Abstract
Aims
The role of calcineurin protein phosphatase 2B (PP2B) in the pathogenesis of human hypertrophic cardiomyopathy (HCM) remains unsettled. We determined potential involvement of calcineurin in the pathogenesis of HCM caused by mutations in myozenin 2 (MYOZ2), an inhibitor of calcineurin.
Methods and results
We generated multiple lines of transgenic mice expressing either Flag-tagged wild-type (WT) (MYOZ2WT) or mutant MYOZ2S48P and MYOZ2I246M, identified in families with HCM, in the heart. To mimic the human genotype, we generated bigenic mice expressing WT and mutant MYOZ2 in the background of hemizygous endogenous MYOZ2 (Myoz2+/−). Transgene proteins constituted 15–48% of the total MYOZ2 protein in the heart. Mutant MYOZ2 mice showed molecular, cellular, and gross cardiac hypertrophy, preserved systolic function, and interstitial fibrosis. Immunofluorescence staining showed co-localization of WT and mutant MYOZ2 proteins with α-actinin at the Z disks. Electron microscopy showed disrupted and mal-aligned Z disks in the mutant mice. Cardiac calcineurin activity, determined by quantifying Rcan1.4 mRNA and protein levels, luciferase activity in triple transgenic Myoz2+/−:NFATc-Luc:MYOZ2I246M and Myoz2+/−:NFATc-Luc:MYOZ2WT mice, and NFATc transcriptional activity assay, was unchanged in the mutant transgenic mice. However, levels of phospho-ERK1/2 and JNK54/46 were altered in the transgenic mice. Likewise, lentiviral-mediated expression of the MYOZ2I246M did not affect RCAN1.4 and calcineurin (PPP3CB) protein levels.
Conclusions
Thus, the cardiac phenotype in HCM caused by MYOZ2 mutations might be independent of calcineurin activity in the heart. Z disk abnormalities might provide the stimulus for the induction of cardiac hypertrophy caused by MYOZ2 mutations.
Keywords: Genetics, Myozenin 2, Calcineurin, Hypertrophy, Sarcomere
1. Introduction
Human hypertrophic cardiomyopathy (HCM) is a relatively common genetic disease caused by mutations in genes encoding sarcomere proteins.1,2 The phenotype is characterized by cardiac and myocyte hypertrophy, disarray, interstitial fibrosis in conjunction with preserved systolic function.2,3 HCM is the most common discernible cause of sudden cardiac death in the young and an important cause of diastolic heart failure.4
Over a dozen causal genes and several hundred mutations have been identified in patients with HCM.2 The known causal genes encode sarcomere including Z disk proteins.2 Molecular mechanistic studies have illustrated the heterogeneity of the responsible mechanisms for the cardiac phenotype in HCM.5–7 We recently identified two mutations (p.S48P and p.I246M) in MYOZ2, which encodes Z disk protein myozenin 2 (MYOZ2) or calsarcin 1 in HCM families.8 The mutations affect highly conserved amino acids.8 Because MYOZ2 is known to inhibit protein phosphatase 2B (PP2B) activity, commonly known as calcineurin,9,10 the findings invoked the calcineurin–nuclear factor of activated T cells (NFAT) pathway in the pathogenesis of HCM.11 Despite the well-established pro-hypertrophic role of the calcineurin–NFATc pathway in the heart,11,12 the role of calcineurin in the pathogenesis of HCM has remained controversial. Contrary to the well-established role of the calcineurin–NFATc pathway in experimental models of cardiac hypertrophy, treatment of organ transplant recipients with calcineurin inhibitors can cause an HCM-like phenotype, which reverses upon discontinuation.13 Similarly, treatment of a mouse model of human MyHC mutation with cyclosporine, a calcineurin inhibitor, caused deleterious effects on cardiac structure and function and led to premature mortality.14
We determine whether cardiac hypertrophy in HCM caused by the MYOZ2 mutations results from an enhanced activity of the calcineurin–NFAT pathway in the heart. We generated transgenic mice that expressed either a wild-type (WT) or a mutant MYOZ2 in the background of endogenous MYOZ2 protein. To mimic the human genotype, we generated mice hemizygous for the endogenous Myoz2 genotype (Myoz2+/−) but expressing the WT or mutant MYOZ2 in the heart. We characterized the phenotype and determined the activation of the calcineurin–NFATc pathway in the heart. In addition, we generated recombinant lentiviruses and expressed MYOZ2WT and MYOZ2I246M in neonatal rat ventricular myocytes (NRVM) and measured a regulator of calcineurin 1 (RCAN1) and calcineurin (PPP3CB) protein levels.
2. Methods
An extended version of Materials and Methods is provided as Supplementary material online.
The investigation conforms with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication, 8th Edition, 2011). The University of Texas Health Science Center Animal Care and Use Committee approved the protocol.
2.1. MYOZ2WT, MYOZ2S48P, and MYOZ2I246M transgenic mice
We generated several lines of transgenic mice (FVB background) expressing either WT or mutant (p.S48P or p.I246M) MYOZ2 in the heart under the transcriptional regulation of a cardiac-restricted α-myosin heavy chain (Myh6) promoter.15 In brief, full-length human WT MYOZ2 cDNA was cloned downstream to a 5.5 kb 5′ genomic fragment of the Myh6 gene. Three sequential FLAG sequences were positioned in frame 5′ to the MYOZ2 cDNA. Mutations were introduced by site-directed mutagenesis. The final transgene constructs were sequenced by the Sanger method in sense and anti-sense directions. The vector-purified transgenes were microinjected into single-cell embryos.
2.2. Double transgenic Myoz2+/−:MYOZ2WT and Myoz2+/−:MYOZ2I246M mice
The Myoz2 null mice have been generated and characterized by Dr Olson's group.9 To simulate the human heterozygous mutations, Myoz2−/− mice were crossed to Myh6-MYOZ2I246M mice to generate chimeric mice that express MYOZ2I246M in the background of heterozygous deficiency of endogenous MYOZ2 (Myoz2+/−). The control group included bigenic mice expressing MYOZ2WT in the background of Myoz2+/−. Because multiple lines of MYOZ2I246M were generated as opposed to a single line of MYOZ2S48P, the former was selected to generate and characterize double transgenic Myoz2+/−:MYOZ2I246M mice.
2.3. Triple transgenic Myoz+/−:MYOZ2WT:NFATc-Luc and Myoz+/−:MYOZ2I246M:NFATc-Luc mice
NFATc-Luc reporter mouse, generated by placing nine copies of an NFATc-binding site 5′ to a minimal promoter from the Myh6 gene (−164 to +16) upstream of the luciferase reporter,16 was a kind gift from Dr Jeffrey Molkentin (University of Cincinnati). Bigenic Myoz2+/−:MYOZ2WT and Myoz2+/−:MYOZ2I246M mice were crossed to NFATc-Luc mice to generate triple transgenic mice that express either MYOZ2WT or MYOZ2I246M in the background of the Myoz2+/− and the NFATc-luciferase reporter, the latter is an indicator of the activation of the calcineurin–NFATc pathway. At the genetic level, the mutant mice resemble the human genotype having one copy of the WT and one copy of the mutant MYOZ2 alleles but also harbour the NFATc-luciferase reporter construct in their genome.
2.4. Mouse genotyping
The transgenes were detected by PCR of genomic DNA extracted from mice tails. The sequences of the primers are shown in Supplementary material online, Table 1. Euthanasia was performed by CO2 inhalation followed by cervical dislocation.
2.5. Echocardiography
Cardiac structure and function in transgenic and non-transgenic (NTG) mice was assessed by the transthoracic M mode, 2D, and Doppler echocardiography using an HP 5500 Sonos echocardiography unit equipped with a 15-MHz linear transducer, as published.17–20 Mice were anaesthetized by ip injection (15 mL/kg) of sodium pentobarbital (4.13 mg/mL, equal to 62 mg/kg). The adequacy of anaesthesia was confirmed by assessing the response to pinching the toes and eyelid reflex. Indices of the cardiac size and function were calculated as described.17–19
2.6. Isolation of neonatal rat ventricular myocytes
NRVM was isolated as described with modest modifications.21 Neonatal rats were euthanized by decapitation. The heart was removed, minced into small pieces and placed in a digestion buffer containing Collagenase Type II. Upon complete digestion, cells were pelleted by centrifugation and re-suspended in a culture media containing 10% foetal bovine serum and antibiotics. The non-attached myocytes were collected and plated into six-well plates for transduction with the recombinant lentiviruses.
2.7. Recombinant lentiviruses
Flag-tagged WT and p.I246M MYOZ2 cDNAs were cloned into the lentiviral expression vector, as described.20 The lentiviral plasmid containing the MYOZ23 cDNAs and the packaging plasmids were transfected into 293T cells to generate the viruses.
2.8. Immunoblotting
Immunoblotting was performed using 30 μg of cardiac or myocytes protein extracts as described.20 To detect the transgene and total MYOZ2 (transgene and endogenous) proteins, the membranes were probed with rabbit polyclonal anti-Flag and goat polyclonal anti-MYOZ2 antibodies, respectively. To detect RCAN1 and PPP3CB (calcineurin) proteins, the membranes were probed with rabbit polyclonal anti-DSCR1 and anti-PPP3CB antibodies, respectively. Likewise, levels of selected mitogen-activated protein kinases (MAPK) involved in cardiac hypertrophic response were detected using antibodies specific against phospho-ERK1/2, total ERK1/2, phospho-JNK, total-JNK, phospho-p38, total-p38, and myosin heavy chain β (MYH7). The secondary antibodies were goat anti-mouse IgG horseradish peroxidase (HRP) conjugated, goat anti-rabbit IgG-HRP, and donkey anti-rabbit IgG-HRP. Signals were detected using chemiluminescence detection reagents.
2.9. Gross phenotype and histomorphometry
Ventricular weight, after the excision of the great vessels and atria, and body weight were measured in 8–21 mice per group. Cardiac histology was examined by H&E staining of thin myocardial sections. The extent of fibrosis was analysed by the quantification of collagen volume fraction (CVF) of Sirius Red stained thin myocardial sections. Images were analysed using the ImageTool 3.0 analysis software (http://ddsdx.uthscsa.edu/dig/itdesc.html). The myocyte cross-sectional area (CSA) was quantified after staining fresh frozen thin myocardial sections with 2 µg/mL wheat germ agglutinin conjugated with Texas-Red®. The sections were mounted in Hard SetTM mounting medium and examined under fluorescence microscopy. Morphometric analysis was performed as published.17,18
2.10. Immunofluorescence staining
Thin myocardial cryosections were fixed with 2% paraformaldehyde, permeabilized, blocked with 5% donkey serum in 0.2% PBS-Triton X, and incubated with monoclonal anti-sarcomeric α-actinin and rabbit polyclonal anti-Flag antibodies. The sections were treated with secondary antibodies conjugated with either Texas-Red or Fluorescein. NRVM were stained for F-actin 48 h of transduction with the recombinant lentiviruses using Phailloidin conjugated to Alexa Fluor® 568. Nuclei were counter-stained with DAPI (Vectorshield™).
2.11. Transmission electron microscopy
Age-matched mice (n = 3 per group) were anaesthetized by ip injection (15 mL/kg) of sodium pentobarbital (4.13 mg/mL). Mice were injected with 100 IU/mL (diluted in PBS) of heparin intraperitoneally. The isolated hearts were perfused retrogradely with calcium-free PBS and perfusion fixed with 2% paraformaldehyde + 2.5% glutaraldehyde in 0.1 M Millonig's phosphate buffer, pH7.4. The hearts were then minced, fixed for 2 days in the same fixation buffer and post-fixed in cold 1% OsO4 in 0.1 M Millonig's phosphate buffer followed by dehydration through a gradient series of ethanol concentrations. The tissue was en bloc stained with saturated uranyl acetate in 50% ethanol and was infiltrated with progressively concentrated mixtures of plastic resin and 100% ethanol. The tissue was then given three changes of pure, freshly made plastic resin, embedded in 00 BEEM capsules and placed in a 63°C oven for 18 h.
Longitudinal thin sections of ∼70 nm were obtained using an RMC MT6000-XL ultramicrotome and a Diatome Ultra45 diamond knife, and collected on 150 hex-mesh copper grids. The sections were counter-stained with Reynold's lead citrate for 4 min. Dry samples were examined on a Hitachi H7500 transmission electron microscope and images were captured using a Gatan US1000 digital camera with the Digital Micrograph v1.82.366 software.
2.12. Quantitative real time PCR
Total RNA was extracted from myocardial tissue using the Qiagen RNeasy Mini Kit. The Rcan1.4 mRNA level was determined by quantitative PCR (qPCR), using specific TaqMan Gene expression assays and normalized to the glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA level. Likewise, mRNA levels of Myh7, Nppb (B-type natriuretic peptide); Nppa (atrial natriuretic peptide), Atp2a2 (SERCA2a), and Acta1 (skeletal α-actin), markers of cardiac hypertrophy, as well as Col1a1 (procollagen 1) and Col3a1 (procollagen III); markers of fibrosis, were determined by qPCR. All reactions were performed in triplicates and in four to six mice per group and in three independent sets of transduced NRVM. Sequence of the probe used is listed in Supplementary material online, Table 1.
2.13. NFATc luciferase activity
Aliquots of 70–80 mg of heart tissues from NFATc-Luc, Myoz2+/−:NFATc-Luc, Myoz2+/−:MYOZ2WT:NFATc-Luc, and Myoz2+/−:MYOZ2I246M:NFATc-Luc were homogenized by sonication in the presence of a Luciferase Assay System Reporter Lysis Buffer and complete protease inhibitor cocktail. Protein concentrations were measured by the Bradford assay. Aliquots of 250 µg of protein extracts were applied to a 96-well plate, aliquots of 100 µL of ONE-Glo Luciferase assay buffer were added and luciferase activity was measured after 3 min using a microplate reader.
2.14. NFATc transcription activity assay
The effects of mutant MYOZ2 proteins (MYOZ2S48P and MYOZ2I246M) on specific activation of the calcineurin-dependent transcription factor NFATc1 were assessed by an ELISA-based method according to manufacturer's recommended conditions. Briefly, nuclear extracts from NTG, MYOZ2WT, MYOZ2S48P, and MYOZ2I246M hearts were prepared using NE-PER® nuclear and cytoplasmic extraction reagents with a proteinase inhibitor and a phosphatase inhibitor. Ten micrograms of nuclear extracts were applied to wells coated with oligonucleotides containing a NFAT consensus-binding site (5′-AGGAAA-3′). After incubation for 1 h at room temperature on a plate shaker an NFATc1-antibody was added to each well followed by incubation with an HRP-conjugated secondary antibody. The reaction was quantified using a microplate reader at 450 ηm with a reference wavelength of 655 ηm. Nuclear extracts from Jurkat T-cells served as a positive control.
2.15. Statistical analysis
Continuous variables were expressed as mean ± SD and differences among the continuous variables that satisfied the normality distribution were compared by the one-way analysis of variance and the Bonferroni correction to multiple comparisons. Variables that were not normally distributed were compared by the Kruskal–Wallis test. Differences in the categorical variables were analysed by the Chi-square test. All statistical analyses were performed using STATA IC v. 10.1 for Macintosh.
3. Results
3.1. Expression of MYOZ2S48P and MYOZ2I246M in the heart
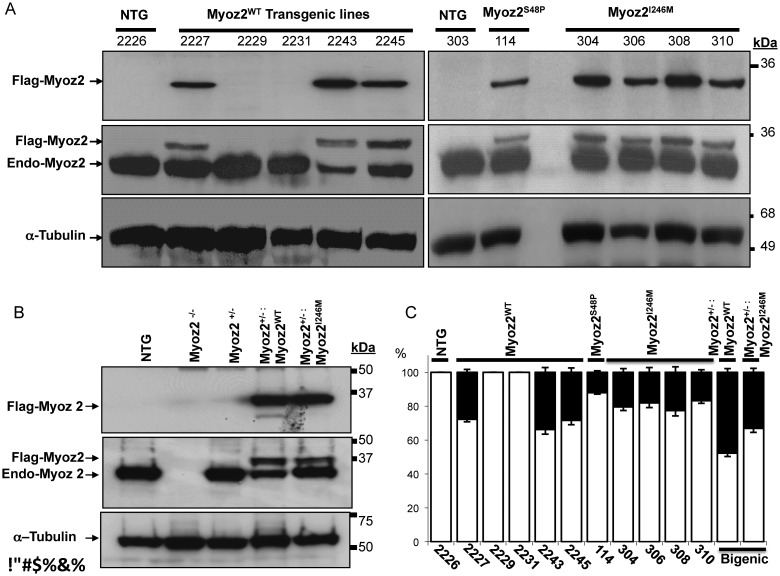
We generated five lines of MYOZ2WT and one line of MYOZ2S48P and four lines of MYOZ2I246M transgenic mice (Figure 1A). Immunoblotting of myocardial protein extracts, probed with antibodies against Flag to detect transgene MYOZ2 only or pan MYOZ2 to detect endogenous as well as Flag-tagged transgene MYOZ2 proteins simultaneously, showed expression of MYOZ2WT in three independent lines and MYOZ2S48P in one line and MYOZ2I246M in all four lines (Figure 1A). Expressions of the transgene MYOZ2WT and MYOZ2I246M in the bigenic Myoz2+/−:MYOZ2WT and Myoz2+/−:MYOZ2I246M mice were detected by immunoblotting using antibodies against Flag and Pan MYOZ2, the latter to detect and quantify transgene and endogenous MYOZ2 (Figure 1B).
Figure 1.
The expression of transgene proteins in the heart. (A) Immunoblots showing the expression of transgenic MYOZ2WT, MYOZ2S48P, and MYOZ2I246M and endogenous MYOZ2 (Endo-MYOZ2) proteins in the heart. Anti-Flag antibody detected transgene MYOZ2 only while a pan specific anti-MYOZ2 antibody detects the endogenous as well as Flag-tagged transgene MYOZ2 in non-transgenic (NTG) and transgenic mice. (B) Immunoblots showing the expression of transgenic MYOZ2WT and MYOZ2I246M in the background of the Myoz2 hemizygous genotype (Myoz2+/−). (C) Quantitative presentation of MYOZ2WT, MYOZ2S48P, and MYOZ2I246M levels as percentages of the total MYOZ2 in the heart in each line of transgenic mice (n = 3–6 mice per line).
Relative expression levels of the transgene protein to that of total MYOZ2 protein, quantified after probing the membranes with a pan MYOZ2 antibody, were 26–36% for MYOZ2WT, 12% for MYOZ2S48P and 18–23% for MYOZ2I246M (Figure 1C). Likewise, expression levels of the MYOZ2WT and MYOZ2I246M in double transgenic mice constituted ∼48 and 33% of the total MYOZ2 level in the heart, respectively (Figure 1C). There were no apparent phenotypic differences among the lines within each genotype and between the corresponding single or double transgenic mice. Accordingly, lines with the highest expression levels were used for subsequently phenotypic characterization.
3.2. Gross morphological and histological phenotypes of transgenic hearts
MYOZ2S48P and MYOZ2I246M mice survived normally up to 2 years of age. Echocardiographic findings are presented in Table 1. The mutant MYOZ2 mice exhibited echocardiographic evidence of left ventricular hypertrophy as indicated by increased interventricular septal thickness and left ventricular mass, when compared with NTG or corresponding MYOZ2WT transgenic mice. Likewise, left ventricular end diastolic diameter was modestly increased in MYOZ2I246M and Myoz2+/−:MYOZ2I246M mice, when compared with NTG mice. The left ventricular systolic function, however, was preserved. Overall, the differences were statistically significant, albeit modest in magnitude.
Table 1.
Echocardiographic phenotype in single transgenic and bigenic MYOZ2 wild-type and mutant mice
| NTG | MYOZ2WT | MYOZ2S48P | MYOZ2I246M | MYOZ2−/− | MYOZ2+/− | MYOZ2+/− :MYOZWT | MYOZ2+/− :MYOZ2I246M | P-value (ANOVA) | |
|---|---|---|---|---|---|---|---|---|---|
| n | 58 | 17 | 8 | 16 | 10 | 11 | 13 | 12 | NA |
| Sex (M/F) | 38/20 | 6/11 | 3/5 | 8/8 | 5/5 | 6/5 | 6/7 | 5/7 | 0.227 |
| Age (months) | 9.8 ± 3.4 | 11.6 ± 7.6 | 16.6 ± 5.9 | 12.3 ± 6.6 | 9.5 ± 2.0 | 9.5 ± 2.8 | 9.5 ± 5.5 | 9.5 ± 3.4 | 0.243* |
| Age (months) | 9.8 ± 3.4 | 11.6 ± 7.6 | 16.6 ± 5.9 | 12.3 ± 6.6 | 9.5 ± 2.0 | 9.5 ± 2.8 | 9.5 ± 5.5 | 9.5 ± 3.4 | 0.243* |
| Body weight (g) | 33.1 ± 4.1 | 34.6 ± 4.2 | 36.6 ± 3.8 | 36.6 ± 5.1 | 36.6 ± 5.3 | 35.3 ± 8.6 | 34.5 ± 4.2 | 35.6 ± 6.5 | 0.003* |
| Heart rate (b.p.m.) | 561.7 ± 86.2 | 650.0 ± 61.6* | 623.8 ± 76.5 | 601.3 ± 62.9 | 528.0 ± 109.3 | 5773 ± 103.6 | 580 ± 77.9 | 586.7 ± 92.8 | 0.010 |
| IVST (mm) | 0.97 ± 0.11 | 1.04 ± 0.14 | 1.18 ± 0.10**,*** | 1.10 ± 0.12** | 0.85 ± 0.10 | 0.90 ± 0.09 | 0.88 ± 0.07 | 1.03 ± 0.05****,***** | <0.0001 |
| PWT (mm) | 0.93 ± 0.12 | 0.93 ± 0.11 | 1.10 ± 0.09**,***,****** | 0.99 ± 0.12 | 0.84 ± 0.08 | 0.89 ± 0.10 | 0.83 ± 0.10 | 0.97 ± 0.10****,***** | <0.0001 |
| LVEDD (mm) | 2.85 ± 0.35 | 3.04 ± 0.31 | 3.12 ± 0.22 | 3.17 ± 0.22** | 3.47 ± 0.22** | 3.40 ± 0.26** | 3.40 ± 0.30** | 3.29 ± 0.33** | <0.0001 |
| LVESD (mm) | 1.02 ± 0.20 | 1.02 ± 0.20 | 1.06 ± 0.17 | 1.01 ± 0.15 | 1.34 ± 0.28** | 1.25 ± 0.19** | 1.13 ± 0.19 | 1.12 ± 0.30 | 0.0004 |
| LV mass (mg) | 87.9 ± 17.0 | 101.9 ± 15.4 | 132.1 ± 15.0**,***,****** | 116.9 ± 7.6**,*** | 100.4 ± 19.8 | 105.5 ± 16.7** | 99.1 ± 13.3 | 117.5 ± 17.2**,***** | <0.0001 |
| FS (%) | 64.3 ± 5.5 | 66.5 ± 5.2 | 66.0 ± 5.01 | 68.0 ± 3.4 | 61.4 ± 7.1 | 63.2 ± 4.9 | 66.7 ± 5.1 | 66.2 ± 7.0 | 0.065 |
| Vcf (mm/s) | 34.6 ± 11.4 | 44.6 ± 12.6 | 41.1 ± 8.1 | 46.1 ± 9.4** | 37.8 ± 9.6 | 41.6 ± 6.7 | 46.6 ± 9.2** | 42.7 ± 10.3 | 0.002 |
| Mitral inflow E/A | 1.17 ± 0.18 | 1.13 ± 0.16 | 1.13 ± 0.21 | 1.00 ± 0.23 | 2.21 ± 0.46** | 1.54 ± 0.26** | 1.44 ± 0.33 | 2.00 ± 0.08**,***** | <0.0001 |
| Aortic outflow velocity (m/s) | 1.11 ± 0.44 | 1.37 ± 0.15 | 1.32 ± 0.23 | 1.11 ± 0.20 | 1.22 ± 0.24 | 1.25 ± 0.22 | 1.18 ± 0.19 | 1.20 ± 0.20 | 0.022* |
n, number; NTG, non-transgenic; MYOZ2, myozenin 2; M/F, male/female; g, gram; b.p.m., beats per minutes; IVST, interventricular septal thickness; PWT, posterior wall thickness; LVEDD, left ventricular end diastolic diameter; LVESD, left ventricular end systolic diameter; LV mass, left ventricular mass; FS, fractional shortening.
*P value by the Kruskal–Wallis rank test because of departure from a normal distribution.
**P < 0.05 by the Bonferroni pairwise multiple comparison test when compared with non-transgenic.
***P < 0.05 vs. Myoz2WT.
****P < 0.05 by the Bonferroni pairwise multiple comparison test between Myoz2+/−:Myoz2I246M and Myoz2+/−:Myoz2WT.
*****P < 0.05 vs. Myoz2+/−:MYOZ2WT.
******P < 0.05 by the Bonferroni pairwise multiple comparison test between Myoz2S48P or Myoz2I246M and Myoz2WT.
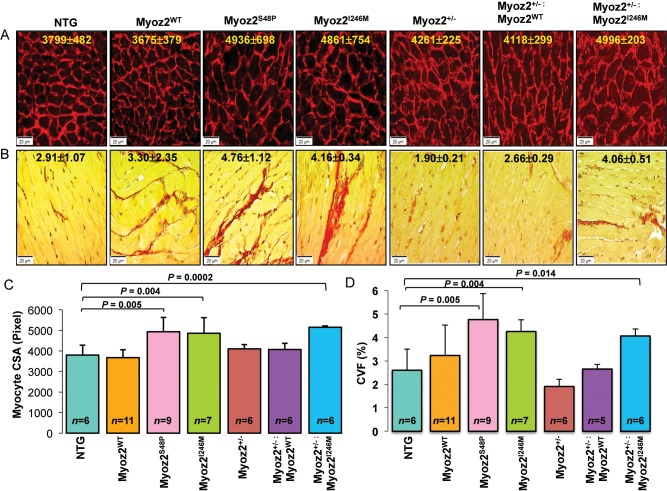
Histological examination showed increased cardiac myocytes CSA in MYOZ2S48P and MYOZ2I246M as well as in Myoz2+/−:MYOZ2I246M mice (Figure 2A). Likewise, interstitial fibrosis, as indicated by the CVF, was increased by approximately two-fold in MYOZ2S48P, MYOZ2I246M, and Myoz2+/−:MYOZ2I246M mice, when compared with NTG (Figure 2B).
Figure 2.
Cardiac myocyte hypertrophy and interstitial fibrosis in NTG and transgenic mice. (A) Texas Red conjugate wheat germ agglutinin (WGA) stained thin myocardial sections depicting myocytes cross-sectional area (CSA) in the experimental groups. The mean and standard deviation of myocytes CSA are shown. (B) Thin myocardial sections stained with Picrosirius Red to detect fibrosis (shown in red). (C) Graphical representation of CSA and (D) collagen volume fraction (CVF) quantified using ImageTool. The number of mice per group is shown in each graph.
The ventricular/body weight ratio, an index of cardiac hypertrophy, was increased in single transgenic mutant MYOZ2 mice as well in bigenic Myoz2+/−:MYOZ2I246M mice, when compared with NTG and the corresponding transgenic MYOZ2WT mice (Supplementary material online, Figure 1A).
3.3. Molecular markers of cardiac hypertrophy and fibrosis in transgenic hearts
The mRNA levels of Myh7 and Nppb, markers of cardiac hypertrophy were increased in the Myoz2+/−:MYOZ2I246M mice, when compared with Myoz2+/−:MYOZ2WT and NTG mice (Supplementary material online, Figure 1B and C). Likewise, The Co1a1 and Col3a1 mRNA level was increased in the mutant bigenic mice when compared with NTG or bigenic MYOZ2WT mice (Supplementary material online, Figure 1D and E).
3.4. Localization of mutant MYOZ2 proteins to Z disks in the myocardium
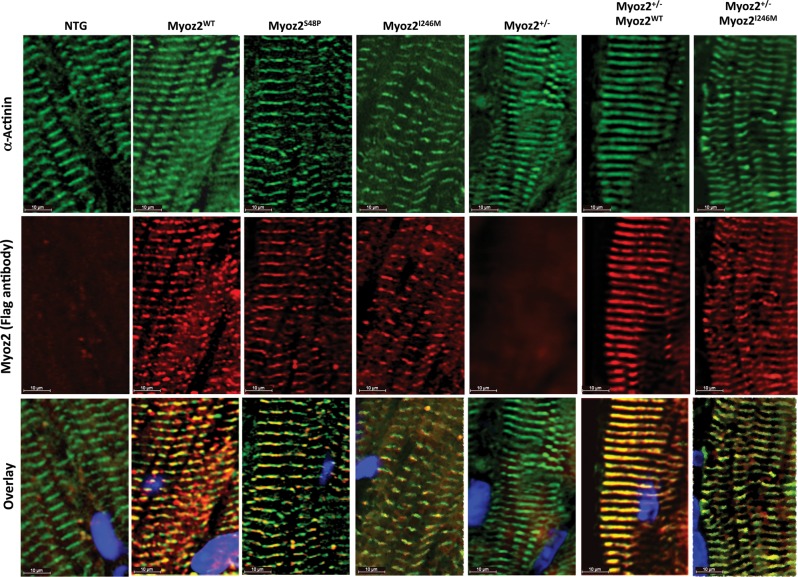
Thin myocardial sections were co-stained with antibodies against Flag to detect transgenes and α-actinin to mark the Z disks. As shown in Figure 3, both mutant proteins, whether expressed in the background of endogenous MYOZ2 or hemizygous endogenous MYOZ2 (Myoz+/−), were co-detected with α-actinin, indicating co-localization to sarcomere Z disk.
Figure 3.
Localization of transgene proteins to Z disk: immunofluorescence panels stained using anti-Flag and anti-α-actinin antibodies on myocardial cryosections showing co-localization of transgenic MYOZ2 and α-actinin at Z disks. MYOZ2 and α-actinin were co-localized in the MYOZ2WT (R = 0.745), MYOZ2S48P (R = 0.702), MYOZ2I246M (R = 0.881), Myoz2+/−:MYOZ2WT (R = 0.914), and Myoz2+/−:MYOZ2I246M (R = 839).
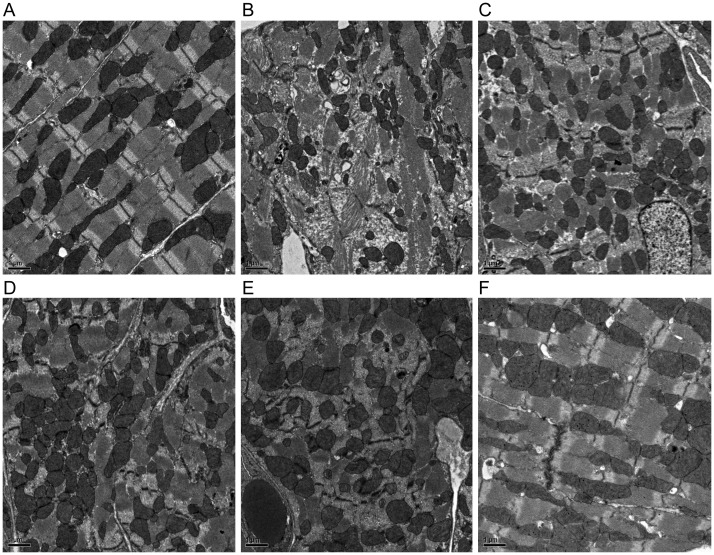
3.5. Z disks structure in the myocardium
Z disks structure analysed by transmission electron microscopy was normal in the Myoz2+/−:MYOZ2WT mice with the exception of occasional misaligned or interrupted Z disks that comprised <4% of ∼300 Z disks that were examined per each mouse heart (Figure 4A). In contrast, various Z disks abnormalities including poorly formed Z disks associated with myofibrillar disarray, interrupted, and discontinuous, bulging and slipped Z disks were observed in the mutant Myoz2+/−:MYOZ2I246M group (Figure 4B–F). The percentage (mean ± SD) of abnormal Z disks in each group, determined after the examination of ∼300 Z disks per each mouse hearts and in three mice per group, was 3.6 ± 0.9% in the MYOZ2WT and 14.3 ± 3.2% in MYOZ2I246M groups (P = 0.003).
Figure 4.
Z disk structural abnormalities: transmission electron microscopy panels showing various structural abnormalities in the Z disks in the Myoz2+/−:MYOZ2I246M bigenic mice. (A) Normal alignment and structure of the Z disk in the heart in the Myoz2+/−:MYOZ2WT mice. (B–F) Various Z disks abnormalities in the MYOZ2I246M mice, including poor Z disk assembly and myofibrillar disorganization, interrupted, poorly aligned, discontinuous, and serrated Z disks along with Z disk slippage and bulging. All magnifications are ×2000. The percentage (mean ± SD) of abnormal Z disks in each group, determined after the examination of ∼300 Z disks per each mouse hearts and in three mice per group, was 3.6 ± 0.9% in the MYOZ2WT and 14.3 ± 3.2% in MYOZ2I246M groups (P = 0.003).
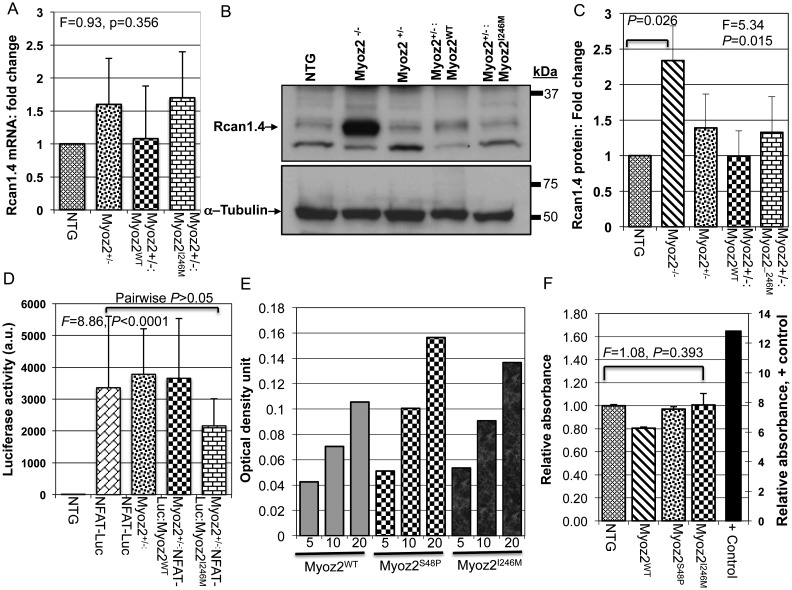
3.6. Rcan1.4 mRNA and protein levels in transgenic hearts
Rcan1.4, also known as MCIP1.4, is considered a reliable marker of the activation of the calcineurin pathway.22 We detected and quantified Rcan1.4 mRNA and protein levels by qPCR and immunoblotting, respectively. The results showed increased Rcan1.4 protein and mRNA levels by more than two-fold in Myoz2−/− mice, which served as a positive control (Figure 5A–C). However, Rcan1.4 mRNA and protein levels were not significantly different among Myoz2+/−, Myoz2+/−:MYOZ2WT Myoz2+/−:MYOZ2I246M mice and were similar to Rcan1.4 mRNA and protein levels in NTG mice (Figure 5A–C).
Figure 5.
The calcineurin–NFATc pathway: (A) quantitative Rcan1.4 mRNA levels as determined by qPCR and normalized to Gapdh mRNA levels (n = 6–8 mice per group). There were no differences in Rcan1.4 mRNA levels among the groups. (B) Representative immunoblots showing the expression of Rcan1. 4 protein in the experimental groups. The Rcan1.4 protein level was increased significantly in Myoz2 null mice, indicating increased calcineurin activity, as a positive control. Rcan1.4 protein levels were not significantly different among NTG, Myoz2+/−:MYOZ2WT, and Myoz2+/−:MYOZ2I246M mice, as quantitatively represented in (C) (n = 4 mice per group). (D) NFATc-luciferase (Luc) activity in NTG (negative control, n = 11), NFATc-Luc mice (positive control, n = 19), Double transgenic NFATc-Lu:Myoz2+/− (n = 7) and triple transgenic Myoz2+/−:NFAT-Lu:MYOZ2WT (n = 6) and Myoz2+/−:NFATc-Lu:MYOZ2I246M (n = 24). ANOVA P-value is shown as well as pairwise comparison P-value. There were no statistically significant different among the luciferase reporter mice. (E) The dose titration assay using increasing amount of nuclear protein extracts showing a dose-dependent increase in NFATc-transcriptional activity in NTG, MYOZ2WT, MYOZ2S48P, and MYOZ2I246M transgenic mice (units are microgram). (F) Levels of NFATc transcriptional activity normalized to that in NTG mice (n = 4 per group). A positive control group is included in the left side of the panel with its own scale. There were no significant differences among the groups.
3.7. NFATc-luciferase reporter assay
To determine whether the expression of mutant MYOZ2 affected activity of the calcineurin–NFATc pathway in vivo, we measured luciferase activity in triple transgenic mice expressing either a WT or mutant MYOZ2 in the background of hemizygous endogenous MYOZ2 and the luciferase reporter, regulated by the NFATc DNA-binding elements.16,23,24 Intra-assay variability was minimal as indicated by a strong correlation between three measurements (correlation coefficient 0.999). Luciferase activity measured in a relatively large number of mice did not differ significantly among the experimental groups (Figure 5D).
3.8. NFAT-c transcriptional activity in transgenic hearts
To further substantiate the observed null findings regarding Rcan1.4 mRNA and protein levels and NFATc-Luciferase activity in the mutant transgenic mice, we detect and quantify calcineurin-dependent activation of transcription factor NFATc1. We tested three different doses of nuclear protein extracts and found a dose-dependent increase in NFATc activity in cardiac protein extracts from NTG, MYOZ2WT, and MYOZ2I246M mice (Figure 5E). We measured NFATc transcriptional activity in four mice per group. The mean NFATc transcriptional activity did not differ significantly among the experimental groups (Figure 5F).
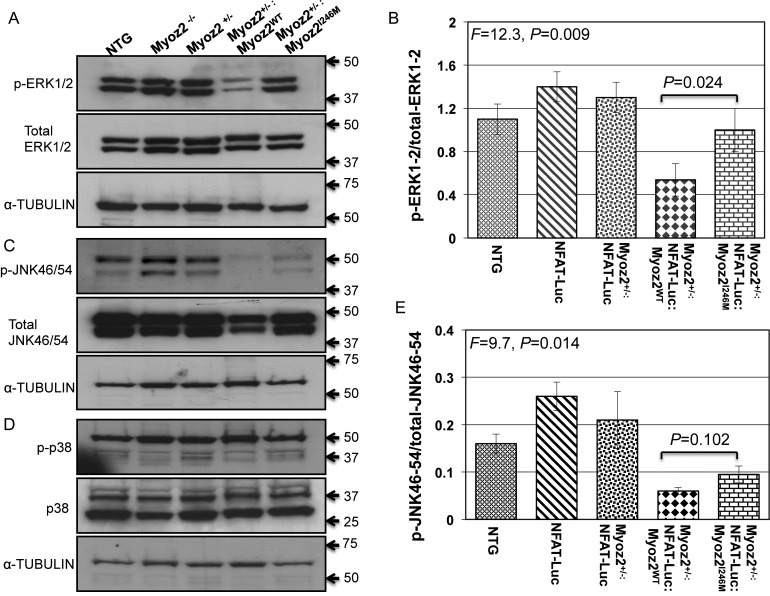
3.9. Selected MAPK levels involved in cardiac hypertrophy in transgenic hearts
Levels of phospho-ERK1/2 were reduced in the heart of the Myoz2+/−:MYOZ2WT mice when compared with NTG mice (Figure 6). However, phospho-ERK1/2 levels were increased in the Myoz2+/−:MYOZ2I246M mice when compared with Myoz2+/−:MYOZ2WT mice. Levels of total ERK1/2 were not significantly different among the experimental groups (Figure 6). In addition, levels of phospho-JNK54/46, but not total JNK54/46 in the heart, were reduced in the Myoz2+/−:MYOZ2WT and Myoz2+/−:MYOZ2I246M mice when compared with the NTG mice. There were no significant differences in the myocardial levels of phospho-p38 and total p38 among the experimental groups.
Figure 6.
The activation of selected mitogen-activated protein kinases involved in cardiac hypertrophy: (A) levels of phospho-ERK1/2 (upper blot), total ERK1/2 (middle blot), and the corresponding blot for α-tubulin (lower blot) in the experimental group. Corresponding quantitative data are shown in (B). As shown phospho-ERK1/2 levels were reduced in the Myoz2+/−:MYOZ2WT as opposed to the corresponding levels in the NTG mice or the Myoz2+/−:MYOZ2I246M mice. (C) Levels of phospho-p38 (upper blot), total p38 (middle blot), and α-tubulin in the experimental groups. There were no significant differences in the ratio of phosphor-p38/total p38 among the experimental groups (D). (E) Levels of phospho-JNK54/46 (upper blot), total JNK54/46 (middle blot), and the corresponding α-tubulin levels (lower blot). A shown (F) relative levels of phospho-JNK54/46 normalized to total JNK54/46 levels were significantly reduced in Myoz2+/−:MYOZ2WT and Myoz2+/−:MYOZ2I246M mice when compared with levels in the NTG mice. However, there were no significant differences between the wild-type and mutant bigenic mice groups.
3.10. Expression of MYOZ2WT and MYOZ2I246M in NRVM
There were no significant difference in mRNA levels of markers of cardiac hypertrophy, namely Myh6, Myh7, Nppb, Nppa, ATP2a, and Acta1 between NRCM transduced with recombinant lentiviruses expressing MYOZWT and MYOZ2I246M (Supplementary material online, Figure 2). Likewise, there were no discernible differences in the protein levels of RCAN1.4, PPP3CB, ERK1/2, as well as in cell morphology, and F-actin distribution between NRVM transduced with the WT or mutant MYOZ2 viruses (Supplementary material online, Figure 3).
4. Discussion
The impetus for the study was the unsettled role of calcineurin (PP2B) in human HCM.13,14 Identification of causal mutations in MYOZ2,8 an inhibitor of calcineurin,9,10 in HCM families led us to propose that MYOZ2 mutations cause HCM in humans through the removal of the inhibitory effect of MYOZ2 on calcineurin–NFATc activity. Consequently, we postulated that the cardiac phenotype in HCM caused by the MYOZ2 mutations results from the activation of the calcineurin–NFATc pathway. Data in the present study show that the expression of mutant MYOZ2 proteins recapitulates an HCM-like phenotype, characterized by cardiac and myocytes hypertrophy, interstitial fibrosis and preserved systolic function. However, MYOZ2 mutations had no discernible effects on calcineurin–NFATc activity, as determined by four complementary methods of detecting Rcan1.4 mRNA and protein levels, a reliable indicator of calcineurin–NFATc activity,22 and measuring calcineurin-activated NFATc transcriptional activity and calcineurin–NFATc-regulated luciferase activity in mice.16,23,24 Similarly, the expression of the mutant MYOZ2I246M in NRVM did not have a significant effect on RCAN1.4 and PPP3CB protein levels. These results reject our primary hypothesis and indicate that the induction of cardiac hypertrophy in mouse models of HCM caused by the MYOZ2 mutations is independent of the effects of MYOZ2 on calcineurin pro-hypertrophic activity.
MYOZ2 is a well-established inhibitor of calcineurin activity.10 The p.S48P and p.I246M, identified in families with HCM, involve highly conserved amino acids, as reported previously.8 However, the mutations do not involve the known MYOZ2 binding domains for calcineurin (amino acids 217–240) and α-actinin (amino acids 153–200).10 We substantiated the null effects of MYOZ2 mutations on calcineurin activity, by multiple complementary methods. The results across methodological platforms were concordant, indicating no discernible effect. Despite the null effect on calcineurin activity, transgenic mice expressing mutant MYOZ2S48P and MYOZ2I246M in the background of endogenous MYOZ2 recapitulated an HCM-like phenotype. Likewise, the expression of the mutant MYOZ2 in the background of hemizygous deletion of endogenous Myoz2 (Myoz+/−), which emulate the genotype of human patients with HCM, also induced a hypertrophic phenotype. However, calcineurin activity was unchanged in these mice.
The results concerning the null effect of the mutant MYOZ2 on calcineurin activity are subject to several potential limitations. The expression level of the mutant MYOZ2 protein was <50% of the total MYOZ2 level in all five independent lines of mutant MYOZ2 mice, which is lower than the expected level in humans with heterozygous MYOZ2 mutations. Likewise, in the bigenic Myoz2+/−:MYOZ2I246M mice, the mutant MYOZ2I246M comprised ∼35% of the total MYOZ2 protein. One might speculate that the lack of a discernible effect on calcineurin–NFATc transcriptional activity reflects <50% level of the mutant MYOZ2 protein in the heart. Although the p.S48P and p.I246M involve conserved amino acids,8 residue 246 differs in humans (isoleucine) and mouse (valine). In addition, several other differences exist between human and mouse MYOZ2 proteins, which have 233/264 (88%) amino acid identify. Thus, species-specific differences between mouse (endogenous) and human MYOZ2 (transgene) might confound the results regarding the effects of the mutations on calcineurin activity. The data also do not exclude possible activation of the calcineurin–NFATc pathway at the level that is beyond the resolution of four different detection methods that were utilized in the present study. In addition, dissociation of the induction of cardiac hypertrophy from the calcineurin–NFATc activity does not exclude potential contribution of the calcineurin–NFATc hypertrophic pathway to the progression of cardiac hypertrophy or the response of cardiac hypertrophy to increased external load, such as exercise or pressure overload. We did not subject these mice to external stress for two main reasons; first, because they exhibited cardiac hypertrophy, albeit mild, in the absence of external stimuli; and secondly, because we intended to model human HCM, which by definition expresses cardiac hypertrophy in the presence of normal loading conditions.
We generated and characterized conventional transgenic mice for two mutations in MYOZ2 but only bigenic mice for the p.I246M mutation (Myoz2+/−:MYOZ2I246M). We did not express mutant MYOZ2S48P in the hemizygous background of the endogenous MYOZ2, as we had generated only one conventional transgenic line of the MYOZ2S48P mutation. In contrast, we had generated and characterized four independent conventional transgenic MYOZ2I246M line. Therefore, we chose the latter, as the expression of a similar cardiac phenotype in independent transgenic lines practically abrogates the potential confounding effects of insertional mutagenesis, which is an inherent concern in a single transgenic line.
The mutant MYOZ2 mice exhibited cardiac hypertrophy as detected at the organ, cellular, and molecular levels. The expression of cardiac hypertrophy, however, was relatively mild. The stimulus for cardiac hypertrophy in these transgenic mice is unlikely to be gross mechanical dysfunction as cardiac systolic function, as assessed by echocardiography, was normal. However, various Z disk abnormalities were present in the mutant MYOZ2 bigenic mice, which might provide the initial impetus for the induction of cardiac hypertrophy, independent of the calcineurin pathway. Z disks are increasingly recognized as signalling hubs for the cardiac hypertrophic response,25 even though the precise molecular events emanating from the Z disks are not fully known. Among the notable changes in the MYOZ2 transgenic mice were comparatively higher levels of phospho-ERK1/2 in the mutant MYOZ2I246M transgenic mice relative to MYOZ2WT transgenic mice. Despite the well-established role of ERK1/2 in cardiac hypertrophy,26 the data in the present manuscript are inadequate to conclude the activation of ERK1/2 as the responsible mechanism for the cardiac phenotype in the mutant MYOZ2 transgenic mice because levels of phospho-ERK1/2 in the Myoz2+/−:MYOZ2WT were reduced. Similarly, levels of phosph-JNK54/46 were also reduced. These changes might reflect functional differences between human (transgene) MYOZ2 and the mouse MYOZ2, particularly in influencing the hypertrophic signalling pathways including the calcineurin–NFATc pathway, which might compound interpretations of the results.
In summary, our data show that the expression of mutant MYOZ2 proteins, responsible for HCM in humans, induces a mild cardiac phenotype that resembles human HCM. Notably these mice exhibit cardiac and myocytes hypertrophy, myofibrillar disarray, Z disk structural abnormalities, interstitial fibrosis, and preserved systolic function. However, despite the well-established role of MYOZ2 as an inhibitor of calcineurin,9,10,22 the induction of the cardiac phenotype in the mutant transgenic mice was independent of the calcineurin–NFATc pathway. These findings imply that calcineurin inhibitors might not be plausible agents for treatment of human patients with HCM caused by MYOZ2 mutations.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported in part by grants from the National Heart, Lung, and Blood Institute (R01-088498), National Institute of Aging (R21 AG038597), Burroughs Wellcome Award In Translational Research (1005907), Greater Houston Community Foundation (TexGen), and George and Mary Josephine Hamman Foundation.
Supplementary Material
Acknowledgements
The authors wish to acknowledge technical help of Ms. Debra Townley at Baylor College of Medicine Integrated Microscopy Core.
Conflict of interest: none declared.
References
- 1.Seidman CE, Seidman JG. Identifying sarcomere gene mutations in hypertrophic cardiomyopathy: a personal history. Circ Res. 2011;108:743–750. doi: 10.1161/CIRCRESAHA.110.223834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marian AJ. Hypertrophic cardiomyopathy: from genetics to treatment. Eur J Clin Invest. 2010;40:360–369. doi: 10.1111/j.1365-2362.2010.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho CY, Lopez B, Coelho-Filho OR, Lakdawala NK, Cirino AL, Jarolim P, et al. Myocardial fibrosis as an early manifestation of hypertrophic cardiomyopathy. N Engl J Med. 2010;363:552–563. doi: 10.1056/NEJMoa1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maron BJ, Doerer JJ, Haas TS, Tierney DM, Mueller FO. Sudden deaths in young competitive athletes: analysis of 1866 deaths in the United States, 1980–2006. Circulation. 2009;119:1085–1092. doi: 10.1161/CIRCULATIONAHA.108.804617. [DOI] [PubMed] [Google Scholar]
- 5.Wen Y, Pinto JR, Gomes AV, Xu Y, Wang Y, Potter JD, et al. Functional consequences of the human cardiac troponin i hypertrophic cardiomyopathy mutation r145g in transgenic mice. J Biol Chem. 2008;283:20484–20494. doi: 10.1074/jbc.M801661200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowey S, Lesko LM, Rovner AS, Hodges AR, White SL, Low RB, et al. Functional effects of the hypertrophic cardiomyopathy r403q mutation are different in an alpha- or beta-myosin heavy chain backbone. J Biol Chem. 2008;283:20579–20589. doi: 10.1074/jbc.M800554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frey N, Brixius K, Schwinger RH, Benis T, Karpowski A, Lorenzen HP, et al. Alterations of tension-dependent atp utilization in a transgenic rat model of hypertrophic cardiomyopathy. J Biol Chem. 2006;281:29575–29582. doi: 10.1074/jbc.M507740200. [DOI] [PubMed] [Google Scholar]
- 8.Osio A, Tan L, Chen SN, Lombardi R, Nagueh SF, Shete S, et al. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2007;100:766–768. doi: 10.1161/01.RES.0000263008.66799.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frey N, Barrientos T, Shelton JM, Frank D, Rutten H, Gehring D, et al. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat Med. 2004;10:1336–1343. doi: 10.1038/nm1132. [DOI] [PubMed] [Google Scholar]
- 10.Frey N, Richardson JA, Olson EN. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc Natl Acad Sci USA. 2000;97:14632–14637. doi: 10.1073/pnas.260501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 13.Turska-Kmiec A, Jankowska I, Pawlowska J, Kalicinski P, Kawalec W, Tomyn M, et al. Reversal of tacrolimus-related hypertrophic cardiomyopathy after conversion to rapamycin in a pediatric liver transplant recipient. Pediatr Transplant. 2007;11:319–323. doi: 10.1111/j.1399-3046.2006.00633.x. [DOI] [PubMed] [Google Scholar]
- 14.Fatkin D, McConnell BK, Mudd JO, Semsarian C, Moskowitz IG, Schoen FJ, et al. An abnormal ca(2+) response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. J Clin Invest. 2000;106:1351–1359. doi: 10.1172/JCI11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulick J, Subramaniam A, Neumann J, Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J Biol Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- 16.Wilkins BJ, Dai YS, Bueno OF, Parsons SA, Xu J, Plank DM, et al. Calcineurin/nfat coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ Res. 2004;94:110–118. doi: 10.1161/01.RES.0000109415.17511.18. [DOI] [PubMed] [Google Scholar]
- 17.Lombardi R, Bell A, Senthil V, Sidhu J, Noseda M, Roberts R, et al. Differential interactions of thin filament proteins in two cardiac troponin t mouse models of hypertrophic and dilated cardiomyopathies. Cardiovasc Res. 2008;79:109–117. doi: 10.1093/cvr/cvn078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marian AJ, Senthil V, Chen SN, Lombardi R. Antifibrotic effects of antioxidant n-acetylcysteine in a mouse model of human hypertrophic cardiomyopathy mutation. J Am Coll Cardiol. 2006;47:827–834. doi: 10.1016/j.jacc.2005.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardi R, da Graca Cabreira-Hansen M, Bell A, Fromm RR, Willerson JT, Marian AJ. Nuclear plakoglobin is essential for differentiation of cardiac progenitor cells to adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ Res. 2011;109:1342–1353. doi: 10.1161/CIRCRESAHA.111.255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen SN, Czernuszewicz G, Tan Y, Lombardi R, Jin J, Willerson JT, et al. Human molecular genetic and functional studies identify trim63, encoding muscle ring finger protein 1, as a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2012;111:907–919. doi: 10.1161/CIRCRESAHA.112.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsybouleva N, Zhang L, Chen S, Patel R, Lutucuta S, Nemoto S, et al. Aldosterone, through novel signaling proteins, is a fundamental molecular bridge between the genetic defect and the cardiac phenotype of hypertrophic cardiomyopathy. Circulation. 2004;109:1284–1291. doi: 10.1161/01.CIR.0000121426.43044.2B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank D, Kuhn C, van Eickels M, Gehring D, Hanselmann C, Lippl S, et al. Calsarcin-1 protects against angiotensin-II induced cardiac hypertrophy. Circulation. 2007;116:2587–2596. doi: 10.1161/CIRCULATIONAHA.107.711317. [DOI] [PubMed] [Google Scholar]
- 23.Braz JC, Bueno OF, Liang Q, Wilkins BJ, Dai YS, Parsons S, et al. Targeted inhibition of p38 mapk promotes hypertrophic cardiomyopathy through upregulation of calcineurin-nfat signaling. J Clin Invest. 2003;111:1475–1486. doi: 10.1172/JCI17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Q, Bueno OF, Wilkins BJ, Kuan CY, Xia Y, Molkentin JD. C-jun n-terminal kinases (jnk) antagonize cardiac growth through cross-talk with calcineurin-nfat signaling. EMBO J. 2003;22:5079–5089. doi: 10.1093/emboj/cdg474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautel M. The sarcomere and the nucleus: functional links to hypertrophy, atrophy and sarcopenia. Adv Exp Med Biol. 2008;642:176–191. doi: 10.1007/978-0-387-84847-1_13. [DOI] [PubMed] [Google Scholar]
- 26.Rose BA, Force T, Wang Y. Mitogen-activated protein kinase signaling in the heart: angels vs. demons in a heart-breaking tale. Physiol Rev. 2010;90:1507–1546. doi: 10.1152/physrev.00054.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.