The authors set out to determine whether dual and antagonistic functionality is a common feature of SR and hnRNP splicing regulators because recent genome-wide analyses called the strict designation of hnRNP-like splicing repressors into question. The results show that this is indeed the case, where SR proteins activate splicing only from within exons but repress exon recognition from intronic positions, and hnRNP splicing factors repress within exons while activating splicing from the intron. Repressive splicing activities do not interfere with E-complex formation but do interfere with the transition from early to later spliceosomal assembly steps.
Keywords: SR protein, hnRNP protein, pre-mRNA splicing, splicing activation, splicing repression
Abstract
Alternative splicing is regulated by splicing factors that modulate splice site selection. In some cases, however, splicing factors show antagonistic activities by either activating or repressing splicing. Here, we show that these opposing outcomes are based on their binding location relative to regulated 5′ splice sites. SR proteins enhance splicing only when they are recruited to the exon. However, they interfere with splicing by simply relocating them to the opposite intronic side of the splice site. hnRNP splicing factors display analogous opposing activities, but in a reversed position dependence. Activation by SR or hnRNP proteins increases splice site recognition at the earliest steps of exon definition, whereas splicing repression promotes the assembly of nonproductive complexes that arrest spliceosome assembly prior to splice site pairing. Thus, SR and hnRNP splicing factors exploit similar mechanisms to positively or negatively influence splice site selection.
INTRODUCTION
Alternative pre-mRNA splicing promotes high proteomic diversity despite the limited number of protein coding genes (Nilsen and Graveley 2010). Intron removal is carried out by the spliceosome and a broad inventory of accessory proteins (Wahl et al. 2009) that recognize the boundaries of exons through sequence-specific interactions. Base-pairing between the 5′ end of the U1 snRNA and the exon/intron junction initiates the assembly of the spliceosome on the pre-mRNA. The efficiency of this process depends on the complementarity between the 5′ splice site (5′ss) and the 5′ end of the U1 snRNA (Kammler et al. 2001) as well as the presence of nearby splicing regulatory elements (SREs) (Blencowe 2006; Nilsen and Graveley 2010).
In general, SREs serve as binding sites for splicing regulatory proteins that either increase or decrease spliceosomal component recruitment (Lin and Fu 2007; Venables 2007). For example, SR proteins associated with exonic SREs to facilitate U1 snRNP recruitment to the 5′ss through their RS domain (Fu and Maniatis 1992; Lin and Fu 2007) or through RNA binding domain interactions with U1 snRNP-specific protein 70K (Cho et al. 2011). However, in some cases SR proteins were shown to interfere with exon selection when bound to intronic positions, potentially due to steric hindrance (Kanopka et al. 1996; Ibrahim et al. 2005; Hicks et al. 2010). hnRNP-mediated repression from exonic positions has been documented (Chen et al. 1999; Jacquenet et al. 2001; Rothrock et al. 2005), and in some cases, it was shown that an intronic hnRNP could activate splicing (Schaub et al. 2007; Wang et al. 2011). Even more, from an evolutionary viewpoint, it appears that intronic hnRNP binding sites have played a major role in shaping the landscape of mammalian SREs (Voelker et al. 2012). In combination with recently defined RNA splicing maps that correlate profiles of RNA–protein interactions with their effects on splicing activation (Ule et al. 2006; Llorian et al. 2010; Huelga et al. 2012), these observations suggest that the activity of splicing regulatory proteins might be highly position dependent.
Here we show that almost all splicing regulatory proteins tested can activate 5′ss selection from defined positions. Surprisingly, all splicing regulatory proteins tested also interfere with spliceosomal recognition by simply relocating their binding sites to the opposite side of the regulated 5′ss. The position-dependent silencing activity of splicing regulatory proteins does not interfere with initial U1 snRNP recruitment, but it induces the formation of nonfunctional complexes incapable of maturing into functional spliceosomes.
RESULTS
The location of a SRE determines if a neighboring 5′ss is used
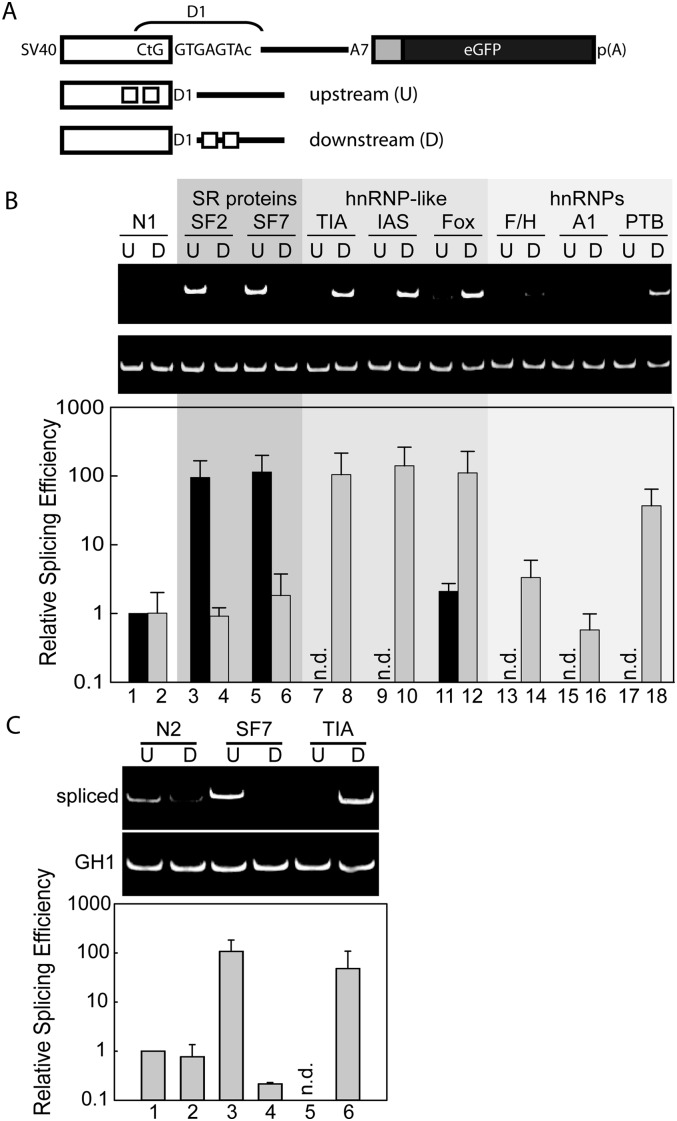
To address how the position of a SRE relative to a 5′ss affects its recognition, a panel of known high-affinity binding sites for splicing regulatory proteins was inserted upstream of or downstream from the 5′ss of a splicing reporter (Fig. 1A; Kammler et al. 2001; Caputi et al. 2004). By use of RT-PCR, we observed that SR proteins only activate splicing when recruited upstream of the 5′ss, while hnRNP proteins activated splicing only when recruited downstream from the 5′ss (Fig. 1B). These results were also validated using splicing-sensitive cell imaging (Supplemental Fig. 1). Relocating the SRSF7 SRE to the downstream position or relocating the TIA-1 SRE to the upstream position did not support 5′ss selection (Fig. 1C, lanes 3–6). In fact, 5′ss usage was repressed when the same constructs were compared with splicing neutral controls (Fig. 1C, lanes 1,2). These results show that mislocalization of SREs to the opposite side of a regulated 5′ss interferes with splice site activation. We conclude that all SREs tested contribute to 5′ss selection in a position-dependent manner, either promoting or repressing splice site recognition.
FIGURE 1.
The positioning of splicing regulatory elements determines their effect on 5′ splice site (5′ss) activation. (A) Schematic drawing of the RNA splicing reporter. Specific binding sites for splicing regulatory proteins are immediately upstream of or downstream from the D1 5′ss, as indicated by boxes. HeLa cells were transiently transfected with each of the reporter constructs and analyzed by semi-quantitative and real-time PCR to determine the position-dependent activity of SREs (B) and to demonstrate that each SRE displays position-dependent activation and repression of splicing (C). N1 and N2 are splicing neutral and near neutral control sequences (Supplemental Fig. 2). U and D refer to upstream and downstream positions relative to the D1 5′ss. n.d. refers to not detected above background. All data were normalized to the constitutively spliced GH1 mRNA.
To evaluate the effects of multiple SREs, we placed SRSF7 and TIA-1 binding sites upstream of and downstream from the 5′ss, or opposing each other on either side of the 5′ss. Multiple SREs located at activating positions promoted splicing of very weak 5′ss's, supporting the notion that the overall ability of the spliceosome to recognize a 5′ss depends on U1 snRNA complementarity and the strength of SREs in its neighborhood (Supplemental Fig. 3). When identical SREs were present at upstream and downstream locations, the exonic activity generally prevailed. Furthermore, if activating and repressive SREs are localized to exonic positions, the SRE closest to the 5′ss dominated splicing decisions (Supplemental Fig. 4).
MS2 tethering of splicing regulatory proteins mimics positional requirements for 5′ss activation
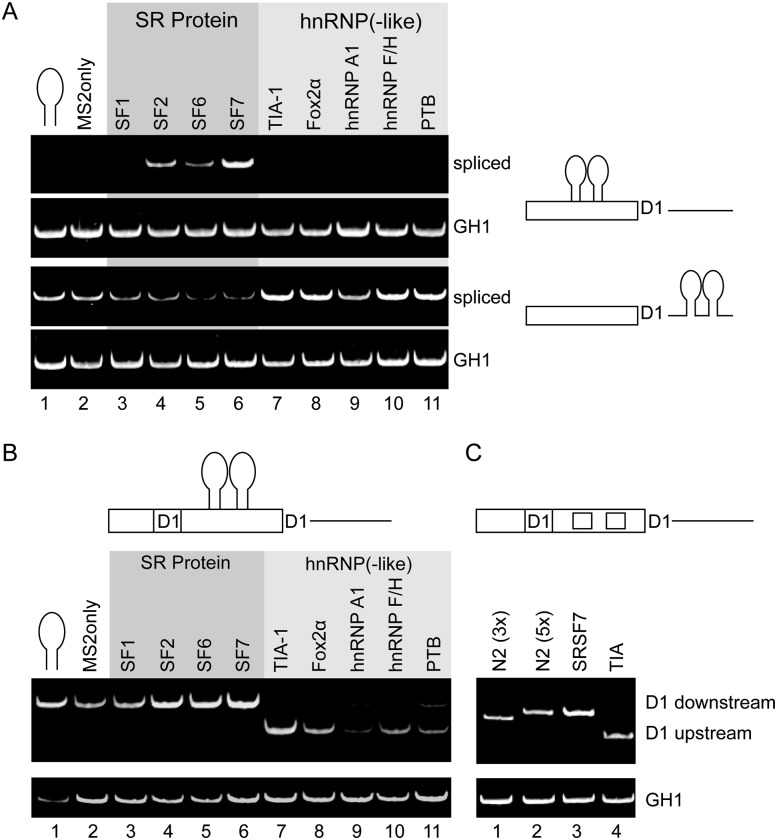
By using naturally occurring SREs, we formally could not exclude the possibility that the SRE itself modulated 5′ss activation or that other factors interact with the SRE. We therefore evaluated position-dependent splicing using an MS2 tethering system. Placing SR protein fusion proteins immediately upstream of the 5′ss promoted splicing, whereas SR fusion recruitment to the downstream intronic position resulted in splicing repression (Fig. 2A). In contrast, hnRNP fusion proteins stimulated splicing only when they were tethered to the downstream intron. When localized to the opposite side of the 5′ss, hnRNP fusion proteins actively repressed intron excision. In summary, the tethering experiments confirm the conclusion that the activities of splicing regulatory proteins are highly position dependent.
FIGURE 2.
MS2 tethering of splicing regulatory proteins mimics positional requirements for 5′ss activation. (A) Expression of MS2 fusion proteins results in position-dependent activation or repression of splicing using reporters that harbor MS2 binding sites upstream of or downstream from the D1 5′ss. Comparable expression of all MS2 fusion proteins was verified by Western blot analysis (Supplemental Fig. 5). (B) Schematic diagram of the splicing reporter in which the two MS2 binding sites are flanked by the identical up- and downstream 5′ss D1. The splicing pattern of each sample was analyzed using semi-quantitative RT-PCR analysis. (C) Identical experiments were performed using splicing reporters where the MS2 binding sites were replaced by high-affinity binding sites for endogenous TIA-1 and SRSF7.
To determine the effects of splicing regulatory proteins on the selection of two alternative 5′ss's, we inserted tandem MS2 RNA hairpins between two identical 5′ss sequences (Fig. 2B). All SR fusion proteins promoted splicing of the downstream 5′ss, whereas all hnRNP factors tested favored the upstream 5′ss (Fig. 2B). Consistent with these results, the natural binding site for SRSF7 showed a strong preference for activating the downstream 5′ss, whereas binding sites for TIA-1 activate the upstream 5′ss (Fig. 2C).
The progression into a functional splicing complex depends on the side from which U1 snRNP is recruited to the 5′ss
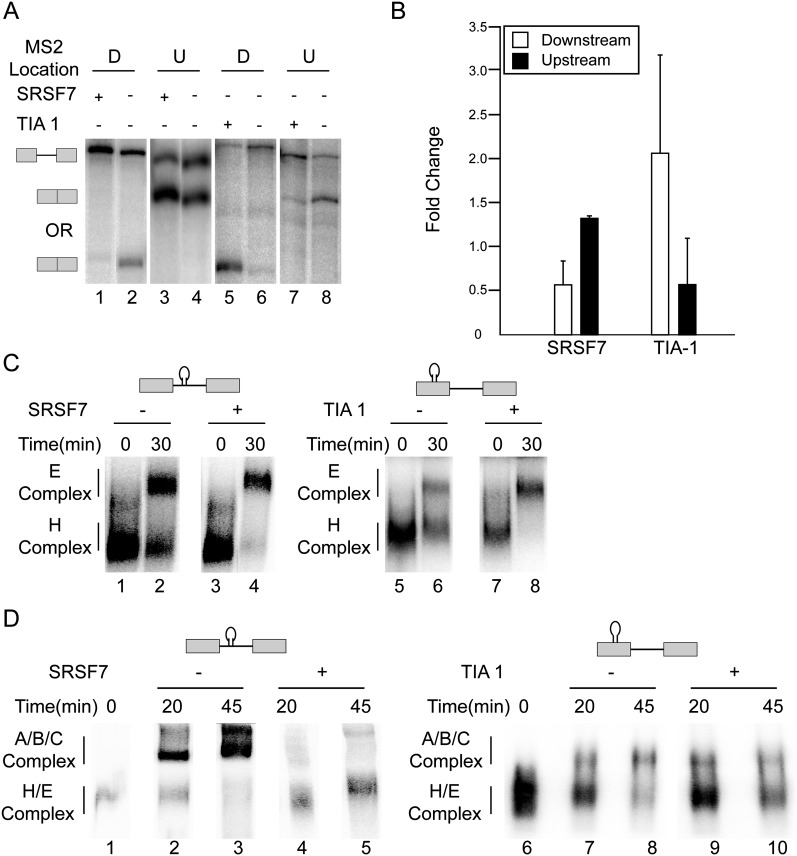
To determine the mechanisms of splicing repression by intronic SR proteins, we measured intron removal efficiency of a β-globin–based pre-mRNA substrate that contains a single MS2 RNA hairpin downstream from its 5′ss. In agreement with the cell culture experiments, splicing of this substrate was impaired when SRSF7 was tethered downstream from the 5′ss (Fig. 3A,B). When placed upstream of the regulated 5′ss, SRSF7 activated splicing. The parallel but opposite effect was observed for TIA-1 (Fig. 3A,B), which repressed from an exonic location but activated from an intronic location. It is worth noting here that the splicing reporters used in the cell transfection and in the in vitro splicing experiments are unrelated, thus supporting the notion that the position-dependent splicing activities are not reporter specific but are perhaps more general in nature.
FIGURE 3.
Binding of SRSF7 to the intron does not interfere with E-complex formation. (A) In vitro splicing reaction time course in the presence or absence of MS2-RS-SRSF7 or MS2-TIA-1 binding either upstream of or downstream from the 5′ss. (B) In vitro splicing reactions were repeated and quantified. (C) ATP-independent E-complex formation in the presence or absence of MS2-RS-SRSF7 or MS2-TIA-1 was visualized using agarose gel electrophoresis. (D) Higher-order, ATP-dependent complex formation in the presence or absence of MS2-RS-SRSF7 or MS2-TIA-1 was visualized using polyacrylamide native gels or agarose gels. Spliceosomal complexes or splicing products formed during the reactions are defined next to each gel.
To determine the mechanisms of the observed splicing repression, we uncoupled the ATP-dependent and ATP-independent steps of splicing through the use of an E-complex pulse chase experiment (Kotlajich et al. 2009). Splicing reactions were enriched for E-complex in the absence of ATP and then chased to spliced products. When MS2-SRSF7 was located within the intron, only 10% of the E-complexes converted into spliced mRNAs. However, the rate of splicing for this small fraction was essentially identical to the rates measured in the absence of MS2-SRSF7 (Supplemental Fig. 6). These results show that only a small portion of splicing-competent E-complexes was formed on the pre-mRNA in the presence of intronic MS2-SRSF7 and that this small fraction behaves kinetically similar to uninhibited pre-mRNAs.
To determine if spliceosomal complex formation was altered when splicing was repressed, we analyzed complex formation in the presence and absence of MS2-SRSF7 and MS2-TIA-1. Neither protein, when localized to their repressive position, interfered with spliceosome assembly at the early E-complex stage, indicating that U1 snRNP is efficiently recruited to the pre-mRNA substrate (Fig. 3C). However, this initial U1 snRNP recruitment appears to be a “dead-end” E-complex because further incubation in the presence of ATP led to less formation of higher-order complexes (A-, B-, and C-complex) (Fig. 3D). These results demonstrate that the presence of SRSF7 or TIA-1 at the repressive positions accumulate splicing-incompetent E-complexes that do not efficiently progress into functional spliceosomes. We conclude that U1 snRNP is recruited efficiently to the pre-mRNA regardless of the presence of an inhibitory SRE. However, the presence of the inhibitory SRE interferes with the progression into later stages of spliceosome assembly.
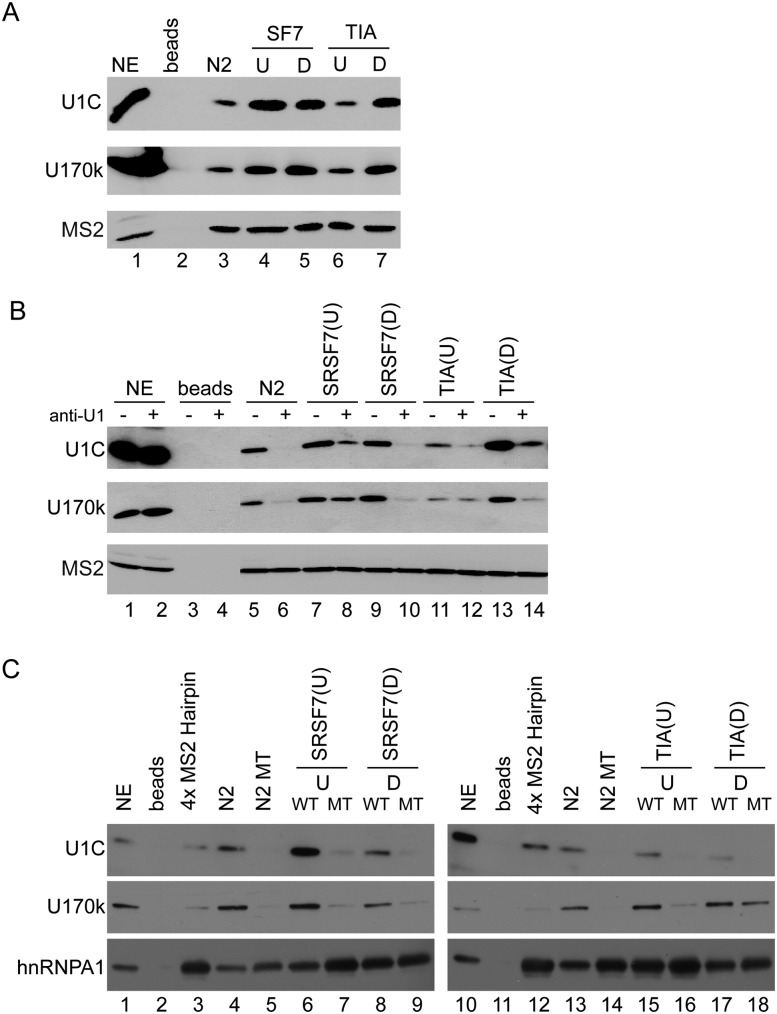
Based on these observations, it is expected that U1 snRNP components interact with the pre-mRNA substrate, independent of SRE location. To confirm this prediction, we tested for the presence of U1-70K and U1-C upon immobilizing in vitro transcribed RNAs containing a 5′ss and SREs at various locations. While the splicing near neutral control precipitated only low levels of U1 snRNP-specific proteins (Fig. 4A, lane 3), we detected high levels of U1-70K and U1-C on RNAs containing the SRSF7 binding site upstream of or downstream from the 5′ss (Fig. 4A, lanes 4,5). These results confirm the prediction that the splicing repressive phenotype of SRSF7 is not caused by an interference to recruit U1 snRNP to the 5′ss. A slightly different U1 snRNP recruitment trend was observed for RNAs containing TIA-1 binding sites. Increased levels of U1-70K and U1-C were detected on RNAs containing the TIA-1 binding site in its activating intronic position (Fig. 4A, cf. lanes 3 and 7). However, when placed into a splicing repressive position, TIA-1 binding did not increase levels of U1 snRNP recruitment. Rather, the presence of TIA-1 in the repressive exonic position does not interfere with the intrinsic binding of U1 snRNP to the D1 5′ss (Fig. 4A, cf. lanes 3 and 6). Together, these results suggest that SR and hnRNP proteins act through similar mechanisms to activate splicing by increasing U1 snRNP recruitment. When located in repressive positions, SR and hnRNP proteins do not interfered with U1 snRNP recruitment. However, higher-order spliceosomal complex formation is impaired at the stage of A-complex formation.
FIGURE 4.
The recruitment of U1 snRNP components to the 5′ss is not decreased by repressive SREs. (A) RNAs were immobilized using Agarose beads and analyzed for the presence of U1 snRNP with specific antibodies directed against U1-70K and U1-C. MS2 coat protein was used as a loading control. N2 represents the splicing near neutral control sequence. (B) HeLa cell nuclear extracts were depleted (D) or mock depleted (U) of functional U1 snRNP using short DNA oligonucleotides and RNase H. The extracts were then used in RNA pulldown assays to demonstrate that U1 snRNA/RNA interactions are necessary for U1 snRNP recruitment. MS2 coat protein and hnRNP A1 were used to control for loading, and N2 represents splicing near neutral control sequences. (C) RNAs containing a muted 5′ss were used in the pulldown assay to demonstrate the effect of a functional or nonfunctional splice site on U1 recruitment.
To determine whether “dead-end” E-complex formation depends on U1 snRNA interactions with the pre-mRNA, we incapacitated U1 snRNA binding by oligonucleotide-directed RNase H digestion of the U1 snRNA 5′ end. In all SRE permutations tested, U1 snRNP recruitment was severely reduced by the loss of the 5′ end of U1 snRNA, demonstrating that U1 snRNP recruitment requires RNA duplex formation (Fig. 4B). Furthermore, identical immobilization experiment using RNAs that harbor a mutated 5′ss showed that a functional 5′ss is required for efficient recruitment of spliceosomal components (Fig. 4C). Together, these results demonstrate that base-pairing between the U1 snRNA and the 5′ss is required for U1 snRNP recruitment to the pre-mRNA but that the ability of the E-complex to progress into spliceosomal A-, B-, and C-complexes is regulated by surrounding SREs. Our findings show that positioning SR proteins to the downstream intron, or hnRNPs to the upstream exon, represses splicing without significantly altering initial recruitment of U1 snRNP to the pre-mRNA.
Based on these results, it is expected that constitutive exons should display a higher frequency of SREs in activating positions compared with alternatively spliced exons. To test this hypothesis, we determined the frequency of SREs in exonic or intronic positions of human exons within a 30-nucleotide (nt) window upstream of and downstream from their 5′ss. Indeed, constitutive exons are more likely to harbor an upstream SRSF7 or a downstream TIA-1 binding site compared with alternative exons (Supplemental Table 1). In contrast, alternative exons display a higher frequency of intronic SRSF7 and exonic TIA-1 binding sites. This analysis can be extended to other hnRNPs, demonstrating that constitutive exons generally display an increased frequency of hnRNP binding sites in the activating intronic position (Supplemental Table 2). Furthermore, knockdown of individual hnRNPs, as recently reported (Huelga et al. 2012), recorded changes in splicing efficiencies of exons that were identified by our motif search as potential hnRNP targets (data not shown).
DISCUSSION
Here we show that the rigid categorization of SREs into enhancers or repressors should be re-evaluated. Based on our results, we propose a model in which the positioning of a SRE relative to a 5′ss determines whether the formation of splicing complexes is productive or not. While a repressing SRE does not interfere with the recruitment of U1 snRNP to the regulated 5′ss, it traps early spliceosomal complexes in an inactive state, potentially resulting in the recruitment of qualitatively distinct U1 snRNPs that may differ in their protein composition (Hernandez et al. 2009) or U1 snRNA (Kyriakopoulou et al. 2006).
SR and hnRNP proteins have activities that differ depending on their position relative to the regulated splice site
For SRE-dependent splice site recognition, a mechanistic picture emerges that indicates surprising similarities between the actions of SR and hnRNP proteins. Both classes of splicing factors demonstrate position-dependent activation or repression. As has been demonstrated previously, SR proteins bound to exons recruit the splicing machinery to splice sites (Graveley 2000; Black 2003; Cho et al. 2011). The observed SR protein splicing repression is based on stalling the progression of spliceosomal assembly after efficient formation of the E-complex. Thus, the recruited components are not able to efficiently establish splice site pairing, a process mediated by RS-domain containing bridging factors Prp5 and Rsd1 (Shao et al. 2012). It is possible that intronic SR proteins may prevent the interaction between pre-mRNA-bound U1 and U2 snRNPs by competing for Prp5 (at the 3′ splice site) or Rsd1 (at the 5′ss) binding. Alternatively, the correct placement of U1 snRNP at the 5′ss could be a measure for rearranging the conformational state of the particle to promote the recruitment of other splicing factors required for further spliceosomal assembly, similar in principle with the demonstration that alternate conformations of U2AF65 promote spliceosome assembly (Mackereth et al. 2011).
The effects of hnRNP proteins on 5′ss activation are mirror images of SR protein activities. When bound to exons, they repress splicing, and when bound to introns, they activate splicing. Interestingly, all hnRNP proteins tested follow this antagonistic activity profile (with the exception of hnRNP A1: No evidence has yet been presented that hnRNP A/B proteins recruit spliceosomal components; this may mean they interact with the spliceosome differently than the other hnRNPs) (Dreyfuss et al. 1993; Mayeda et al. 1998). Activating hnRNP positions promote E-complex assembly; however, splicing repression from an exonic position did not interfere with initial 5′ss recognition. Thus, splicing suppression is achieved after initial splice site recognition regardless of whether the repression is exerted by exonic hnRNPs or intronic SR proteins. These observations are consistent with studies demonstrating that splicing silencer motifs impair the commitment to splice site pairing (Yu et al. 2008). Splice site activation by hnRNPs follows the mechanisms worked out for classical exonic enhancers, increased recruitment of U1 snRNP to the 5′ss. In summary, SR and hnRNP splicing regulatory proteins appear to target the same steps to promote or interfere with spliceosomal assembly.
While the splicing promoting and repressing activities of SR proteins can be explained by RS-domain interactions, it is unclear how hnRNP splicing regulators increase U1 snRNP recruitment or interfere with prespliceosomal complex formation. Activation by intronic TIA-1 has been demonstrated to target the U1-C component of U1 snRNP (Forch et al. 2002), suggesting that other intronically bound hnRNPs may use similar interactions. Exonic hnRNPs may stabilize cross–exon interactions, thus reducing the efficiency of splice site pairing (Motta-Mena et al. 2010).
The rescue of very weak 5′ss when embedded within a context of activating SRE positions highlights the shortcomings of present in silico approaches that usually ignore the sequence environment of 5′ss for prediction of their functionality. Our studies illustrate that the position of SREs relative to a 5′ss defines their splicing phenotype, providing a mechanistic framework for exonic and intronic splicing repression/activation. Integrating these new insights into current splicing code algorithms will likely improve the success rate of splice pattern predictions.
MATERIALS AND METHODS
Plasmids
All transfection constructs are based on the HIV-1 glycoprotein/eGFP expression plasmid (Kammler et al. 2001). All upstream and downstream splicing elements (Supplemental Table 3) were cloned into the reporter using PCR-product using a specific forward and reverse primer pairs (please contact corresponding authors for primer sequences). MS2 fusion protein expressing plasmids were generated according to the method described previously (Singh et al. 2010).
Cell culture and RT-PCR analysis
HeLa cells were maintained in DMEM supplemented with 10% fetal calf serum. Total RNA was collected 30 h after transfection, reverse transcribed, and analyzed by qPCR or semi-quantitative PCR. All experiments were repeated three times.
In vitro splicing and complex formation
Reactions were carried out in 30% nuclear extract containing 32P-labeled RNA transcripts with or without saturating amounts of recombinant MS2-RS-SRSF7 (250 nM) or MS2-TIA-1 (500 nM), according to the method previously described (Hicks et al. 2005). Pulse chase experiments were carried out according to the method previously described (Konarska and Sharp 1986; Kotlajich et al. 2009). ATP-independent complex formation reactions were carried out like the splicing reactions except no ATP or creatine phosphate was added to the reaction. Following incubation, reactions were loaded onto a 1.5% low-melt Agarose gel and analyzed by PhosphorImager analysis (Das and Reed 1999; Hicks et al. 2005). ATP-dependent complex formation was analyzed according to the method previously described (Konarska and Sharp 1986; Kotlajich et al. 2009).
Immobilization of RNA on Agarose beads and RNA affinity assays
Substrate RNAs were covalently linked to adipic acid dihydrazide-Agarose beads according to the method previously described (Kammler et al. 2006). Immobilized RNAs were incubated in 30% HeLa cell nuclear extract/buffer D for 30 min at 30°C. Beads were then washed, and proteins were eluted by the addition of 2× protein sample buffer. Proteins were resolved by SDS-PAGE and were subjected to immunoblot analysis (see Protein Analysis section).
Protein analysis
Transfected cells were lysed in RIPA buffer. Proteins were separated by SDS-PAGE and subjected to immunoblotting. Membranes were probed with rabbit antibody against HA (H6908), mouse antibody against β-actin (A2228), rat antibody against U1-C (4H12) (all Sigma-Aldrich), goat antibody against U1-70K (Santa Cruz Biotechnology, C-18), mouse antibody against hnRNP A1 (Santa Cruz Biotechnology, 9H10), and rabbit antibody against MS2 coat protein (Tetracore, TC-7004).
Computational analysis
To determine the frequency of potential SRE binding sites upstream of and/or downstream from known splice sites within the human genome, a list of constitutive (99885) and alternative 5′ss (5554) exons was compiled based on known mRNA isoforms and EST information (GRCh37/hg19) (Kent et al. 2002). To calculate frequencies of potential TIA-1 and SRSF7 binding sites, the surrounding 30-nt sequence space was scanned for occurrences of either TTTTT or at least two occurrences of TTT in the case of TIA-1 (Dember et al. 1996) and for occurrences of [A/T]C[A/T][A/T]C in the case of SRSF7 (Cavaloc et al. 1999). P-values were calculated using Fisher's exact test, comparing the frequency of each motif combination (in both groups) with the sum of the frequencies of any other combination. To determine the effect of hnRNP knockdowns on directional splicing events, we used a list of exons that change their splicing behavior upon knockdown of hnRNP A1 and hnRNP F, respectively, which were kindly provided by Gene Yeo (Huelga et al. 2012). For all exons in our list, we extracted the sequence of 30 nt upstream of and downstream from the 5′ss. Those sequences were scanned for occurrences of potential binding sites for hnRNP A1 (TTAGGG) and hnRNP F (AGGGA), respectively.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank John Conboy, Juan Valcárcel, Massimo Caputi, Brenton Graveley, Chris Smith, Alain Cochrane, and David Peabody for providing plasmids. We thank Björn Wefers, Eva Leimbach, Christin Bruchhagen, Christiane Breuer, Tomasz Ochman, and Sarah Rosin for excellent technical assistance. This work was supported by grants from the Stiftung für AIDS-Forschung, Düsseldorf (H.S.), the Jürgen-Manchot-Stiftung (H.S.), Boehringer Ingelheim Fonds (travel allowance to S.E.), NIH (RO1 GM62287 and R21 CA149548 to K.J.H.), and a fellowship within the Postdoc Programme of the German Academic Exchange Service, DAAD (A.B.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.037044.112.
REFERENCES
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ 2006. Alternative splicing: new insights from global analyses. Cell 126: 37–47 [DOI] [PubMed] [Google Scholar]
- Caputi M, Freund M, Kammler S, Asang C, Schaal H 2004. A bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu, and nef gene expression. J Virol 78: 6517–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Bourgeois CF, Kister L, Stevenin J 1999. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5: 468–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Kobayashi R, Helfman DM 1999. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat β-tropomyosin gene. Genes Dev 13: 593–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G 2011. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci 108: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Reed R 1999. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5: 1504–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember LM, Kim ND, Liu KQ, Anderson P 1996. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem 271: 2783–2788 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG 1993. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 62: 289–321 [DOI] [PubMed] [Google Scholar]
- Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J 2002. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J 21: 6882–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T 1992. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc Natl Acad Sci 89: 1725–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez H, Makarova OV, Makarov EM, Morgner N, Muto Y, Krummel DP, Robinson CV 2009. Isoforms of U1-70k control subunit dynamics in the human spliceosomal U1 snRNP. PLoS ONE 4: e7202 doi: 10.1371/journal.pone.0007202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks MJ, Lam BJ, Hertel KJ 2005. Analyzing mechanisms of alternative pre-mRNA splicing using in vitro splicing assays. Methods 37: 306–313 [DOI] [PubMed] [Google Scholar]
- Hicks MJ, Mueller WF, Shepard PJ, Hertel KJ 2010. Competing upstream 5′ splice sites enhance the rate of proximal splicing. Mol Cell Biol 30: 1878–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S et al. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1: 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T 2005. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci 102: 5002–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquenet S, Mereau A, Bilodeau PS, Damier L, Stoltzfus CM, Branlant C 2001. A second exon splicing silencer within human immunodeficiency virus type 1 Tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J Biol Chem 276: 40464–40475 [DOI] [PubMed] [Google Scholar]
- Kammler S, Leurs C, Freund M, Krummheuer J, Seidel K, Tange TO, Lund MK, Kjems J, Scheid A, Schaal H 2001. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA 7: 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammler S, Otte M, Hauber I, Kjems J, Hauber J, Schaal H 2006. The strength of the HIV-1 3′ splice sites affects Rev function. Retrovirology 3: 89 doi: 10.1186/1742-4690-3-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanopka A, Muhlemann O, Akusjarvi G 1996. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381: 535–538 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Sharp PA 1986. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46: 845–855 [DOI] [PubMed] [Google Scholar]
- Kotlajich MV, Crabb TL, Hertel KJ 2009. Spliceosome assembly pathways for different types of alternative splicing converge during commitment to splice site pairing in the A complex. Mol Cell Biol 29: 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulou C, Larsson P, Liu L, Schuster J, Soderbom F, Kirsebom LA, Virtanen A 2006. U1-like snRNAs lacking complementarity to canonical 5′ splice sites. RNA 12: 1603–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Fu XD 2007. SR proteins and related factors in alternative splicing. Adv Exp Med Biol 623: 107–122 [DOI] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. 2010. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol 17: 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth CD, Madl T, Bonnal S, Simon B, Zanier K, Gasch A, Rybin V, Valcarcel J, Sattler M 2011. Multi-domain conformational selection underlies pre-mRNA splicing regulation by U2AF. Nature 475: 408–411 [DOI] [PubMed] [Google Scholar]
- Mayeda A, Munroe SH, Xu RM, Krainer AR 1998. Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA 4: 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Heyd F, Lynch KW 2010. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol Cell 37: 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothrock CR, House AE, Lynch KW 2005. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J 24: 2792–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MC, Lopez SR, Caputi M 2007. Members of the heterogeneous nuclear ribonucleoprotein H family activate splicing of an HIV-1 splicing substrate by promoting formation of ATP-dependent spliceosomal complexes. J Biol Chem 282: 13617–13626 [DOI] [PubMed] [Google Scholar]
- Shao W, Kim HS, Cao Y, Xu YZ, Query CC 2012. A U1-U2 snRNP interaction network during intron definition. Mol Cell Biol 32: 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Erkelenz S, Rattay S, Dehof AK, Hildebrandt A, Schulze-Osthoff K, Schaal H, Schwerk C 2010. Human SAP18 mediates assembly of a splicing regulatory multiprotein complex via its ubiquitin-like fold. RNA 16: 2442–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586 [DOI] [PubMed] [Google Scholar]
- Venables JP 2007. Downstream intronic splicing enhancers. FEBS Lett 581: 4127–4131 [DOI] [PubMed] [Google Scholar]
- Voelker RB, Erkelenz S, Reynoso V, Schaal H, Berglund JA 2012. Frequent gain and loss of intronic splicing regulatory elements during the evolution of vertebrates. Genome Biol Evol 4: 659–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang E, Mueller WF, Hertel KJ, Cambi F 2011. G Run-mediated recognition of proteolipid protein and DM20 5′ splice sites by U1 small nuclear RNA is regulated by context and proximity to the splice site. J Biol Chem 286: 4059–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW 2008. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell 135: 1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]