Abstract
Nearly all vertebrate cells have been shown to express heparan sulfate proteoglycans (HSPGs) at the cell surface. The HSPGs bind to many secreted signaling proteins, including numerous growth factors, cytokines, and morphogens, to affect their tissue distribution and signaling. The heparan sulfate (HS) chains may have variable length and may differ with regard to both degree and pattern of sulfation. As the sulfation pattern of HS chains in most cases will determine if an interaction with a potential ligand will take place, as well as the affinity of the interaction, a key to understanding the function of HSPGs is to clarify how HS biosynthesis is regulated in different biological contexts. This review provides an introduction to the current understanding of HS biosynthesis and its regulation, and identifies research areas where more knowledge is needed to better understand how the HS biosynthetic machinery works.
Keywords: heparan sulfate, biosynthesis, regulation, enzyme, specificity, GAGosome, Golgi
Heparan sulfate proteoglycans (HSPGs) are indispensible during embryonic development and have important regulatory functions in adulthood as well as affect several common pathophysiological conditions (Hacker et al. 2005; Lindahl and Li 2009; Iozzo and Sanderson 2011). The structurally complex HSPGs modulate signaling, turnover, and tissue distribution of many secreted signaling molecules such as growth factors and morphogens. In addition, they participate in uptake of triglyceride-rich lipoproteins (Foley and Esko 2010) and interact with cell adhesion molecules (Xian et al. 2010). After shedding into the extracellular matrix, their function as co-receptors can be lost, and they may then instead work as secreted antagonists to inhibit signaling events at the cell surface (Kreuger et al. 2004).
One well-studied and important function of HSPGs is to modulate signaling by shaping and conferring robustness to secreted protein gradients (Hufnagel et al. 2006) and by mediating the formation of ligand-receptor complexes, for example, the ternary FGF-HS-FGFR complexes (Schlessinger et al. 2000). However, due to the complex nature of proteoglycans and the heterogeneity of the HS glycosaminoglycan (GAG) chains and their large interactome, HSPGs may at the same time modulate the functions of multiple protein ligands. Despite this complexity, characterization of the phenotypes of knockout mice, where the expression of HS biosynthetic enzymes has been perturbed, points to some distinct processes that seem to be particularly sensitive to reduced HS biosynthesis. These include cartilage and bone formation, as well as lung, kidney, eye, brain, lacrimal gland, and mammary gland development (Merry and Wilson 2002; Inatani et al. 2003; Crawford et al. 2010; Habuchi and Kimata 2010; Li 2010; Ringvall and Kjellen 2010; Qu et al. 2011).
Evidence for Regulated HS Biosynthesis
Investigations of the HS biosynthetic machinery have been and still are hampered by the lack of analytical methods for high-capacity, high-resolution sequence analysis of full-length HS chains. Partial sequence analysis has successfully been performed of purified, shorter HS oligosaccharides and heparin derivatives using different techniques (Turnbull et al. 1999; Venkataraman et al. 1999; Kreuger et al. 2001; Yang et al. 2011). A promising mass spectrometry methodology was also recently used to determine the predominant sequence of the single chondroitin sulfate chain of bikunin (Ly et al. 2011). However, it is not presently possible to identify all the structurally different HS chains expressed at the surface of a single cell.
Still, some very important information on HS structure and biosynthesis has indeed been obtained using global HS disaccharide analysis, a variety of chromatographic methods, and enzymatic assays with defined substrates. Although the expression of HS as judged by global disaccharide composition analysis seems to be highly conserved between organs and different cell types (Ledin et al. 2004), the variability at the level of single chains may be such that very few, if any, HS chains have the same structure. The fine structure of HS could in theory accommodate an infinite number of (protein-binding) epitopes. For example, it has been calculated that over 1,000,000 structurally different epitopes are possible in only an octasaccharide fragment (Sasisekharan and Venkataraman 2000). However, the regulation of enzyme expression and substrate specificities of the HS biosynthetic enzymes will greatly restrict the number of HS epitopes expressed (Rudd and Yates 2012). All available data indicate that the expression of HS (as well as proteoglycan core proteins) is tightly regulated during development. Structural analyses of HS isolated from different mammalian tissues have pointed to the existence of tissue-specific HS composition in support of the stringent regulation of HS biosynthesis (Maccarana et al. 1996; Ledin et al. 2004; Lawrence et al. 2008). Furthermore, immunohistochemical analyses using antibodies selectively recognizing different HS epitopes have revealed reproducible patterns of individual HS motifs within tissues (van Kuppevelt et al. 1998). At which different levels might then HS biosynthesis be regulated?
Core Protein Formation and Processing
It is possible that the amount of core protein in some situations may be the limiting factor for HS biosynthesis or that core proteins could compete for enzymes involved in HS biosynthesis in the Golgi compartment. In this way, different types and amounts of HS may be presented at the cell surface (e.g., attached to syndecans and glypicans) and/or in the extracellular matrix (e.g., attached to agrin, perlecan, and collagen XVIII). Furthermore, it has been shown that domains in the core protein other than the GAG attachment site can have regulatory functions. For example, in glypicans, removal of the globular domain resulted in decreased HS decoration and increased chondroitin sulfate (CS) substitution of the core protein (Chen and Lander 2001).
Formation of the Linkage Region
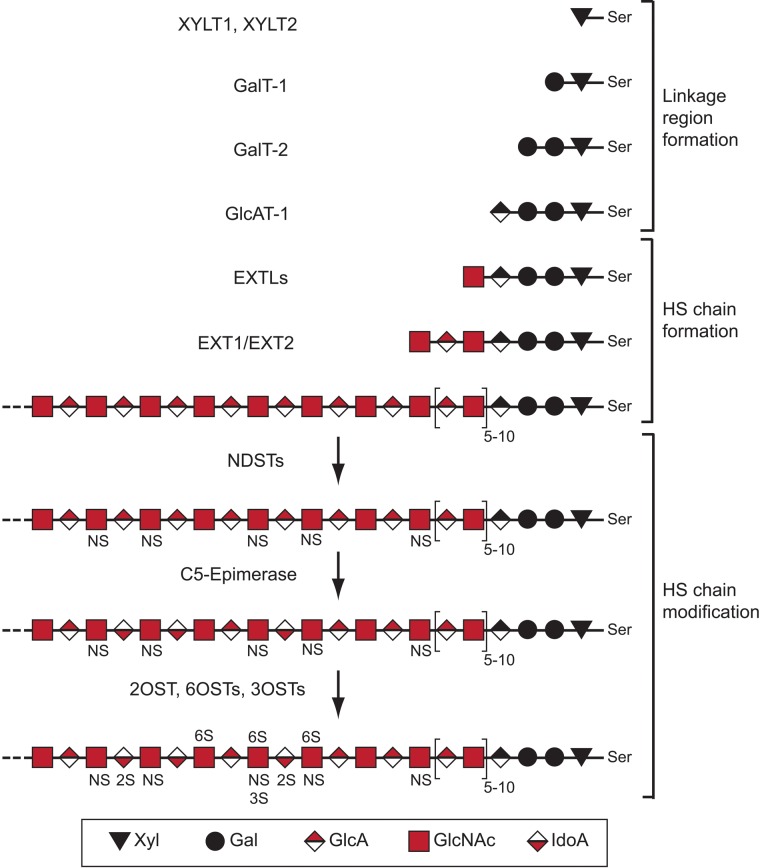
It should be noted that all biosynthetic enzymes (in humans and mice) directly involved in the making of HS chains, with the exception of a 3-O-sulfotransferase, have been reported to be transmembrane proteins exerting their catalytical activities inside the Golgi compartment. In zebrafish, however, three of the eight 3-O-sulfotransferases lack transmembrane domains (Cadwallader and Yost 2006). Initiation of HS biosynthesis starts with the formation of a glucuronic acid-galactose-galactose-xylose tetrasaccharide linkage region (Fig. 1). The xylose (Xyl) residue is attached to a serine next to a glycine residue flanked by acidic and hydrophobic amino acids (Zhang et al. 1995; Esko and Zhang 1996) in the core protein by the action of one of the two xylosyltransferases XYLT1 or XYLT2. Two galactose (Gal) residues are thereafter added in sequence by the galactosyltransferases GalT-1 and GalT-2, respectively. The formation of the linkage region is finally completed by the addition of a glucuronic acid (GlcA) unit by the GlcAT-1 transferase.
Figure 1.
Heparan sulfate (HS) structure and biosynthesis scheme. Shown is a simplified scheme outlining the different steps of HS biosynthesis involving specific enzymes or enzyme families. The structure of HS is variable, and a hypothetical example is shown. The saccharide units corresponding to symbols used are defined below the scheme. The abbreviations related to structure are as follows: NS, N-sulfated GlcN; 6S, 6-O-sulfated GlcN; 2S, 2-O-sulfated IdoA; 3S, 3-O-sulfated GlcN; Ser, serine. For additional information, see Figure 2 and the main text.
The linkage region saccharides may be modified by phosphorylation of the Xyl unit and by sulfation of the two Gal units. These modifications have been shown to affect downstream enzymatic polymerization activities, such that multiple modifications mainly inhibit or restrict enzymes involved in the formation of the linkage region (Gulberti et al. 2005; Tone et al. 2008). It has further been suggested that 4-O-sulfation of the second Gal residue in the link region is associated with CS biosynthesis (Ueno et al. 2001). Thus, phosphorylation and sulfation of the linkage region may affect both HS and CS biosynthesis.
HS Polymer Formation
The members of the EXTL family of glycosyltransferases initiate HS chain formation by attaching an N-acetylglucosamine (GlcNAc) residue to the non-reducing end of the acceptor linkage tetrasaccharide region. Importantly, the linkage region may also serve as a primer for biosynthesis of the closely related polysaccharide CS. In the case of CS biosynthesis, a β-N-acetylgalactosamine (β-GalNAc) residue is transferred to the linkage region by a CSGALNAC-transferase.
EXTL1, EXTL2, and EXTL3 have all been shown to possess α-GlcNAc transferase activity, thus being capable of assisting in HS polymerization (Kim et al. 2001). Although EXTL3 seems to be the main enzyme catalyzing the initiation of HS biosynthesis in vivo (Han et al. 2004; Holmborn et al. 2012), suppression of both EXTL2 and EXTL3 leads to reduced HS biosynthesis (Kaidonis et al. 2010). It has also been demonstrated that lowered levels of EXTL3 lead to increased HS chain length (Busse et al. 2007), pointing to a complex regulation. Notably, EXTL2 has been shown to transfer either GlcNAc or GalNAc to the link region. When GlcNAc is added, it will serve as a starting point for HS biosynthesis, whereas transfer of α-GalNAc by EXTL2 has been suggested to block CS biosynthesis, as this residue does not serve as an acceptor for further CS polymerization (Kitagawa et al. 1999). Accordingly, there may be competition or regulation of the balance between HS and CS biosynthesis at the level of HS/CS initiation; however, the relevance of this concept remains to be investigated in vivo. Because the HS biosynthetic enzymes seem to reside in the proximal parts of the Golgi compartment while the CS enzymes locate more to the distal stacks (Presto et al. 2008), it is possible that CS in some situations will be synthesized onto HSPG core proteins carrying linkage regions that, for some reason, escaped HS biosynthesis.
After the EXTL-mediated initiation, the HS chain is extended by the action of the functional HS-polymerase complex, consisting of the EXT1 and EXT2 enzymes, which transfers alternating GlcA and GlcNAc residues to the growing polymer. In vitro results suggest that sulfation of the growing HS backbone stimulates polymerization and leads to increased chain length (Lidholt and Lindahl 1992). In this context, the finding of an interaction between EXT2 and N-deacetylase/N-sulfotransferase-1 (NDST1; see below) is interesting. Finally, EXT-mutant cells that do not synthesize HS instead produce increased amounts of CS (Lidholt et al. 1992; Stickens et al. 2005; Le Jan et al. 2012). This interplay between HS and CS biosynthesis will be discussed further below.
HS Chain Modification
NS-Domain Formation
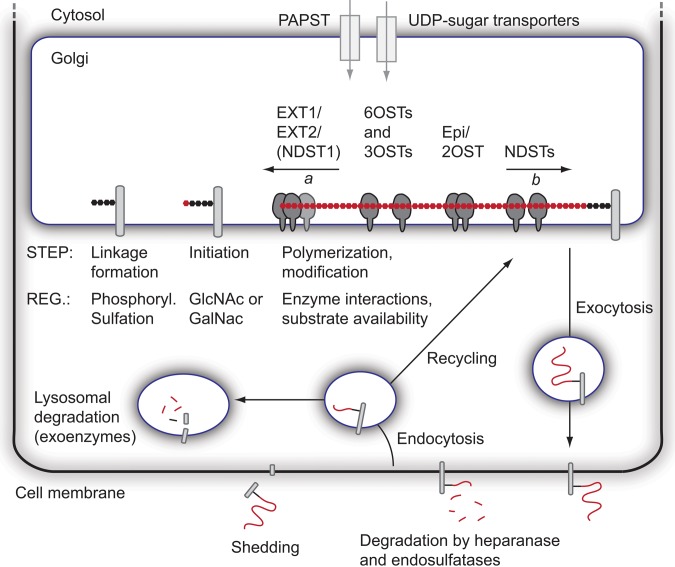
The modification of the HS backbone is thought to most often start with N-deacetylation/N-sulfation of GlcNAc residues by the NDST enzymes. This process is incomplete, such that the final HS products display a highly variable domain-type arrangement with N-sulfated regions separated by N-acetylated regions (Esko and Lindahl 2001). Both the N-sulfated and N-acetylated regions may be of different lengths, but it is not understood how and to what extent the N-sulfation pattern is regulated. However, it has been shown in vitro that the presence of the sulfate donor 3′-phosphoadenosine-5′-phosphosulfate (PAPS) makes the NDST enzymes work in a processive manner, so that the lengths of the N-sulfated domains correlate to the concentration of PAPS (Carlsson et al. 2008). Recently, it was suggested that the direction of NDST enzyme action is opposite to that of the EXT1/EXT2 polymerase complex (Sheng et al. 2011) (Fig. 2). NDST1 has the capacity to bind to EXT2, and the N-sulfation degree is affected by the level of EXT1 and EXT2 expression (Presto et al. 2008).
Figure 2.
Formation and fate of heparan sulfate (HS). The formation of HS takes place in the Golgi network, where most of the biosynthetic enzymes are anchored to the Golgi membrane. Biosynthetic precursors (3′-phosphoadenosine-5′-phosphosulfate [PAPS] and UDP-sugars) are formed in the cytosol and transported into the Golgi. Prior to HS polymerization, the linkage region is formed attached to a serine residue in a core protein. Next, the EXT1/EXT2 polymerase complex adds alternating units of GlcNAc and GlcA to the non-reducing end of the growing chain (arrow a indicates the direction of polymerization). The polymerization is followed by a series of modification reactions, likely to begin with N-deacetylation/N-sulfation, followed by epimerization and 2-O-sulfation, and finally 6-O- and 3-O-sulfation. Notably, it has recently been proposed that the direction of N-deacetylation/N-sulfation is opposite to that of polymerization (arrow b). Known interactions between enzymes are indicated, but additional protein interactions as well as larger GAGosome complexes encompassing many enzymes may exist. After completion of the modification process, the core proteins are transported to the cell membrane, where they are exocytosed. HS chains of both membrane-intercalated and secreted proteoglycans (PGs) can be trimmed by the actions of heparanase and endosulfatases, and surface-bound PGs can also be shed. Finally, endocytosis of PGs leads to degradation of HS by exoenzymes in lysosomes or, alternatively, to recycling and possibly additional rounds of HS biosynthesis/modification onto recycled core proteins. Some regulatory steps (Reg.) during the biosynthetic process are indicated.
The NDSTs clearly play a key role in the formation of ligand-binding domains because many of the other modification reactions occur at the sites of N-sulfation (Esko and Lindahl 2001). Protein ligands may interact with single N-sulfated (NS) domains rich in O-sulfate groups (e.g., growth factors of the fibroblast growth factor [FGF] family) or with two NS-domains separated by N-acetylated (NA) disaccharide residues, so-called SAS domains (Kreuger et al. 2002), as has been shown for the angiogenic growth factor VEGFA165 (Robinson et al. 2006), interleukin-8 (Spillmann et al. 1998), interferon-γ (Lortat-Jacob et al. 1995), and platelet factor 4 (Stringer and Gallagher 1997).
Heparin, produced exclusively by connective tissue–type mast cells attached to serglycin core proteins, contains the same polysaccharide backbone as HS but exhibits a much higher degree of modification (i.e., epimerization and sulfation). Although no heparin is produced in mast cells of the NDST2 knockout mice (Forsberg et al. 1999; Humphries et al. 1999), the structure of HS isolated from different tissues of the same mice appears to be normal (Ledin et al. 2004). Thus, although NDST2, just like NDST1, is expressed by most cells in the body, it is apparently only essential for heparin biosynthesis.
Epimerization and 2-O-Sulfation
A single C5-epimerase converts many but not all GlcA units positioned next to glucosamine units into IdoA (Li et al. 1997). Epimerization is followed by 2-O-sulfation of a majority of the IdoA units by a 2-O-sulfotransferase (2OST). Thus, the pattern of N-sulfation will dictate the position of the highly modified NS-domains. The C5-epimerase and the 2OST co-localize in the Golgi apparatus and may interact (Pinhal et al. 2001). However, although some overlap occurs, the expression of the 2OST and the C5-epimerase is not always coordinately regulated (Cadwallader and Yost 2007).
6-O- and 3-O-Sulfation
As the capacity of HS to interact with protein ligands is dependent on the occurrence of O-sulfate groups, it is striking to note that, although there is only a single 2-O-sulfotransferase, there are three 6-O-sulfotransferases (6OST1-3) and seven 3-O-sulfotransferases. A lot of the functionally relevant regulation of HS biosynthesis resulting in the generation of protein binding sites may thus lie at the level of 6-O and 3-O sulfation, given the evolution and preservation of the corresponding ten O-sulfotransferases. The substrate specificities of the 6-O-sulfotranferases differ slightly, and although all three enzymes may act on both GlcNAc and GlcNS residues (Smeds et al. 2003), 6-O sulfation has been reported to preferentially occur on GlcNS residues flanked by 2-O-sulfated IdoA units (Jemth et al. 2003).
Special attention should be given to the seven 3-O-sulfotransferases. They do have distinct temporal and spatial expression patterns (Cadwallader and Yost 2006) and have been shown to be involved in the formation of the still relatively few HS motifs that interact in a selective manner with protein ligands. Examples of such motifs include the antithrombin-binding pentasaccharide motif in heparin/HS (Petitou et al. 2003), the gD herpes simplex virus type 1–binding octasaccharide motif (Shukla et al. 1999), and the chemokine cyclophilin B–binding epitope (Vanpouille et al. 2007). Interestingly, mouse knockouts for HS3OST-1, central to endothelial cell production of anticoagulant HS, exhibited lethality only on a specific genetic background and showed intrauterine growth retardation but showed no obvious coagulopathy (HajMohammadi et al. 2003). Reduction of 3OST activity in Drosophila resulted in compromised Notch signaling with resultant neurogenic phenotypes (Kamimura et al. 2004). Thus, 3OST activity seems to modulate several distinct developmental processes.
Degradation of Surface-Bound and Extracellular HS
Two endosulfatases have been identified, SULF1 and SULF2, which can remove 6-O-sulfate groups from HS in the extracellular space (Ai et al. 2006). In addition, the secreted enzyme heparanase degrades extracellular HS, resulting in trimming of the HS chains attached to core proteins and the release of smaller and potentially bioactive HS fragments (Gong et al. 2003). An alternative mechanism for the release of HS from the cell surface is by shedding of the proteoglycan. Syndecans, for example, are shed by the action of matrix metalloproteinases (Fitzgerald et al. 2000), and glypicans can be cleaved by the action of the notum protein and phospholipases, leading to release of the HS-substituted extracellular domain (Kreuger et al. 2004; Traister et al. 2008).
Intracellular Degradation and Recycling
Proteoglycans have also been shown to recycle, with the same core protein being internalized and subsequently routed back to the cell surface (Fig. 2). Recycling brings about the possibility of partial intracellular degradation or trimming of HS chains, possibly together with new rounds of HS biosynthesis, where after core proteins with altered HS chains can be re-exocytosed to the cell membrane (Fransson et al. 2004). The final degradation of HS occurs intracellularly in the lysosomes through the action of several exoenzymes (Freeman and Hopwood 1992), and malfunction of the exoenzymes leads to different mucopolysaccharidosis lysosomal storage diseases (Clarke 2008).
Substrates for HS Biosynthesis: Nucleotide Sugars and PAPS
The biosynthesis of HS requires UDP-sugars as well as the sulfate donor PAPS (Berninsone and Hirschberg 2000; Caffaro et al. 2006). All these precursors are synthesized in the cytosol and transported into the Golgi compartment. Two PAPS synthases, PAPSS1 and PAPSS2 (Fuda et al. 2002; Stelzer et al. 2007), and two PAPS transporters, PAPST1 and PAPST2, have been identified (Kamiyama et al. 2006). As mentioned above, PAPS is required for the processive action of NDSTs and as a sulfate donor for the NDSTs as well as the different O-sulfotransferases. It should be noted that altered synthesis and/or altered transport of these precursors could affect HS biosynthesis.
Regulation of Enzyme Expression at the DNA and RNA Levels
Not much is known regarding the transcriptional and translational regulation of the HS biosynthetic enzymes. Recent studies report on the transcriptional regulation of SULFs (Langsdorf et al. 2011), NDST2 (Morii et al. 2001), and the PAPS synthases (Shimizu et al. 2002). Furthermore, the Runx2 transcription factor has been shown to increase the expression of EXT1 and heparanase (Teplyuk et al. 2009). Epigenetic regulation of EXT1 has been shown to lead to a loss of HS biosynthesis (Teplyuk et al. 2009), and the levels of some 3OSTs are also determined by this type of regulation (Bui et al. 2010). Evidence for translational regulation of HS biosynthesis enzymes has also been presented (Bornemann et al. 2008; Grobe and Esko 2002), and it was also recently shown that miRNAs have the potential to downregulate the levels of enzymes related to HS biosynthesis (Small et al. 2010).
The Hypothetical GAGosome Complex
The GAGosome model proposed in 2002 by Jeffrey Esko and Scott Selleck proposes that some of the HS biosynthetic enzymes may act together in a physical complex (Esko and Selleck 2002). The ability of the enzymes to associate with each other and the stoichiometry and presence of enzyme isoforms with different catalytic activities will likely affect GAGosome function. Compatible with the GAGosome hypothesis, the HS polymerases EXT1 and EXT2 are known to function as a complex (Kobayashi et al. 2000; McCormick et al. 2000; Senay et al. 2000), and an interaction between EXT2 and NDST1, regulating NDST1 activity, has also been reported (Presto et al. 2008). Here it was shown that the expression levels of the EXT polymerases affected the NDST1 protein levels. Interactions have also been observed between XylT and GalT-1 (Schwartz 1975) and between the C5-epimerase and 2OST (Pinhal et al. 2001). The rapid production of HS/heparin, with a complete heparin chain being produced in ~1 min (Hook et al. 1975; Lidholt et al. 1989), could perhaps be explained by the existence of highly efficient GAGosomes.
Compartmentalization of Enzymes
Previous work where the expression of green fluorescent protein (GFP)–tagged biosynthesis enzymes have been studied indicates that enzymes responsible for the biosynthesis of the linkage region as well as the EXT1/EXT2 co-polymerase, NDST1, 2OST, the C5-epimerase, and 6OSTs all reside in the cis/medial stacks of the Golgi compartment (McCormick et al. 2000; Crawford et al. 2001; Pinhal et al. 2001; Nagai et al. 2004). In contrast, the CS biosynthesis enzymes appear to localize more to the trans-Golgi/trans-Golgi network (Velasco et al. 1988; Uhlin-Hansen and Yanagishita 1993; Kolset et al. 2002). More refined techniques, such as the one described by Multhaupt and Couchman (2012), will make it possible to get a more detailed picture of how the different enzymes localize in different sub-Golgi compartments. Of note, it is possible that the compartmentalization of HS and CS biosynthesis varies between cell types.
The Interplay between HS and CS Biosynthesis
It has been shown that inhibition of HS biosynthesis may affect CS biosynthesis (Holmborn et al. 2012; Wei et al. 2000). Accordingly, EXT1-deficient embryoid bodies were recently shown to have doubled their production of CS in the absence of HS biosynthesis, and it was suggested that this increase in CS production allowed formation of capillary-like vascular structures (Le Jan et al. 2012). These findings may indicate that there is a direct link between HS and CS production and that the two polysaccharides even may have partially overlapping functions. How, then, could reduced HS biosynthesis lead to increased and/or altered CS biosynthesis? It is possible that a reduction in HS biosynthesis allows for CS substitution of linkage regions that normally should carry HS. Also, a lack of HS production would conceivably lead to larger pools of available PAPS and UDP-sugar precursors that now could be used for CS biosynthesis. Several other mechanisms, including a response to altered surface levels of HS and CS, could of course also affect both HS and CS biosynthesis.
Future Perspectives
One of the main challenges in this research field is to establish the functionalities of the postulated enzyme complexes in the Golgi and to investigate how dynamic or stable these complexes are. We also need to find out more about the transcriptional and translational regulation of the different biosynthetic enzymes. In addition, methods to analyze the structure of extended HS chains at high resolution need to be further developed and become widely accessible. It will also be important to better understand how primary HS sequences relate to three-dimensional structures recognized by various protein ligands.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: L.K. is supported by the Swedish Research Council, the Swedish Cancer Society, and Polysackaridforskning AB. J.K. is supported by the Swedish Research Council, the Swedish Cancer Society, the Swedish Childhood Cancer Foundation, the Swedish Foundation for Strategic Research (project no A3 05:207g), Polysackaridforskning AB, and Uppsala University.
References
- Ai X, Do AT, Kusche-Gullberg M, Lindahl U, Lu K, Emerson CP., Jr 2006. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J Biol Chem. 281:4969–4976 [DOI] [PubMed] [Google Scholar]
- Berninsone PM, Hirschberg CB. 2000. Nucleotide sugar transporters of the Golgi apparatus. Curr Opin Struct Biol. 10:542–547 [DOI] [PubMed] [Google Scholar]
- Bornemann DJ, Park S, Phin S, Warrior R. 2008. A translational block to HSPG synthesis permits BMP signaling in the early Drosophila embryo. Development. 135:1039–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui C, Ouzzine M, Talhaoui I, Sharp S, Prydz K, Coughtrie MW, Fournel-Gigleux S. 2010. Epigenetics: methylation-associated repression of heparan sulfate 3-O-sulfotransferase gene expression contributes to the invasive phenotype of H-EMC-SS chondrosarcoma cells. FASEB J. 24:436–450 [DOI] [PubMed] [Google Scholar]
- Busse M, Feta A, Presto J, Wilen M, Gronning M, Kjellen L, Kusche-Gullberg M. 2007. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J Biol Chem. 282:32802–32810 [DOI] [PubMed] [Google Scholar]
- Cadwallader AB, Yost HJ. 2006. Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: I. The 3-O-sulfotransferase family. Dev Dyn. 235:3423–3431 [DOI] [PubMed] [Google Scholar]
- Cadwallader AB, Yost HJ. 2007. Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: III. 2-O-sulfotransferase and C5-epimerases. Dev Dyn. 236:581–586 [DOI] [PubMed] [Google Scholar]
- Caffaro CE, Hirschberg CB, Berninsone PM. 2006. Independent and simultaneous translocation of two substrates by a nucleotide sugar transporter. Proc Natl Acad Sci U S A. 103:16176–16181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson P, Presto J, Spillmann D, Lindahl U, Kjellen L. 2008. Heparin/heparan sulfate biosynthesis: processive formation of N-sulfated domains. J Biol Chem. 283:20008–20014 [DOI] [PubMed] [Google Scholar]
- Chen RL, Lander AD. 2001. Mechanisms underlying preferential assembly of heparan sulfate on glypican-1. J Biol Chem. 276:7507–7517 [DOI] [PubMed] [Google Scholar]
- Clarke LA. 2008. The mucopolysaccharidoses: a success of molecular medicine. Expert Rev Mol Med. 10:e1. [DOI] [PubMed] [Google Scholar]
- Crawford BE, Garner OB, Bishop JR, Zhang DY, Bush KT, Nigam SK, Esko JD. 2010. Loss of the heparan sulfate sulfotransferase, Ndst1, in mammary epithelial cells selectively blocks lobuloalveolar development in mice. PLoS ONE. 5:e10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford BE, Olson SK, Esko JD, Pinhal MA. 2001. Cloning, Golgi localization, and enzyme activity of the full-length heparin/heparan sulfate-glucuronic acid C5-epimerase. J Biol Chem. 276:21538–21543 [DOI] [PubMed] [Google Scholar]
- Esko JD, Lindahl U. 2001. Molecular diversity of heparan sulfate. J Clin Invest. 108:169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. 2002. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 71:435–471 [DOI] [PubMed] [Google Scholar]
- Esko JD, Zhang L. 1996. Influence of core protein sequence on glycosaminoglycan assembly. Curr Opin Struct Biol. 6:663–670 [DOI] [PubMed] [Google Scholar]
- Fitzgerald ML, Wang Z, Park PW, Murphy G, Bernfield M. 2000. Shedding of syndecan-1 and -4 ectodomains is regulated by multiple signaling pathways and mediated by a TIMP-3-sensitive metalloproteinase. J Cell Biol. 148:811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EM, Esko JD. 2010. Hepatic heparan sulfate proteoglycans and endocytic clearance of triglyceride-rich lipoproteins. Prog Mol Biol Transl Sci. 93:213–233 [DOI] [PubMed] [Google Scholar]
- Forsberg E, Pejler G, Ringvall M, Lunderius C, Tomasini-Johansson B, Kusche-Gullberg M, Eriksson I, Ledin J, Hellman L, Kjellen L. 1999. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 400:773–776 [DOI] [PubMed] [Google Scholar]
- Fransson LA, Belting M, Cheng F, Jonsson M, Mani K, Sandgren S. 2004. Novel aspects of glypican glycobiology. Cell Mol Life Sci. 61:1016–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman C, Hopwood J. 1992. Lysosomal degradation of heparin and heparan sulphate. Adv Exp Med Biol. 313:121–134 [DOI] [PubMed] [Google Scholar]
- Fuda H, Shimizu C, Lee YC, Akita H, Strott CA. 2002. Characterization and expression of human bifunctional 3′-phosphoadenosine 5′-phosphosulphate synthase isoforms. Biochem J. 365:497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Jemth P, Escobar Galvis ML, Vlodavsky I, Horner A, Lindahl U, Li JP. 2003. Processing of macromolecular heparin by heparanase. J Biol Chem. 278:35152–35158 [DOI] [PubMed] [Google Scholar]
- Grobe K, Esko JD. 2002. Regulated translation of heparan sulfate N-acetylglucosamine N-deacetylase/n-sulfotransferase isozymes by structured 5′-untranslated regions and internal ribosome entry sites. J Biol Chem. 277:30699–30706 [DOI] [PubMed] [Google Scholar]
- Gulberti S, Lattard V, Fondeur M, Jacquinet JC, Mulliert G, Netter P, Magdalou J, Ouzzine M, Fournel-Gigleux S. 2005. Phosphorylation and sulfation of oligosaccharide substrates critically influence the activity of human beta1,4-galactosyltransferase 7 (GalT-I) and beta1,3-glucuronosyltransferase I (GlcAT-I) involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J Biol Chem. 280:1417–1425 [DOI] [PubMed] [Google Scholar]
- Habuchi H, Kimata K. 2010. Mice deficient in heparan sulfate 6-O-sulfotransferase-1. Prog Mol Biol Transl Sci. 93:79–111 [DOI] [PubMed] [Google Scholar]
- Hacker U, Nybakken K, Perrimon N. 2005. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 6:530–541 [DOI] [PubMed] [Google Scholar]
- HajMohammadi S, Enjyoji K, Princivalle M, Christi P, Lech M, Beeler D, Rayburn H, Schwartz JJ, Barzegar S, de Agostini AI, et al. 2003. Normal levels of anticoagulant heparan sulfate are not essential for normal hemostasis. J Clin Invest. 111:989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Khodoun M, Tauchi M, Lin X. 2004. Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development. 131:1563–1575 [DOI] [PubMed] [Google Scholar]
- Holmborn K, Habicher J, Kasza Z, Eriksson AS, Filipek-Gorniok B, Gopal S, Couchman JR, Ahlberg PE, Wiweger M, Spillmann D, et al. 2012. On the roles and regulation of chondroitin sulfate and heparan sulfate in zebrafish pharyngeal cartilage morphogenesis. J Biol Chem. 287:33905–33916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook M, Lindahl U, Hallen A, Backstrom G. 1975. Biosynthesis of heparin. Studies on the microsomal sulfation process. J Biol Chem. 250:6065–6071 [PubMed] [Google Scholar]
- Hufnagel L, Kreuger J, Cohen SM, Shraiman BI. 2006. On the role of glypicans in the process of morphogen gradient formation. Dev Biol. 300:512–522 [DOI] [PubMed] [Google Scholar]
- Humphries DE, Wong GW, Friend DS, Gurish MF, Qiu WT, Huang C, Sharpe AH, Stevens RL. 1999. Heparin is essential for the storage of specific granule proteases in mast cells. Nature. 400:769–772 [DOI] [PubMed] [Google Scholar]
- Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. 2003. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 302:1044–1046 [DOI] [PubMed] [Google Scholar]
- Iozzo RV, Sanderson RD. 2011. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 15:1013–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemth P, Smeds E, Do AT, Habuchi H, Kimata K, Lindahl U, Kusche-Gullberg M. 2003. Oligosaccharide library-based assessment of heparan sulfate 6-O-sulfotransferase substrate specificity. J Biol Chem. 278:24371–24376 [DOI] [PubMed] [Google Scholar]
- Kaidonis X, Liaw WC, Roberts AD, Ly M, Anson D, Byers S. 2010. Gene silencing of EXTL2 and EXTL3 as a substrate deprivation therapy for heparan sulphate storing mucopolysaccharidoses. Eur J Hum Genet. 18:194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura K, Rhodes JM, Ueda R, McNeely M, Shukla D, Kimata K, Spear PG, Shworak NW, Nakato H. 2004. Regulation of Notch signaling by Drosophila heparan sulfate 3-O sulfotransferase. J Cell Biol. 166:1069–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama S, Sasaki N, Goda E, Ui-Tei K, Saigo K, Narimatsu H, Jigami Y, Kannagi R, Irimura T, Nishihara S. 2006. Molecular cloning and characterization of a novel 3′-phosphoadenosine 5′-phosphosulfate transporter, PAPST2. J Biol Chem. 281:10945–10953 [DOI] [PubMed] [Google Scholar]
- Kim BT, Kitagawa H, Tamura J, Saito T, Kusche-Gullberg M, Lindahl U, Sugahara K. 2001. Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode alpha 1,4- N-acetylglucosaminyltransferases that likely are involved in heparan sulfate/heparin biosynthesis. Proc Natl Acad Sci U S A. 98:7176–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H, Shimakawa H, Sugahara K. 1999. The tumor suppressor EXT-like gene EXTL2 encodes an alpha1, 4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region: the key enzyme for the chain initiation of heparan sulfate. J Biol Chem. 274:13933–13937 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Morimoto K, Shimizu T, Takahashi M, Kurosawa H, Shirasawa T. 2000. Association of EXT1 and EXT2, hereditary multiple exostoses gene products, in Golgi apparatus. Biochem Biophys Res Commun. 268:860–867 [DOI] [PubMed] [Google Scholar]
- Kolset SO, Prydz K, Fjeldstad K, Safaiyan F, Vuong TT, Gottfridsson E, Salmivirta M. 2002. Effect of brefeldin A on heparan sulphate biosynthesis in Madin-Darby canine kidney cells. Biochem J. 362:359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuger J, Matsumoto T, Vanwildemeersch M, Sasaki T, Timpl R, Claesson-Welsh L, Spillmann D, Lindahl U. 2002. Role of heparan sulfate domain organization in endostatin inhibition of endothelial cell function. EMBO J. 21:6303–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuger J, Perez L, Giraldez AJ, Cohen SM. 2004. Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Dev Cell. 7:503–512 [DOI] [PubMed] [Google Scholar]
- Kreuger J, Salmivirta M, Sturiale L, Gimenez-Gallego G, Lindahl U. 2001. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J Biol Chem. 276:30744–30752 [DOI] [PubMed] [Google Scholar]
- Langsdorf A, Schumacher V, Shi X, Tran T, Zaia J, Jain S, Taglienti M, Kreidberg JA, Fine A, Ai X. 2011. Expression regulation and function of heparan sulfate 6-O-endosulfatases in the spermatogonial stem cell niche. Glycobiology. 21:152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence R, Olson SK, Steele RE, Wang L, Warrior R, Cummings RD, Esko JD. 2008. Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J Biol Chem. 283:33674–33684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Jan S, Hayashi M, Kasza Z, Eriksson I, Bishop JR, Weibrecht I, Heldin J, Holmborn K, Jakobsson L, Soderberg O, et al. 2012. Functional overlap between chondroitin and heparan sulfate proteoglycans during VEGF-induced sprouting angiogenesis. Arterioscler Thromb Vasc Biol. 32:1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledin J, Staatz W, Li JP, Gotte M, Selleck S, Kjellen L, Spillmann D. 2004. Heparan sulfate structure in mice with genetically modified heparan sulfate production. J Biol Chem. 279:42732–42741 [DOI] [PubMed] [Google Scholar]
- Li J, Hagner-McWhirter A, Kjellen L, Palgi J, Jalkanen M, Lindahl U. 1997. Biosynthesis of heparin/heparan sulfate. cDNA cloning and expression of D-glucuronyl C5-epimerase from bovine lung. J Biol Chem. 272:28158–28163 [DOI] [PubMed] [Google Scholar]
- Li JP. 2010. Glucuronyl C5-epimerase an enzyme converting glucuronic acid to iduronic acid in heparan sulfate/heparin biosynthesis. Prog Mol Biol Transl Sci. 93:59–78 [DOI] [PubMed] [Google Scholar]
- Lidholt K, Kjellen L, Lindahl U. 1989. Biosynthesis of heparin. Relationship between the polymerization and sulphation processes. Biochem J. 261:999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidholt K, Lindahl U. 1992. Biosynthesis of heparin. The D-glucuronosyl- and N-acetyl-D-glucosaminyltransferase reactions and their relation to polymer modification. Biochem J. 287(Pt 1):21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidholt K, Weinke JL, Kiser CS, Lugemwa FN, Bame KJ, Cheifetz S, Massague J, Lindahl U, Esko JD. 1992. A single mutation affects both N-acetylglucosaminyltransferase and glucuronosyltransferase activities in a Chinese hamster ovary cell mutant defective in heparan sulfate biosynthesis. Proc Natl Acad Sci U S A. 89:2267–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl U, Li JP. 2009. Interactions between heparan sulfate and proteins-design and functional implications. Int Rev Cell Mol Biol. 276:105–159 [DOI] [PubMed] [Google Scholar]
- Lortat-Jacob H, Turnbull JE, Grimaud JA. 1995. Molecular organization of the interferon gamma-binding domain in heparan sulphate. Biochem J. 310(Pt 2):497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly M, Leach FE, Laremore TN, Toida T, Amster IJ, Linhardt RJ. 2011. The proteoglycan bikunin has a defined sequence. Nat Chem Biol. 7:827–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarana M, Sakura Y, Tawada A, Yoshida K, Lindahl U. 1996. Domain structure of heparan sulfates from bovine organs. J Biol Chem. 271:17804–17810 [DOI] [PubMed] [Google Scholar]
- McCormick C, Duncan G, Goutsos KT, Tufaro F. 2000. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci U S A. 97:668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry CL, Wilson VA. 2002. Role of heparan sulfate-2-O-sulfotransferase in the mouse. Biochim Biophys Acta. 1573:319–327 [DOI] [PubMed] [Google Scholar]
- Morii E, Ogihara H, Oboki K, Sawa C, Sakuma T, Nomura S, Esko JD, Handa H, Kitamura Y. 2001. Inhibitory effect of the mi transcription factor encoded by the mutant mi allele on GA binding protein-mediated transcript expression in mouse mast cells. Blood. 97:3032–3039 [DOI] [PubMed] [Google Scholar]
- Multhaupt HA, Couchman JR. 2012. Heparan sulfate biosynthesis: methods for investigation of the heparanosome. J Histochem Cytochem. 60:908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai N, Habuchi H, Esko JD, Kimata K. 2004. Stem domains of heparan sulfate 6-O-sulfotransferase are required for Golgi localization, oligomer formation and enzyme activity. J Cell Sci. 117:3331–3341 [DOI] [PubMed] [Google Scholar]
- Petitou M, Casu B, Lindahl U. 2003. 1976-1983, a critical period in the history of heparin: the discovery of the antithrombin binding site. Biochimie. 85:83–89 [DOI] [PubMed] [Google Scholar]
- Pinhal MA, Smith B, Olson S, Aikawa J, Kimata K, Esko JD. 2001. Enzyme interactions in heparan sulfate biosynthesis: uronosyl 5-epimerase and 2-O-sulfotransferase interact in vivo. Proc Natl Acad Sci U S A. 98:12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presto J, Thuveson M, Carlsson P, Busse M, Wilen M, Eriksson I, Kusche-Gullberg M, Kjellen L. 2008. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc Natl Acad Sci U S A. 105:4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XX, Carbe C, Tao CQ, Powers A, Lawrence R, van Kuppevelt TH, Cardoso WV, Grobe K, Esko JD, Zhang X. 2011. Lacrimal gland development and Fgf10-Fgfr2b signaling are controlled by 2-O- and 6-O-sulfated heparan sulfate. J Biol Chem. 286:14435–14444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringvall M, Kjellen L. 2010. Mice deficient in heparan sulfate N-deacetylase/N-sulfotransferase 1. Prog Mol Biol Transl Sci. 93:35–58 [DOI] [PubMed] [Google Scholar]
- Robinson CJ, Mulloy B, Gallagher JT, Stringer SE. 2006. VEGF165-binding sites within heparan sulfate encompass two highly sulfated domains and can be liberated by K5 lyase. J Biol Chem. 281:1731–1740 [DOI] [PubMed] [Google Scholar]
- Rudd TR, Yates EA. 2012. A highly efficient tree structure for the biosynthesis of heparan sulfate accounts for the commonly observed disaccharides and suggests a mechanism for domain synthesis. Mol Biosyst. 8:1499–1506 [DOI] [PubMed] [Google Scholar]
- Sasisekharan R, Venkataraman G. 2000. Heparin and heparan sulfate: biosynthesis, structure and function. Curr Opin Chem Biol. 4:626–631 [DOI] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. 2000. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 6:743–750 [DOI] [PubMed] [Google Scholar]
- Schwartz NB. 1975. Biosynthesis of chondroitin sulfate: immunoprecipitation of interacting xylosyltransferase and galactosyltransferase. FEBS Lett. 49:342–345 [DOI] [PubMed] [Google Scholar]
- Senay C, Lind T, Muguruma K, Tone Y, Kitagawa H, Sugahara K, Lidholt K, Lindahl U, Kusche-Gullberg M. 2000. The EXT1/EXT2 tumor suppressors: catalytic activities and role in heparan sulfate biosynthesis. EMBO Rep. 1:282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Liu R, Xu Y, Liu J. 2011. The dominating role of N-deacetylase/N-sulfotransferase 1 in forming domain structures in heparan sulfate. J Biol Chem. 286:19768–19776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu C, Fuda H, Lee YC, Strott CA. 2002. Transcriptional regulation of human 3′-phosphoadenosine 5′-phosphosulphate synthase 2. Biochem J. 363:263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 99:13–22 [DOI] [PubMed] [Google Scholar]
- Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. 2010. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res. 107:1336–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeds E, Habuchi H, Do AT, Hjertson E, Grundberg H, Kimata K, Lindahl U, Kusche-Gullberg M. 2003. Substrate specificities of mouse heparan sulphate glucosaminyl 6-O-sulphotransferases. Biochem J. 372:371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillmann D, Witt D, Lindahl U. 1998. Defining the interleukin-8-binding domain of heparan sulfate. J Biol Chem. 273:15487–15493 [DOI] [PubMed] [Google Scholar]
- Stelzer C, Brimmer A, Hermanns P, Zabel B, Dietz UH. 2007. Expression profile of Papss2 (3′-phosphoadenosine 5′-phosphosulfate synthase 2) during cartilage formation and skeletal development in the mouse embryo. Dev Dyn. 236:1313–1318 [DOI] [PubMed] [Google Scholar]
- Stickens D, Zak BM, Rougier N, Esko JD, Werb Z. 2005. Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development. 132:5055–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer SE, Gallagher JT. 1997. Specific binding of the chemokine platelet factor 4 to heparan sulfate. J Biol Chem. 272:20508–20514 [DOI] [PubMed] [Google Scholar]
- Teplyuk NM, Haupt LM, Ling L, Dombrowski C, Mun FK, Nathan SS, Lian JB, Stein JL, Stein GS, Cool SM, et al. 2009. The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts. J Cell Biochem. 107:144–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y, Pedersen LC, Yamamoto T, Izumikawa T, Kitagawa H, Nishihara J, Tamura J, Negishi M, Sugahara K. 2008. 2-O-phosphorylation of xylose and 6-O-sulfation of galactose in the protein linkage region of glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis. J Biol Chem. 283:16801–16807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traister A, Shi W, Filmus J. 2008. Mammalian Notum induces the release of glypicans and other GPI-anchored proteins from the cell surface. Biochem J. 410:503–511 [DOI] [PubMed] [Google Scholar]
- Turnbull JE, Hopwood JJ, Gallagher JT. 1999. A strategy for rapid sequencing of heparan sulfate and heparin saccharides. Proc Natl Acad Sci U S A. 96:2698–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Yamada S, Zako M, Bernfield M, Sugahara K. 2001. Structural characterization of heparan sulfate and chondroitin sulfate of syndecan-1 purified from normal murine mammary gland epithelial cells: common phosphorylation of xylose and differential sulfation of galactose in the protein linkage region tetrasaccharide sequence. J Biol Chem. 276:29134–29140 [DOI] [PubMed] [Google Scholar]
- Uhlin-Hansen L, Yanagishita M. 1993. Differential effect of brefeldin A on the biosynthesis of heparan sulfate and chondroitin/dermatan sulfate proteoglycans in rat ovarian granulosa cells in culture. J Biol Chem. 268:17370–17376 [PubMed] [Google Scholar]
- van Kuppevelt TH, Dennissen MA, van Venrooij WJ, Hoet RM, Veerkamp JH. 1998. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J Biol Chem. 273:12960–12966 [DOI] [PubMed] [Google Scholar]
- Vanpouille C, Deligny A, Delehedde M, Denys A, Melchior A, Lienard X, Lyon M, Mazurier J, Fernig DG, Allain F. 2007. The heparin/heparan sulfate sequence that interacts with cyclophilin B contains a 3-O-sulfated N-unsubstituted glucosamine residue. J Biol Chem. 282:24416–24429 [DOI] [PubMed] [Google Scholar]
- Velasco A, Hidalgo J, Perez-Vilar J, Garcia-Herdugo G, Navas P. 1988. Detection of glycosaminoglycans in the Golgi complex of chondrocytes. Eur J Cell Biol. 47:241–250 [PubMed] [Google Scholar]
- Venkataraman G, Shriver Z, Raman R, Sasisekharan R. 1999. Sequencing complex polysaccharides. Science. 286:537–542 [DOI] [PubMed] [Google Scholar]
- Wei G, Bai X, Gabb MM, Bame KJ, Koshy TI, Spear PG, Esko JD. 2000. Location of the glucuronosyltransferase domain in the heparan sulfate copolymerase EXT1 by analysis of Chinese hamster ovary cell mutants. J Biol Chem. 275:27733–27740 [DOI] [PubMed] [Google Scholar]
- Xian X, Gopal S, Couchman JR. 2010. Syndecans as receptors and organizers of the extracellular matrix. Cell Tissue Res. 339:31–46 [DOI] [PubMed] [Google Scholar]
- Yang B, Solakyildirim K, Chang Y, Linhardt RJ. 2011. Hyphenated techniques for the analysis of heparin and heparan sulfate. Anal Bioanal Chem. 399:541–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, David G, Esko JD. 1995. Repetitive Ser-Gly sequences enhance heparan sulfate assembly in proteoglycans. J Biol Chem. 270:27127–27135 [DOI] [PubMed] [Google Scholar]