GW182 family proteins, core components in miRNA gene silencing, interact with Argonaute (Ago) proteins and localize in processing bodies, which are cytoplasmic foci involved in mRNA degradation and storage. Here, the authors demonstrate that a human GW182 paralog, TNRC6A, is a nuclear–cytoplasmic shuttling protein that can bring Ago proteins into the nucleus via its Ago-interacting motif(s). Furthermore, miRNAs also colocalized with nuclear TNRC6A-Ago and exhibited gene silencing activity. These results indicate that TNRC6A plays an important role in navigating Ago protein into the nucleus to lead miRNA-mediated gene silencing.
Keywords: Argonaute, microRNA-mediated gene silencing, nuclear export signal, nuclear localization signal, TNRC6A
Abstract
GW182 family proteins play important roles in microRNA (miRNA)-mediated gene silencing. They interact with Argonaute (Ago) proteins and localize in processing bodies, which are cytoplasmic foci involved in mRNA degradation and storage. Here, we demonstrated that human GW182 paralog, TNRC6A, is a nuclear–cytoplasmic shuttling protein, and its subcellular localization is conducted by a nuclear export signal (NES) and a nuclear localization signal (NLS) identified in this study. TNRC6A with mutations in its NES region was predominantly localized in the nucleus in an Ago-independent manner. However, it was found that TNRC6A could bring Ago protein into the nucleus via its Ago-interacting motif(s). Furthermore, miRNAs were also colocalized with nuclear TNRC6A-Ago and exhibited gene silencing activity. These results proposed the possibility that TNRC6A plays an important role in navigating Ago protein into the nucleus to lead miRNA-mediated gene silencing.
INTRODUCTION
GW182 family proteins are components of microRNA (miRNA)-induced silencing complexes (miRISCs) in animal cells and play important roles in miRNA-mediated gene silencing (for review, see Ding and Han 2007; Eulalio et al. 2009a). They interact with Argonaute (Ago) proteins, core components of miRISC, and mainly localize in processing (P) bodies, which are cytoplasmic foci that contain proteins involved in diverse post-transcriptional processes such as mRNA degradation, storage, and translational repression (Ding et al. 2005; Jakymiw et al. 2005; Liu et al. 2005; Meister et al. 2005; Behm-Ansmant et al. 2006; Zhang et al. 2007; Landthaler et al. 2008). There are three GW182 paralogs in vertebrates (TNRC6A/GW182, TNRC6B, and TNRC6C), one ortholog in insects (GW182), and two orthologs in Caenorhabditis elegans (AIN-1 and AIN-2) (Eulalio et al. 2009a). Among them, the vertebrate and insect proteins share a common structural organization characterized by the following five regions: multiple glycine-tryptophan (GW) repeats in the N-terminal region, a central ubiquitin-associated (UBA) domain, a glutamine-rich (Q-rich) region, a poly(A)-binding protein (PABP) binding motif 2 (PAM2), and a C-terminal RNA recognition motif (RRM) (Fig. 1A).
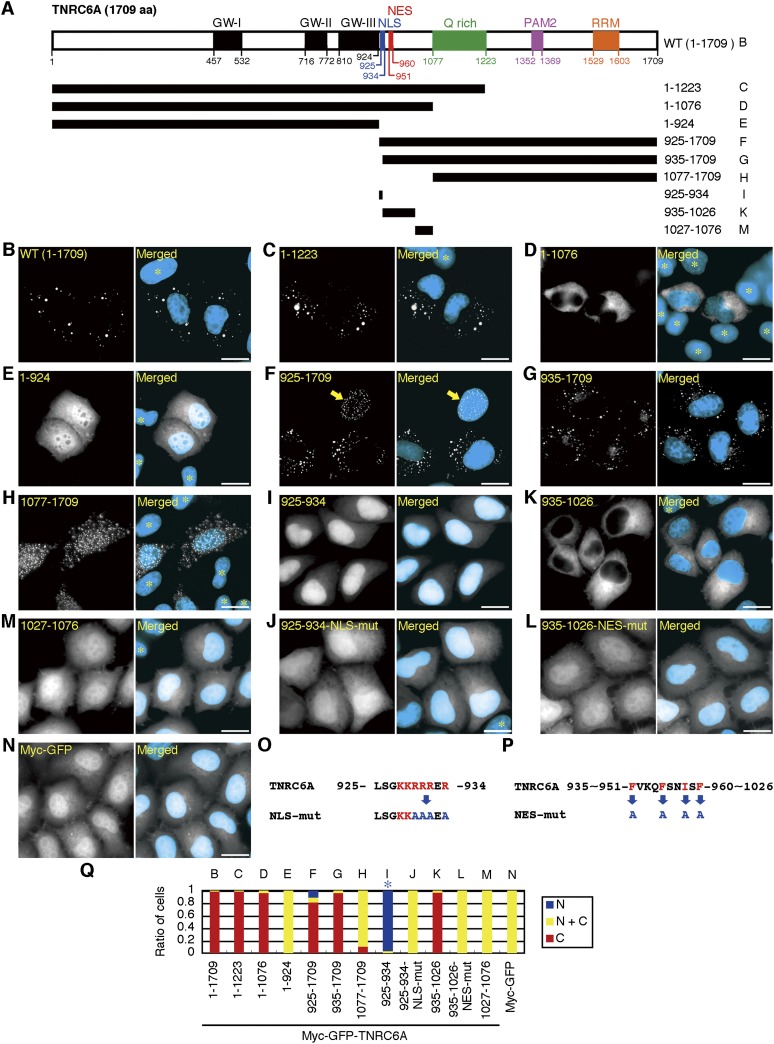
FIGURE 1.
Subcellular localization of TNRC6A fragments and identification of NES and NLS. (A) Schematic representation of TNRC6A deletion mutants, which were expressed as fusion proteins with myc-GFP. Numbers denote the amino acid position relative to the N terminus. GW-I, -II, and -III (black boxes) indicate GW-repeated Ago-binding motifs. NLS (blue box) and NES (red box) were identified in this study. B–I, K, and M at the right side indicate the fragments used in B–I, K, and M. (WT) Wild type. (B–N) Subcellular localization of the indicated myc-GFP-tagged TNRC6A fragments (B–M) and the control myc-GFP (N). Fluorescent microscopy images of GFP are shown in the left panels, and their merged images with DAPI are shown in the right panels, in which the GFP signal is shown in white and the DAPI signal in cyan. An arrow in F indicates the cell in which myc-GFP-TNRC6A-(925–1709) signals are exclusively observed in the nucleus. (*) Cells not transfected with each expression construct. Bars, 20 μm. (O,P) Sequences of the TNRC6A NLS motif and its mutant (NLS-mut) (O), and TNRC6A NES and its mutant (NES-mut) (P). The red letters represent basic amino acids or hydrophobic residues. The blue letters indicate substituted amino acids. (Q) The ratio of cells expressing GFP exclusively in the nucleus (N, blue), cytoplasm (C, red), or both (N+C, yellow). B–N at the upper side indicate the fragments used in B–N. A blue asterisk indicates that the expression of GFP is strongly observed in the nucleus, although a negligible signal is also observed in the cytoplasm.
Many recent reports have revealed the molecular function of GW182 family proteins in miRNA-mediated gene silencing in the cytoplasm. The human and Drosophila GW182 family proteins have multiple Ago interaction sites in an N-terminal GW-repeat-containing region (Till et al. 2007; Eulalio et al. 2009b; Lazzaretti et al. 2009; Lian et al. 2009; Takimoto et al. 2009). The C-terminal region containing the PAM2 motif and RRM possesses silencing activity (Chekulaeva et al. 2009; Eulalio et al. 2009b; Lazzaretti et al. 2009; Zipprich et al. 2009). This region of human TNRC6A, TNRC6B, and TNRC6C and Drosophila GW182 (dGW182) interacts with cytoplasmic poly(A)-binding protein 1 (PABPC1) (Fabian et al. 2009; Zekri et al. 2009; Huntzinger et al. 2010). The most recent reports show that two regions of GW182 proteins interact with PABPC1: the first contains the PAM2 motif, which is a direct binding site for PABPC1 (Jinek et al. 2010; Kozlov et al. 2010), and the second contains the M2 and C-terminal sequences, which mediate indirect binding to PABPC1 (Huntzinger et al. 2010). Furthermore, the interaction between Ago and GW182 triggers mRNA decay by inducing accelerated biphasic deadenylation mediated by Pan2-Pan3 and Ccr4-Caf1 deadenylase complexes (Chen et al. 2009).
The components of miRISC containing miRNAs, Ago and GW182 family proteins are localized in the cytoplasm, but several instances of evidence indicating their nuclear localization were reported recently (Meister et al. 2004; Hwang et al. 2007; Till et al. 2007; Rüdel et al. 2008; Politz et al. 2009; Liao et al. 2010; Taft et al. 2010; Yoo et al. 2010; Jeffries et al. 2011). In addition, several reports demonstrated that miRISC components are functional in the nucleus as well. The 7SK RNA level in the nuclear fraction is reduced by siRNAs targeting 7SK RNA in combination with Ago2 in the nucleus (Robb et al. 2005). In human cells, transcriptional gene silencing (TGS) induced by exogeneous siRNAs and miRNAs targeting to promoter-overlapping RNAs is dependent on Ago proteins (Kim et al. 2006; Han et al. 2007; Younger and Corey 2011). Furthermore, Ago2 high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP) data shows that 12% of Ago-mRNA HITS-CLIP tags are mapped to introns (Chi et al. 2009). These reports imply that miRNAs associated with Ago protein interact with RNAs in the nucleus. However, the precise mechanisms defining subcellular localization of the nuclear miRISC components containing Ago protein consolidating their nuclear function remain unresolved.
In this study, we identified a functional nuclear export signal (NES) and a nuclear localization signal (NLS) in the central region of human GW182 family protein(s) and demonstrate that TNRC6A is a nuclear–cytoplasmic shuttling protein and navigates Ago2 to translocate into the nucleus. Furthermore, miRNAs were found to be colocalized with Ago-TNRC6A in the nucleus and showed gene silencing activity.
RESULTS
Motif analyses of a member of human GW182 family proteins, TNRC6A
To investigate motifs involved in the subcellular localization of human GW182 family proteins, we generated the expression constructs encoding N-terminal myc- and GFP-tagged TNRC6A (AY035864), named pmyc-GFP-TNRC6A, and their deletion mutants (Fig. 1A). These constructs were transfected into human HeLa cells, and the subcellular localization of their GFP signals was observed under a fluorescent microscope (Fig. 1B–N) and quantified by classifying the cells into three groups: those with GFP signals (1) exclusively in the nucleus, (2) exclusively in the cytoplasm, and (3) in both the nucleus and the cytoplasm (Fig. 1Q). Myc-GFP-tagged wild-type TNRC6A protein (myc-GFP-TNRC6A-[1–1709]) was localized in cytoplasmic foci in almost all of the transfected cells (Fig. 1B), as previously reported for endogenous TNRC6A and GFP-TNRC6A (Eystathioy et al. 2002). The N-terminal fragment of myc-GFP-TNRC6A-(1–924) and the C-terminal fragment of myc-GFP-TNRC6A-(1077–1709) were distributed in both the cytoplasm and the nucleus in >90% of the transfected cells (Fig. 1E,H,Q), although myc-GFP-TNRC6A-(1–924) was distributed uniformly (Fig. 1E) and myc-GFP-TNRC6A-(1077–1709) showed dotted foci (Fig. 1H). The other fragments of myc-GFP-TNRC6A-(1–1223), -(1–1076), -(925–1709), and -(935–1709) were localized predominantly in the cytoplasm in >80% of the cells (Fig. 1C,D,F,G,Q). That the detected GFP signals were mainly derived from the transfected constructs was verified by Western blot (Supplemental Fig. S1A). These results indicate that myc-GFP-tagged TNRC6A fragments lacking the 925–1076 amino acids, which are situated between GW-III and a Q-rich domain, were distributed in both the cytoplasm and the nucleus, but the other fragments containing the 925–1076 amino acids were preferentially localized in the cytoplasm.
Identification of NES and NLS sequences in human GW182 family proteins
In contrast to the exclusive cytoplasmic localization of myc-GFP-TNRC6A-(935–1709) proteins (Fig. 1G,Q), we noticed that myc-GFP-TNRC6A-(925–1709) proteins were localized exclusively in the nucleus in about 10% of the total cells counted (Fig. 1F,Q), suggesting that 10 amino acids in the 925–934 region mediate nuclear transport. The amino acid alignment of human GW182 family proteins showed that both TNRC6B (NP_055903) and TNRC6C (NP_061869) have conserved regions with 935–1026 amino acids in TNRC6A, although the corresponding region with the 925–934 amino acids in TNRC6A was not found (Supplemental Fig. S2A). Myc-GFP-TNRC6B-(997–1084) and myc-GFP-TNRC6C-(928–1020) were localized almost exclusively in the cytoplasm (Supplemental Fig. S2D,F,H), indicating that the 997–1084 amino acids in TNRC6B and 928–1020 in TNRC6C might be involved in cytoplasmic localization.
To examine the detailed function of the region between GW-III and a Q-rich domain (Fig. 1A; Supplemental Fig. S2A), the expression patterns of the following three myc-GFP-tagged TNRC6A fragments were examined: the 925–934 fragment containing a TNRC6A-specific region, the 935–1026 fragment containing a region commonly conserved in three human GW182 family proteins, and the 1027–1076 fragment containing a region deleted in TNRC6B (Supplemental Fig. S2A). In most of the transfected cells, the myc-GFP-TNRC6A-(1027–1076) fragment was detected in the cytoplasm and the nucleus (Fig. 1M,Q). However, strong nuclear localization was observed in almost all cells transfected with myc-GFP-TNRC6A-(925–934) (Fig. 1I,Q). On the contrary, in >98% of the cells, myc-GFP-TNRC6A-(935–1026) was detected exclusively in the cytoplasm (Fig. 1K,Q). These results suggest that the 925–934 and 935–1026 regions of TNRC6A function as the NLS and NES, respectively. The 925–934 region is rich in basic amino acids (Fig. 1O), which represents a characteristic feature of the NLS. To confirm the NLS function of the 925–934 region, myc-GFP-TNRC6A-(925–934) was mutated by replacing four arginine residues with alanines (TNRC6A-(925–934)-NLS-mut) (Fig. 1O). This mutated product showed diffused distribution in the cytoplasm (Fig. 1J), which is similar to the control myc-GFP protein (Fig. 1N), indicating that the 925–934 region containing 10 amino acids is a functional NLS. The NES consensus sequences, Φ-X2-3-Φ-X2-3-Φ-X-Φ (Φ = L, I, V, F, M; X is any amino acid) (Kutay and Güttinger 2005), were found in the 951–960 region of TNRC6A, 1009–1018 of TNRC6B, and 943–952 of TNRC6C (Fig. 1P; Supplemental Fig. S2B,C). Their capabilities of nuclear export were examined by substituting four amino acid residues with alanines in these putative NES fragments (Fig. 1P; Supplemental Fig. S2B,C). All of the NES-mutated proteins (myc-GFP-TNRC6A-[935–1026]-NES-mut, myc-GFP-TNRC6B-[997–1084]-NES-mut, and myc-GFP-TNRC6C-[928–1020]-NES-mut) accumulated in the nuclei in addition to the cytoplasm in most of the cells (Fig. 1L,Q; Supplemental Fig. S2E,G,H). These results indicate that these regions are functional NESs.
Identified NLS and NES are functional in the full-length TNRC6A
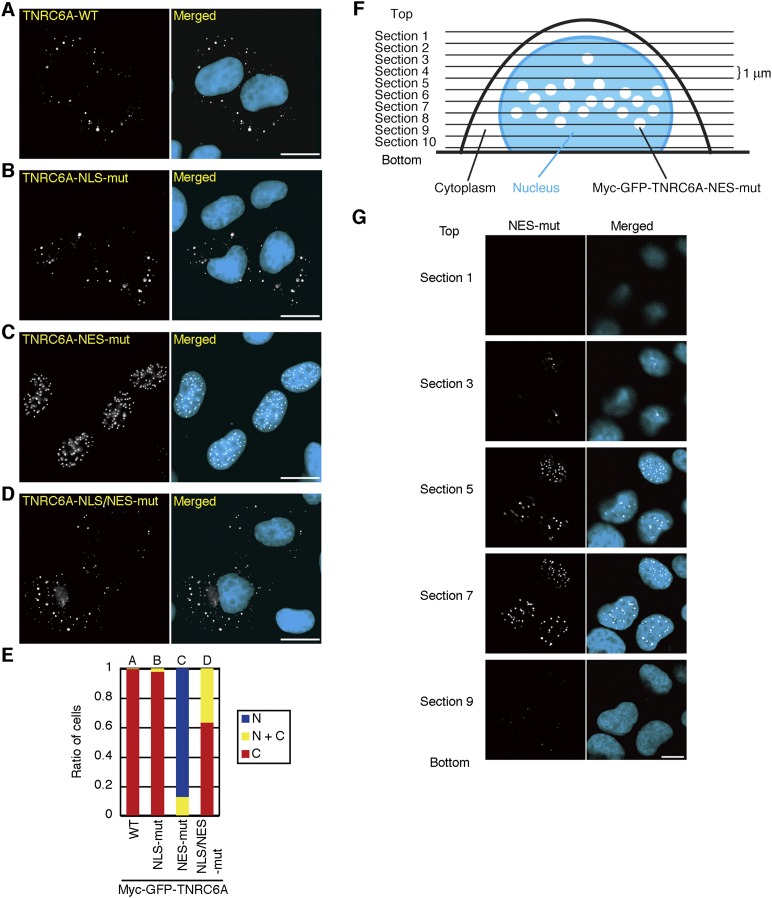
To investigate whether the identified NLS and NES is functional in the full-length context of TNRC6A, the same amino acids shown in Figure 1, O and P, were substituted in a full-length TNRC6A (myc-GFP-TNRC6A-NLS-mut and myc-GFP-TNRC6A-NES-mut). The NES mutant showed punctate localization exclusively in the nucleus (Fig. 2C,E). This was also confirmed under a confocal microscope. The NES mutant proteins were distributed in the middle area in the nucleus, and they were not associated with the nuclear membrane (Fig. 2F,G). However, the subcellular localization of the NLS mutant was not distinguished from that of wild type (Fig. 2A,B,E). An NLS mutation was then additionally introduced into the NES mutant (myc-GFP-TNRC6A-NLS/NES-mut). The product was preferentially distributed in the cytoplasm (Fig. 2D,E), demonstrating that the NLS and NES of TNRC6A are functional in the full-length context.
FIGURE 2.
Subcellular localization of NES and NLS mutants of TNRC6A in the full-length context. (A–D) Subcellular localization of the myc-GFP-TNRC6A (WT; A), myc-GFP-TNRC6A-NLS-mut (B), myc-GFP-TNRC6A-NES-mut (C), and myc-GFP-TNRC6A-NLS/NES-mut (D) proteins. Bars, 20 μm. (E) The ratio of the cells transfected with pmyc-GFP-TNRC6A and its mutants by classification according to their subcellular localization. Protein expression was verified by Western blot (Supplemental Fig. S1B). (F) The schematic diagram of observation of the cells expressing myc-GFP-TNRC6A-NES-mut by confocal microscopy. (G) Confocal images of the myc-GFP-TNRC6A-NES-mut at each section shown in F. Bar, 10 μm.
However, cells transfected with 500 ng/well of TNRC6A and its mutant expression constructs resulted in protein product expression levels approximately 5–13 times higher than the endogenous levels (data not shown). To exclude the possibility that the NLS- and NES-regulated subcellular localization of TNRC6A was due to their high expression levels, the subcellular localization was examined in cells transfected with 25 ng/well of expression constructs, representing a similar or lower level of protein expression compared to endogenous TNRC6A levels. However, GFP signals were not detected in these experiments due to the small amount of protein expressed by the transfected expression constructs (Supplemental Fig. S3, GFP). Therefore, these cells were stained using an anti-GFP antibody (Supplemental Fig. S3, anti-GFP). The subcellular localizations of TNRC6A and its NLS and/or NES mutant proteins at endogenous levels were essentially consistent with those observed under the overexpression conditions at 500 ng/well, although clear dotted foci were not detected (Supplemental Figs. S3, S4). These results suggest that subcellular localization of endogenous TNRC6A is also conducted by the NLS and NES identified in this study. Additionally, the dotted foci of TNRC6A and its mutant proteins in both cytoplasm and nucleus might be formed when they are expressed excessively.
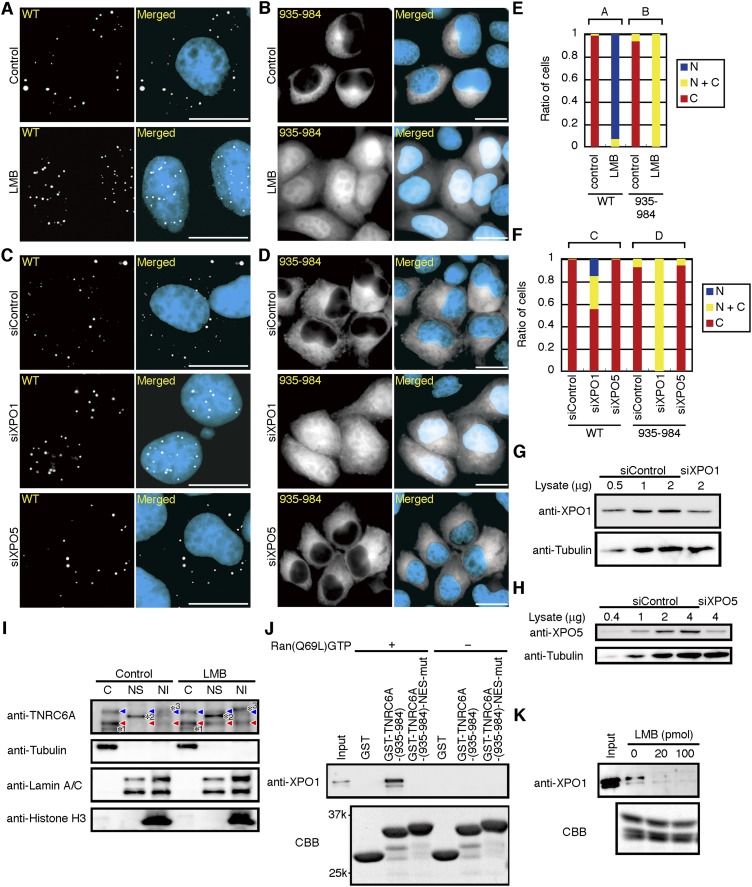
TNRC6A is exported from the nucleus by Exportin 1
Because Exportin 1 is an export receptor for various NESs (Kutay and Güttinger 2005), we investigated whether TNRC6A is exported from the nucleus by Exportin 1. Following transfection of the plasmids encoding myc-GFP-TNRC6A or its deletion mutants into HeLa cells, the cells were treated with Leptomycin B (LMB), a specific inhibitor of Exportin 1-mediated protein export (Kudo et al. 1999). Four h after LMB treatment, most of the dotted GFP signals of wild-type myc-GFP-TNRC6A were detected exclusively in the nucleus (Fig. 3A,E). Nuclear translocation of a fragment containing NES, myc-GFP-TNRC6A-(935–984), was also observed by LMB treatment (Fig. 3B,E). However, the signals in the cytoplasm remained weak, probably because this fragment does not contain the NLS sequence.
FIGURE 3.
Nuclear export of TNRC6A by Exportin 1. (A–D) Fluorescent images of HeLa cells transfected with pmyc-GFP-TNRC6A (WT; A,C) or pmyc-GFP-TNRC6A-(935–984) (B,D), treated with LMB (A,B), or siXPO1 and siXPO5 (C,D). Bars, 20 μm. (E,F) The ratio of cells expressing GFP exclusively in the nucleus (N, blue), cytoplasm (C, red), or both (N+C, yellow). A and B (E), and C and D (F) at the upper side indicate the results shown in A and B, and C and D, respectively. (G,H) Western blot of Exportin 1 (G) or Exportin 5 (H) in HeLa cells transfected with siControl and siXPO1, or siControl and siXPO5, respectively. Anti-tubulin antibody was used as a loading control. (I) HeLa cells treated with and without LMB for 8 h were harvested, and cell fractionation was performed. Ten μg of the cytoplasmic fraction (C), 5 μg of the nuclear soluble fraction (NS), or 10% of the nuclear insoluble fraction (NI) were analyzed by Western blot using an anti-TNRC6A antibody. Blue and red arrowheads indicate the positions corresponding to ∼210-kDa TNRC6A (NP_055309) and ∼182-kDa TNRC6A (AAK62026) proteins, respectively. Anti-tubulin, anti-Lamin A/C, and anti-Histone H3 antibodies were used as a cytoplasmic marker, a nuclear soluble and insoluble marker, and a nuclear insoluble marker, respectively. Asterisks (*1, *2, *3) indicate nonspecific bands. (J) GST pulldown assay of Exportin 1 and TNRC6A fragments. Equal amounts of His-GST, His-GST-TNRC6A-(935–984), or His-GST-TNRC6A (935–984)-NES-mut were immobilized on glutathione-sepharose beads and incubated with recombinant Exportin 1 with or without Ran(Q69L)GTP. Bound fractions were analyzed by Western blot using an anti-XPO1 antibody. The lower panel is the pattern of Coomassie brilliant blue (CBB) staining, which indicates that each protein is similarly immobilized. (K) GST-TNRC6A-(935–984) was immobilized on glutathione-sepharose beads and incubated with the indicated amount of LMB treated-Exportin 1 in the presence of Ran(Q69L)GTP. Bound fractions were analyzed by Western blot using anti-XPO1. The lower panel shows the CBB staining pattern.
To confirm Exportin 1-dependent cytoplasmic transport of TNRC6A, an RNAi experiment targeting Exportin 1 was conducted. The siRNA against Exportin 1 (siXPO1) efficiently reduced the expression of Exportin 1 protein (Fig. 3G). Following the transfection of siXPO1 with the plasmids encoding myc-GFP-TNRC6A or myc-GFP-TNRC6A-(935–984) into HeLa cells, notable nuclear transport of myc-GFP-TNRC6A and myc-GFP-TNRC6A-(935–984) proteins was observed in >40% and ∼100% of the cells, respectively (Fig. 3C,D,F). Another export receptor, Exportin 5, has been shown to be involved in the nuclear export of pre-miRNA (Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004). Transfection of siXPO5 showed no significant change in the subcellular localization of myc-GFP-TNRC6A or myc-GFP-TNRC6A-(935–984) (Fig. 3C,D,F), although Exportin 5 was efficiently knocked down (Fig. 3H). Thus, Exportin 1 might be a specific mediator of TNRC6A export from the nucleus to the cytoplasm.
This result was further confirmed for endogenous TNRC6A. Because the antibodies against endogenous TNRC6A protein tested so far were not available for immunofluorescence experiments (probably due to a low expression level of endogenous TNRC6A protein or for technical reasons), we performed cell fractionation experiments followed by Western blot analysis to determine the subcellular localization of endogenous TNRC6A protein. Following LMB treatment, the amount of endogenous TNRC6A was clearly increased in the nuclear soluble and insoluble fractions (Fig. 3I). This result strongly suggests that endogenous TNRC6A is also exported from the nucleus by Exportin 1, as in the case with exogenous myc-GFP-TNRC6A protein.
Direct binding of Exportin 1 to the NES of TNRC6A was examined by a glutathione S-transferase (GST) pulldown experiment using a recombinant His- and GST-tagged NES fragment containing 935–984 amino acids (His-GST-TNRC6A-[935–984]). Control His-GST, His-GST-TNRC6A-(935–984), or His-GST-TNRC6A-(935–984)-NES-mut was immobilized on glutathione-sepharose, and the purified His-tagged Exportin 1 (His-XPO1) was added in the presence or absence of His-tagged Ran(Q69L), a GTPase-deficient Ran mutant locked in the GTP-bound state, to stimulate binding of the NES to Exportin 1 (Fornerod et al. 1997). His-XPO1 bound to His-GST-TNRC6A-(935–984) in the presence of His-Ran(Q69L)GTP but not to that with the NES mutation (Fig. 3J). This indicates that Exportin 1 binds the NES of TNRC6A. Furthermore, binding of His-XPO1 to the NES fragment immobilized on the glutathione-sepharose was eliminated by the pretreatment of His-XPO1 with LMB (Fig. 3K), which binds directly to Exportin 1 and inhibits its function (Kudo et al. 1999). These results revealed that TNRC6A is exported from the nucleus by direct binding to Exportin 1 in a RanGTP-dependent manner.
TNRC6A has two strong Ago2 binding motifs and a weak one
Human TNRC6A has been shown to interact with Ago proteins by using its three Ago-interacting motifs (Eulalio et al. 2009a; Takimoto et al. 2009). We confirmed that TNRC6A has three GW-repeated Ago-binding motifs, named as GW-I (amino acid residues 457–532), -II (716–772), and -III (810–924) in this study (Supplemental Fig. S5A,B). These regions overlapped with the previously reported regions and showed high homology with those involved in TNRC6B and TNRC6C (Supplemental Fig. S5C).
To confirm that GW-I, -II, and -III are functional Ago2-binding sites in a full-length TNRC6A, pmyc-GFP-TNRC6A expression constructs lacking each of or combinations of GW-I, -II, and -III motifs (pmyc-GFP-TNRC6A-del.GW-I, -II, -III, -II+III, -I+III, -I+II, or -I+II+III) were generated. These constructs were transfected into HeLa cells, and binding of their products with endogenous Ago2 was examined by immunoprecipitation with an anti-GFP antibody. The results of Western blot showed that little or no Ago2 binding with myc-GFP-TNRC6A-del.GW-I+II+III was observed, indicating that the three GW motifs are principal Ago2 binding sites in a full-length context (Supplemental Fig. S6). Myc-GFP-TNRC6A-del.GW-II+III, -I+III, and -I+II are considered to bind Ago2 by the GW-I, -II, and -III motifs, respectively. Endogenous Ago2 bound to myc-GFP-TNRC6A-del.GW-II+III and -I+III in clearly detectable levels, but myc-GFP-TNRC6A-del.GW-I+II showed a weak and negligible level of Ago2 interaction (Supplemental Fig. S6). These results indicate that GW-I and -II are strong Ago2-binding motifs with similar binding activities, but GW-III is a weak one. Myc-GFP-TNRC6A-del.GW-I, -II, and -III are considered to bind Ago2 using two of three Ago2 binding motifs, GW-II+III, -I+III, and -I+II motifs, respectively. However, their binding activities with Ago2 were similar and indistinguishable from those of myc-GFP-TNRC6A-del.GW-II+III, -I+III and wild-type myc-GFP-TNRC6A (Supplemental Fig. S6), suggesting that endogenous Ago2 binds to either of GW-I or -II motif in a full-length TNRC6A with similar binding affinity in HeLa cells.
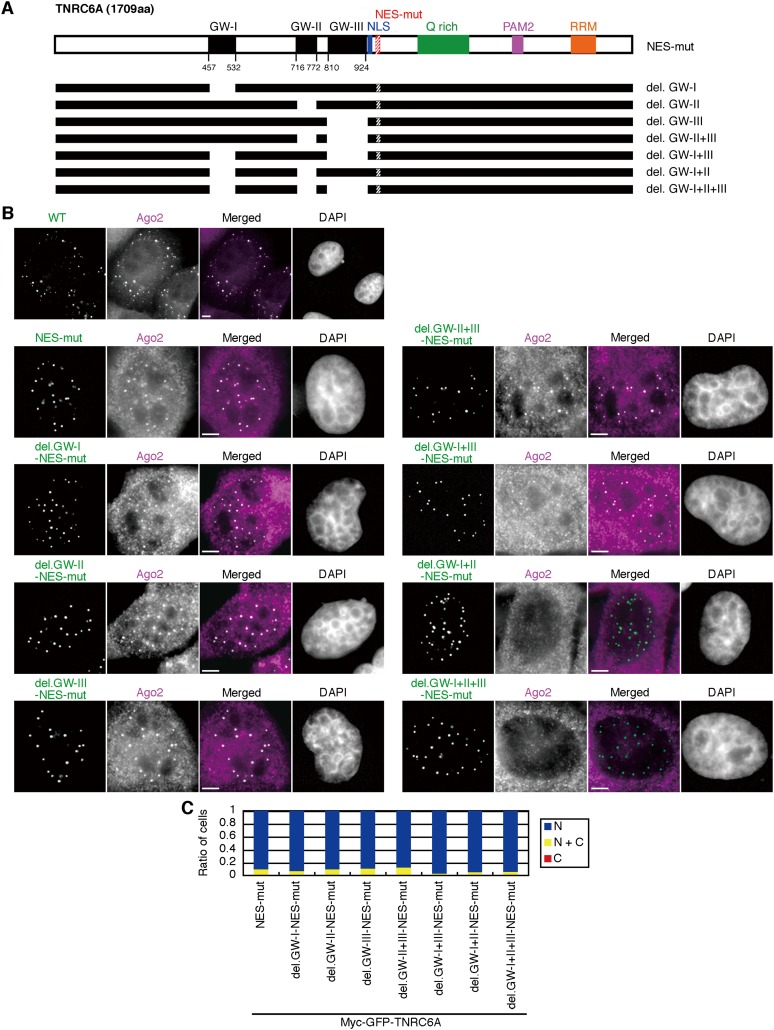
TNRC6A translocates into the nucleus in an Ago2-independent manner
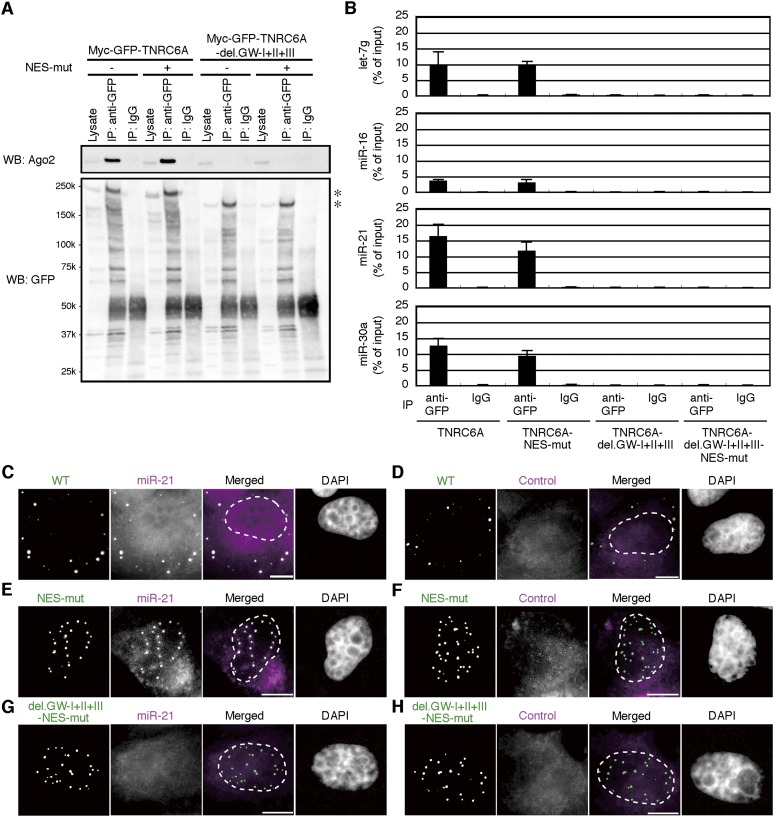
We examined whether TNRC6A localized in the nucleus can interact with Ago2 by detecting endogenous Ago2 using anti-Ago2 antibody. The endogenous Ago2 signals were detected slightly, if at all, by anti-Ago2 antibody in the nucleus of normal HeLa cells (Supplemental Fig. S7A). These signals were confirmed as Ago2 signals, since these signals were greatly reduced by Ago2 knockdown (Supplemental Fig. S7B,C). In the cells transfected with the wild-type TNRC6A expression construct, GFP signals of myc-GFP-TNRC6A were mainly observed in the cytoplasmic foci where endogenous Ago2 was clearly colocalized (Fig. 4B). Following the transfection with pmyc-GFP-TNRC6A-NES-mut, noticeable Ago2 signals emerged in both the nucleus and the cytoplasm, and myc-GFP-TNRC6A-NES-mut and Ago2 were colocalized well in nucleoplasmic foci (Fig. 4B). A similar colocalization of TNRC6A and Ago2 in the nucleus was observed in the cells transfected with wild-type pmyc-GFP-TNRC6A followed by LMB treatment (Supplemental Fig. S7D). Then, Ago2-interacting sites in TNRC6A in the nucleus were examined using NES-mutated TNRC6A expression constructs lacking either GW-I, -II, or -III motifs, or combinations of them (Fig. 4A). All the NES-mutated TNRC6As were localized exclusively in the nucleus in about 90% of cells (Fig. 4B,C). However, the subcellular localization of Ago2 was different according to the deleted GW motif(s) in TNRC6A. When myc-GFP-TNRC6A-del.GW-I, -II, -III, -II+III, or -I+III with NES mutation were expressed in the cells, no apparent change in the nuclear localization of endogenous Ago2 was observed (Fig. 4B). However, when pmyc-GFP-TNRC6A-del.GW-I+II+III-NES-mut lacking all functional GW motifs was transfected, Ago2 was not colocalized with TNRC6A in the nucleus (Fig. 4B). An almost identical result was obtained when myc-GFP-TNRC6A-del.GW-I+II-NES-mut was used (Fig. 4B), probably due to weak Ago2-binding ability of GW-III. These results demonstrate that TNRC6A can actively translocate into the nucleus regardless of the interaction of Ago2.
FIGURE 4.
Nuclear transport of TNRC6A is Ago2-independent. (A) Schematic representation of myc-GFP-TNRC6A-NES-mut with GW motif deletions. Note that NES regions were mutated. (B) Fluorescent images of HeLa cells transfected with the expression constructs of myc-GFP-TNRC6A (WT) or myc-GFP-TNRC6A-NES-mut with GW motif deletions and stained by an anti-Ago2 antibody, followed by Cy5-conjugated anti-mouse IgG. Fluorescent images are shown, from the left: GFP signals of TNRC6A fragments; Cy5 signals of the second antibody against anti-Ago2 antibody; the merged images of GFP and Cy5, in which GFP is shown in green, the Cy5 in magenta, and the overlapped GFP/Cy5 in white; and DAPI signals. Bars, 5 μm. Protein expression was verified by Western blot (Supplemental Fig. S1C). (C) The ratio of cells expressing GFP exclusively in the nucleus (N, blue), cytoplasm (C, red), or both (N+C, yellow).
Next, we examined the translocation of Ago2 protein in the TNRC6A knockdown cells with or without LMB treatment (Supplemental Fig. S8A,B), as quantitatively represented in Supplemental Figure S8C. In cells transfected with siControl, the endogenous Ago2 signals were apparently translocated into the nucleus in the presence of LMB treatment. However, the amount of Ago2 signals translocated into the nucleus in the LMB-treated TNRC6A knockdown cells was significantly small. This result suggests that Ago2 translocation into the nucleus is dependent on TNRC6A. The small increase in Ago2 signals might have been caused by the residual endogenous TNRC6A that had not been knocked down (Supplemental Fig. S8D). Alternatively, the translocation of Ago2 might be regulated by factors other than TNRC6A.
Nuclear TNRC6A is colocalized with miRNAs
The result that TNRC6A can interact with Ago2 in the nucleus as well as in the cytoplasm raises the question of whether Ago2 associates with miRNAs in the nuclear foci. To clarify this point, pmyc-GFP-TNRC6A and its NES mutants with or without Ago2-binding motifs were transfected into HeLa cells, and immunoprecipitation with an anti-GFP antibody was carried out. Then, miRNAs in the purified RNA from the immunoprecipitates were detected by RT-PCR. As expected, both of myc-GFP-TNRC6A and myc-GFP-TNRC6A-NES-mut were immunoprecipitated with Ago2, but TNRC6A without Ago2 binding motifs, myc-GFP-TNRC6A-del.GW-I+II+III and myc-GFP-TNRC6A-del.GW-I+II+III-NES-mut were not (Fig. 5A). All of the four miRNAs examined in this study (let-7g, miR-16, miR-21, and miR-30a) were clearly detected in RNAs purified from the immunoprecipitates from the cells expressing myc-GFP-TNRC6A and myc-GFP-TNRC6A-NES-mut (Fig. 5B). However, little or none of miRNAs were detected in the immunoprecipitates from the cells expressing TNRC6A without GW motifs.
FIGURE 5.
Nuclear TNRC6A is colocalized with miRNAs. (A) Cell lysates from HeLa cells expressing myc-GFP-TNRC6A and myc-GFP-TNRC6A-del.GW-I+II+III with or without NES mutation were immunoprecipitated with an anti-GFP antibody or rabbit normal IgG. Then, the immunoprecipitates (IP) and cell lysates were analyzed by Western blot using a monoclonal anti-Ago2 antibody (upper panel) and an anti-GFP antibody (lower panel). Asterisks indicate products of myc-GFP-TNRC6A and myc-GFP-TNRC6A-del.GW-I+II+III with or without NES mutation. (B) The amount of miRNAs contained in the immunoprecipitates was quantified by real-time PCR. Data are the mean and standard deviation of three independent experiments. (C–H) HeLa cells expressing myc-GFP-TNRC6A (WT; C,D), myc-GFP-TNRC6A-NES-mut (NES-mut; E,F), or myc-GFP-TNRC6A-del.GW-I+II+III-NES-mut (del.GW-I+II+III-NES-mut; G,H) were stained by digoxigenin-labeled LNA probe targeting miR-21 (C,E,G) or control probe (D,F,H). Fluorescent images are shown, from the left: GFP signals of TNRC6A; Rhodamine signals of the antibody against miRNA probes; the merged images of GFP and Rhodamine, in which GFP is shown in green and the Rhodamine in magenta; and DAPI signals. In the merged images, the dashed lines represent nuclear outlines. Bars, 10 μm.
To confirm the nuclear localization of miRNAs, we performed an in situ hybridization analysis using a locked nucleic acid (LNA) probe specific to miR-21 (Fig. 5C–H). In HeLa cells expressing wild-type myc-GFP-TNRC6A, miR-21 signals were detected as dotted foci overlapped with GFP signals in the cytoplasm (Fig. 5C), although such apparent dotted structures of miR-21 were not detected in the cells without overexpression of myc-GFP-TNRC6A (data not shown). In contrast, no definite signals were detected by a scrambled LNA miRNA probe (Fig. 5D). In the cells expressing TNRC6A with NES mutation, miR-21 but not control LNA probe signals were observed in the nuclear foci containing GFP signals (Fig. 5E,F). Such nuclear localization of miR-21 was not observed in the cells transfected with TNRC6A without any GW motif (Fig. 5G,H). Altogether, our results indicate that miRNAs might be associated with Ago2 protein, which is tethered by TNRC6A in the nuclear foci as well as in the cytoplasmic P bodies.
miRNA-mediated gene silencing is induced by TNRC6A with NES and/or NLS mutations
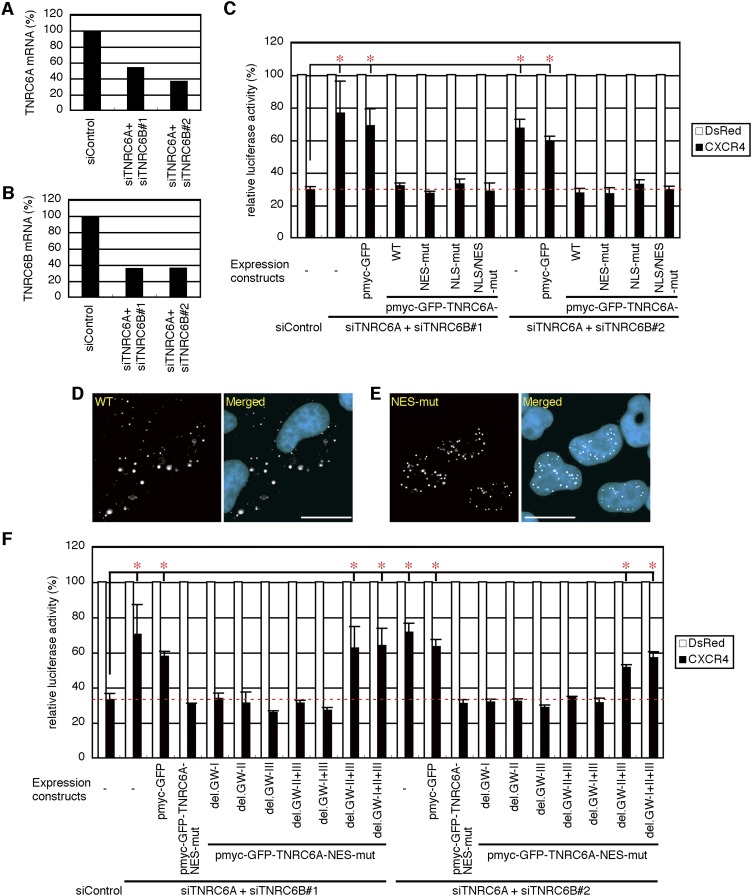
TNRC6A has been shown to be necessary for miRNA-mediated gene silencing (Liu et al. 2005). To investigate whether NES and/or NLS mutations in TNRC6A affect miRNA-mediated gene silencing, we used a CXCR4 miRNA-mediated gene silencing assay system (Doench et al. 2003). However, since TNRC6A, TNRC6B, and TNRC6C are known to be functionally redundant (Eulalio et al. 2009a), absolute mRNA quantification of TNRC6s was carried out. Endogenous TNRC6A and TNRC6B were found to be expressed abundantly in HeLa cells (160 and 112 copies per ng total RNA, respectively), but the amount of TNRC6C was very low (12 copies per ng total RNA). Then, to eliminate the effects of endogenous TNRC6A and TNRC6B, they were knocked-down by the simultaneous transfection of siRNAs against TNRC6A (siTNRC6A) and TNRC6B (siTNRC6B#1 or siTNRC6B#2) targeting their 3′ UTRs. Both combinations of siTNRC6A/siTNRC6B#1 and siTNRC6A/siTNRC6B#2 effectively reduced the expression levels of endogenous TNRC6A and TNRC6B transcripts at ∼35%–50% (Fig. 6A,B). The reduction of endogenous TNRC6A and TNRC6B remarkably repressed miRNA-mediated gene silencing activity (Fig. 6C), indicating that TNRC6s are essential for miRNA-mediated gene silencing, as shown in the previous reports (Liu et al. 2005; Eulalio et al. 2009a; Zipprich et al. 2009). Under these conditions, the introduction of myc-GFP-TNRC6A and its NES and/or NLS mutants showed substantial silencing activities (Fig. 6C). However, control myc-GFP exhibited almost no silencing activity. The protein expression of myc-GFP, myc-GFP-TNRC6A and its mutants was verified by Western blot (Supplemental Fig. S9A). Moreover, under the same conditions shown in Figure 6C, the unambiguous changes in subcellular localization of TNRC6A or TNRC6A-NES-mut were not observed (Fig. 6D,E). In addition, whereas NES-mutated TNRC6A having at least the GW-I or GW-II motif exhibited miRNA-mediated gene silencing activities, little or no silencing activities were observed by NES-mutated TNRC6A without any GW motifs or with the weakest GW-III motif (Fig. 6F; Supplemental Fig. S9B), indicating that interaction of TNRC6A with Ago2 might be essential for silencing activity in the nucleus.
FIGURE 6.
miRNA-mediated gene silencing activity of TNRC6A with NES and/or NLS mutations. (A,B) TNRC6A (A) and TNRC6B (B) mRNA levels in HeLa cells transfected with siTNRC6A/siTNRC6B#1 or siTNRC6A/siTNRC6B#2. (C) One d after the transfection with siControl, siTNRC6A/siTNRC6B#1 or siTNRC6A/siTNRC6B#2, cells were further transfected with a mixture of the following four plasmids: psiCHECK-CXCR4 target, pGL3-Control, pSUPER-CXCR4 or control pSUPER-DsRed, and myc-GFP or the indicated TNRC6A expression constructs. Two d after second transfection, luciferase activities were measured. The activities were shown as the mean and standard deviation of three or four independent experiments. P-values were determined by Student's t-test. (*) P < 0.05. (D,E) Fluorescent microscopy images of the cells expressing WT (D) or NES-mut (E) under the same conditions shown in C. WT was exclusively localized in the cytoplasm, but NES-mut was in the nucleus. Bars, 20 μm. (F) Relative luciferase activities by the transfection of NES-mutated TNRC6As lacking various combinations of GW motifs.
TNRC6A with NES mutation enhances RNAi activity against a nuclear-specific noncoding RNA, MALAT-1
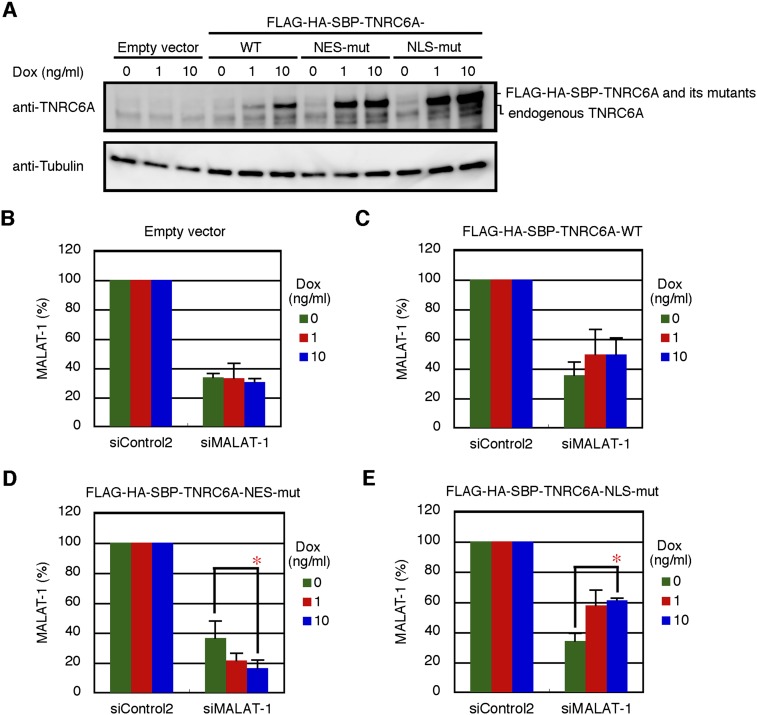
To confirm that RNA silencing in the nucleus is associated with the nuclear Ago2, navigated by TNRC6A with NES mutation, we generated TNRC6A- and TNRC6A NES mutant-inducible cells using the Flp-In T-REx system in HEK293 cells and examined the effects of their overexpression. Under control of the tetracycline-inducible promoter, expression of FLAG-HA-SBP-tagged TNRC6A (FLAG-HA-SBP-TNRC6A) or its NES mutant was induced by doxycycline (dox) treatment in HEK293 cells. Then, we examined the RNAi activity against metastasis-associated in lung adenocarcinoma transcript 1 (MALAT-1), a nuclear-specific noncoding RNA that is known to be stably retained in the nucleus (Miyagawa et al. 2012). FLAG-HA-SBP-TNRC6A and its mutants were induced by dox in a dose-dependent manner (Fig. 7A). In cells transfected with siRNA against MALAT-1 (siMALAT-1), a nearly identical level of MALAT-1 repression was observed as that found in cells transfected with either TNRC6A or its mutant protein expression constructs in the absence of dox (Fig. 7B–E). Induction of FLAG-HA-SBP-TNRC6A-NES-mut increased RNAi activity for MALAT-1 (Fig. 7D). In contrast, FLAG-HA-SBP-TNRC6A-NLS-mut decreased this activity (Fig. 7E). However, the empty vector or TNRC6A-WT did not show any remarkable effects (Fig. 7B,C). The results suggest that the overexpression of TNRC6A with NES mutation navigates many Ago2 proteins into the nucleus, resulting in the increase in RNAi activity for nuclear RNA, whereas TNRC6A with NLS mutation retains Ago2 in the cytoplasm, thereby decreasing RNAi activity in the nucleus.
FIGURE 7.
Effects of overexpression of TNRC6A and its NES or NLS mutants on RNAi activity against MALAT-1. (A) The induction of FLAG-HA-SBP-TNRC6A or its mutant proteins by the treatment of the indicated concentration of dox for 24 h in HEK293 cells stably transfected with either empty vector, FLAG-HA-SBP-TNRC6A-WT, FLAG-HA-SBP-TNRC6A-NES-mut, or FLAG-HA-SBP-TNRC6A-NLS-mut expression constructs was verified by Western blot using anti-TNRC6A antibody. Anti-tubulin antibody was used as a loading control. (B–E) Stable HEK293 cells with pcDNA5/FRT/TO/FLAG-HA-SBP (empty vector; B), and expressing FLAG-HA-SBP-TNRC6A (C), FLAG-HA-SBP-TNRC6A-NES-mut (D), and FLAG-HA-SBP-TNRC6A-NLS-mut (E) were transfected with siControl2 or siMALAT-1 and cultured in the presence of the indicated concentration of dox for 24 h. MALAT-1 RNA levels were shown as the mean and standard deviation of three independent experiments. P-values were determined by Student's t-test. (*) P < 0.05.
Colocalization of Dcp1 and p54 with cytoplasmic TNRC6A but not with nuclear TNRC6A
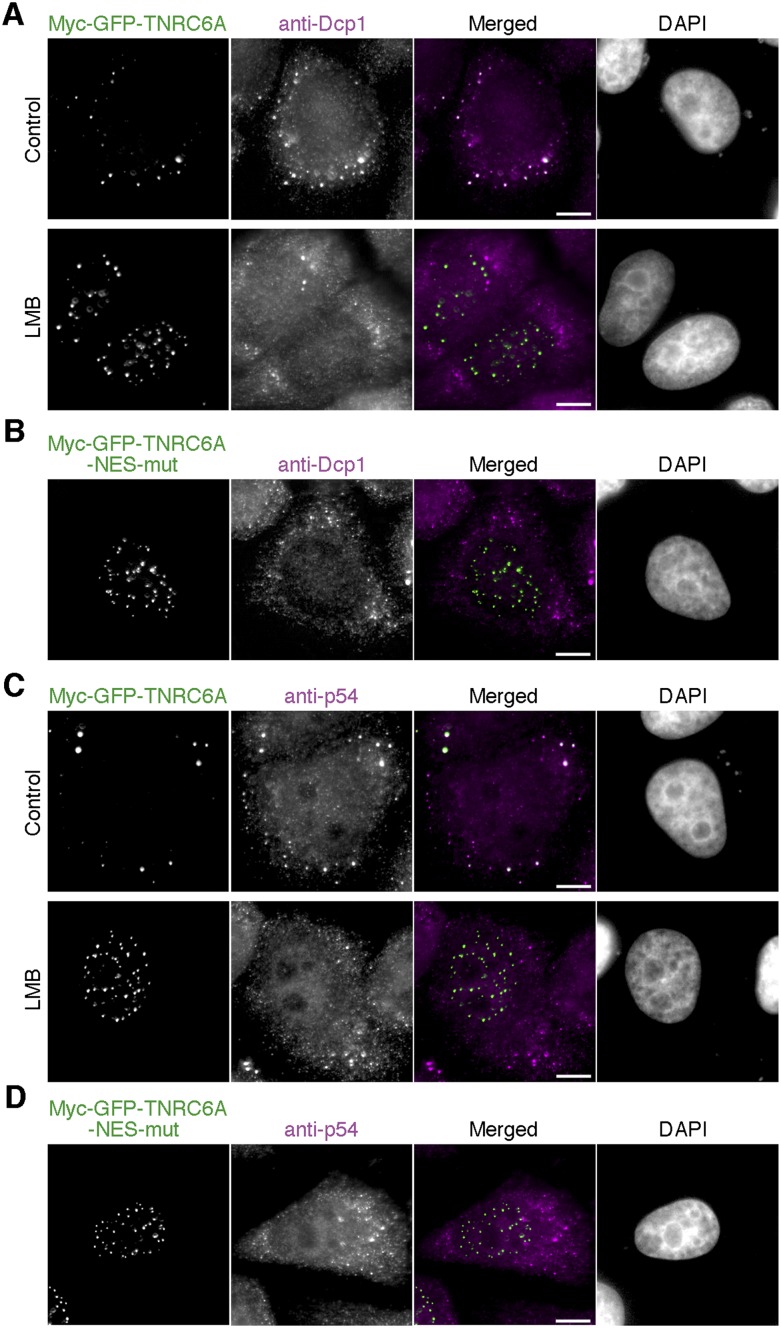
We investigated the subcellular localization of the marker proteins of the cytoplasmic P bodies Dcp1 and p54, which are known to be colocalized with TNRC6A or Ago2 in the cytoplasm (Eystathioy et al. 2003; Chu and Rana 2006). Dcp1 and p54 proteins were clearly colocalized with wild-type myc-GFP-TNRC6A in the cytoplasmic foci, but such colocalization was not observed in the nuclear foci formed by the expression of myc-GFP-TNRC6A-NES-mut or wild-type myc-GFP-TNRC6A after LMB treatment (Fig. 8). Thus, the components of cytoplasmic and nuclear foci differed from one another.
FIGURE 8.
Dcp1 and p54 were colocalized with cytoplasmic but not with nuclear TNRC6A. (A,C) HeLa cells transfected with pmyc-GFP-TNRC6A were treated with or without LMB, and stained with anti-Dcp1 (A) or anti-p54 (C) antibody. Fluorescent images are shown, from the left: GFP signals of TNRC6A; Cy5 signals of the second antibody against anti-Dcp1 (A) or anti-p54 (C) antibody; the merged images of GFP and Cy5, in which GFP is shown in green and the Cy5 in magenta; and DAPI signals. Bars, 10 μm. (B,D) HeLa cells transfected with pmyc-GFP-TNRC6A-NES-mut were stained with anti-Dcp1 (B) or anti-p54 (D) antibody. Fluorescent images are shown, from the left: GFP signals of TNRC6A; Cy5 signals of the second antibody against anti-Dcp1 (B) or anti-p54 (D) antibody; the merged images of GFP and Cy5, in which GFP is shown in green and the Cy5 in magenta; and DAPI signals. Bars, 10 μm.
DISCUSSION
In this study, we found that human TNRC6A protein is a nuclear–cytoplasmic shuttling protein, and its subcellular localization is conducted by its NES and NLS sequences. There are functional NESs in the central regions between GW-III and Q-rich domains in TNRC6A as well as in TNRC6B and TNRC6C (Supplemental Fig. S2A). These NES sequences in human TNRC6A–C are completely conserved in their mouse counterparts, MmTNRC6A–C, and their consensus sequences are also found in C. elegans AIN-1 and AIN-2, and Drosophila dGW182 (Supplemental Fig. S10), suggesting that the NES is functionally essential in diverse organisms. In human GW182 family proteins, the NES motif is included in the previously reported UBA domain (Supplemental Fig. S2A). However, because four Drosophila putative NESs in dGW182 did not overlap its UBA domain, the UBA domains of human and mouse GW182 family proteins might have an alternative function other than the NES. A recent report revealed that each of UBA domains of human TNRC6A and TNRC6C can interact with a UBA domain of mammalian hyperplastic discs protein EDD, which is an E3 ubiquitin ligase and participates in miRNA-mediated gene silencing by recruiting downstream effectors (Su et al. 2011). As shown in Supplemental Figure S2A, the UBA domain in TNRC6A is overlapped with the identified NES. The NES mutant of TNRC6A could potentially destroy the function of the UBA domain. Thus, it is not negligible that the miRNA-mediated gene silencing activity via EDD is disturbed.
TNRC6A also has a NLS in the region between GW-III and a Q-rich region (Supplemental Fig. S2A), but neither TNRC6B nor TNRC6C has NLS sequence in this corresponding region. No NLS-like region has been found in the GW182 family proteins of C. elegans or Drosophila melanogaster. Thus, the NLS might be not essential for all of the GW182 family proteins. However, Till et al. (2007) reported that GFP-TNRC6B iso3 (NP_001020014; ∼110 kDa) substantially accumulates in the nucleus by LMB treatment, indicating that TNRC6B iso3 is a nuclear–cytoplasmic shuttling protein. Since any proteins larger than ∼40 kDa require NLS sequences for their nuclear import (Davis 1995), TNRC6B or its interacting protein(s) might have a NLS. The best understood pathway of nuclear transport is conducted by a heterodimeric import receptor consisting of the β-karyopherin importin β, which mediates interactions with the nuclear pore complex, and the adaptor protein importin α, which directly binds to the NLS motif (Lange et al. 2007). Weinmann et al. (2009) reported that Importin 8 (Imp8) binds to Ago in P bodies and shuttles Ago protein to the nucleus. Therefore, it is probable that Ago translocates into the nuclei independent of TNRC6A. However, they described that, even when Imp8 was depleted, a significant portion of Ago remained in the nucleus, suggesting that alternative nuclear import pathways for Ago proteins may exist. Although we could not identify an import receptor of TNRC6A in this study, TNRC6A might be a strong candidate for an Ago-navigator protein into the nucleus by some other type of importin receptor that is able to bind to the NLS sequence.
Human GW182 family proteins have three GW motifs. Takimoto et al. (2009) demonstrated that GW182 could associate with multiple Ago proteins, but each of them can independently bind to Ago2. Our results indicate that binding activities of GW-I, -II, and -III with Ago2 were different in the full-length context (Supplemental Fig. S6). The results of nuclear translocation with Ago2 indicated that the GW-I and GW-II motifs in TNRC6A showed strong binding activities, but the GW-III motif has little or negligible activity (Fig. 4B). Furthermore, NES-mutated TNRC6A having at least GW-I and/or GW-II motif(s) showed substantial silencing activities, but the mutants lacking both the GW-I and GW-II motifs exhibited no or little silencing activities (Fig. 6F). Thus, at least one of the GW-I or GW-II motifs in TNRC6A might be functionally sufficient for nuclear–cytoplasmic shuttling of endogenous Ago2 and gene silencing activity in the nucleus.
We investigated the effects of NES and/or NLS mutations in TNRC6A using the CXCR4 miRNA-mediated gene silencing assay system (Fig. 6). Under knockdown conditions of the endogenous GW182 family proteins, the expression of NES-mutated TNRC6A showed adequate silencing activity, and this activity was eliminated when Ago2-binding motifs were deleted. The result strongly suggested that TNRC6A exerts miRNA-mediated gene silencing activity with Ago2 in the nucleus. However, because a small amount of NES-mutated TNRC6A protein was observed in the cytoplasm (Supplemental Fig. S4C), the possibility that the NES-mutated TNRC6A exhibits silencing activity in the cytoplasm could not be excluded. Therefore, we carried out an RNAi assay targeting nuclear-specific MALAT-1 RNA (Fig. 7). RNAi activity against MALAT-1 was increased by the induction of NES-mutated TNRC6A expression, although little or no effect was observed by the induction of wild-type TNRC6A proteins. The result suggests that the Ago2 protein is navigated into the nucleus by binding with TNRC6A to enhance the RNAi activity in the nucleus.
Under the overexpression condition of wild-type TNRC6A, clear dotted foci were observed in the nucleus by Exportin 1 knockdown or LMB treatment (Fig. 3). Furthermore, not only TNRC6A but also TNRC6C was proved to produce nuclear foci, and both proteins were shown to be colocalized in the same nuclear foci (L Badertscher and W Filipowicz, pers. comm.). As shown previously (Eystathioy et al. 2003; Chu and Rana 2006), we confirmed that the subcellular localization of the P-body marker proteins Dcp1 and p54 were colocalized with the wild-type myc-GFP-TNRC6A in the cytoplasmic foci, but such colocalization was not observed in the nuclear foci formed by the expression of myc-GFP-TNRC6A-NES-mut or wild-type myc-GFP-TNRC6A after LMB treatment (Fig. 8). Thus, we revealed that the components of cytoplasmic and nuclear foci were different from one another, suggesting that the miRNA-mediated silencing mechanism in the nucleus might not be parallel to that in the cytoplasm, although the possibility that the nuclear foci are formed due to the overexpression of TNRC6A could not be excluded. So far, conceivable silencing mechanisms in the nucleus are as follows: (1) Target mRNAs are tethered by miRNA-Ago-TNRC6 complex in the nucleus to avoid translocation into cytoplasm for translation; (2) TNRC6A destabilizes poly(A) RNAs in the nucleus, since the components involved in miRNA-mediated deadenylation, PABPC1, Ccr4-Caf1, and Pan2-Pan3 (Chen et al. 2009; Fabian et al. 2009), were shown to be nuclear–cytoplasmic shuttling proteins (Afonina et al. 1998; Yamashita et al. 2005); and (3) miRNA associates with promoter RNAs to induce miRNA-mediated TGS or heterochromatin formation (Kim et al. 2008; Younger and Corey 2011).
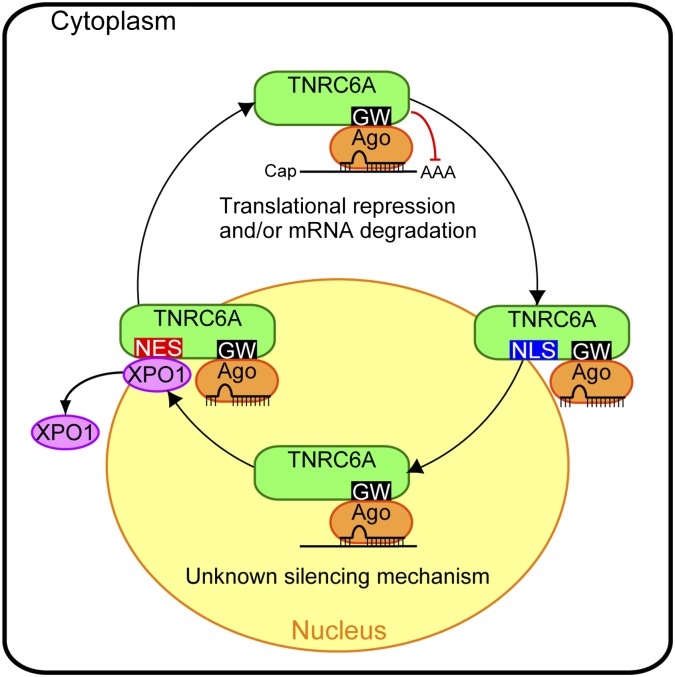
In summary, we found that: (1) nuclear–cytoplasmic transport of human TNRC6A is conducted by its NLS and NES (Fig. 9); (2) its nuclear export is mediated by Exportin 1; (3) TNRC6A can translocate into the nucleus with or without Ago2 interaction; (4) in the nucleus, miRNAs are found to be colocalized with TNRC6A which interacts with Ago2; and (5) small RNA-containing Ago2 protein navigated into the nucleus by TNRC6A might cleave MALAT-1 RNA or hold target RNAs in the nucleus. Our results proposed the possibility that GW182 family proteins could regulate miRNA-mediated gene silencing in the nucleus as well as in the cytoplasm by navigating Ago proteins, although the silencing mechanism in the nucleus remains unknown.
FIGURE 9.
TNRC6A regulates miRNA-mediated gene silencing in the nucleus and cytoplasm by navigating Ago protein. TNRC6A translocates from the cytoplasm to the nucleus via its own NLS sequence. Its nuclear export is mediated by Exportin 1 (XPO1), which directly binds to its NES sequence. In the cytoplasm, interaction between Ago and TNRC6A triggers translational repression and/or mRNA degradation by a known mechanism (see Introduction). In the nucleus, the miRNA-Ago-TNRC6A complex might induce miRNA-mediated gene silencing targeting nuclear RNA by an unknown mechanism. See the text for details.
MATERIALS AND METHODS
Plasmid construction
The expression plasmids of TNRC6A, B, and C and their derivatives, Exportin 1 and Ran(Q69L) were constructed as follows: pIRESneo-FLAG/HA-Ago2 (Meister et al. 2004), kindly provided by Dr. Thomas Tuschl through Addgene, was used as pFLAG/HA-Ago2 in this study. All PCR primers used for plasmid construction are shown in Supplemental Table S1.
Myc-GFP-TNRC6A expression construct, pmyc-GFP-TNRC6A: A fragment encoding EGFP was amplified using pEGFP-N3 (Clontech) as a template by PCR with a forward primer containing the myc-tag and a reverse primer. An amplified fragment was digested with NheI and HindIII and inserted into the NheI/HindIII-digested pcDNA3.1-myc-Dicer (Provost et al. 2002). A 5130-bp fragment encoding TNRC6A, which was amplified using pCMV-SPORT/FLAG-GW182 (Wakiyama et al. 2007) by PCR, was digested with HindIII and XbaI and inserted into pcDNA3.1-myc-GFP-Dicer by two-step ligation: (1) A 1994-bp fragment was inserted into pcDNA3.1-myc-GFP-Dicer digested with HindIII and XbaI; and (2) a 3136-bp fragment was inserted into the HindIII site. The orientation of the insert was determined by nucleotide sequence analysis.
Myc-GFP-TNRC6B expression construct, pmyc-GFP-TNRC6B: Fragment encoding TNRC6B was amplified using cDNA derived from HEK293 cells. A 5172-bp fragment of TNRC6B was digested with HindIII and XbaI and inserted into the HindIII/XbaI-digested pcDNA3.1-myc-GFP-Dicer.
Myc-GFP-TNRC6C expression construct, pmyc-GFP-TNRC6C: A 5073-bp fragment of TNRC6C amplified using cDNA from HEK293 cells was digested with HindIII and XbaI, and inserted into pcDNA3.1-myc-GFP-Dicer by two-step ligation: (1) A 3202-bp fragment was inserted into pcDNA3.1-myc-GFP-Dicer digested with HindIII and XbaI; and (2) a 1871-bp fragment was inserted into the XbaI site. Deleted constructs of TNRC6A, B, and C were amplified using pmyc-GFP-TNRC6A, B, and C, respectively, as templates. They were digested with HindIII and XbaI, and inserted into the HindIII/XbaI-digested pcDNA3.1-myc-GFP-Dicer. Amino acid-substituted constructs and several deletion constructs of TNRC6A, B, and C were generated using the corresponding pmyc-GFP-TNRC6A, B, and C by site-directed mutagenesis using PCR.
pET-Exportin 1 and pET-Ran(Q69L): A fragment encoding Exportin 1 or Ran was amplified by PCR using cDNA derived from total RNA of HEK293 cells. They were then digested with NheI and NotI, and inserted into NheI/NotI sites of pET28a (Novagen). pET-Ran(Q69L) was generated from pET-Ran by site-directed mutagenesis using PCR.
pET-GST, pET-GST-TNRC6A-(935–984) and pET-GST-TNRC6A-(935–984)-NES-mut: A fragment encoding GST was amplified by PCR using pGEX-6P2 (GE Healthcare) as a template. It was digested with NheI and HindIII, and inserted into the NheI/HindIII sites of pET28a. A fragment encoding GFP-TNRC6A-(935–984) or TNRC6A-(935–984)-NES-mut was amplified from pmyc-GFP-TNRC6A derivatives. They were digested with NheI and NotI, and inserted into the NheI/NotI sites of pET28a. A fragment encoding GST was amplified by PCR using pGEX-6P2. It was then digested with NheI and HindIII, and inserted into pET-GFP-TNRC6A-(935–984) or pET-GFP-TNRC6A-(935–984)-NES-mut digested with NheI/HindIII.
pcDNA5/FRT/TO/FLAG-HA-SBP-TNRC6A and its mutant expression constructs: A fragment encoding TNRC6A, which was amplified from pCMV-SPORT/FLAG-GW182 by PCR, was cloned into pcDNA5/FRT/TO/FLAG-HA-SBP (Okada-Katsuhata et al. 2012) using an In-Fusion HD Cloning Kit (Clontech). Amino acid-substituted constructs were generated using pcDNA5/FRT/TO/FLAG-HA-SBP-TNRC6A by site-directed mutagenesis using PCR.
The siRNA expression constructs: The expression constructs of chemokine receptor CXCR4 siRNA (Doench et al. 2003) or control DsRed siRNA, named pSUPER-CXCR4 or pSUPER-DsRed, were constructed by inserting annealed synthetic oligonucleotides consisting of two strands of single-stranded 67-mer oligonucleotides (Supplemental Table S1) into BamHI/HindIII sites in the pSUPER.retro.Puro vector (OligoEngine). From the inserted oligonucleotides, the transcript was encoded in the following order: (1) a 21-nt siRNA guide strand; (2) a human miR-23a loop; and (3) a 19-nt passenger strand of the identical siRNA lacking a 3′ overhang.
The siRNA target-containing construct: The plasmid expressing the CXCR4 siRNA target site (Doench et al. 2003) was constructed by inserting oligonucleotides with a central bulge (Supplemental Table S1) into the XhoI/EcoRI sites in the 3′ UTR of psiCHECK-1 (Promega), which encodes the Renilla luciferase gene (psiCHECK-CXCR4 target).
Cell culture and LMB treatment
HeLa and HEK293 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen) with 10% fetal bovine serum (FBS; Sigma) at 37°C with 5% CO2. Transfection was conducted using Lipofectamine 2000 (Invitrogen). For LMB (Calbiochem) treatment, 2 d after transfection, HeLa cells were washed twice with DMEM with 10% FBS and incubated in DMEM with 10% FBS containing 10 ng/mL LMB or the same volume of solvent for 4 h, except for Figure 3I.
Fluorescence microscopy
HeLa cells were inoculated on a glass coverslip in a well of a 12-well culture plate at 1 × 105 cells/mL/well at 1 d before transfection. Various GFP-tagged TNRC6A expression constructs (500 ng/well) were transfected into the cells. At 2 d after transfection, cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde at room temperature for 15 min. The cells were mounted in ProLong Gold antifade reagent with DAPI (Invitrogen). GFP and DAPI signals were observed under a Zeiss Axiovert 200 fluorescence microscope, using a 63× objective lens, or Olympus Fluoview FV1000 confocal microscopy system, using a 60× objective lens. In confocal imaging, optical sections were taken at 1-μm intervals. Quantification of fluorescence microscopy images was performed using Image J software (NIH). The antibody staining was performed as previously reported (Nishi and Saigo 2007). Briefly, the fixed cells permeabilized with PBS containing 0.2% Triton X-100 were incubated with the first antibody (1:200–400 dilution) in PBS, and this was followed by treatment with the second antibody (1:400 dilution) in PBS. They were mounted with DAPI and observed under fluorescence microscope. In subcellular localization assays, the numbers of counted cells were listed in Supplemental Table S2. The first antibodies used for staining the cells were as follows: Living Colors Full-Length Aequorea victoria polyclonal GFP antibody (Clontech), anti-human AGO2 monoclonal antibody, 4G8 (Wako), mouse monoclonal antibody to Dcp1a (Abcam; ab57654), and anti-Rck/p54 polyclonal antibody (MBL). The second antibodies were as follows: Cy5-conjugated goat anti-rabbit IgG (Jackson Immunoresearch Laboratories), and goat anti-mouse IgG conjugated with Cy5 (Amersham Pharmacia Biotech).
Western blot
Cells were lysed with ice-cold lysis buffer (10 mM Hepes-NaOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 140 mM NaCl, 0.5 mM DTT, 1 mM EDTA, 1 mM Na3VO4, 10 mM NaF, 0.5% NP40, and 1× complete protease inhibitor cocktail [Roche]) on ice for 30 min and centrifuged. The supernatants were mixed with same volume of 2× SDS-PAGE sample buffer and boiled for 5 min. Except for Western blot of TNRC6A, samples were separated by SDS-PAGE, and the gels were transferred to BioTrace PVDF membrane (Pall Corp.) with an iBlot Dry Blotting System (Invitrogen). The membranes were blocked for 1 h in TBST (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.2% Triton X-100 or 0.1% Tween-20) supplemented with 5% skim milk (Difco) and washed once with TBST. Following incubation with the first antibody (1:200–2000 dilution) in TBST containing 1% liquid block (GE Healthcare) at 4°C overnight, the membranes were washed three times with TBST and treated with the second antibody (1:10,000 dilution) in TBST containing 1% liquid block. After being washed three times with TBST, the membranes were incubated with CDP-Star detection reagent (GE Healthcare), and signals were detected by an LAS3000 imaging system (FUJIFILM). For Western blot of TNRC6A, samples were separated by SDS-PAGE, and the gels were transferred to PVDF membranes using a Trans-Blot Turbo Transfer System (Bio-Rad). The membranes were blocked for 1 h in TBST (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.1% Tween-20) supplemented with 2% ECL advance blocking agent (GE Healthcare). Following incubation with the first antibody (1:2000 dilution) in TBST containing 2% ECL advance blocking agent at 4°C overnight, the membranes were washed three times with TBST and treated with the second antibody (1:10,000 dilution) in TBST containing 2% ECL advance blocking agent. After being washed three times with TBST, the membranes were reacted with an ECL advance Western blotting detection kit (GE Healthcare), and signals were detected by an LAS3000 imaging system.
The first antibodies used for Western blot were as follows: polyclonal anti-Ago2 antibody (Upstate), myc-tag rabbit antibody (Cell Signaling Technology), anti-human AGO2 monoclonal antibody, 4G8, Living Colors Full-Length Aequorea victoria polyclonal GFP antibody, anti-CRM1 (Exportin 1) rabbit polyclonal IgG, H-300 (Santa Cruz Biotechnology), anti-Exportin 5 rabbit polyclonal IgG, H-300 (Santa Cruz Biotechnology), Monoclonal Anti-α Tubulin antibody (ICN/CAPPEL Biomedicals), mouse anti-Lamin A/C monoclonal antibody (BD Transduction Laboratories), Anti-Histone H3 antibody (Abcam; ab1791), and Rabbit anti-GW182 (TNRC6A) Antibody (Bethyl; A302-329A). The second antibodies were as follows: alkaline phosphatase-conjugated goat affinity purified antibody to rabbit IgG (ICN/CAPPEL Biomedicals), alkaline phosphatase-conjugated goat affinity purified antibody to mouse IgG (ICN/CAPPEL Biomedicals), and ECL Anti-rabbit IgG, Horseradish Peroxidase linked whole antibody (GE Healthcare).
Knockdown of Exportin 1 and Exportin 5
The siRNAs against human Exportin 1 (siXPO1) and Exportin 5 (siXPO5) were designed by our functional siRNA selection algorithm (Ui-Tei et al. 2004, 2008) using siDirect2.0 (Naito et al. 2009) and purchased from Sigma. Their sequences are described in Supplemental Table S3. HeLa cells inoculated on a glass coverslip at 1 × 105 cells/mL/well in wells of 12-well culture plate were transfected twice at 24-h intervals with 50 nM siRNA. At the second transfection, 500 ng/well pmyc-GFP-TNRC6A or its mutant constructs were simultaneously transfected with each siRNA. One day after the second transfection, cells were fixed and observed under the fluorescent microscope or harvested for Western blot.
Cell fractionation
Nuclear soluble and cytoplasmic fractions were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagent (Thermo scientific) according to the manufacturer's recommendation. The remaining pellets were sonicated in 2× SDS sample buffer and used as nuclear insoluble fractions.
Purification of recombinant proteins
His-GST, His-GST-TNRC6A-(935–984), His-GST-TNRC6A-(935–984)-NES-mut, His-Exportin 1, and His-Ran(Q69L) were expressed from the pET constructs (see “Plasmid construction” section) in Escherichia coli Rosetta (DE3) pLysS (Novagen). Cells were first grown in L Broth with 50 μg/mL kanamycin and 34 μg/mL chloramphenicol at 37°C overnight. The culture was diluted 1:20 in the same medium and grown to A600 = 0.6–1.0 at room temperature, and isopropyl β-D-thiogalactoside was added to a final concentration of 0.5 mM. After an additional 12 h of culture, the cells were collected by centrifugation at 4°C. The pellets were frozen and stored at −20°C. Cells were resuspended in lysis buffer (50 mM sodium phosphate [pH 8.0], 10 mM imidazole, and 300 mM NaCl) and disrupted by sonication. The lysate was centrifuged at 12,000g for 1 h at 4°C. The supernatant was loaded onto a Ni-NTA column (Qiagen) equilibrated with lysis buffer. It was washed with lysis buffer and then wash buffer (50 mM sodium phosphate [pH 8.0], 20 mM imidazole, and 300 mM NaCl), and finally eluted with the elution buffer (50 mM sodium phosphate [pH 8.0], 250 mM imidazole, and 300 mM NaCl). Eluted proteins were dialyzed against PBS containing 10% glycerol on a PD10 column (GE Healthcare). Purified protein was stored at −80°C.
In vitro binding assay
His-GST, His-GST-TNRC6A-(935–984), His-GST-TNRC6A-(935–984)-NES-mut, His-Exportin 1, and His-Ran-GTP(Q69L) were prepared as described in the “Purification of recombinant proteins” section. A total of 600 pmol GST fusion proteins were bound to 20 μL of glutathione-Sepharose 4B (GE Healthcare) in PBS containing 10% glycerol for 2 h at 4°C. They were washed three times with PBS containing 10% glycerol, and 500 μL PBS containing 20 pmol Exportin 1, 190 pmol Ran(Q69L), 10 nmol GTP, and 10% glycerol was added. After incubation for 2 h at 4°C, they were washed three times with PBS containing 500 mM NaCl, 2 μM GTP, 0.2% Triton X-100, and 10% glycerol and resuspended in 2× SDS sample buffer. Bound proteins were eluted by boiling the beads for 5 min and separated by SDS-PAGE. To detect the inhibitory effect of LMB on Exportin 1 binding to TNRC6A NES, 20 pmol Exportin 1 protein was treated with 20 or 100 pmol LMB or the solvent in PBS for 1 h on ice before the binding assay.
Immunoprecipitation
HEK293 cells were plated in a 9-cm dish at 2 × 106 cells/10 mL 1 d before transfection. Three μg of pFLAG/HA-Ago2 were transfected into the cells with 3 μg of pmyc-GFP-TNRC6A or its mutant expression constructs. Two days after transfection, cells were washed once with PBS and lysed with ice-cold lysis buffer (10 mM Hepes-NaOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 140 mM NaCl, 0.5 mM DTT, 1 mM EDTA, 1 mM Na3VO4, 10 mM NaF, 0.5% NP40, and 1× complete protease inhibitor cocktail [Roche]). Lysates were centrifuged at 20,000g for 10 min at 4°C, and the supernatants were centrifuged at 48,000g for 30 min at 4°C. The supernatants were incubated with 30 μL anti-c-Myc agarose conjugate beads (Sigma) with constant rotation for 2 h at 4°C. The beads were washed six times with the lysis buffer supplemented with 300 mM NaCl, and resuspended in 30 μL 2× SDS sample buffer. Bound proteins were eluted by boiling the beads for 5 min and separated by SDS-PAGE (SuperSep 5%–20% [Wako]).
HeLa cells were plated in a 9-cm dish at 2 × 106 cells/10 mL 1 d before transfection. The cell lysates from the cells transfected with 5 μg of pmyc-GFP-TNRC6A or its mutants were prepared as described above. Thirty μL of protein A agarose (MILLPORE) was equilibrated with 500 μL of lysis buffer and incubated with 2.5 μg of anti-GFP antibody in lysis buffer for 1 h at 4°C. Then, the cell lysates were added to the antibody-bound protein A agarose and incubated with constant rotation for 2 h at 4°C. The beads were washed twice with the lysis buffer containing 300 mM NaCl, once with lysis buffer, and resuspended in 30 μL 2× SDS sample buffer. Bound proteins were eluted by boiling the beads for 5 min and separated by SDS-PAGE.
Knockdown of Ago2 or TNRC6A
The sequences of siRNAs against Ago2 (siAgo2) and TNRC6A (siTNRC6A) are described in Supplemental Table S3. For verification of immunofluorescence signals of Ago2, HeLa cells inoculated on a glass coverslip at 1 × 105 cells/mL/well in wells of 12-well culture plate were transfected twice at 24-h intervals with 50 nM siAgo2. One day after the second transfection, cells were fixed, stained with an anti-Ago2 antibody, and observed under the fluorescence microscope or harvested for Western blot. For TNRC6A knockdown, HeLa cells were transfected once with 50 nM siTNRC6A. Two days after the transfection, cells were treated with LMB, stained with an anti-Ago2 antibody, and observed.
miRNA quantification
Quantification of miRNAs contained in immunoprecipitates prepared from HeLa cells transfected with pmyc-GFP-TNRC6A or its mutant expression constructs was carried out. Sixty μL of beads carrying the immunoprecipitates were resuspended in 60 μL lysis buffer. Eighty μL of suspension was used for RNA extraction, and the remaining 40 μL suspension was used for protein analysis.
RNA was extracted by using the mirVana miRNA Isolation Kit (Ambion). MiRNAs were quantified using a modified procedure of the stem–loop RT-PCR reported previously (Chen et al. 2005). The reverse transcriptase reactions were performed in a 10-μL reaction mixture containing 2.5 μL of eluted RNA, 1.5 pmoles of RT primers specific for each of the miRNAs, 1× RT buffer (RETROscript Kit; Ambion), 0.25 mM dNTPs, 35 U of MultiScribe reverse transcriptase (Applied Biosystems), and 20 U of RNase OUT (Invitrogen). The mixture was incubated at 16°C for 30 min, 42°C for 30 min, 85°C for 5 min, and then held at 4°C. A mixture of the synthesized cDNA, FastStart Universal SYBR Green Master (Roche), and respective primers for each miRNA was incubated at 95°C for 10 min. The 40-cycle PCR reaction was then carried out at 95°C for 15 sec and 60°C for 1 min in triplicate. The levels of PCR products were monitored with an ABI PRISM 7000 sequence detection system (Applied Biosystems). The stem–loop RT primers and PCR primers are listed in Supplemental Table S4.
Fluorescence in situ hybridization
Fluorescence in situ hybridization was performed essentially according to the procedure reported by Nathans et al. (2009) with slight modifications. LNA hybridization probes with 5′ digoxigenin modification complementary to miR-21 (5′-TCAACATCAGTCTGATAAGCTA-3′) and control scramble-miR (5′-GTGTAACACGTCTATACGCCCA-3′) were purchased from Exiqon. Briefly, HeLa cells fixed with 4% paraformaldehyde were rehydrated in PBS for 10 min and prehybridized for 15 min with 40% formamide in 2× SSC. Each probe (20 ng) was mixed with E. coli tRNA (5 μg) and salmon sperm DNA (5 μg) in 40 μL of 80% formamide, heated for 5 min at 95°C, and immediately mixed with an equal volume of hybridization buffer (2 mg/mL BSA, 20% dextran sulfate in 4× SSC). Cells were incubated with 60 μL of probe in hybridization buffer for 3 h at 37°C, and washed. Cells were then incubated with 1:100 diluted Anti-Digoxigenin-Rhodamine Fab fragments (Roche) in PBS at 4°C overnight, and washed three times with PBS. They were visualized under a fluorescence microscope.
Gene silencing activity assay
HeLa cells were plated at 0.6 × 105 cells/well in a well of a 24-well plate. Cells were transfected with 50 nM control siRNA (siControl), 25 nM siTNRC6A and 25 nM siRNA#1 against TNRC6B (siTNRC6B#1), or 25 nM siTNRC6A and siTNRC6B#2. Their sequences are described in Supplemental Table S3. Twenty-four h after siRNA transfection, the cells were simultaneously transfected with psiCHECK-CXCR4 target (10 ng), pGL3-Control (100 ng; Promega) encoding firefly luciferase gene, pSUPER-CXCR4 or pSUPER-DsRed (100ng), and pmyc-GFP-TNRC6A or its mutant expression constructs (500 ng). The cells were harvested 48 h after the second transfection, and the relative Renilla/firefly luciferase activity was determined using a Dual-Luciferase Reporter Assay System (Promega).
Two d after siRNA transfection, knockdown efficiency of TNRC6A and TNRC6B by each siRNA was quantified by real-time RT-PCR using total RNA isolated using an RNeasy Mini kit (Qiagen). The expression of TNRC6A and TNRC6B was normalized to that of GAPDH. The primers' sequences are listed in Supplemental Table S4.
Absolute quantification of TNRC6s' mRNAs
cDNA was synthesized from 1 μg of the total RNA of HeLa cells by SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). The PCR primers used are listed in Supplemental Table S4. The copy numbers of mRNA was calculated from serial dilutions of known copy numbers of pCMV-SPORT/FLAG-GW182, pmyc-GFP-TNRC6B, or pmyc-GFP-TNRC6C.
Generation of stable HEK293 cells and knockdown experiment for MALAT-1
Flp-In T-REx 293 cells (Invitrogen) were transfected with pcDNA5/FRT/TO/FLAG-HA-SBP, pcDNA5/FRT/TO/FLAG-HA-SBP-TNRC6A, or its mutant expression constructs along with pOG44 (Invitrogen) using Polyethylenimine “Max” (Polysciences). One d after transfection, cells were selected with hygromycin B (50 μg/mL) for 7–8 wk. Obtained stable HEK293 cells were maintained in DMEM supplemented with 10% Tet System Approved FBS (Clontech).
Stable HEK293 cells were plated at 1.3 × 105 cells/well in a well of a 24-well plate. Cells were transfected with 50 nM control siRNA (siControl2) or 50 nM siMALAT-1. Their sequences are described in Supplemental Table S3. After transfection, cells were cultured in the presence or absence of dox for 24 h. Knockdown efficiency of MALAT-1 was quantified by real-time RT-PCR. The expression of MALAT-1 was normalized to that of GAPDH. The primers' sequences are listed in Supplemental Table S4.
Statistical analysis
All statistical analyses were performed using the two-tailed unpaired Student's t-test.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article (10 figures and four tables).
ACKNOWLEDGMENTS
We thank Drs. Patrick Provost, Shigeyuki Yokoyama, Thomas Tuschl, and Akio Yamashita for kindly providing plasmids, and Dr. Tetsuya Kojima for conducting the confocal microscopy imaging. This work was partially supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) and the Cell Innovation Project (MEXT) to K.U.-T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.034769.112.
REFERENCES
- Afonina E, Stauber R, Pavlakis GN 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem 273: 13015–13021 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W, Parker R 2009. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA 15: 794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res 33: e179 doi: 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Zheng D, Xia Z, Shyu AB 2009. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol 16: 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol 4: e210 doi: 10.1371/journal.pbio.0040210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI 1995. The nuclear pore complex. Annu Rev Biochem 64: 865–896 [DOI] [PubMed] [Google Scholar]
- Ding L, Han M 2007. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol 17: 411–416 [DOI] [PubMed] [Google Scholar]
- Ding L, Spencer A, Morita K, Han M 2005. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell 19: 437–447 [DOI] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA 2003. siRNAs can function as miRNAs. Genes Dev 17: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, Izaurralde E 2009a. The GW182 protein family in animal cells: New insights into domains required for miRNA-mediated gene silencing. RNA 15: 1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E 2009b. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA 15: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ 2002. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell 13: 1338–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Séraphin B, Cougot N, Fritzler MJ 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Han J, Kim D, Morris KV 2007. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci 104: 12422–12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Braun JE, Heimstädt S, Zekri L, Izaurralde E 2010. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J 29: 4146–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT 2007. A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100 [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol 7: 1267–1274 [DOI] [PubMed] [Google Scholar]
- Jeffries CD, Fried HM, Perkins DO 2011. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA 17: 675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA 2010. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol 17: 238–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ 2006. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol 13: 793–797 [DOI] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O Jr, Rossi JJ 2008. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci 105: 16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Safaee N, Rosenauer A, Gehring K 2010. Structural basis of binding of P-body associated proteins GW182 and Ataxin-2 by the Mlle domain of poly(A)-binding protein. J Biol Chem 285: 13599–13606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci 96: 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Güttinger S 2005. Leucine-rich nuclear-export signals: Born to be weak. Trends Cell Biol 15: 121–124 [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T 2008. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14: 2580–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH 2007. Classical nuclear localization signals: Definition, function, and interaction with importin α. J Biol Chem 282: 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaretti D, Tournier I, Izaurralde E 2009. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA 15: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian SL, Li S, Abadal GX, Pauley BA, Fritzler MJ, Chan EK 2009. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA 15: 804–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH 2010. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE 5: e10563 doi: 10.1371/journal.pone.0010563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR III, Parker R, Hannon GJ 2005. A role for the P-body component GW182 in microRNA function. Nat Cell Biol 7: 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Lührmann R, Tuschl T 2005. Identification of novel argonaute-associated proteins. Curr Biol 15: 2149–2155 [DOI] [PubMed] [Google Scholar]
- Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T, et al. 2012. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 18: 738–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Yoshimura J, Morishita S, Ui-Tei K 2009. siDirect 2.0: Updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics 10: 392 doi: 10.1186/1471-2105-10-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell 34: 696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K, Saigo K 2007. Cellular internalization of green fluorescent protein fused with herpes simplex virus protein VP22 via a lipid raft-mediated endocytic pathway independent of caveolae and Rho family GTPases but dependent on dynamin and Arf6. J Biol Chem 282: 27503–27517 [DOI] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S 2012. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 40: 1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Hogan EM, Pederson T 2009. MicroRNAs with a nucleolar location. RNA 15: 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Rådmark O 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 21: 5864–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb GB, Brown KM, Khurana J, Rana TM 2005. Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol 12: 133–137 [DOI] [PubMed] [Google Scholar]
- Rüdel S, Flatley A, Weinmann L, Kremmer E, Meister G 2008. A multifunctional human Argonaute2-specific monoclonal antibody. RNA 14: 1244–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Meng S, Lu Y, Trombly MI, Chen J, Lin C, Turk A, Wang X 2011. Mammalian hyperplastic discs homolog EDD regulates miRNA-mediated gene silencing. Mol Cell 43: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJ, Rasko JE, et al. 2010. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol 17: 1030–1034 [DOI] [PubMed] [Google Scholar]
- Takimoto K, Wakiyama M, Yokoyama S 2009. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA 15: 1078–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG 2007. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol 14: 897–903 [DOI] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K 2004. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res 32: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Nishi K, Juni A, Saigo K 2008. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res 36: 7100–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S 2007. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev 21: 1857–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann L, Höck J, Ivacevic T, Ohrt T, Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G 2009. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136: 496–507 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063 [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger ST, Corey DR 2011. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res 39: 5682–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo NJ, Hur SY, Kim MS, Lee JY, Lee SH 2010. Immunohistochemical analysis of RNA-induced silencing complex-related proteins AGO2 and TNRC6A in prostate and esophageal cancers. APMIS 118: 271–276 [DOI] [PubMed] [Google Scholar]
- Zekri L, Huntzinger E, Heimstädt S, Izaurralde E 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol 29: 6220–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, Liu X, Yates JR 3rd, Han M 2007. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell 28: 598–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W 2009. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA 15: 781–793 [DOI] [PMC free article] [PubMed] [Google Scholar]