar]
- Rose T, Lambotte O, Pallier C, Delfraissy J-F, Colle J-H 2009. Identification and biochemical characterization of human plasma soluble IL-7R: lower concentrations in HIV-1-infected patients. J Immunol 182: 7389–7397 [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH 2009. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res 69: 9422–9430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell 2: 761–771 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952 [DOI] [PubMed] [Google Scholar]
- Tian B, Pan Z, Lee JY 2007. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res 17: 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD, Nave K-A 2008. Multiple sclerosis: an immune or neurodegenerative disorder? Annu Rev Neurosci 31: 247–269 [DOI] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev 14: 403–413 [PMC free article] [PubMed] [Google Scholar]
- Vorlová S, Rocco G, LeFave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L 2011. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell 43: 927–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-S, Cooper TA 2007. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8: 749–761 [DOI] [PubMed] [Google Scholar]
- Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA 2011. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol 8: 1173–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H-H, Kovanen PE, Rise-Masisom CA, Berg M, Radovich MF, Brady JN, Leonard WJ 2002. IL-2 negatively regulates IL-7 receptor α chain expression in activated T lymphocytes. Proc Natl Acad Sci 99: 13759–13764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Yajima T, Tanabe M, Fukui K, Okada E, Okamoto R, Oshima S, Nakamura T, Kanai T, Uehira M, et al. 2003. Mucosal T cells expressing high levels of IL-7 receptor are potential targets for treatment of chronic colitis. J Immunol 171: 1556–1563 [DOI] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang I, Rusch M, et al. 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukemia. Nature 481: 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]