SUMMARY
During infection of the nervous system, alphaherpesviruses, including pseudorabies virus (PRV) use retrograde axonal transport to travel towards the neuronal cell body and anterograde transport to traffic back to the cell periphery upon reactivation from latency. The PRV protein Us9 plays an essential but unknown role in anterograde viral spread. To determine Us9 function, we identified viral and host proteins that interact with Us9 and explored the role of KIF1A, a microtubule-dependent kinesin-3 motor involved in axonal sorting and transport. Viral particles are co-transported with KIF1A in axons of primary rat superior cervical ganglion neurons and overexpression or disruption of KIF1A function respectively increases and reduces anterograde capsid transport. Us9 and KIF1A interact early during infection with the aid of additional viral protein(s) but exhibit diminished binding at later stages when capsids typically stall in axons. Thus, alphaherpesviruses repurpose the axonal transport and sorting pathway to spread within their hosts.
INTRODUCTION
Alphaherpesviruses are pathogens of the nervous systems of their mammalian hosts. Well-studied alphaherpesviruses include human herpes simplex virus types 1 and 2 (HSV-1 and HSV-2), varicella zoster virus (VZV), as well as the veterinary pathogen pseudorabies virus (PRV) (Pellett and Roizman, 2007). Neuronal spread of infection requires bidirectional transport of viral particles over long distances in axons (Smith, 2012). Virions enter termini of the peripheral nervous system (PNS) and undergo retrograde transport towards the cell body to establish lifelong latent infection. Following reactivation from latency, newly assembled virions are sorted into axons and transported in the anterograde direction back out towards the periphery (Smith, 2012). For PRV, the highly conserved Us9 protein is required for anterograde spread of infection both in vitro and in vivo (Ch’ng and Enquist, 2005). However, the molecular mechanism underlying Us9-dependent anterograde spread has remained elusive.
Prior to axonal sorting, PRV nucleocapsids and tegument proteins bud into a membrane vesicle derived from the trans-Golgi network (TGN) through a process known as secondary envelopment. This produces an enveloped virion surrounded by a transport vesicle that contains viral and host transmembrane proteins (Smith, 2012). Us9 is a type II tail-anchored membrane protein that is incorporated within these particles and is oriented such that its functional motifs can interact with cytoplasmic proteins (Brideau et al., 1998). By live cell imaging of rat superior cervical ganglion (SCG) neurons infected with PRV strains expressing functional GFP-tagged Us9 fusion proteins, we demonstrated that GFP-Us9 co-transports with anterograde directed viral particles in axons (Taylor et al., 2012). In the absence of Us9, viral particles are assembled in cell bodies but are unable to undergo axonal sorting (Lyman et al., 2007). Combined, these findings suggest that Us9 functions by engaging previously unidentified viral or host proteins that mediate axonal sorting and transport of intracellular virions.
Here, we used mass spectrometry to identify proteins that specifically co-purified with GFP-Us9 from infected cells. Among these was KIF1A, a microtubule-dependent kinesin-3 motor that has a well-defined role in axonal sorting and transport of other axonal cargoes (Klopfenstein et al., 2002; Lo et al., 2011). We demonstrate that viral particles are co-transported with KIF1A and that overexpression of dominant negative KIF1A proteins disrupts anterograde transport of viral particles. Our results suggest that alphaherpesviruses repurpose the synaptic vesicle sorting pathway for efficient spread within their hosts.
RESULTS
Identification of GFP-Us9 interactions with viral and host proteins
To identify Us9-associated proteins, we isolated GFP-Us9 from PRV infected cells by immunoaffinity purification (IP). We used neuronal growth factor (NGF) differentiated PC12 cells, which can be readily cultured in sufficient quantities for these experiments. Differentiated PC12 cells possess many of the characteristics of sympathetic neurons, including long polarized neurites that label with classic axonal markers (Lyman et al., 2008). Importantly, the Us9-null phenotype observed in SCG neurons is recapitulated in differentiated PC12 cells (Lyman et al., 2007; Lyman et al., 2008). Since Us9 must be incorporated into lipid rafts in order to facilitate axonal sorting (Lyman et al., 2008), protein extraction conditions were chosen to preserve protein interactions that depend on Us9’s presence within these membrane domains (see Experimental Procedures). Differentiated PC12 cells were infected with PRV strains that express GFP-Us9 (PRV 340) or GFP (PRV 151) as a control. At 20 hours post infection (hpi), IPs were performed with anti-GFP antibodies (Figure 1A). The efficiency of GFP-Us9 purification was assessed by Coomassie staining and western blot analysis (Figures 1B and 1C). GFP-Us9 consistently migrated as three bands on 10% SDS polyacrylamide gels; these bands reacted with antibodies against GFP and Us9 by western blot (Figure 1C). Untagged Us9, when resolved on 12.5% SDS polyacrylamide gels, also migrates as three bands (Brideau et al., 1998). We attribute this migration pattern to Us9’s phosphorylation state (Brideau et al., 2000) or a common result of membrane protein biochemistry (Rath et al., 2009). Overall, we conclude that GFP-Us9 was efficiently purified from infected cells (Figures 1B and 1C).
Figure 1. Identification of GFP-Us9 interacting proteins.
(A) Schematic diagram for parallel IPs of GFP (PRV 151) and GFP-Us9 (PRV 340) from PRV infected PC12 cells. (B) Representative Coomassie-stained gel of a GFP-Us9 IP (4–12% polyacrylamide gel). (C) Western blot analysis of GFP and GFP-Us9 IP samples with anti-GFP and anti-Us9 antibodies (10% polyacrylamide gel). (D) Visualization of putative Us9-associated proteins at 20 hpi. Proteins were manually categorized based on their biological function or subcellular localization and color-coded according to their enrichment value. Proteins highlighted with blue borders were depleted or not detected in at least one of the functionally defective GFP-Us9 IP samples compared to wild type GFP-Us9 as presented in Figure S2. See also Figure S1 and Table S1.
To identify Us9 interactions by mass spectrometry, immunoisolates of GFP-Us9 and GFP were resolved by SDS-PAGE, subjected to in-gel trypsin digestion, and analyzed by nLC-tandem MS (MS/MS) using an LTQ-Orbitrap Velos mass spectrometer (Figure 1A). The specificity of protein interactions was assessed using a label-free spectral counting approach to calculate the relative enrichment of each protein in the GFP-Us9 samples compared to the GFP controls (Tsai et al., 2012). Proteins with an average enrichment of at least twofold and a minimum of eight spectra in both biological replicates were considered putative Us9-associated proteins (see Experimental Procedures). Overall, 63 proteins met these criteria (Figure 1D and Table S1). As expected, Us9 was the most enriched protein. Ten additional viral proteins were enriched in the GFP-Us9 samples, eight of which represent structural proteins belonging to the capsid (UL17), tegument (Us2, EP0, UL42), and envelope (UL43, as well as glycoproteins gE, gC, gB, and gM) layers of PRV extracellular virions (Kramer et al., 2011).
Among the cellular proteins that co-isolated with Us9, one of the most notable was KIF1A, a member of the kinesin-3 family of microtubule-dependent molecular motor (Figure 1D). Previous studies have established that KIF1A mediates axonal sorting and long distance transport of axonal cargoes, such as synaptic vesicle precursors and dense core vesicles (Klopfenstein et al., 2002; Lo et al., 2011). In addition to KIF1A, four other proteins known to be involved in synaptic vesicle dynamics were detected. These proteins include the vesicle SNARE (v-SNARE) VAMP2, as well as the target SNARE (t-SNARE) subunits SNAP-25, VTI1B, and syntaxin-6. We also identified 14 proteins that are involved in an assortment of intracellular and intercellular signaling pathways, such as regulators of GTPase activity, G-protein coupled receptors, neurotransmitters, and neuronal growth and morphogenesis (Figure 2). Two of the most highly enriched proteins within this category are the GTPase activating proteins RABGAP1 and RABGAP1-like. We also detected the E3 ubiquitin ligase NEDD4, which has been shown to interact with the HSV-2 membrane protein UL56 (Ushijima et al., 2008). In agreement with our findings during PRV infection, HSV-2 UL56 has also been shown to interact with KIF1A by yeast two-hybrid and in vitro purifications (Koshizuka et al., 2005).
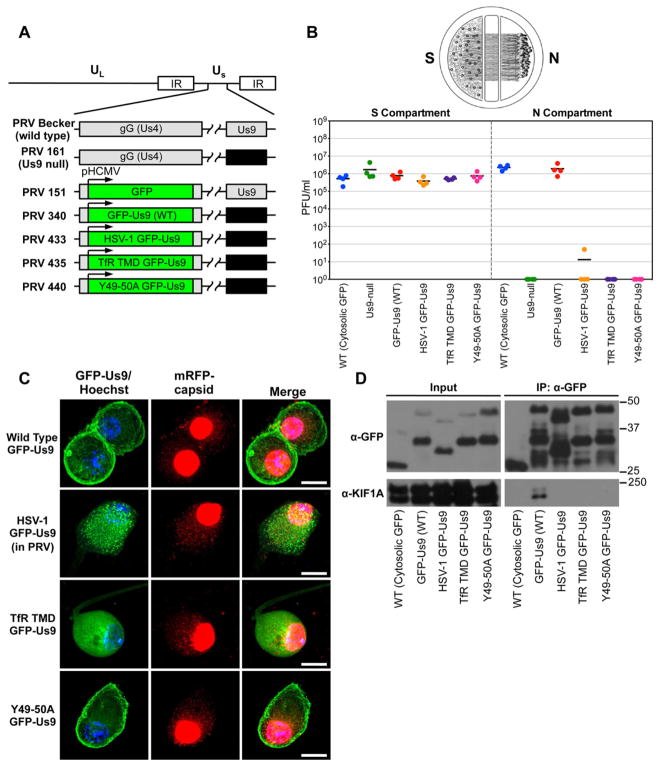
Figure 2. Characterization of PRV strains expressing wild type and functionally defective GFP-Us9 proteins.
(A) Schematic representation of the genomes of PRV Becker (wild type), PRV 161 (Us9 null), PRV 151 (diffusible GFP), PRV GFP-Us9 (wild type), PRV HSV-1 GFP-Us9 (HSV-1 Us9 in PRV), PRV TfR TMD GFP-Us9 (the transmembrane domain of Us9 was swapped for that of the transferrin receptor, thus making it non-lipid raft associated), PRV Y49-50A GFP-Us9 (mutated PRV Us9 in which the tyrosine residues at positions 49–50 have been substituted with alanines). The GFP and GFP-Us9 proteins are expressed ectopically from the HCMV promoter in the non-essential gG locus of the PRV genome. (B) Quantification of the efficiency of anterograde axonal spread using a chambered neuronal culture system. Cell bodies in the S compartment were infected at a high MOI with the indicated PRV strains. Four chambers were used for each viral strain. At 24 hpi, the entire content of the S and N compartments were harvested separately and titered on PK15 cells using a standard plaque assay. (C) Confocal microscopy of SCG neuron cell bodies infected with PRV strains expressing wild type and functionally defective GFP-Us9 proteins at 20 hpi (scale bars = 15 μm). (D) Western blot analysis of IPs from differentiated PC12 cells that are infected (at 20 hpi) with PRV strains that express the indicated GFP-tagged proteins. See also Figure S2 and Table S2.
Temporal analysis of GFP-Us9 interactions over the course of infection
We next monitored the dynamics of viral and host proteins associated with Us9 over the course of infection (Figure S1). Western blot analysis showed that KIF1A co-purified most efficiently with GFP-Us9 at 8 and 12 hpi, and then was detectable in diminishing amounts at 18 and 24 hpi (Figure S1A). The reduced amount of co-purified KIF1A at later times post infection is consistent with previous measurements showing that the number of stalled capsids in axons increases over the course of infection (Smith et al., 2001). Reduced co-purification of KIF1A with GFP-Us9 is likely correlated with depletion of KIF1A in the input cell lysates over time in both PC12 cells and SCG neurons (Figures S1A and S1B). Furthermore, efficient reduction of KIF1A protein levels is dependent on Us9 expression (Figure S1B).
Mass spectrometry-based proteomics and spectral counting analyses were used to identify proteins that were reproducibly detected (Table S1). Our results suggest that most proteins consistently co-purify with GFP-Us9 over the course of infection (Figure S1C-F). Additionally, by mass spectrometry we detected phosphorylation of five Us9 serine residues, including two previously identified sites (Ser51 and 53) (Brideau et al., 2000), and three additional sites (Ser38, 46, and 59) (Figure S1 G–I).
Analysis of mutant PRV strains expressing functionally defective GFP-tagged Us9 proteins
To validate these proteomic findings and gain insight into their functional relationship with Us9, we took advantage of three mutant viral strains that express functionally defective GFP-tagged Us9 proteins (Figure 2A). These included HSV-1 Us9 (HSV-1 GFP-Us9), non-lipid raft associated Us9 (TfR TMD GFP-Us9), and Us9 with mutations in an essential dityrosine motif (Y49-50A GFP-Us9). To confirm that these PRV mutants are defective in anterograde neuronal spread, we utilized a previously described compartmentalized neuronal culture system (Ch’ng and Enquist, 2005) (Figure 2B). The anterograde axonal spread defect of these mutants was similar to that observed during infection with a PRV Us9 null strain.
In neuron cell bodies, wild type GFP-Us9 localized to the plasma membrane as well as intracellular vesicular puncta (Figure 2C). The Y49-50A GFP-Us9 displayed a similar localization pattern. In contrast, the HSV-1 GFP-Us9 appeared to localize more prominently to intracellular punctate structures and less to the plasma membrane. Finally, the TfR TMD GFP-Us9 displayed a diffuse or reticular localization pattern that was unlike any of the other three GFP-tagged Us9 proteins.
We next performed IPs from PC12 cells infected with the GFP-tagged Us9 mutants. Western blot analysis confirmed equivalent expression levels of GFP-tagged Us9 proteins from these strains and similar IP efficiencies (Figure 2D). Mass spectrometry-based spectral counting analysis of the IP samples was used to identify and compare proteins that co-purify with the wild type and functionally defective GFP-Us9. Overall, we identified 110 proteins enriched versus the GFP-Us9 control that were detected in at least one of the samples (Figure S2 and Table S2). Notably, KIF1A was the only protein exclusively detected in the wild type GFP-Us9 sample (Figure S2). This was confirmed by western blot analysis (Figure 2D). This conclusion is consistent with the inability of these proteins to facilitate anterograde spread of PRV infection, and confirms that the interaction between wild type Us9 and KIF1A confers both physical and functional specificity.
KIF1A co-transports with anterograde-directed viral particles in axons of primary SCG neurons
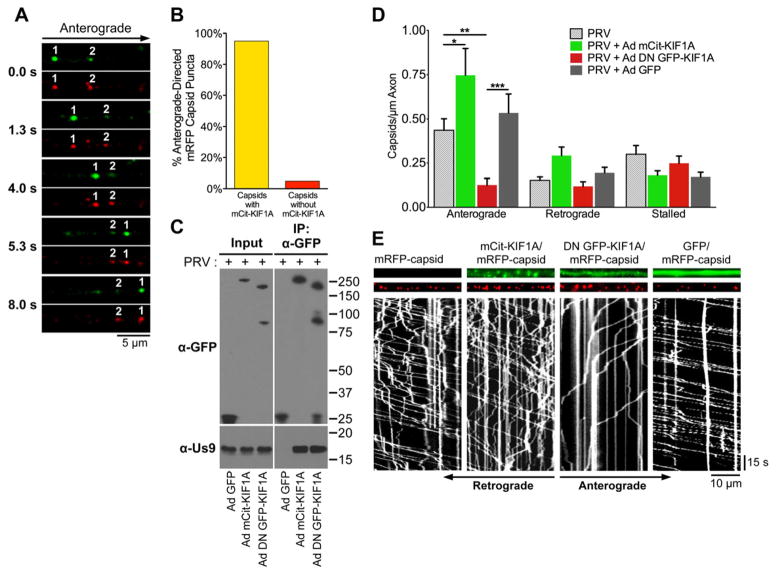
We next used live cell imaging to directly test whether viral particles are co-transported with KIF1A in axons. Since primary SCG neurons are challenging to transfect, we utilized a previously characterize non-replicating adenoviral transduction vector (AdEasy) that is widely used for protein expression in many cell types (Luo et al., 2007). SCG neurons were transduced with an adenovirus vector that expresses mCitrine-tagged KIF1A (Ad mCit-KIF1A) and co-infected with a PRV strain that expresses mRFP-tagged capsid proteins (mRFP-VP26). Visualization of these two fluorescent proteins by live cell imaging revealed that anterograde-directed but not retrograde-directed capsid puncta co-transport with KIF1A in axons (Figure 3A and Movie S1). KIF1A was detectable on over 95% of anterograde-directed capsids (Figure 3B). KIF1A puncta were also seen moving independently of capsids, but did not associate with retrograde-directed capsid puncta. Similarly, we also found that mCherry-tagged viral membrane glycoprotein gM (gM-mCherry) puncta co-transport with KIF1A in axons (Movie S1). To confirm that mCitrine-KIF1A interacts with Us9, we performed reciprocal co-IP experiments from differentiated PC12 cells that were transduced with Ad mCit-KIF1A and co-infected with wild type PRV (PRV Becker). Us9 specifically co-purified with mCitrine-KIF1A but not with a parallel control in which cells were transduced with an adenoviral vector that expresses diffusible GFP (Ad GFP; Figure 3C).
Figure 3. KIF1A mediates anterograde-directed transport of viral particles in axons.
(A) Live cell imaging of KIF1A (green) and capsid puncta (red) in axons of SCG neurons. Neurons were transduced with Ad mCit-KIF1A. At 4 days post transduction, neurons were infected with PRV mRFP-capsids and imaged between 8–12 hours post PRV infection. (B) Quantification of the percentage of anterograde-directed mRFP-tagged capsid puncta (mRFP-VP26) that co-transport with mCitrine-KIF1A in axons of SCG neurons. n = 555 capsids in 21 axons from 4 independent experiments. (C) PC12 cells were transduced with Ad GFP, Ad mCit-KIF1A, or Ad DN GFP-KIF1A and subsequently infected with PRV Becker. At 12 hpi (and 4 days post adenoviral transduction), cell lysates were prepared and subject to immunoaffinity purification using anti-GFP antibodies. (D) SCG neurons were transduced with Ad mCit-KIF1A, Ad DN GFP-KIF1A, Ad GFP, or mock. At 4 days post transduction, neurons were infected with PRV mRFP-capsids and visualized by live cell imaging between 8–12 hours post PRV infection. The number of anterograde, retrograde, and stalled particles was manually quantified and normalized to the measured axon segment length. Data is from: 12 axons containing 972 capsids for PRV mRFP-VP26 only; 9 axons containing 938 particles for Ad mCit-KIF1A and PRV mRFP-VP26, 14 axons containing 620 capsids for Ad DN GFP-KIF1A and PRV mRFP-VP26; 12 axons containing 999 capsids for Ad GFP and PRV mRFP-VP26. For each of these conditions, data was acquired from at least two independent replicates. Error bars indicate the mean ± SEM. * is p < 0.05, ** is p < 0.01, and *** is p < 0.001. (E) Representative kymographs of mRFP-capsids in axons of neurons transduced with the indicated adenoviral vectors. See also Movie S1.
To determine whether KIF1A is functionally required for mediating anterograde axonal transport of viral particles, we constructed an adenovirus vector that expresses dominant negative KIF1A proteins (Ad DN GFP-KIF1A) in which the N-terminal motor domain of KIF1A was substituted with GFP (Klopfenstein et al., 2002). Us9 co-purifies with DN GFP-KIF1A, indicating that this construct functions by inhibiting Us9’s capacity to recruit endogenous cellular KIF1A. SCG neurons were transduced with Ad mCit-KIF1A, Ad DN GFP-KIF1A, Ad GFP, or mock transduced, and then infected with PRV expressing mRFP-capsids. Expression of dominant negative GFP-KIF1A resulted in a significant reduction of anterograde directed capsids in axons compared to neurons that were transduced with GFP, mCitrine-KIF1A, or mock. Moreover, expression of mCitrine-KIF1A resulted in a modest but statistically significant increase in anterograde-directed capsids (Figure 3D and 3E). In contrast, the number of stalled or retrograde-directed capsids was not significantly altered following transduction with any of the tested constructs (Figure 3D). We conclude that KIF1A is functionally required for mediating anterograde-directed transport of viral particles in axons.
The interaction between Us9 and KIF1A requires other viral proteins
We next constructed an adenoviral vector that expresses GFP-Us9 (Ad GFP-Us9). When expressed independently of PRV infection, GFP-Us9 localizes to puncta within cell bodies of SCG neurons (Figure 4A). While GFP-Us9 was enriched in these structures in all imaged cells, it was occasionally also detected at the plasma membrane. Using our trichamber neuronal culture system, we demonstrated that Ad GFP-Us9 efficiently trans-complements the anterograde spread defect of a Us9 null PRV mutant (Figure 4B). In neurons that were transduced with Ad GFP-Us9 and co-infected with wild type PRV, GFP-Us9 was consistently localized both to the plasma membrane and intracellular puncta (Figure 4A).
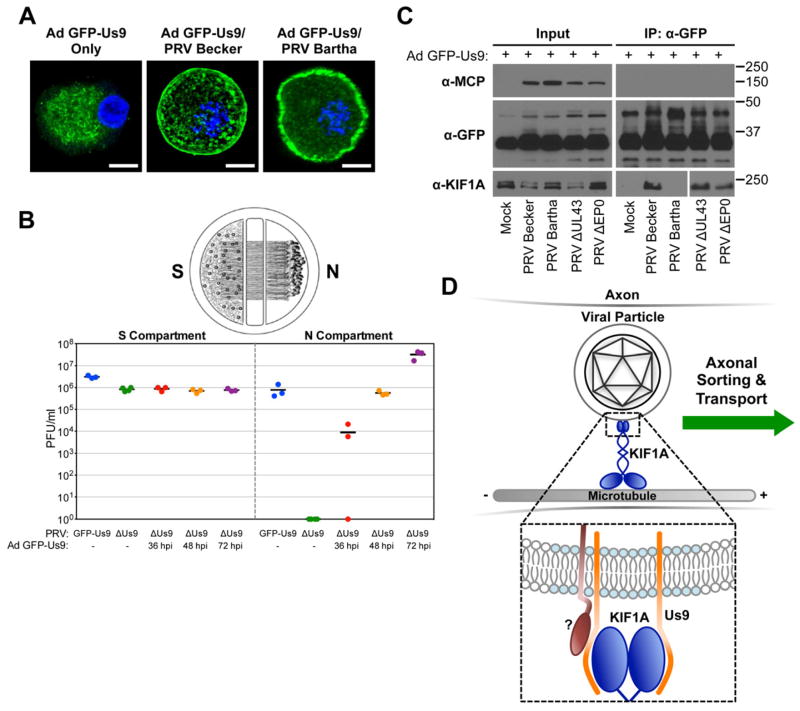
Figure 4. Us9 is necessary but not sufficient for interacting with KIF1A during PRV infection.
(A) SCG neurons were transduced with an adenoviral vector expressing GFP-Us9 (Ad GFP-Us9). At 5 days post transduction, neurons were infected with PRV Becker, PRV Bartha, or mock infected (scale bars = 10 μm). (B) Neurons grown in a trichamber were transduced with Ad GFP-Us9 in the S compartment. Us9 null PRV (PRV 161) was added at the indicated times post transduction. For comparison, we also tested PRV Us9 null and PRV expressing GFP-Us9 (PRV 340) without adenoviral transduction. The S and N compartments of all chambers were harvested at 24 hours post PRV infection and titered using a standard plaque assay. The total times of Ad GFP-Us9 transduction, including the PRV infection period, are the indicated times plus 24 hours. (C) PC12 cells were transduced with Ad GFP-Us9 for 4 days and then infected with the indicated PRV strains or mock infected. Cells were lysed at 12 hours post PRV infection and subject to immunoaffinity purification using anti-GFP antibodies as previously described. (D) Schematic model of Us9-dependent axonal sorting and transport of viral particles. Our data indicate that the membrane protein Us9 (orange) recruits KIF1A (dark blue) to intracellular viral particles contained within transport vesicles. Us9 must be incorporated within membrane microdomains known as lipid rafts (light blue) to interact with KIF1A. At least one additional viral protein (dark red) is required to facilitate the interaction between Us9 and KIF1A.
To test whether Us9 is sufficient to interact with KIF1A, we purified GFP-Us9 from Ad GFP-Us9 transduced PC12 cells in the presence or absence of wild type PRV infection. We conclude that at least one additional PRV protein is required for GFP-Us9 to interact with KIF1A (Figure 4C). We also performed similar IP experiments with cells transduced with Ad GFP-Us9 and co-infected with PRV strains that are mutant for one or more of these proteins. Three PRV mutants were tested that contained mutations in genes that are non-essential for viral replication in cell culture. One of these was the attenuated vaccine strain PRV Bartha. The PRV Bartha genome contains multiple mutations, including a large deletion in the coding region for gI, gE, Us9, and Us2 (Szpara et al., 2011). Since PRV Bartha does not express Us9, this strain is defective for anterograde spread of infection in neurons both in vitro and in vivo (Ch’ng and Enquist, 2005). Furthermore, both gE and gI have a previously reported but poorly understood role in promoting anterograde spread (Ch’ng and Enquist, 2005). We also tested PRV mutants that are null for the virion proteins UL43 or EP0. Both of these proteins co-purified with GFP-Us9 (Figure 1), but have no known function in mediating anterograde spread.
Overall, our results show that KIF1A co-isolated with GFP-Us9 in the absence of UL43 and EP0 expression during PRV infection, but not from cells that were infected with PRV Bartha (Figure 4C). During PRV Bartha infection, GFP-Us9 was detected in both the plasma membrane and intracellular puncta (Figure 4A). However, we observed that at this time point (12 hours post PRV infection, 4 days post Ad GFP-Us9 transduction), GFP-Us9 was enriched at the plasma membrane during PRV Bartha infection compared to PRV Becker infection. The mutations in PRV Bartha may disrupt the interaction between Us9 and KIF1A due to mislocalization of GFP-Us9. Thus, we conclude that while the Us9-KIF1A interaction is necessary for anterograde transport of viral particles, one or more of the viral proteins that are mutated or null in Bartha (besides Us9) is required to facilitate this interaction.
DISCUSSION
Anterograde axonal transport of viral particles is necessary for efficient spread of infection within and between hosts. A longstanding hypothesis has been that the PRV Us9 protein promotes anterograde transport by directly or indirectly interacting with a kinesin motor (Smith, 2012). However, identification of the specific motors that mediate viral particle transport has been challenging, in part because mammalian genomes encode up to 45 different kinesins (Lo et al., 2011). Here, we demonstrate that PRV Us9 functions by recruiting the kinesin-3 motor KIF1A to viral particles for efficient anterograde axonal sorting and transport (Figure 4D). Our results suggest that viral particles utilize the synaptic vesicle secretory pathway for efficient axonal sorting and transport. We propose that this represents the molecular basis for anterograde spread of alphaherpesvirus infection in neurons.
We found that KIF1A co-purified with wild type GFP-Us9, but not with the GFP control or any of the functionally defective GFP-Us9 mutants. This confirmed that the interaction between KIF1A and wild type GFP-Us9 is specific and strongly suggests that Us9 functions to recruit KIF1A. Our experiments on mutant GFP-Us9 proteins also provide insight into how the interaction between Us9 and KIF1A may be regulated. For example, failure of the TfR TMD GFP-Us9 to interact with KIF1A is consistent with previous work from our lab showing that Us9 must be incorporated within lipid rafts in order to function (Lyman et al., 2008). Furthermore, failure of KIF1A to co-purify with Y49-50A GFP-Us9 indicates that KIF1A is not required for plasma membrane targeting of Us9 and suggests that this motif may mediate Us9-KIF1A protein-protein interactions.
By isolating GFP-Us9 at different times post infection, we found that more KIF1A co-purified at 8 and 12 hpi than at later times post infection (Figure S1A). This is likely due to depletion of intracellular KIF1A over the course of infection. Previously, Kumar et al. demonstrated that after binding to and transporting cargo, KIF1A is targeted for degradation by the ubiquitin-proteasome pathway at synaptic regions (Kumar et al., 2010). In agreement with this, KIF1A is not efficiently depleted during infection if Us9 is not expressed (Figure S1B) or cannot interact with KIF1A (as during PRV Bartha infection; Figure 4C). We speculate that the E3 ubiquitin ligases EP0 and NEDD4 may target KIF1A for degradation (Ushijima et al., 2008; Van Sant et al., 2001).
Besides KIF1A, we identified 61 viral and host proteins that co-purified with GFP-Us9 at 20 hpi (Figure 1D). For the viral proteins we detected, additional experiments are needed to determine whether these interactions are required for virion assembly or viral spread. For example, virion structural components (e.g. UL17, Us2, and gM) may co-purify with GFP-Us9 as a result of direct or indirect protein-protein interactions that link Us9 to the capsid, tegument, and envelope layers during virion assembly (Kramer et al., 2011). Also, we found evidence that PRV utilizes the neuronal secretory pathway for targeted membrane fusion along axons. Specifically, GFP-Us9 co-purified with the v-SNARE VAMP2, which has been previously shown to co-transport with anterograde-directed PRV and HSV- 1 viral particles in axons (Antinone et al., 2010). We also detected proteins that are part of t-SNARE complexes, including SNAP-25, VTI1B, and syntaxin-6. We hypothesize that incorporation of Us9 and these SNARE proteins within the same membrane microdomains is functionally required for efficient spread of infection. Furthermore, we recently demonstrated that alphaherpesvirus infection disrupts mitochondrial motility in axons (Kramer and Enquist, 2012). Additional work is needed to determine whether the dynamics of synaptic vesicles and other cellular axonal cargoes may be altered.
Involvement of KIF1A in directional viral particle transport process fits well with several known properties of KIF1A function and viral particle dynamics. First, KIF1A is highly expressed in neuronal tissues and is enriched in axons (Klopfenstein et al., 2002). Second, KIF1A has a well-established role in mediating axonal sorting and transport of cargoes such as synaptic vesicle precursors and dense core vesicles (Lo et al., 2011). Loss of KIF1A leads to a reduced number of synaptic vesicle precursors in axons and accumulation of these organelles in cell bodies (Pack-Chung et al., 2007). The Us9 null phenotype is strikingly similar; viral particles are assembled in the cell body, but fail to sort into axons (Lyman et al., 2007; Lyman et al., 2008). Finally, KIF1A is a relatively fast kinesin motor with average velocities of 1.2–2.5 μm/s, depending on the model system (Klopfenstein et al., 2002). The measured average velocity of anterograde-directed PRV capsids in axons is within this range (1.97 μm/s) (Smith et al., 2001).
Future studies will focus on identifying whether other alphaherpesviruses utilize KIF1A for axonal transport and whether Us9 is involved in mediating this interaction. We demonstrate that KIF1A does not co-purify with HSV-1 GFP-Us9 expressed during PRV infection (Figure 3D), but also found that at least one additional PRV protein is required for facilitating the interaction between Us9 and KIF1A (Figure 4C). Thus, we postulate that a virus-specific protein co-factor is required for various Us9 homologues to interact with KIF1A. For both PRV and HSV-1, gE and gI, which function as a heterodimer, are involved in promoting anterograde spread of infection (Ch’ng and Enquist, 2005). Since the genes that encode these proteins are deleted in PRV Bartha (Szpara et al., 2011), they likely act in concert with Us9 to promote recruitment of KIF1A through an unknown mechanism. As deletion of gE/gI severely limits but does not completely ablate the anterograde spread capacity of PRV in vitro (Ch’ng and Enquist, 2005), these proteins may function as accessory proteins to stabilize the interaction between Us9 and KIF1A (Figure 4D). Alternatively, gE/gI may function by targeting Us9 to the proper subcellular compartment so that it can interact with KIF1A. Our unpublished results show that gI and Us9, but not KIF1A, co-purify with gE-GFP from PRV infected PC12 cells. Finally, further is needed determine how other alphaherpesvirus particles recruit KIF1A, or potentially or other motors, for efficient transport within their hosts.
Overall, our results suggest that alphaherpesviruses hijack and repurpose KIF1A for efficient axonal transport of viral particles in order to spread within the nervous system of their hosts. This finding is important for understanding the molecular nature of viral spread and disease in the nervous system.
EXPERIMENTAL PROCEDURES
Virus strains
Viral stocks were propagated in PK15 cells grown with Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 2% fetal bovine serum and 1% penicillin/streptomycin (all from HyClone, Logan, UT). A complete list of viral strains used in this study is available in the Supplemental Experimental Procedures.
PC12 cell culture and infection
A detailed protocol for culturing and differentiating PC12 cells has been described previously (Ch’ng et al., 2005). PC12 cells were differentiated in the presence of neuronal growth factor (NGF) for 10–12 days. Differentiated PC12 cells were infected with 108 plaque forming units (PFUs) of the indicated PRV strains per 10 cm dish or 2×108 PFUs per 15 cm dish. For adenoviral transductions, PC12 cells were incubated with 108 fluorescent foci units (FFUs) per 10 cm dish and subsequently infected with PRV at the indicated times post transduction. Additional details are available in the Supplemental Experimental Procedures.
Primary neuronal culture and trichamber system
Embryonic superior cervical ganglia (SCG) were isolated from Sprague-Dawley rats (Hill-Top Labs Inc., Scottdale, PA) at embryonic day 17, dissociated into individual neurons, and cultured as described previously (Ch’ng et al., 2005). Neurons used for experiments were grown between 14–28 days in culture. For PRV infection of SCG neurons, cells were incubated with 106 PFUs of the indicated viral strains. These conditions have been previously demonstrated to allow for a synchronous infection of all of the neuron cell bodies in the culture dish (Ch’ng et al., 2005). For adenoviral transductions, SCG neurons were incubated with 107 FFUs in neuronal medium and subsequently infected with PRV at the indicated times post transduction. The trichamber system for assaying anterograde spread of infection has been described previously (Curanovic et al., 2009). Neuron cell bodies in the S compartment were infected with the indicated viral strains and incubated until 24 hours post PRV infection. The S and N compartments were harvested separately by scraping each compartment and titered on epithelial cells.
Immunoaffinity purifications, mass spectrometry, and data analysis
Immunoaffinity purifications were performed on magnetic beads as previously described (Cristea and Chait, 2011a, b) with modifications. Differentiated PC12 cells were infected with the indicated viral strains and resuspended in immunoprecipitation lysis buffer designed to preserve Us9’s incorporation within lipid rafts (see Supplemental Experimental Procedures). Insoluble debris was pelleted by centrifugation and the supernatant was incubated with Dynal M-270 epoxy magnetic beads (Invitrogen, Carlsbad, CA) coated with custom made high affinity rabbit polyclonal anti-GFP antibodies. Protein complexes were eluted in 1X SDS-PAGE loading buffer (Invitrogen). For mass spectrometry analysis, GFP-Us9 affinity purification samples were resolved by SDS-PAGE and digested in-gel with trypsin. NanoLC-MS/MS analysis of tryptic peptide fractions was performed on a Dionex Ultimate 3000 RSLC (Dionex, Amsterdam, the Netherlands) coupled online to an Orbitrap Velos mass spectrometer (Thermo-Fisher Scientific, San Jose, CA). Label-free spectral counting analyses were performed as previously described (Tsai et al., 2012), with slight modification.
Microscopy and image analysis
Imaging experiments were performed on a Leica SP5 laser scanning confocal microscope (Leica Microsystems, Wetzlar, Germany) and a Nikon Ti-Eclipse inverted epifluorescence microscope (Nikon Instruments, Tokyo, Japan). Additional details are provided in the Supplemental Experimental Procedures.
Statistical analysis
One-way ANOVA with Tukey’s post-test was performed using GraphPad Prism 5.0d for Max OS X (GraphPad Software, San Diego, CA, www.graphpad.com).
Supplementary Material
ARTICLE HIGHLIGHTS.
Identification of proteins that co-purify with the pseudorabies virus (PRV) protein Us9
KIF1A binds to Us9 and mediates Us9-dependent axonal transport of viral particles
The Us9-KIF1A interaction requires at least one additional viral protein
PRV utilizes the synaptic vesicle sorting pathway for anterograde spread of infection
Acknowledgments
We thank K.J. Verhey, B. Vogelstein, S.J. Flint, and P. Ryan for generously providing reagents, and R. Kratchmarov for assistance with virus construction. This work was supported by National Institutes of Health grants to L.W.E. (R37 NS33506, R01 NS060699, and P40 RR18604) and I.M.C. (DP1 DA026192), a Human Frontiers Science Program Organization award to I.M.C (RGY0079/2009- C), a National Science Foundation graduate research fellowship to T.K. (DGE- 0646086), and an American Cancer Society postdoctoral research fellowship to M.P.T. (PF-1005701-MPC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antinone SE, Zaichick SV, Smith G. Resolving the assembly state of herpes simplex virus during axon transport by live-cell imaging. J Virol. 2010 doi: 10.1128/JVI.01296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau AD, Banfield BW, Enquist LW. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J Virol. 1998;72:4560–4570. doi: 10.1128/jvi.72.6.4560-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau AD, Eldridge MG, Enquist LW. Directional transneuronal infection by pseudorabies virus is dependent on an acidic internalization motif in the Us9 cytoplasmic tail. J Virol. 2000;74:4549–4561. doi: 10.1128/jvi.74.10.4549-4561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng TH, Enquist LW. Neuron-to-cell spread of pseudorabies virus in a compartmented neuronal culture system. J Virol. 2005;79:10875–10889. doi: 10.1128/JVI.79.17.10875-10889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng TH, Flood EA, Enquist LW. Culturing primary and transformed neuronal cells for studying pseudorabies virus infection. Methods Mol Biol. 2005;292:299–316. doi: 10.1385/1-59259-848-x:299. [DOI] [PubMed] [Google Scholar]
- Cristea IM, Chait BT. Affinity purification of protein complexes. Cold Spring Harb Protoc. 2011a;2011:pdb prot5611. doi: 10.1101/pdb.prot5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea IM, Chait BT. Conjugation of magnetic beads for immunopurification of protein complexes. Cold Spring Harb Protoc. 2011b;2011:pdb prot5610. doi: 10.1101/pdb.prot5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curanovic D, Ch’ng TH, Szpara M, Enquist L. Compartmented neuron cultures for directional infection by alpha herpesviruses. Curr Protoc Cell Biol. 2009;Chapter 26(Unit 26):24. doi: 10.1002/0471143030.cb2604s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshizuka T, Kawaguchi Y, Nishiyama Y. Herpes simplex virus type 2 membrane protein UL56 associates with the kinesin motor protein KIF1A. J Gen Virol. 2005;86:527–533. doi: 10.1099/vir.0.80633-0. [DOI] [PubMed] [Google Scholar]
- Kramer T, Enquist LW. Alphaherpesvirus infection disrupts mitochondrial transport in neurons. Cell Host Microbe. 2012;11:504–514. doi: 10.1016/j.chom.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer T, Greco TM, Enquist LW, Cristea IM. Proteomic characterization of pseudorabies virus extracellular virions. J Virol. 2011;85:6427–6441. doi: 10.1128/JVI.02253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Choudhary BC, Metpally R, Zheng Q, Nonet ML, Ramanathan S, Klopfenstein DR, Koushika SP. The Caenorhabditis elegans Kinesin-3 motor UNC-104/KIF1A is degraded upon loss of specific binding to cargo. PLoS Genet. 2010;6:e1001200. doi: 10.1371/journal.pgen.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Kuzmin A, Unger SM, Petersen JD, Silverman MA. KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett. 2011;491:168–173. doi: 10.1016/j.neulet.2011.01.018. [DOI] [PubMed] [Google Scholar]
- Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- Lyman M, Feierbach B, Curanovic D, Bisher M, Enquist LW. Pseudorabies virus Us9 directs axonal sorting of viral capsids. J Virol. 2007;81:11363–11371. doi: 10.1128/JVI.01281-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman MG, Curanovic D, Enquist LW. Targeting of pseudorabies virus structural proteins to axons requires association of the viral Us9 protein with lipid rafts. PLoS Pathog. 2008;4:e1000065. doi: 10.1371/journal.ppat.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack-Chung E, Kurshan PT, Dickman DK, Schwarz TL. A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat Neurosci. 2007;10:980–989. doi: 10.1038/nn1936. [DOI] [PubMed] [Google Scholar]
- Pellett PE, Roizman B. The Family: Herpesviridae A Brief Introduction. 5. Vol. 2. Philadelphia, PA: Lippincott, Williams, and Wilkins; 2007. [Google Scholar]
- Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci USA. 2009;106:1760–1765. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. Herpesvirus transport to the nervous system and back again. Annu Rev Microbiol. 2012;66:153–176. doi: 10.1146/annurev-micro-092611-150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G, Gross SP, Enquist LW. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc Natl Acad Sci USA. 2001;98:3466–3470. doi: 10.1073/pnas.061029798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpara ML, Tafuri YR, Parsons L, Shamim SR, Verstrepen KJ, Legendre M, Enquist LW. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS pathogens. 2011;7:e1002282. doi: 10.1371/journal.ppat.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MP, Kramer T, Lyman MG, Kratchmarov R, Enquist LW. Visualization of an alphaherpesvirus membrane protein that is essential for anterograde axonal spread of infection in neurons. MBio. 2012:3. doi: 10.1128/mBio.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics. 2012;11:M111 015156. doi: 10.1074/mcp.M111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima Y, Koshizuka T, Goshima F, Kimura H, Nishiyama Y. Herpes simplex virus type 2 UL56 interacts with the ubiquitin ligase Nedd4 and increases its ubiquitination. J Virol. 2008;82:5220–5233. doi: 10.1128/JVI.02515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sant C, Hagglund R, Lopez P, Roizman B. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2001;98:8815–8820. doi: 10.1073/pnas.161283098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.