Abstract
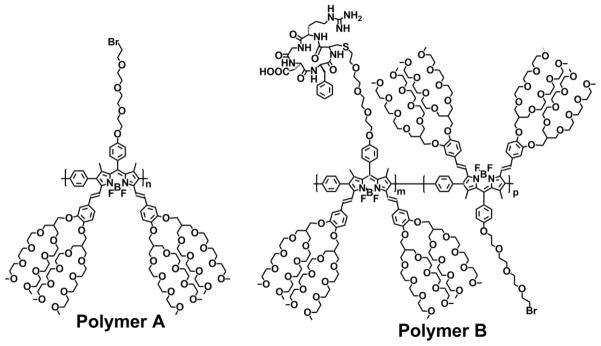
Near-infrared emissive BODIPY polymeric dye bearing cancer-homing cyclic arginine-glycine-aspartic acid (RGD) peptide residues (polymer B) was prepared by post-polymerization functionalization of BODIPY polymeric dye bearing bromo groups through tetra(ethylene glycol tethered spacers (polymer A) with thiol-functionalized RGD cancer-homing peptide through thioether bonds under a mild basic condition. Polymer B possesses excellent water solubility, good photostability, biocompatibility and resistance to nonspecific interactions to normal endothelial cells, and can efficiently detect breast tumor cells through specific cooperative binding of cancer-homing RGD peptides to αVβ3 integrins of cancer cells while its parent polymer Awith outRGD residues fails to target cancer cells.
Keywords: BODIPY dye, Conjugated polymer, Cancer imaging, Fluorescence, RGD peptide
1. Introduction
Fluorescence-based optical imaging offers a convenient approach to monitor multiple biological processes simultaneously and in real time with high sensitivity, low tissue autoabsorption and autofluorescence, low light scattering, operational simplicity, relatively low cost, potential miniaturization and mobility.[1–4] The demand for near-infrared (NIR) fluorescent dyes for noninvasive and simple diagnostic techniques such as in vivo imaging has been drastically growing as NIR dyes with emission wavelengths in the region between 700 nm and 900 nm can propagate through several centimeters of living tissues and may enable deep tissue imaging.[1–4] Functional bio-conjugated quantum dots (CdSe) could lead to a new generation of nanoparticle imaging probes for in vivo tumor imaging at high sensitivity and specificity,[5] but their limited tissue penetration, lack of spatial resolution in tumor depth and potential toxicity concerns still restrict their clinical application. As a result, it is very important to develop a novel, highly water-soluble, near-infrared emissive agent with low cost, good biocompatibility, high intensity and photostability. In addition, high specificity for tumor cells, and tissue penetration and quantitative visualization for real time, in vivo detection of tumor will be highly desirable features in such a fluorescent dye. Numerous long-wavelength fluorophores such as 4,4′-difluoro-4-bora-3a,4a-diaza-s-indacene (BODIPY) dyes,[6] rhodamine,[7] squaraine,[8] or cyanine dyes[9] have been recently reported. BODIPY dyes have received renewed interest for promising applications as imaging agents because of their many distinctive and desirable properties such as high absorption coefficients, narrow absorption bands, sharp emissions, high fluorescence quantum yields, and excellent chemical and photostabilities.[6] Functionalization of BODIPY dyes by the number and nature of the substituents at different positions (Scheme 1) has been used to tune to near-infrared emission. However, most reported studies have focused on small molecules of BODIPY dyes and only a few reported near-infrared emissive BODIPY dyes are soluble in aqueous solution.[6, 10, 11] Very recently, we and other groups reported some deep-red and near-infrared emissive polymeric BODIPY dyes.[12–16] However, all these BODIPY polymeric dyes are insoluble in aqueous solution.[12–16]
Scheme 1.
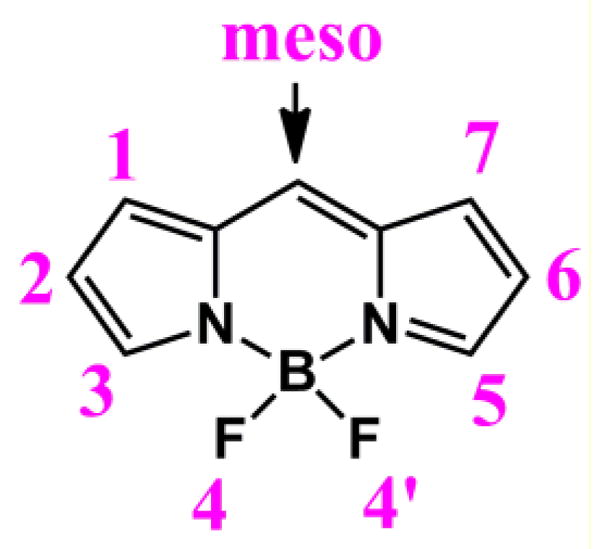
Chemical structure of BODIPY core.
In this paper, we report a facile, convenient and versatile approach to prepare highly water-soluble, near-infrared emissive BODIPY polymeric dye bearing RGD cancer-homing peptide residues for cancer imaging application (Scheme 2). We prepared highly water-soluble BODIPY polymeric dye (polymer A) through the palladium-catalyzed Suzuki polymerization of highly water-soluble 2,6-diiodo-3,5-distyryl-BODIPY dye bearing bromide residue at the meso-position with 1,4-phenyldiboronic acid because this polymer is expected to display the relatively weak π-π stacking interactions among different polymer backbones. Incorporation of new branched oligo(ethylene glycol)methyl ether residues to distyryl units of BODIPY core at 3,5-positions effectively enhances enthalpic interactions of BODIPY dye with water, significantly increases water solubility of BODIPY polymeric dyes and prevents potential non-specific interactions of BODIPY dyes with other proteins through intermolecular interactions. RGD cancer-homing peptide residues were covalently attached to BODIPY cores at the meso positions through tetra(ethylene glycol) tethered spacers. This BODIPY polymeric dye functions as intrinsic near-infrared fluorophore with high water solubility, good photostability, biocompatibility and resistance to nonspecific interactions to biomolecules, targeting efficacy to tumor cells and easy-to-use during imaging detection.
Scheme 2.
Chemical structures of highly water-soluble, near-infrared BODIPY polymeric dyes.
2. Experimental Section
2.1 Materials
Unless otherwise indicated, all reagents and solvents were obtained from commercial suppliers (Aldrich, Sigma, Fluka, Acros Organics, Fisher Scientific, Lancaster) and used without further purification. Air- and moisture-sensitive reactions were conducted in oven-dried glassware using a standard Schlenk line or drybox techniques under an inert atmosphere of dry nitrogen.
BODIPY dye 3
The aldehyde 3 (2.0 g, 5.56 mmol) and 2,4-dimethylpyrrole (1.16 g, 12.23 mmol) were dissolved in 1000 mL of dry CH2Cl2 in a 2000-mL three-neck flask. Five drops of trifluoroacetic acid (TFA) were added to the reaction mixture, and the resulting mixture was stirred in the dark for 12 h under a nitrogen atmosphere at room temperature. After the complete consumption of aldehyde 3 (which was monitored by TLC), DDQ (2,3-dichloro-5,6-dicyano-1,4-benzoquinone) (1.26 g, 5.56 mmol) was added to the reaction mixture. When the mixture was stirred for 40 min, 15 mL of diisopropylethylamine (DIPEA) and 15 mL of BF3·OEt2 were added to the mixture. After the mixture was further stirred for 40 min, it was concentrated to 200 mL and filtered. The filtrate was washed twice with water and brine solution, dried over anhydrous MgSO4, and concentrated under reduced pressure. The crude product was purified by column chromatography to obtain 4 as a red-orange oil (1.35 g, 42%). 1H NMR (400 MHz, CDCl3): δ 7.12 (d, J = 8.8 Hz, 2H), 6.99 (d, J = 8.8 Hz, 2H), 5.94 (s, 2H), 4.16 (t, J = 4.4 Hz, 2H), 3.88 (t, J = 4.8 Hz, 2H), 3.1–3.65 (m, 10H), 3.45 (t, J = 6.4 Hz, 2H), 2.52 (s, 6H), 1.39 (s, 6H). 13C NMR (100 MHz, CDCl3): δ 159.6, 155.4, 143.4, 142.0, 132.0, 129.4, 127.4, 121.3, 115.4, 71.4, 71.1, 70.9, 70.8, 70.7, 69.9, 67.7, 30.5, 14.8. IR (cm−1): 2867, 1609, 1542, 1508, 1468, 1409, 1363, 1304, 1285, 1245, 1191, 1155, 1106, 1083, 1047, 973, 833, 810, 764, 704. HRMS(FAB) calcd for C27H34BBrF2N2O4[M] +, 578.1763; found, 578.1757.
2,6-Diiodo-BODIPY dye (5)
When iodic acid (0.54 g, 3.065 mmol) in 3 mL of water was added dropwise (30 mins) to the ethanol solution (20 mL) containing compound 4 (896 mg, 1.55 mmol) and iodine (0.43 g, 3.36 mmol), the mixture was stirred for 4 h. After the completion of the reaction, the mixture was concentrated under reduced pressure, dissolved in CH2Cl2, and washed twice with water and saturated saline solution. The organic layer was collected, dried over anhydrous Na2SO4, and concentrated under reduced pressure. The crude product was purified by column chromatography to yield 5 as red crystals (1.20 g, 93%). 1H NMR (400 MHz, CDCl3): δ 7.11 (d, J = 8.8 Hz, 2H), 7.02 (d, J = 8.8 Hz, 2H), 4.18 (t, J = 4.4 Hz, 2H), 3.91 (t, J = 4.8 Hz, 2H), 3.81 (t, J = 6.4 Hz, 2H), 3.77–3.68 (m, 8H), 3.46 (t, J = 6.4 Hz, 2H), 2.62 (s, 6H), 1.42 (s, 6H). 13C NMR (100 MHz, CDCl3): δ 160.0, 156.8, 145.6, 141.8, 131.9, 129.3, 127.1, 115.7, 71.4, 71.1, 70.9, 70.8, 70.7, 69.9, 67.8, 30.5, 17.4, 16.2. IR (cm−1): 2862, 1607, 1522, 1454, 1396, 1342, 1304, 1247, 1173, 1114, 1091, 1059, 994, 961, 913, 834, 763, 704. HRMS(FAB) calcd for C27H32BBrF2I2N2O4 [M]+, 829.9696; found, 829.9690.
2,5-Diiodo-3,5-distyryl BODIPY dye (7)
2,6-Diiodo-BODIPY dye 5 (1.66 g, 2.0 mmol), and compound 6 (5.4 g, 6.0 mmol) were refluxed in a mixture of benzene (150 mL), piperidine (3.5 mL) and AcOH (3.2 mL). Any water formed during the reaction, was removed azeotropically by heating the mixture in a Dean-Stark apparatus. After 2.5 h, the mixture was concentrated in vacuo, diluted with EtOAc, and then washed with water and brine, respectively. The organic phase was dried over Na2SO4 and concentrated in vacuo. The crude product was simply purified by silica gel column chromatography (hexane/EtOAc/CH2Cl2/EtOH, 5/1/3/0.5, v/v to hexane/EtOAc/CH2Cl2/EtOH, 5/1/3/1, v/v) to obtain distyryl BODIPY 7 (1.23 g, 24%) as green oil. 1H NMR (400 MHz, CDCl3): δ 8.00 (d, J = 16.8 Hz, 2H), 7.46 (d, J = 16.4 Hz, 2H), 7.25 (d, J = 5.6 Hz, 2H), 7.10 (d, J = 8.4 Hz, 2H), 7.02 (s, 2H), 6.99 (d, J = 5.2 Hz, 4H), 6.87 (d, J = 8.4 Hz, 2H), 4.17 (t, J = 4.4 Hz, 2H), 4.04 (t, J = 5.6 Hz, 6H), 3.88 (t, J = 5.6 Hz, 2H), 3.77 (t, J = 6.4 Hz, 2H), 3.73–3.41 (m, 104H), 3.32–3.29 (s× 8CH3, 24H), 2.42–2.36 (m, 4H), 1.45 (s, 6H). 13C NMR (100 MHz, CDCl3): δ 160.0, 150.6, 150.5, 149.2, 145.8, 139.7, 138.7, 133.5, 130.0, 129.8, 127.6, 120.5, 117.0, 115.6, 113.9, 113.7, 83.3, 77.5, 72.1, 71.4, 71.1, 70.9, 70.8, 70.8, 70.7, 70.7, 70.6, 69.9, 69.5, 69.4, 67.8, 67.3, 67.1, 63.7, 59.2, 41.4, 40.2, 30.5, 17.9. IR (cm−1): 2867, 1594, 1509, 1463, 1409, 1350, 1265, 1250, 1178, 1095, 1012, 958, 849, 806, 769, 711. HRMS(MAIDL) calcd for C113H184BBrF2I2N2O40 [M+Na]+, 2613.9656; found, 2613.9688.
BODIPY dye A
2,5-Diiodo-3,5-distyryl BODIPY dye 7 (80 mg, 0.03 mmol), phenylboronic acid (12.2 mg, 0.1 mmol), Pd(dppf)2Cl2 (1.1 mg) and Na2CO3 (32 mg, 0.30 mmol) were added to a 50-mL flask under a nitrogen atmosphere. When toluene (6 mL), EtOH (4 mL) and H2O (3 mL) were added to the flask, the mixture was stirred at 80 °C under a nitrogen atmosphere overnight. After removal of the solvent, the residue was dissolved in ethyl acetate and washed with water twice. The organic layer was collected, dried over anhydrous Na2SO4, and filtered. The filtrate was concentrated under reduced pressure and purified using TLC plate (hexanes/CH2Cl2/EtOAc/MeOH, 5/3/1/0.7, v/v) to attain BODIPY dye A as green oil (74 mg, 83%). 1H NMR (400 MHz, CDCl3): δ 7.52 (d, J = 16.4 Hz, 2H), 7.41–7.33 (m, 6H), 7.24–7.21 (m, 6H), 7.03 (dd, J = 8.4, 1.6 Hz, 2H), 6.99 (d, J = 8.8 Hz, 2H), 6.79 (d, J = 8.4 Hz, 2H), 6.48 (d, J = 1.6Hz, 2H), 6.42 (d, J = 16.4 Hz, 2H), 4.14 (t, J = 4.4 Hz, 2H), 3.98 (d, J = 5.6 Hz, 4H), 3.91 (d, J = 5.6 Hz, 4H), 3.85 (t, J = 4.4 Hz, 2H), 3.76 (t, J = 6.0 Hz, 2H), 3.72–3.47 (m, 120H), 3.42 (t, J = 6.0 Hz, 2H, -CH2Br), 3.33–3.32 (s, 8CH3, 24H), 2.38–2.34 (m, 4H), 1.24 (s, 6H). 13C NMR (100 MHz, CDCl3): δ 159.6, 150.3, 150.2, 149.0, 139.6, 138.9, 138.5, 135.6, 133.8, 133.7, 131.4, 130.7, 130.4, 129.9, 128.9, 128.5, 128.4, 128.2, 127.6, 120.0, 117.3, 115.4, 114.0, 113.6, 77.5, 72.1, 71.4, 71.1, 70.9, 70.8, 70.7, 70.6, 69.9, 69.5, 67.7, 67.3, 67.1, 59.2, 40.2, 40.2, 30.5, 12.9. IR (cm−1): 2868, 1594, 1576, 1509, 1449, 1398, 1364, 1348, 1268, 1246, 1185, 1090, 1029, 1009, 996, 964, 849, 808, 707. ESI MS(m/z): 1248.0 (M + 2H+).
Polymer A
2,6-Diiodo3,5-distyryl BODIPY dye 7 (600 mg, 0.23 mmol), benzene-1,4-diboronic acid (42 mg, 0.253 mmol), Pd(dppf)2Cl2 (19.0 mg, 0.023 mmol) and Na2CO3 (244 mg, 2.3 mmol) were added to a 50 mL flask under a nitrogen atmosphere. When toluene (40 mL), EtOH (10 mL) and H2O (20 mL) were added to the flask, the mixture was stirred at 80 °C under a nitrogen atmosphere for 2 days. After removal of the solvent by under vacuum, the residue was dissolved in ethyl acetate and washed with water twice. The organic layer was collected, dried over anhydrous Na2SO4, and filtered. The filtrate was concentrated under reduced pressure and purified using TLC plate (hexanes/CH2Cl2/EtOAc/MeOH, 5/3/1/1, v/v) to attain polymer A as green oil (426 mg). 1H NMR (400 MHz, CDCl3): δ 7.55 (d, J = 14.4, 2H), 7.32–7.23 (m, 6H), 7.05 (br, 2H), 6.91 (br, 2H), 6.75 (br, 2H), 6.63 (s, 2H), 6.60–6.56 (m, 2H), 4.18 (br, 2H), 4.07 (br, 2H), 3.98 (br, 4H), 3.89 (br, 4H), 3.78–3.24 (m, 148H), 2.42–2.34 (m, 4H), 1.23 (s, 6H). IR (cm−1): 2867, 1594, 1575, 1506, 1447, 1416, 1362, 1349, 1268, 1247, 1180, 1087, 1021, 1006, 962, 841, 808, 770, 709. GPC (THF, polystyrene standard), Mn: 38,700 g/mol; polydispersity: 2.1.
Polymer B
Polymer A (20 mg, 0.00865 mmol, repeated unit), c(RGDfC) (5 mg, 0.00865 mmol) and Na2CO3 (9 mg, 0.0865 mmol) dissolved in absolute solvents of DMF (3 mL) and H2O (0.5 mL). After the mixture was stirred at room temperature under nitrogen atmosphere for 2 days, the solvents were removed under reduced pressure. The residue was dissolved in CH2Cl2 and filtered. The filtrate was concentrated under a reduce pressure and precipitated by using Et2O and CH2Cl2. The solid was collected and dried under reduced pressure to attain polymer B as green solid (7 mg). IR (cm−1): 3304 (N-H), 2869, 1747 (C=O), 1594, 1509, 1449, 1414, 1362, 1349, 1268, 1248, 1183, 1024, 964, 842, 770, 709. Elem. Anal. Found: N, 6.77; S, 0.96. Functionalization degree with RGD peptide residuesis ~ 20 % according to elemental analysis of nitrogen and sulfur atoms.
2.2 Instrumentation
1H NMR and 13C NMR spectra were taken on a 400 MHz Varian Unity Inova spectrophotometer instrument. 1H and 13C NMR spectra were recorded in CDCl3, chemical shifts (δ) are given in ppm relative to solvent peaks (1H: δ 7.26; 13C: δ 77.3) as internal standard. Absorption spectra were taken on a Perkin Elmer Lambda 35 UV-Vis spectrometer. Fluorescence spectra were recorded on a Jobin Yvon Fluoromax-4 spectrofluorometer. The excitation and emission slit width was set to 1 nm and all the samples were scanned with an increment of 1 nm. Fluorescence lifetimes were measured on GL-3300 Nitrogen Laser laserstrobe, PTI instrument and analyzed using FeliX32 software. Fluorescein dye (φn = 85% in 0.1 N NaOH aqueous solution) was used as a standard to determine the fluorescence quantum yields of BODIPY dye 4 and 5.[17] Sulforhodamine 101 dye (φn = 95% with excitation wavelength at 577 nm in ethanol) was used as fluorescence standard to determine fluorescence quantum yields of BODIPY dye 7, A and polymers A and B.[18] Both samples and reference dye were prepared under identical conditions and the absorption, excitation and emission spectra were acquired under same experimental conditions. The areas for all emission spectra were calculated by integration of peak. Each plot of integrated area versus absorbance contained the corresponding reference data at that wavelength as well as the data for the test sample. A linear fit was then employed to determine the slope of the line (gradient) for the reference and the sample. The fluorescence quantum yield of each sample was obtained by using the following equation: [18]
Where the subscripts ‘st’ and ‘x’ represent standard and test samples, respectively, Φ is the fluorescence quantum yield, Grad stands for the gradient from the plot of integrated fluorescence intensity versus absorbance, and ηis the refractive index of the solvent.
Molecular weights of the polymers were determined by gel permeation chromatography (GPC) by using a Waters Associates Model 6000A liquid chromatograph. Mobile phase was HPLC grade THF which was filtered and degassed by vacuum filtration through a 0.5 μm Fluoropore filter prior to use. The polymer was detected by a Waters Model 2410 refractive index detector. Molecular weight was measured relative to polystyrene standards.
2.3 Procedures
2.3.1 Cell Culture and Fluorescence Imaging
MDA-MB-231 and HUVEC-C cell lines were procured from ATCC®. MDA-MB-231 cells were cultured in Dulbecco’s Modified Eagle’s Medium/Nutrient Mixture F12 Ham’s Liquid Media (DMEM/F-12) supplemented with 10% fetal bovine serum (FBS, GIBCO), penicillin (100 U / ml) and streptomycin (100 μg/ ml) (1X PenStrep, GIBCO), and HUVEC cells were cultured in medium 199 with 2mM L-glutamine, 10% FBS, heparin (100 μg/ml), endothelial cell growth supplements (50 μg/ml) and 1X PenStrep at 37 °C in humidified incubator with 5% CO2. The cells were plated on 12-well culture plates at a density of 1 × 105 cells/ ml for live cell imaging. After 24-hour incubation at 37 °C, 5% CO2 incubator, the media was removed and rinsed with 1X PBS and fresh serum free media with polymer A or polymer B was added at 50 μg/ml concentration. Polymer A (or polymer B) were incubated with cells for 4 hours and then washed with 1X PBS before imaging. The cell imaging was performed with fluorescence microscope (Zeiss Axiovert 200) using DAPI and Cy5 filters for Hoechst stain and polymer A/B dye respectively. The fluorescence images were obtained with 20× magnification objective and the exposure times for each filter were kept constant for each image series.
2.3.2 Photostability experiment
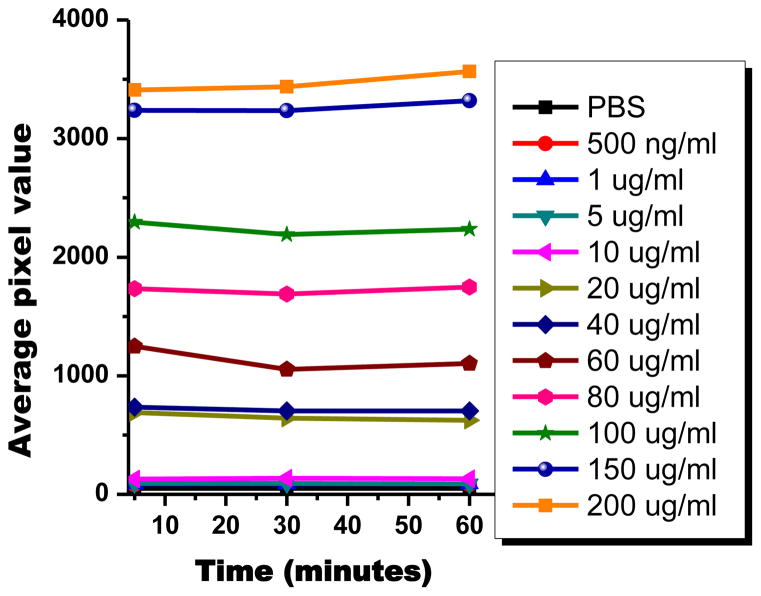
To test the photostability (bleaching) of polymer Bin cell culture media polymer B was dissolved at a concentration of 0.5 and 200 μg/mL in 1 mL media and placed on an inverted microscope (NIKON TE 2000U). In a 96 well plate, a 200 μL aliquot of each of the concentrations was dispensed with the control being a 200 μL aliquot of media solution. Each concentration was tested one at a time using the Nikon CY5 LP filter to excite the polymer in solution. Fluorescence intensity of the polymer was then quantified. The intensity was then measured (×20 objective) up to 60 minutes without switching off the excitation light with an exposure time of 30 sec. Images were captured using a cooled CCD camera (Coolsnap HQ2, Roper Scientific) and post processed using imaging software (NIKON Elements, Melville, NY). ImageJ software (NIH) was used to determine the average pixel intensity, representing the fluorescence emission brightness, of each fluorescent image. The light intensity used is comparable to that used in microscopy for cancer imaging. The absorbance of the polymer solution is between 0 and 0.3.
3. Results and Discussion
3.1 Synthesis of BODIPY Polymeric Dyes
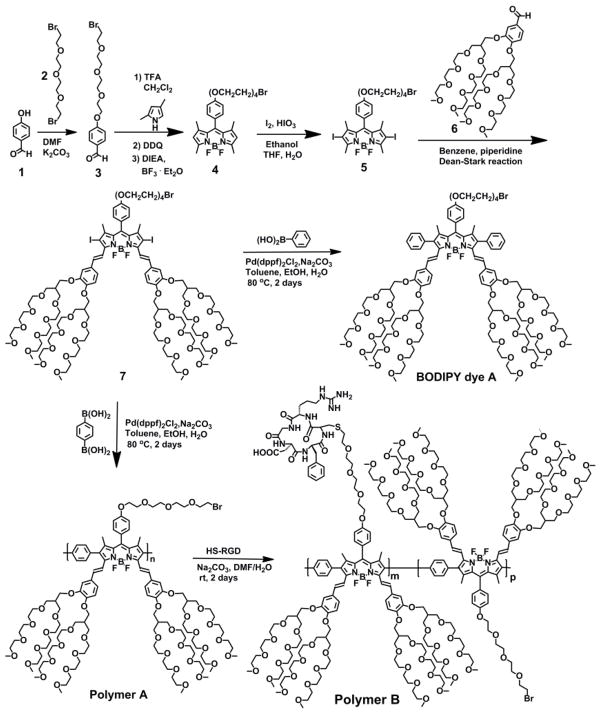
We employed a simple, convenient, versatile post-polymerization functionalization method to prepare BODIPY copolymeric dye bearing RGD cancer-homing peptide residues as we have demonstrated its feasibility to functionalize bromide-bearing conjugated fluorescent polymers such as polythiophenes and fluorene-based conjugated polymers with thiol-functionalized carbohydrates through via thioether bonds under a mild basic condition with almost 100% degree post-polymerization functionalization of the bromide groups with the thiol groups.[19–21] In order to introduce a bromide group through tetra(ethylene glycol) spacer to BODIPY dye at the meso position, we prepared bromide-functionalized aldehyde derivative (3) by reaction of 1-bromo-2-(2-(2-bromoethoxy)ethoxy)ethoxy)ethane (2) with 4-hydroxybenzaldehyde (1) in DMF under a basic condition (Scheme 3). BODIPY dye bearing bromide group through a tetra(ethylene glycol) tethered spacer at the meso position (4) was prepared through the condensation of the benzaldehyde derivative (3) with 2,4-dimethylpyrrole in the presence of a catalytic amount of trifluoroacetic acid (TFA), followed by oxidation with 2,3-dichloro-5,6-dicyano-1,4-benzoquinone (DDQ) and chelation with BF3-etherate in the presence of N,N-diisopropylethylamine (DIPEA).[11] 2,6-Diiodo-BODIPY dye (5) was prepared by iodination of BODIPY dye 4 at 2,6-positions. In order to prepare highly water-soluble 2,6-diiodo-3,5-distyryl-BODIPY monomeric dye with longer emission (7), we incorporated branched oligo(ethylene glycol)methyl ether into distyryl groups at 3,5-positions by the Knoevenagel condensation of methyl groups of 2,6-diiodo-BODIPY dye (5) at 3,5-positions with aldehyde derivative (6).[11, 22] In order to confirm the feasibility to prepare distyryl 2,6-diiodo-3,5-distyryl-BODIPY dye (7) bearing a bromide group through a tetra(ethylene glycol) tethered spacer at the meso position via the Knoevenagel reaction, we prepared BODIPY dye bearing bromide group through tetra(ethylene glycol) at the meso position (A) by the palladium-catalyzed Suzuki coupling of 2,6-diiodo-3,5-distyryl-BODIPY dye (7) with 1-phenylboronic acid (Scheme 3). This is because the proton peak of methylene protons adjacent to bromide group in 2,6-diiodo-3,5-distyryl-BODIPY dye 7 was expected to overlap with those proton peaks from branched oligo(ethylene glycol)ether methyl groups in 1H NMR spectrum. Comparison to 1H NMR spectra and high-resolution MS data of BODIPY dyes 5, 7 and A clearly indicate that bromide group of 2,6-diiodo-BODIPY dye 5 remains intact during the Knoevenagel reaction in the presence of piperidine and AcOH in benzene solution under a reflux condition (Figure S1 in supporting information). Triplet peaks corresponding to methylene protons adjacent to bromide group were observed at 3.46 ppm in 2,6-diiodo-BODIPY dye 5 (Figure S1 in supporting information). After the Knoevenagel reaction, those peaks corresponding to methylene protons adjacent to bromide group were overlapped with proton peaks of branched oligo(ethylene glycol)methyl ether in 2,6-diiodo-3,5-distyryl-BODIPY dye 7. However, after the palladium-catalyzed Suzuki coupling of 2,6-diiodo-3,5-distyryl-BODIPY dye (7) with 1-phenylboronic acid, triplet peaks corresponding to methylene protons adjacent to bromide group show up at 3.43 ppm in BODIPY dye A (Figure S1 in supporting information), which indicates that the bromide groups in BODIPY dyes 5 and 7 are intact during the Knoevenagel condensation reaction and the palladium-catalyzed Suzuki coupling reaction. Highly water-soluble, near-infrared emissive BODIPY polymeric dye bearing bromide groups (polymer A) was prepared by the palladium-catalyzed Suzuki polymerization of 2,6-diiodo-3,5- distyryl-BODIPY dye 7 with 1,4-phenyldiboronic acid. Highly water-soluble, near-infrared emissive BODIPY polymeric dye bearing RGD peptide residues (polymer B) was prepared by postpolymerization functionalization of polymer A with thiol-functionalized RGD peptide through thioether bonds under a mild basic condition. The post-polymerization functionalization degree of polymer A with thiol-functionalized RGD peptide was estimated as 20% according to the elemental analysis (sulfur and nitrogen atoms) of polymer B.
Scheme 3.
Synthetic route to highly water-soluble, near-infrared BODIPY polymeric dyes bearing bromide and RGD peptide residues (polymers Aand B).
3.2 Optical properties of BODIPY-based polymeric dyes
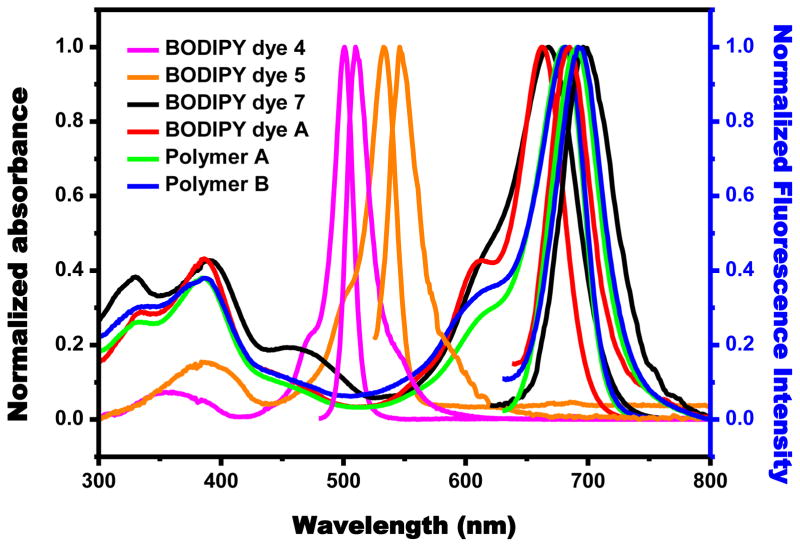
BODIPY dye (4) in methylene chloride shows two absorption peaks which are attributed to a strong S0→S1 (π-π*) transition at 501 nm and a weaker broad band around 350 nm resulting from the S0→S2 (π-π*) transition (Table 1). Introduction of 2,6-diiodo substituents to the BODIPY dye 4 to give BODIPY dye 5 causes large red shifts (32 nm and 36 nm) in both the absorption and fluorescence maxima, respectively, because 2,6-diiodo substituents serve as auxochromes (Table 1). 2,6-Diiodo-3,5-distyryl-BODIPY dye 7 shows absorption and emission peaks at 675 nm and 706 nm, respectively, because of its significantly extended π-conjugation compared with 2,6-diiodo-BODIPY dye 5. It shows slightly red shifts by 8 nm and 9 nm in absorption and emission spectra from methylene chloride to aqueous solution, respectively. BODIPY dye A exhibits absorption and emission peaks at 665 nm and 702 nm, respectively, indicating that two phenyl units and BODIPY core are not coplanar because of steric hindrance among phenyl protons with vinyl protons at 3,5-positions and methyl protons at 1,7-positions. Polymer A displays slightly red shifts by only 22 nm and 8 nm in absorption and emission spectra in aqueous solution compared with BODIPY dye A (Table 1), indicating that phenyl units and BODIPY cores in the polymer backbone are not coplanar because of the same kind of steric hindrance as BODIPY dye A. The lack of significant shifts in emission spectra of polymer A and BODIPY dye A could be due to phenylene moiety itself without any donor groups such as OMe or NMe2. However, the weak π-conjugation of polymer A significantly enhances solubility of the polymer in aqueous solution by preventing potential π-π stacking interactions among polymer backbones. Polymer B shows slightly red shifts by 2 nm and 1 nm in absorption and emission spectra in aqueous solution compared with polymer A. BODIPY dye A, and polymers Aand Bare highly soluble with at least 20 mg/mL in aqueous solutio n.
Table 1.
Absorption and emission peaks, fluorescence quantum yields, and fluorescence lifetime of BODIPY monomeric and polymeric dyes in methylene chloride and aqueous solution.
| BODIPY dye | Absorption Peak (nm) | Emission Peak (nm) | tlifetime (ns) | Φf (%) | Solvent |
|---|---|---|---|---|---|
| 4 | 501 | 510 | 3.8 | 81 | CH2Cl2 |
|
| |||||
| 5 | 533 | 546 | 4.5 | 3.6 | CH2Cl2 |
|
| |||||
| 667 | 697 | 3.4 | 2 | CH2Cl2 | |
| 7 | 635, 675 | 706 | 0.6 | 0.1 | Water |
|
| |||||
| A | 614, 663 | 685 | 7.7 | 18 | CH2Cl2 |
| 622, 665 | 702 | 2.6 | 0.7 | Water | |
|
| |||||
| Polymer A | 680 | 691 | 1.0 | 2.3 | CH2Cl2 |
| 687 | 711 | 0.4 | 0.8 | Water | |
|
| |||||
| Polymer B | 682 | 694 | 1.0 | 2.8 | CH2Cl2 |
| 689 | 712 | 0.6 | 0.9 | Water | |
Figure 2 shows the fluorescence intensity plot of the polymer at different concentrations. As expected, the average pixel value significantly depends on the different polymer concentration. The measurement of average pixel value was conducted at different times as 5 minutes, 30 minutes, and 60 mnutes. The percent change seen among the three time points is less than 5%. The results show that the polymer exhibits very good photostability without any significant decline in intensity due to photobleaching.
Figure 2.
Average light intensity versus time of different concentrations of polymer B in media under light illumination.
3.3 Application of BODIPY polymeric dye bearing cancer-homing RGD peptides in near-infrared imaging of cancer cells
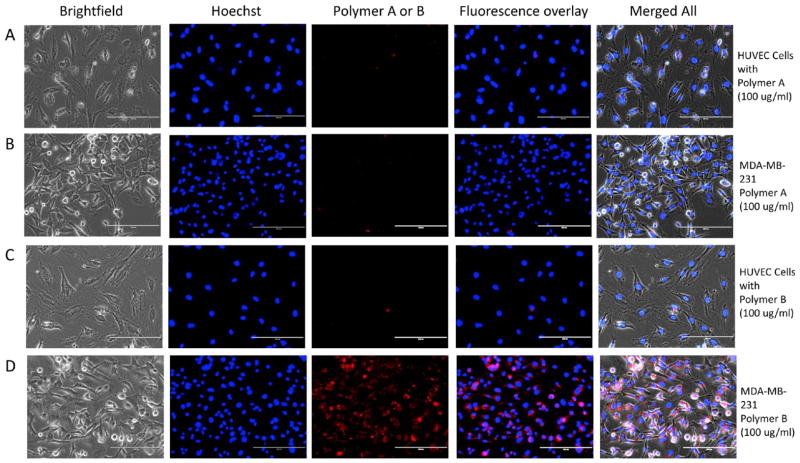
The specificity of RGD-bearing polymeric BODIPY dye (polymer B) was evaluated for tumor cell targeting using MDA-MB-231 breast cancer cell lines and compared with polymer A (lacking RGD peptide). The normal endothelial cells (HUVEC) were used as control cells. We observed a strong fluorescence signal with polymer B at 100 μg/mL concentration (Figure 3) as well as at 50 μg/mL (Figure S21 in supporting information) in MDA-MB-231 cells. The fluorescence signal was very poor with polymer A in both MDA-MB-231 and HUVEC cells, suggesting very little or no incorporation of free polymer A in these cells. Whereas, polymer B showed increased fluorescence signal for MDA-MB-231 cells in the peri-nuclear region (Figure 3 and Figure S22 in supporting information), indicating that RGD-receptor mediated specific transport and incorporation of polymer B to the breast tumor cells through αVβ3 integrins that are overexpressed in MDA-MB-231 cells but are known to be expressed at very low concentrations on normal cells such as the HUVEC-C cells [23]. The sensitivity of the RGD peptide-bearing BODIPY polymeric dye for targeting MDA-MB-231 cells is 40 μg/mL (Figure S23 in supporting information). The high affinity and selectivity of the polymer B arise from the cyclic RGD peptides that bind αVβ3integrins that are over-expressed in MDA-MB-231 breast cells.[23]
Figure 3.
Bright-field and fluorescence images of HUVEC-C cells (A) and (C), and MDA-MB-231 cells (B) and (D) after incubation with 100 μg/ml of polymer A(control BODIPY dye) or polymerB (RGD -bearing polymeric BODIPY dye) for 4 hours and then washing the cells with PBS before imaging. The bright-field images were acquired at 20× magnification with transmitted light and fluorescence images were acquired using DAPI (for Hoechst stain) andCy5 filters (for polymers Aor B). Hoechst stain was used for nuclear localization and is shown in the middle panel (blue). Fluorescence overlay (Hoechst and polymer Aor B) and merged all (Brightfield + fluorescence overlay) shows peri-nuclear localization of polymer Bin MDA -MB-231 cells with little or no incorporation in HUVEC cells.Please see close -up image of panels C and Din figure S22 in supporting information.
4. Conclusion
Conjugation of cancer-homing cyclic RGD peptide to BODIPY polymeric dye not only further enhances water solubility of the polymer, but also significantly increases binding affinity and specificity of the polymer to breast cancer cells via specific cooperative bindings of RGD peptide residues to αvβ3 integrins on cancer cells. Combination of BODIPY polymeric dye with other specific caner-homing peptides such as NGR, GFE, F3, and LyP-1 instead of RGD peptide is expected to significantly extend this application in in vitro and in vivo detection of different cancer cells and tissues.
Supplementary Material
Figure 1.
Absorption and Emission spectra of BODIPY dyes (4, 5, 7, A) and polymers A and B in methylene chloride solution.
Highlights.
Highly water-soluble near-infrared emissive BODIPY polymeric dye bearing bromide groups (polymer A) was prepared.
BODIPY polymeric dye bearing RGD peptides (polymer B) was prepared by post-polymerization functionalization.
Polymer B was used for specific near-infrared fluorescence imaging of breast cancer cells.
Acknowledgments
H. Liu and J. Wei acknowledges NIH SBIR grant (contract number: 1R43CA134039-01) and NSF grant for financial support. A. Tiwari acknowledges MTU (new faculty startup funds) for financial support. We would like to acknowledge Professor Sarah Green for her help in measurement of fluorescence lifetime of all BODIPY dyes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shah N, Cerussi A, Eker C, Espinoza J, Butler J, Fishkin J, Hornung R, Tromberg B. Proc Natl Acad Sci U S A. 2001;98:4420. doi: 10.1073/pnas.071511098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weissleder R. Nat Biotechnol. 2001;19:316. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- 3.Hawrysz DJ, Sevick-Muraca EM. Neoplasia. 2000;2:388. doi: 10.1038/sj.neo.7900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo SL, Zhang EL, Su YP, Cheng TM, Shi CM. Biomaterials. 2011;32:7127. doi: 10.1016/j.biomaterials.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 5.Cai WB, Shin DW, Chen K, Gheysens O, Cao QZ, Wang SX, Gambhir SS, Chen XY. Nano Lett. 2006;6:669. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 6.Loudet A, Burgess K. Chem Rev. 2007;107:4891. doi: 10.1021/cr078381n. [DOI] [PubMed] [Google Scholar]
- 7.Sakanoue J, Ichikawa K, Nomura Y, Tamura M. J Biochem (Tokyo) 1997;121:29. doi: 10.1093/oxfordjournals.jbchem.a021565. [DOI] [PubMed] [Google Scholar]
- 8.Lee YD, Lim CK, Kim S, Kwon IC, Kim J. Adv Funct Mater. 2010;20:2786. [Google Scholar]
- 9.Berezin MY, Guo KV, Akers W, Livingston J, Solomon M, Lee H, Liang KX, Agee A, Achilefu S. Biochem (Mosc) 2011;50:2691. doi: 10.1021/bi2000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bura T, Ziessel R. Org Lett. 2011;13:3072. doi: 10.1021/ol200969r. [DOI] [PubMed] [Google Scholar]
- 11.Zhu SL, Zhang JT, Vegesna G, Luo FT, Green SA, Liu HY. Org Lett. 2011;13:438. doi: 10.1021/ol102758z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donuru VR, Vegesna GK, Velayudham S, Meng G, Liu HY. J Polym Sci, Part A: Polym Chem. 2009;47:5354. [Google Scholar]
- 13.Donuru VR, Vegesna GK, Velayudham S, Green S, Liu HY. Chem Mater. 2009;21:2130. [Google Scholar]
- 14.Yoshii R, Nagai A, Chujo Y. J Polym Sci, Part A: Polym Chem. 2010;48:5348. [Google Scholar]
- 15.Donuru VR, Zhu SL, Green S, Liu HY. Polymer. 2010;51:5359. [Google Scholar]
- 16.Thivierge C, Loudet A, Burgess K. Macromolecules. 2011;44:4012. doi: 10.1021/ma200174w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brannon JH, Magde D. J Phys Chem. 1978;82:705. [Google Scholar]
- 18.Velapoldi RA, Tonnesen HH. J Fluoresc. 2004;14:465. doi: 10.1023/b:jofl.0000031828.96368.c1. [DOI] [PubMed] [Google Scholar]
- 19.Xue CH, Donuru VRR, Liu HY. Macromolecules. 2006;39:5747. [Google Scholar]
- 20.Xue CH, Luo FT, Liu HY. Macromolecules. 2007;40:6863. [Google Scholar]
- 21.Xue C, Velayudham S, Johnson S, Saha R, Smith A, Brewer W, Murthy P, Bagley ST, Liu H. Chem Eur J. 2009;15:2289. doi: 10.1002/chem.200801875. [DOI] [PubMed] [Google Scholar]
- 22.Zhu SL, Zhang JT, Vegesna GK, Pandey R, Luo FT, Green SA, Liu HY. Chem Commun. 2011;47:3508. doi: 10.1039/c0cc05303a. [DOI] [PubMed] [Google Scholar]
- 23.Desgrosellier JS, Cheresh DA. Nat Rev Cancer. 2010;10:890. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.