Summary
Human papillomaviruses (HPV) cause diseases ranging from benign warts to invasive tumours. A subset of these viruses termed “high risk” infects the cervix where persistent infection can lead to cervical cancer. Although many HPV genomes have been sequenced, knowledge of virus gene expression and its regulation is still incomplete. This is due in part to lack, until recently, of suitable systems for virus propagation in the laboratory. HPV gene expression is polycistronic initiating from multiple promoters. Gene regulation occurs at transcriptional, but particularly post-transcriptional levels, including RNA processing, nuclear export, mRNA stability and translation. A close association between the virus replication cycle and epithelial differentiation adds a further layer of complexity. Understanding HPV mRNA expression and its regulation in the different diseases associated with infection may lead to development of novel diagnostic approaches and will reveal key viral and cellular targets for development of novel antiviral therapies.
Keywords: Human papillomavirus, gene expression, transcription, RNA processing, translation, diagnostics, antiviral therapy
Introduction
Human papillomaviruses (HPV) are the causative agents of a range of diseases of the cutaneous and mucosal epithelia. These diseases range from benign warts, for example common warts on the hands, to invasive cancers, for example cervical cancer (Table 1) [1]. There are over 120 HPV types identified to date characterised by full genome sequencing. HPV can be classified into two main phylogenetic genera, the α-HPV and the β-HPV, although [2]. These correspond broadly to the mucosally infective and cutaneous infective HPV respectively. The former genus include HPV 16 and 18 that cause 70% of cases of cervical cancer and are transmitted directly through sexual contact. Mucosal HPV can be further categorised into “low” and “high” risk types depending on whether they cause benign lesions or cause lesions that may progress to malignant tumours. The cutaneous types of the β-genus, for example HPV5 associated with non-melanoma skin cancer in immunosuppressed and immunocompromised individuals, can be transmitted directly through skin contact. Around 40 HPV infect the anogenital region. There are a number of types that cause genital warts but HPV6 and 11 predominate. Genital warts are a common condition that has a major health impact with significant social and economic implications. However, genital warts rarely cause lesions that progress to cancer. In contrast, the more abundant “high risk” (HR) anogenital infective types cause intraepithelial neoplasia, or flat warts, of various anogenital sites, which left untreated can lead to invasive cancers, including cervical, vulvar, anal and penile cancers.
Table 1.
List of diseases caused by human papillomavirus infection.
| Site | Disease |
|---|---|
| Skin | Deep plantar warts |
| Common warts | |
| Butchers’ warts | |
| Periungal warts | |
| Carcinoma and keratosis in epidermodysplasia verruciformis | |
| Anogenital tract | Anogenital warts |
| Cervical, vulvar, perianal intraepithelial neoplasia | |
| Carcinoma of the cervix, vulva, anus, penis | |
| Respiratory tract | Recurrent respiratory papillomatosis |
| Squamous cell carcinoma of the larynx, tonsils, tongue base, sinuses, lung | |
| Other | Conjunctival papillomatosis |
| Oesophageal cancer | |
| Various carcinomas associated with immunodeficiencies and HIV-related. | |
| Breast cancers? |
The most prevalent HR-HPV in the world today is HPV type 16 (HPV16). HPV16 causes around 55% of cases of cases of cervical cancer. The remaining cervical cancer cases are caused by other related high risk types such as HPV18, 31 and 45. Cervical cancer is the second most common cancer in women worldwide. Cervical screening (Papanicolau (Pap) smear test) has led to around a 50% reduction in cases of cervical cancer in the developed world. However, HPV infection rates are increasing and cervical disease remains a major health issue especially in the third world with around 500,000 new cases diagnosed and up to 290,00 deaths per annum worldwide. Similar to the situation in the rest of the developed world, a vaccine against HPV types 16 and 18 was rolled out in 2008 [3] in the UK, where uptake is proving excellent at over 90%. However, it will be several decades before efficacy of the vaccine can be determined fully. Moreover, the vaccine does not protect against the 30% of cervical cancers caused by other HR HPV types so cases of disease will still arise and require treatment. It is of paramount importance to continue research efforts to understand the basic biology of these viruses so that novel antiviral therapies may be developed.
The HPV genome and proteins
All HPV are non-enveloped double stranded DNA viruses. Their genomes are circular and approximately 8 kilobase pairs in size. Most encode eight major proteins, 6 located in the “early” region 2 in the “late” region (Table 2). The “early” proteins are regulatory in function. For example, they play roles in HPV genome replication and transcription, cell cycle, cell signalling and apoptosis control, immune modulation and structural modification of the infected cell. Most of these proteins are expressed throughout the infectious cycle perhaps with reduced expression at late times. E4 is the first virus protein expressed late in infection [4] followed by the late proteins, L1 and L2, which are specifically expressed in the granular layer of the epithelium. For the mucosal epithelium, this is the outermost epithelial layer. L1 and L2 comprise the virus capsid required for virus transmission, spread and survival in the environment [5]. To date functional studies of virus proteins have focussed on these eight proteins primarily in the HR HPV (Table 2). However, there is evidence for other virus proteins that may have important functions. For example, HPV16 and 18 express more than one isoform of the E6 cell regulatory oncoprotein. Four mRNA isoforms (FLE6, E6*I, E6*II, E6*X) have been observed in HPV16 infected cervical epithelial cells [6] and two in HPV18 infection [7]. A role for the E6*I isoform in antagonising FLE6 function has been suggested [7], as has opposing roles of FLE6 and E6*I in regulation of procaspase 8 in the extrinsic apoptotic pathway [8]. More recently, a stand-alone function of the E6*I isoform has been determined in cellular protein degradation [9]. Several isoforms of the virus replication/transcription factor E2 have also been noted for a number of HPV. E2 has an N-terminal domain that mediates protein-protein interactions, a flexible hinge region and a C-terminal DNA binding domain. Truncated E2 proteins may be translated from alternatively spliced E2 RNAs to generate E1^E2 and E8^E2 protein isoforms present in HPV16 and 31-infected cells [10-13]. These E2 isoforms may act in a dominant-negative manner to modulate the function of full length E2 [10,12,13]. For example, a full length E2/E8^E2 dimer may bind DNA but fail to recruit E1 to initiate virus replication. Similarly, such a dimer may be unable to interact with cellular transcription factors to alter virus genome transcription.
Table 2.
Major roles of proteins expressed by high risk HPV.
| Protein | Role in the virus life cycle |
|---|---|
| E1 | Genome replication: ATP-dependent DNA helicase. |
| E2 | Genome replication, transcription, segregation, encapsidation. Regulation of cellular gene expression. Cell cycle and apoptosis regulation. |
| E4 | Remodels cytokeratin network. Cell cycle arrest. Virion assembly. |
| E5 | Control of cell growth and differentiation. Immune modulation |
| E6 | Oncoprotein. Inhibits apoptosis and differentiation. Regulates cell shape, polarity, mobility and signalling. |
| E7 | Cell cycle control. Controls centrosome duplication. |
| L1 | Major capsid protein |
| L2 | Minor capsid protein. Recruits L1. Virus assembly. |
HPV replication and epithelial differentiation
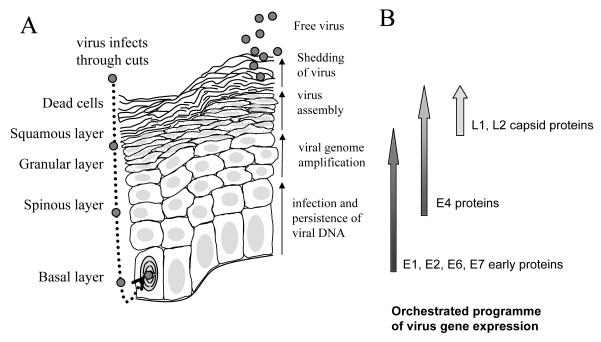
The replication cycle of HPV is tightly linked to differentiation of the epithelium it infects. HPV may access dividing basal epithelial cells by falling down microabrasions in the epithelium and attaching to cells using common cell surface molecules such as heparin sulphate proteoglycans [14,15]. α6-integrin may be used as the virus receptor [16,17], at least in some cases, and virus entry is through either calveolar or clathrin-mediated endocytosis [14,18]. Once inside the cell, uncoating occurs and the circular virus genome is transported to the nucleus. Upon division of the infected cell the nuclear episomal genome is replicated to 20-100 copies and maintained in daughter cells upon segregation through attachment of virus genomes to host chromosomes via the virus replication/ transcription factor E2 following the pattern established for BPV-1 [19]. One idea is that during the first phase of infection of basal layer cells only the virus replication proteins E1 and E2 are expressed in order to establish and maintain the low copy number genomes [20]. However, further work needs to be carried out to establish this as fact. Although the E6 and E7 oncoproteins are also expressed soon after infection, the exact temporal lineage of early events in the virus life cycle is difficult to evaluate. This is because it is still technically very difficult to infect epithelial cells and follow the course of infection with HPV. Infection may persist in the infected stem cell, and its daughter cells, in the basal layer of the epithelium over a long period of time, up to several years. Persistence is considered the major risk factor underlying cervical tumour progression [21]. However, a normal infection requires that division of a basal stem cell sometimes yields a daughter transit amplifying cell with the ability to differentiate to upper epithelia layers. Normally, upon differentiation epithelial cells stop dividing and commence terminal differentiation. Importantly, in HPV-infected epithelial cells the concerted action of the E6 and E7 oncoproteins and their partner proteins p53 and pRb respectively, reactivates cell division, inhibits apoptosis and abrogates epithelial differentiation [22]. Unscheduled division of infected cells in the differentiating epithelial layers gives rise to the physical manifestation of infection as a wart. Reactivated cell division allows further amplification of virus genomes to yield many thousands of copies. Finally, upon differentiation to the granular epithelial layer, the virus capsid proteins L1 and L2 are expressed and encapsidate the newly synthesised virus genomes [23]. For release, new virions may rely on mechanical breakage of infected cells sloughed off from the upper later of the epithelium. However evidence exists that the E4 protein has an active role in priming the infected differentiated cell by disrupting the keratin filamentous network [24] and inhibiting formation of the cornified envelope [25,26]. Figure 1 summarises the above description that is relevant for the anogenital infective HPV. Although the same general scheme may apply to HPV of the β-genus, some important differences do exist. For example, cutaneous HPV E6 and E7 proteins do not appear to operate by the same mechanisms as anogenital HPV in regulating the cell cycle, differentiation and apoptosis of the infected cell. The temporal organisation of the virus replication cycle is also different between different HPV types perhaps reflecting differences in sites of infection and in different modes of transmission [27].
Figure 1. The replication cycle of high risk HPV in a differentiating epithelium.
A. The different epithelial layers are indicated on the left hand side of the diagram. Virus is show as small grey circles. The nucleus of the infected, dividing basal epithelial cell is indicated with curved lines representing condensed chromosomes. All nuclei are shaded in light grey. The granular layer is shown with dotted cytoplasm. The key events in the virus replication cycle are indicated to the right hand side of the diagram of the epithelium. B. A schematic diagram of the gene expression program of the virus within the infected epithelium. Shading on the arrows represents the quantity of expression of each protein subset during the virus replication cycle.
HPV gene expression
Transcription
Transcription of the circular virus genome occurs from only one DNA strand, is initiated from more than one promoter region and is polycistronic, yielding multiple mRNAs with several open reading frames (Figure 2). A major problem with RNAs that potentially encode several proteins is that only the first open reading frame in each mRNA will be translated efficiently [28]. Expression of multiple mRNAs from the virus genome may be necessary to ensure that each open reading frame is first in at least one mRNA.
Figure 2. The gene expression profile of HPV16 in W12 cervical epithelial cells.
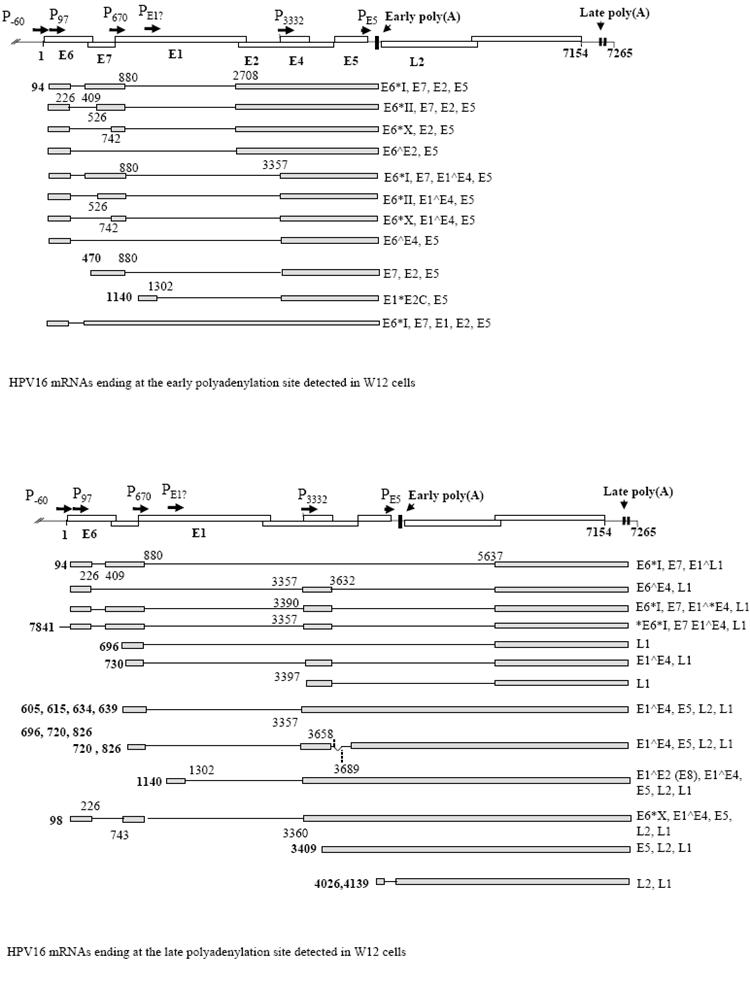
A. At the top is shown a diagram of the HPV16 genome linearised at the first nucleotide. Open boxes, open reading frames with proteins encoded indicated with bold text. Promoters are indicated with arrows and “P”. The early and the two late polyadenylation sites are indicated with arrowheads and vertical bars on the genome map. At the bottom is a diagram of the W12 early mRNAs discovered to date [48], (Milligan and Graham, unpublished data). Grey boxes, exons; black lines, introns. Numbers in bold indicate transcription start sites. The remaining numbers locate splice donor and acceptor sites. The coding potential of each mRNA is indicated to the right hand side. B. A diagram of the late mRNAs detected in W12 cells [45] with similar annotation as in part (A).
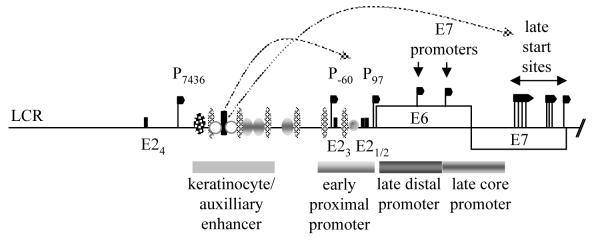
For the α-HPV there are two virus promoters that have been well characterised, the early promoter (P97 for HPV16 and 31, P105 for HPV18) in the virus long control region (LCR) that appears to be constitutively active throughout the virus replication cycle [29-32] and a late differentiation-regulated promoter within the E7 open reading frame (P670 for HPV16, P742 for HPV31) [33]. Early promoters have also been characterised for HPV6 and 11. For HPV6 there are three, P90, P270, and P680, with HPV11 initiation sites similarly located [34-39]. The P97 promoter of HPV16 is perhaps the best understood (Figure 3). It is controlled by the virus E2 protein but also by an array of cellular transcription factors linking virus gene expression into regulation by the infected cell [40]. There are four binding sites for E2 in the promoter; two in the proximal promoter adjacent to the TATA box, one in the distal promoter region and one upstream of the keratinocyte enhancer. The keratinocyte enhancer acts synergistically with an auxiliary enhancer to specify tissue tropism [41]. In a normal infection, the P97 promoter is transcriptionally repressed by E2 in order that expression levels of the E6 and E7 oncoproteins remain low and normally do not activate tumour progression. Repression seems to involve a combination of E2-mediated steric hindrance of TFIID abrogating TATA binding protein location its binding site, the TATA box, and binding of cellular transcriptional repressor proteins, such as Sp1, to the virus LCR. It is clear that the LCR binds many cellular transcription activators. Although all their roles are not clear at present, it is expected that their binding pattern must confer keratinocyte specificity to transcription. Dynamic changes in the profile of the proteins bound and strength of binding occur upon epithelial differentiation [42] and likely mediate some transcriptional changes necessary for completion of the virus life cycle. It is possible that E2 interaction with cellular proteins that bind the LCR might lead to a more repressive promoter conformation or could finely tune repression in response to epithelial differentiation. Truncated E2 isoforms that might act as dominant negative transcription factors leading to repression of P97 have also been characterised for HPV16 and 18 [10,12,13] although the mechanism by which these operate is unclear because the transactivation domain of E2 necessary for transcriptional repression is missing from the isoforms [43,44].
Figure 3. Regulation of promoters at the start of the HPV16 genome.
A schematic diagram of the long control region and the start of the early coding region of the HR HPV genome showing the position of various transcription start sites (black flags), the early proximal promoter region, the keratinocyte/auxiliary enhancer region and the late core and distal promoter regions in the E6/E7 coding region. Data are taken from results with HR HPV16 and 31. LCR, long control region. E2, E2 binding sites indicated by black bars. Ovals and circles indicate approximate regions of transcription factor binding. Only a limited number of binding sites are illustrated. There are very many known binding sites making it impossible to illustrate all transcription factors and their precise binding sites. In addition, most of the transcription factors regulating the E7 promoters and the late promoter have not yet been identified. The box indicates the transcription factors known to bind the HPV16 early region [40].
The dominant promoter used for transcription initiation late in the virus replication cycle, termed the “late” promoter, is located within the E7 open reading frame and is dependent upon keratinocyte differentiation. For HPV16 and 31, many late promoter-regulated transcription initiation sites have been mapped over a 200 nucleotide region [33,45,46]. For example, HPV31 has over 30 initiation sites stretching from genome position 605 to 779 [46]. This complexity in transcription initiation, plus a clear contribution from cis-acting control elements in the LCR, has led to difficulty in mapping cis-acting control elements for the late promoter. The HPV31 P742 core promoter has been mapped to a 150 nucleotide region upstream of the region of most late transcription initiation sites. Furthermore, two differentiation responsive elements, DRE1 and DRE2 have been delineated in the E6/E7 gene region [47]. However, the mode of action of these is not necessarily at the level of control of transcription initiation. One or both elements may regulate stability of certain late mRNAs if they reside in the 5′ untranslated region. It is clear that chromatin remodelling regulates the late promoter in a differentiation stage-specific manner as DNase I hypersensitive sites are uncovered upon differentiation of virus genome-containing keratinocytes [46]. Interestingly, the keratinocyte enhancer in the long control region contributes to regulation of the late promoter. Further studies are required to examine the relative contributions of the different regulatory regions to differentiation stage-specific transcription of the virus genome.
Apart from the major early and late promoters, the existence of other promoters has been reported in various HPV genomes. It seems likely that each HPV has a similar array of promoters in the early region of the genome to that found for BPV-1 [48]. Several novel transcription start sites upstream of the P97 promoter have been observed in a number of HPV [49,50]. At least one located in the LCR is active in expression of mRNAs late in infection [51]. A promoter at the start of the E1 open reading frame may also exist at least for HPV16, but this has not been proven definitively [45]. Several studies have verified the existence of a late promoter at the start of the E4 open reading frame [45,51,52]. This promoter is particularly interesting as it may express the mRNA from which L1 capsid protein is the first open reading frame, thus ensuring efficient translation. Finally, a promoter in the E5 open reading frame has been demonstrated for HPV6 and 16 [37,45] that can transcribe mRNAs encoding L2 as a first open reading frame. Regulation of these transcription starts has not yet been elucidated and, like the “late” promoter, may prove complex involving multiple genome regions.
Post-transcriptional events
Although HPV gene expression is regulated at the level of transcription initiation and in response to epithelial differentiation signals, it is clear that post-transcriptional regulation is also very important. The HPV genome is compact. Polycistronic transcription, overlapping open reading frames, regulated read-through of the early polyadenylation site and alternative splicing ensure maximum coding potential but mean that regulation of RNA processing in the nucleus and translation in the cytoplasm must be key to regulation of virus gene expression. For example, an early observation that L1-encoding mRNAs could be detected in lower epithelial layers where the capsid proteins are not expressed indicated post-transcriptional regulation of virus late gene expression [36,53-55]. There are multiple possibilities for virus post-transcriptional regulation. Those studied to date are discussed below.
Polyadenylation
Upon transcription, 3′ end formation of virus RNAs may occur either at the early or late polyadenylation sites (Figure 2). The early polyadenylation site, as studied for HPV16 and 31, is “weak” meaning that it should be utilised poorly. A consensus AAUAAA polyadenylation signal that binds cleavage and polyadenylation specificity factor (CPSF) is present between the E5 and L2 open reading frames. Co-binding of CPSF with cleavage stimulatory factor (CstF) regulates efficiency of 3′ end formation. Three different G/U-rich regions located downstream of the early cleavage site were found to bind CstF only weakly, and so this could be responsible for inefficient use of HPV early poly(A) sites. The presence of degenerate CPSF binding sites and more than one G/U-rich CstF binding site may also explain the observed heterogeneity in early 3′ end formation [56]. Clearly, the early polyadenylation site is active in formation of viral early mRNAs (Figure 2A, Early poly(A)). However, its inherent inefficiency may be important late in the virus replication cycle to allow production of late mRNAs that are transcribed through the early polyadenylation region and which terminate within the 5′ portion of the LCR downstream of the L1 open reading frame (Figure 2B, Late poly(A)) [56] [57]. Sequences in the HPV16 E4 open reading frame, an upstream polyadenylation element in the early 3′ untranslated region and a polyadenylation element in the first 174 nucleotides of the L2 open reading frame may act to enhance efficiency of early polyadenylation in an epithelial differentiation-specific manner at least in the context of reporter gene assays [58-60]. The L2 element possesses G-rich motifs that bind CstF 64, a component of CstF polyadenylation complex and hnRNP H that may stimulate polyadenylation complex assembly [59]. Elements that repress activity of the HPV31 early polyadenylation site during the virus replication cycle have been mapped within the first 800 nucleotides of the L2 open reading frame but their mechanism of action is unknown [57]. For HPV16 there are two late polyadenylation sites located in the late 3′ untranslated region just downstream of the L1 open reading frame (Figure 2B). Each possesses good consensus polyadenylation signals and HPV16 mRNAs terminating at each site have been detected in infected cervical epithelial cells [45].
Splicing
Although HPV do express some unspliced mRNAs (Figure 2A), most HPV mRNAs are the products of constitutive and alternative splicing. Constitutive splicing is where every intron in the primary RNA transcript is removed as transcription progresses along the gene. Alternative splicing is where exon or intron skipping can occur. For HPV, several main splice events can occur (Figure 2). The first is generated by splicing from a splice donor site in the E6 open reading frame and the second from one at the end of the E4 open reading frame each onto a range of splice acceptor sites located in the E6, E7, E4 and L1 regions of the genome. (Figure 2A,B). Both splice events occur in some mRNAs (Figure 2B).
Regulation of splicing is complex and occurs through efficient recognition of cis-acting splicing signals and RNA/protein complexes known as spliceosomes that form across the exon-intron junction [61,62]. Splicing efficiency is regulated by other cis-acting signal sequences within exons, termed exonic sequence enhancers (ESE) or silencers (ESS) or within introns termed intronic sequence enhancers (ISE) or silencers (ISS) [61]. The enhancer elements bind members of the serine-arginine (SR)-rich protein family of which there are 9 classical members and a number of other related factors [63]. Conversely, silencers bind members of the hnRNP protein family [64]. SR and hnRNP proteins antagonise the action of each other [65] and complex combinations of these factors may often be required to regulate splicing [66].
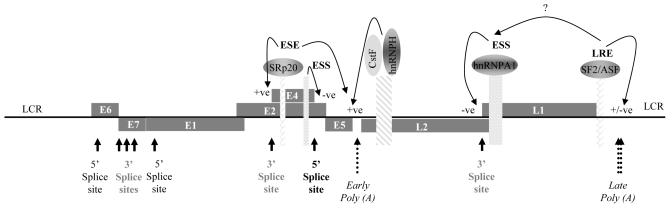
No studies to date have examined HPV splicing regulation in the infected epithelial cell. However, some splicing regulatory sequences on the HPV genome have been defined through analysis of splicing reporter expression plasmids. In the case of HPV16, an ESE required for efficient use of the splice acceptor site 3358 at the start of the E4 open reading frame and an ESS regulating efficiency of the splice donor site at the 3′ end of the E4 open reading frame have been demonstrated [58](Figure 4). This ESE binds the smallest SR protein, SRp20. By analogy with the situation for BPV1, SRp20 may control selection of the splice acceptor site required for synthesis of fully processed virus L1 capsid mRNAs. This ESE also controls early polyadenylation site efficiency. This dual role is unsurprising as exonic regulatory sequences are frequent players in defining exons by cross talk with downstream polyadenylation complexes [67]. Proteins that bind, and mechanism of action of the E4 ESS, are unidentified as yet. ESS are found within the L1 open reading frame and bind hnRNP A1 to inhibit expression of L1 capsid mRNA (Figure 4). The silencing effect may be abrogated by an ESE in the L1 region [68] demonstrating the potential complexity of splicing control on RNA expressed from an intact virus genome (Figure 4). Future studies must focus on global control of HPV splicing regulation and charting which mRNAs are expressed in conditions of overexpression and knock down of individual splicing regulatory proteins.
Figure 4. HPV16 RNA processing regulation; ESE, ESS and polyadenylation control elements and their binding proteins.
A schematic diagram of the HPV16 genome showing open reading frames (grey rectangles containing gene names), splice donor (5′) and splice acceptor (3′) splice sites (black arrows), polyadenylation sites (dotted arrows). When transcribe to RNA, cis-acting RNA processing regulatory elements (patterned oblongs) and the proteins these bind (annotated ovals) are indicated. Arrowed lines indicate the target and type of regulation of each RNA/protein interaction. LCR, long control region.
Importantly, HPV regulate expression of a subset of SR proteins that control viral RNA splicing of viral. Levels of the three smallest SR proteins, SRp20, SC35 and SF2/ASF are specifically increased in the mid to upper layers of HR HPV-infected cervical epithelia [69]. The HPV E2 transcription factor controls expression of these SR proteins during infection. For SF2/ASF at least, the mechanism is by trans-activation of the promoter of its gene [70]. E2-mediated SF2/ASF upregulation during the late phase of the virus replication cycle is necessary for expression of the virus capsid proteins [71] (Graham, unpublished observations) and therefore necessary for completion of the virus replication cycle.
RNA stability
HPV gene regulation is also controlled at other post-transcriptional levels. In particular, expression of the virus capsid proteins L1 and L2 is regulated largely through changes in stability of their mRNAs. A regulatory element located at the end of the L1 open reading frame and extending into the late 3′ untranslated region has been delineated for HPV1, 16 and 31 [72-82]. These elements preclude expression of the virus capsid proteins in cells in the lower epithelial layers by rendering any mRNAs transcribe there unstable [83]. This is important because the capsid proteins are highly immunogenic and premature expression in the lower epithelial layers would result in generation of a robust immune response and clearance of the virus. The HPV1 and 16 late regulatory elements are the best understood and have been shown to bind a number of cellular proteins including HuR, a key RNA stability regulator [84]. For HPV16, HuR overexpression in undifferentiated HPV-infected epithelial cells results in an increase in L1 capsid protein expression. Conversely, siRNA depletion of HuR in differentiated HPV-infected cells results in decreased L1 capsid protein expression [80] demonstrating the importance of this cellular factor in facilitating HPV late gene expression.
Translation
Few studies have addressed any contribution of virus mRNA translation regulation for efficient virus replication. One notable exception is the identification of cis-acting RNA elements located in the HPV16 L2 open reading frame that bind hnRNP K and poly(rC) binding proteins 1 and 2 to inhibit translation of L2 mRNA in vitro [85,86]. A translation regulatory element has also been characterised in the late 3′ untranslated region of HPV1 [87]. This is the same element that regulates stability of HPV1 late mRNAs. A dual function for this element is expected as mRNA stability in the cytoplasm and translation are linked processes [88], so it is possible that other HPV late regulatory elements control translation as well as mRNA stability.
Another aspect of translational control involves efficiency of translation related to codon usage. HPV mRNAs use rare codons, presumably as an adaptation to efficient translation in the face of ongoing host cell translation [89,90]. Studies have demonstrated a keratinocyte differentiation-dependent alteration of papillomavirus capsid protein translation [91] using transient transfection. However, studies using mutated virus genomes stably transfected into keratinocytes are really required to assess any effect of codon usage on HPV gene expression during infection.
CONCLUSIONS
Temporal expression of HPV in response to epithelial differentiation
The pattern of HPV gene expression in infected cervical epithelia is that early gene expression begins in the lower epithelial layers. Next E4 is the first protein expressed at the beginning of the late phase of the life cycle in the suprabasal epithelial layers with the L1 and L2 capsid proteins being expressed last of all in the upper granular layer. This is a carefully orchestrated programme of expression that ensures smooth progression of the key events in the virus replication cycle together with immune evasion [92]. HPV gene expression relies on a complex interplay of differential promoter use, selection of the appropriate polyadenylation site, generation of alternative splice isoforms of polycistronic RNAs transcribed from the genome, regulated stability or decay and translation efficiency of the mRNAs generated. All these processes are controlled by cellular DNA or RNA-binding factors, some of which are known to be expressed in a differentiation stage-specific manner [79,80,93,94]. One key challenge for future HPV research will be to elucidate the host cell factors that are required for virus gene expression and its control. Such factors could prove to be good biomarkers of disease and targets for novel antiviral therapies. There have been no in depth studies of HPV-induced changes in cellular gene expression upon epithelial differentiation. Such analyses may be important in assessing what cellular factors may be required to allow completion of the HPV life cycle. Next generation sequencing approaches and new speedy proteomics should make such studies easy to carry out, perhaps even on an individual patient basis in future. In addition, new high through-put technologies to define protein-protein interactions and the components of large protein complexes have now opened up the possibility of generating a comprehensive database of virus-host cell interactions [95].
FUTURE PERSPECTIVE
Diagnosis
A major issue of high relevance for treatment of HPV-related disease in the clinic is accurate diagnosis of severity of disease [21]. In particular for cervical lesions, current approaches use cytological assessment of Pap smear material which is low sensitivity and subjective in nature, yielding false positive and negative results. Detection of HPV DNA by PCR is exquisitely sensitive and specific and is used in certain settings. For example the Roche linear array HPV PCR detection kit detects 37 different anogenital-infective HPV. However, again results can prove misleading as DNA detection can indicate the present of viral sequences but not whether the virus is actively expressing the proteins that cause disease. Understanding virus gene expression patterns in disease and during disease progression will aid development of novel accurate diagnostic tests. Improved accuracy in diagnosis will inform treatment regimes and save money in terms of patient treatment and may alleviate anxiety in patients diagnosed with HPV infection but whose disease may not progress to tumour. Improvements in diagnosis will arise from identification of hallmarks of active virus infection, present in low grade cervical lesions, and specific biomarkers of high grade disease. Analysis of HPV protein expression is difficult due to extremely low levels of most viral proteins expressed during infection. Moreover, antibodies currently available work inefficiently. mRNA profiling could be used to determine if virus-infected tissue is expressing virus proteins and which proteins are being expressed. mRNA profiling gives a depiction of viral activity and therein a potential handle on the clinical significance of the infection. Currently, quantitative PCR to detect E6 and E7 mRNA expression has recently been developed and is prove to be one of the most promising tests to detect high grade lesions. For example, “Pre-tect HPV proofer” (Norchip) detects the HR-HPV E6 and E7 mRNAs and may specifically detect persistent infections. Completion of HPV transcription profiles at different stages of cervical disease may prove essential in developing novel diagnostic tests [96] that distinguish between transient and persistent infection.
Antiviral therapies
The virus early proteins have diverse regulatory functions during virus replication making them potentially excellent drug targets. The only enzyme expressed by HPV is E1, an ATP-dependent helicase essential for initiation of viral DNA replication. However, currently there are no non-toxic small molecule inhibitors of E1 available. Besides, such inhibitors could also prove inhibitory to DNA helicases involved in cellular DNA replication leading to cell death and toxicity. E1 requires E2 to locate to the origin of replication in the HPV LCR, so E2 is also essential for replication of the HPV genome. The other roles of E2 as the virus transcription and genome segregation factor and an inducer of apoptosis also make it an attractive therapeutic target. So far, an E2 fusion protein with the herpes simplex virus (HSV-1) VP22 protein that is able to penetrate cell membranes has been shown to induce cell death [97]. This type of approach could be developed for topical application. E1 and E2 are expressed only during a productive virus replication cycle so any inhibitors would be useful only in treatment of low grade cervical lesions. Conversely, only the E6 and E7 oncoproteins are expressed in cervical tumour cells so therapies targeting these may be applicable to cervical cancer. In addition, as E6 and E7 are required in a normal infectious life cycle, anti-E6/E7 therapies may also be beneficial in cases of low grade disease. Considerably less is known about the functions of E4 and E5 making them less likely drug targets at present. Virus transcript mapping in HPV-infected tissues is required to reveal if protein isoforms other than those previously detected, e.g. E6 isoforms, may be expressed as these could also present suitable antiviral targets.
There are several options for antiviral therapies in future. Chemically stabilised siRNAs are proving promising therapeutic reagents [98] in a range of diseases. For example, siRNA against the nucleocapsid of respiratory syncitial virus (RSV) is in phase II clinical trials. siRNA directed against the early and late 3′ untranslated regions of HPV mRNAs would be predicted to ablate all virus transcripts and halt infection. In cervical tumours siRNA against E6/E7 mRNAs may suppress tumour growth and reduce cancer progression. Therapeutic nucleic acids also include antisense oligodeoxy-ribonucleotides and ribozymes and have been demonstrated to be effective against HR HPV oncoprotein expression [99]. Finally, intrabodies, therapeutic antibodies capable of blocking protein/protein interaction even when those proteins do not have active sites have been proposed as an alternative route of therapeutic treatment for cervical disease [100].
The virus structural proteins, L1 and L2 may be targets for therapy but only for inhibition of reinfection. This type of approach should not be underestimated as virus shed from an infected cervix often results in reinfection in the vicinity of the original lesion leading to new lesions requiring further treatment. Our improved understanding of HPV late gene expression and its regulation may allow development of novel topically applied antiviral therapies designed to inhibit not only capsid protein expression but also other virus proteins expressed in the upper epithelial layers, for example, E2 [101].
Interestingly, indole derivative compounds (IDCs) have been demonstrated to inhibit the activity of SR proteins during retrovirus infection [102]. IDC-16 has been shown to inhibit SF2/ASF-mediated splicing of RNAs encoding HIV-1 regulatory proteins and thereby interfere with virion assembly while having no detectable effect on cell viability or cellular RNA processing [103]. Moreover, IDC13 and 78 inhibit production of the murine leukaemia virus envelope protein in a splicing-dependent manner resulting in inhibition of virus-induced pathogenesis in vivo [104]. As HPV capsid protein expression has a requirement for SR protein activity, exploration of the applicability of IDCs to inhibit the HPV replication cycle may open up new avenues in HPV anti-viral therapy. Similarly, small molecule inhibitors of HuR, also involved in regulating virus late gene expression, are available [105]. So if these could be applied topically either alone or in combination with other drugs such as IDCs to HPV-infected epithelia they may inhibit capsid protein production and reduce the efficiency of virus transmission and spread.
Development of a topically applied reagent to inhibit HPV replication cycle would be ideal. The upper epithelial layers are normally quite metabolically inactive as their main function is to provide a barrier to the environment. This means that toxicity of any treatment may be less important than if the therapy had to be delivered systemically. Topical application of novel therapies, for example therapeutic RNAs, requires considerable further research as it may not be easy to achieve [106]. Recent developments in gel design and application, for example, for the cervix, use of cervical rings, may prove important [107].
The next decade in HPV research should be geared up to fill in the gaps in knowledge of HPV gene expression and its regulation. This, coupled with the application of next generation sequencing and proteomic technologies, screening of new, more extensive small molecule libraries to look for molecules inhibitory to either the virus replication cycle or to tumour formation and development into the clinic of promising new avenues for drug development, for example, RNA-based therapies should advance the prospects of new anti-HPV treatments to alleviate disease.
Executive summary.
HPV replication cycle
Required a three dimensional epithelium.
Highly organised replication cycle that responds to differentiation signals from the infected host cell.
Virus replication cycle is completed in the upper epithelial layers from where newly synthesised virions are released.
HPV gene expression
The ~8.0 kbp double stranded DNA genome is transcribed in one direction from one DNA strand.
Transcription is polycistronic yielding RNAs that may encode more than one protein and that are alternatively spliced.
At least 6 early (regulatory proteins) and 2 late proteins (structural) are expressed.
Regulation of HPV gene expression
A complex interplay of control mechanisms regulated by virus and host cell proteins leads to correct spatial and temporal expression of the various virus proteins within the infected epithelium.
Transcriptional regulation is important. An array of gene promoters in the HPV early region may be actively engaged in transcription of the genome and controlled in response to epithelial differentiation.
There is evidence for post-transcriptional regulation making a significant contribution to epithelial differentiation stage-specific gene expression. Regulation is at the level of RNA processing, nuclear export, mRNA stability and translation of viral RNAs.
Using information on HPV gene expression and its regulation in design of novel diagnostic tests and anti-viral therapies
Different stages of HPV-induced disease, especially cervical disease, can be characterised by virus mRNA expression profiles. Development of viral mRNA detection as a diagnostic test of virus activity is ongoing.
Understanding virus gene regulation may lead to development of novel anti-viral therapies designed to inhibit viral and cellular proteins essential for completion of the virus replication cycle.
The epithelium is an easily accessible organ and infected epithelia may prove suitable for topical treatment. For example, upper epithelial layers are relatively metabolically inactive so inhibitors of cellular proteins essential for virus replication may not have the same toxic effects as systemic treatments.
Development of novel delivery systems is required, however some already show promise.
Table 3.
HR HPV transcription initiation sites.
| HPV TYPE | GENE REGION | GENOME POSITION OF PROMOTER |
|---|---|---|
| HPV16 | E6 | P97, P441 (multiple initiation sites), P482 (multiple initiation sites), P542 |
| E7 | P670 (multiple initiation sites) | |
| E1 | P1140? | |
| E4 | P3397 | |
| E5 | P4062 | |
| Early noncoding region | P4139 | |
| Long control region | P7841, P22 | |
| HPV31 | E6 | P97 |
| E7 | P742 (multiple initiation sites) | |
| E4 | P3320 | |
| L2 | P5600 | |
| Early noncoding region | P4139 | |
| Long control region | P7375, P7855, P7790, P77, P53 | |
| HPV18 | E6 | P105 |
| E1 | P2598 | |
| E2 | P3036 | |
| Early noncoding region | PURR |
Acknowledgements
The author would like to thank John Doorbar for useful discussions, and Alasdair MacDonald and Melanie McFarlane for critical reading of the manuscript. The author would like to point out that the literature relating to this field of research is vast and would like to apologise to those colleagues whose work could not be included in this review due to word limits.
BIBILIOGRAPHY
- 1.zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virol. 2009;384:260–5. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 2.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virol. 2004;324:17–24. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 3.Stanley MA. Human papillomavirus vaccines. Rev. Med. Virol. 2006;16:139–49. doi: 10.1002/rmv.498. [DOI] [PubMed] [Google Scholar]
- 4.Doorbar J, Foo C, Coleman N, et al. Characterisation of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virol. 1997;238:40–52. doi: 10.1006/viro.1997.8768. [DOI] [PubMed] [Google Scholar]
- 5.Graham SV. Late events in the life cycle of human papillomaviruses. In: Campo MS, editor. Papillomavirus research: from natural history to vaccines and beyond. Caister Academic Press; Wymondham, Norfolk: 2006. pp. 193–212. [Google Scholar]
- 6.Tang S, Tao M, McCoy JP, Zheng Z-M. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 2006;80(9):4249–63. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pim D, Massimi P, Banks L. Alternatively spliced HPV-18 E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene. 1997;15:257–64. doi: 10.1038/sj.onc.1201202. [DOI] [PubMed] [Google Scholar]
- 8.Filippova M, Johnson MM, Bautista M, et al. The large and small isoforms of human papillomavirus type 16 E6 bind to and differentially affect procaspase 8 stability and activity. J. Virol. 2007;81(8):4116–29. doi: 10.1128/JVI.01924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pim D, Tomaic V, Banks L. The human papillomavirus (HVP) E6* proteins from high-risk mucosal HPVs can direct degradation of cellular proteins in the absence of full length E6 protein. J. Virol. 2009;83(19):9863–74. doi: 10.1128/JVI.00539-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stubenrauch F, Hummel M, Iftner T, Laimins LA. The E8^E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 2000;74(3):1178–86. doi: 10.1128/jvi.74.3.1178-1186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alloul N, Sherman L. Transcription-modulatory activities of differentially spliced cDNAs encoding the E2 protein of human papillomavirus type 16. J. Gen. Virol. 2006;80(9):2461–70. doi: 10.1099/0022-1317-80-9-2461. [DOI] [PubMed] [Google Scholar]
- 12.Lace MJ, Anson JR, Thomas GS, Turek LP, Haugen TH. The E8^E2 gene product of human papillomavirus type 16 represses early transcription and replication but is dispensible for viral plasmid persistence in keratinocytes. J. Virol. 2008;82(3):10841–53. doi: 10.1128/JVI.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ammermann I, Bruckner M, Matthes F, Iftner T, Stubenrauch F. Inhibition of transcription and DNA replication by the papillomavirus E8^E2C protein is mediated by interaction with corepressor molecules. J. Virol. 2008;82(11):5127–36. doi: 10.1128/JVI.02647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patterson NA, Smith JL, Ozbun MA. Human papillomavirus type 31b infection of human keratinocytes does not require heparan sulfate. J. Virol. 2005;79(11):6838–47. doi: 10.1128/JVI.79.11.6838-6847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shafti-Keramat S, Handisurya A, Kriehuberm E, Slupetzky K, Kirnbauer R. Different heparan sulfate proteoglycans serve as cellular receptors for human papillomaviruses. J. Virol. 2003;77(24):13125–35. doi: 10.1128/JVI.77.24.13125-13135.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evander M, Frazer IH, Payne E, et al. Identification of the alpha 6 integrin as a candidate receptor for papillomaviruses. J. Virol. 1997;71(3):2449–56. doi: 10.1128/jvi.71.3.2449-2456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMillan NA, Payne E, Frazer IH, Evander M. Expression of the α6 integrin confers papillomavirus binding upon receptor-negative B-cells. Virol. 1999;261(2):271–9. doi: 10.1006/viro.1999.9825. [DOI] [PubMed] [Google Scholar]
- 18.Smith JL, Campos SK, Ozbun MA. Human papillomavirus type 31 uses a caveolin 1- and dynamin 2-mediated entry pathway for infection of human keratinocytes. J. Virol. 2007;81(18):9922–31. doi: 10.1128/JVI.00988-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.You J, Croyle JL, Nishimura A, Ozato K, Howley PM. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell. 2004;117(3):349–60. doi: 10.1016/s0092-8674(04)00402-7. [DOI] [PubMed] [Google Scholar]
- 20.Ozbun MA. Human papillomavirus type 31b infection of human keratinocytes and the onset of early transcription. J. Virol. 2002;76(2):11291–300. doi: 10.1128/JVI.76.22.11291-11300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodman CBJ, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Cancer. 2007;7:11–22. doi: 10.1038/nrc2050. An extremely comprehensive bioclinical review of HPV infection and tumour progression.
- 22.Hamid NA, Brown C, Gaston K. The regulation of cell proliferation by the papillomavirus early proteins. Cell. Mol. Life Sci. 2009;66(10):1700–17. doi: 10.1007/s00018-009-8631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florin L, Sapp C, Streeck RE, Sapp M. Assembly and translocation of papillomavirus capsid proteins. J. Virol. 2002;76(19):10009–14. doi: 10.1128/JVI.76.19.10009-10014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Griffin HM, Southern S, et al. Functional analysis of the human papillomavirus type 16E1^E4 protein provides a mechanism for in vivo and in vitro keratin filament reorganization. J. Virol. 2004;78(2):821–33. doi: 10.1128/JVI.78.2.821-833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryan J, Brown DR. Association of the human papillomavirus type 11 E1^E4 protein with cornified cell envelopes derived from infected genital epithelium. Virol. 2000;277(2):262–9. doi: 10.1006/viro.2000.0599. [DOI] [PubMed] [Google Scholar]
- 26.Lehr E, Jarnik M, Brown DR. Human papillomavirus type 11 alters the transcription and expression of loricrin, the major cell envelope protein. Virol. 2002;298(2):240–7. doi: 10.1006/viro.2002.1445. [DOI] [PubMed] [Google Scholar]
- 27.Middleton K, Peh W, Southern SA, et al. Organisation of the human papillomavirus productive cycle during neoplastic progression provides a basis for the selection of diagnostic markers. J. Virol. 2003;77(19):10186–201. doi: 10.1128/JVI.77.19.10186-10201.2003. A definitive study of the HPV life cycle in relation to the infected epithelium.
- 28.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 1991;15(4):887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smotkin D, Wettstein FO. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc. Natl. Acad. Sci. USA. 1986;83(13):4680–4. doi: 10.1073/pnas.83.13.4680. An important early study of HPV16 transcription that underlies all subsequent studies.
- 30.Ozbun MA, Meyers C. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J. Virol. 1998;72(4):2715–22. doi: 10.1128/jvi.72.4.2715-2722.1998. One of the first descriptions of multiple promoters active during HR-HPV infection.
- 31.Thierry F, Howley PM. Functional analysis of E2-mediated repression of the HPV18 p105 promoter. New Biol. 1991;3(1):90–100. [PubMed] [Google Scholar]
- 32.Thierry F, Heard JJ, Dartmann K, Yaniv M. Characterisation of a transcriptional promoter of human papillomavirus 18 and modulation of its expression by simian virus 40 and adenovirus early antigens. J. Virol. 1987;61(1):134–42. doi: 10.1128/jvi.61.1.134-142.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grassman K, Rapp B, Maschek H, Petry KU, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J. Virol. 1996;70(4):2339–49. doi: 10.1128/jvi.70.4.2339-2349.1996. The first paper describing the HPV16 differentiation-specific promoter.
- 34.Chow LT, Nasseri M, Wolinsky SM, Broker TR. Human papillomavirus types 6 and 11 mRNAs from genital condylomata acuminata. J. Virol. 1987;61(8):2581–8. doi: 10.1128/jvi.61.8.2581-2588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smotkin D, Prokoph H, Wettstein FO. Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J. Virol. 1989;63(3):1441–7. doi: 10.1128/jvi.63.3.1441-1447.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoler MH, Wolinsky SM, Whitbeck A, Broker TR, Chow LT. Differentiation-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridisation with message-specific RNA probes. Virol. 1989;172(1):331–40. doi: 10.1016/0042-6822(89)90135-9. [DOI] [PubMed] [Google Scholar]
- 37.Karlen S, Offord EA, Beard P. Functional promoters in the genome of human papillomavirus type 6b. J. Gen. Virol. 1996;77(1):11–6. doi: 10.1099/0022-1317-77-1-11. [DOI] [PubMed] [Google Scholar]
- 38.Zhao W, Chow LT, Broker TR. Transcription activities of human papillomavirus type 11 E6 promoter-proximal elements in raft and submerged culture of foreskin keratinocytes. J. Virol. 1997;71(11):8832–40. doi: 10.1128/jvi.71.11.8832-8840.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiLorenzo TP, Steinberg BM. Differential regulation of human papillomavirus type 6 and 11 early promoters in cultures cells derived from laryngeal papillomas. J. Virol. 1995;69(11):6865–72. doi: 10.1128/jvi.69.11.6865-6872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carson A, Khan SA. Characterisation of transcription factor binding to human papillomavirus type 16 DNA during cellular differentiation. J. Virol. 2006;80(9):4356–62. doi: 10.1128/JVI.80.9.4356-4362.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thierry F. Transcriptional regulation of the papillomavirus oncogenes by cellular and viral transcription factors in cervical cancer. Virol. 2009;384(2):375–9. doi: 10.1016/j.virol.2008.11.014. A comprehensive review of HR-HPV transcription regulation.
- 42.Woolridge T, Laimins LA. Regulation of human papillomavirus type 31 gene expression during the differentiation-dependent life cycle through histone modifications and transcription factor binding. Virol. 2008;374(2):371–80. doi: 10.1016/j.virol.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodwin EC, Naeger LK, Breiding DE, Androphy EJ, DiMaio D. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J. Virol. 1998;72(5):3925–34. doi: 10.1128/jvi.72.5.3925-3934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimura A, Ono T, Ishimoto A, et al. Mechanisms of human papillomavirus E2-mediated repression of viral oncogene expression and cervical cancer cell growth inhibition. J. Virol. 2000;74(8):3752–60. doi: 10.1128/jvi.74.8.3752-3760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milligan SG, Veerapraditsin T, Ahamat B, Mole S, Graham SV. Analysis of novel human papillomavirus type 16 late mRNAs in differentiated W12 cervical epithelial cells. Virol. 2007;360:172–81. doi: 10.1016/j.virol.2006.10.012. A comprehensive description of HPV16 lare gene expression in W12 cervical epithelial cells.
- 46.Del Mar Pena L, Laimins LA. Differentiation-dependent chromatin rearrangement coincides with activation of human papillomavirus type 31 late gene expression. J. Virol. 2001;75(20):10005–13. doi: 10.1128/JVI.75.20.10005-10013.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bodily JM, Meyers C. Genetic analysis of the human papillomavirus type 31 differentiation-dependent late promoter. J. Virol. 2005;79(6):3309–21. doi: 10.1128/JVI.79.6.3309-3321.2005. Imnportant study characterising the HPV 31 late promoter region.
- 48.Zheng Z-M, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front. Biosci. 2006;11:2286–302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glahder JA, Hansen CN, Vinther J, Madsen BS, Norrild B. A promoter within the E 6 ORF of human papillomavirus type 16 contributes to the expression of the E7 oncoprotein from a monocistronic mRNA. J. Gen. Virol. 2003;84(12):3429–41. doi: 10.1099/vir.0.19250-0. [DOI] [PubMed] [Google Scholar]
- 50.Hansen CN, Nielsen L, Norrild B. Activities of E7 promoters in the human papillomavirus type 16 genome during cell differentiation. Virus Res. 2010;150(1-2):34–42. doi: 10.1016/j.virusres.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 51.Ozbun MA, Meyers C. Characterisation of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 1997;71(7):5161–72. doi: 10.1128/jvi.71.7.5161-5172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doorbar J, Parton A, Hartley K, et al. Detection of novel splicing patterns in a HPV-16-containing keratinocyte cell line. Virol. 1990;178(1):254–62. doi: 10.1016/0042-6822(90)90401-c. [DOI] [PubMed] [Google Scholar]
- 53.Stoler MH, Rhodes CR, Whitbeck A, et al. Human papillomavirus type 16 and 18 gene expression in cervical neoplasia. Human Pathol. 1992;23(2):117–28. doi: 10.1016/0046-8177(92)90232-r. An important early study of HR HPV gene expression in cervical tumours.
- 54.Beyer-Finkler E, Stoler MH, Girardi F, Pfister HJ. Cell differentiation-related gene expression of human papillomavirus 33. Med. Microbiol. Immunol. 1990;179(4):185–92. doi: 10.1007/BF00195249. [DOI] [PubMed] [Google Scholar]
- 55.Crum CP, Barber S, Symbula M, et al. Coexpression of the human papillomavirus type 16 E4 and L1 open reading frames in early cervical neoplasia. Virol. 1990;178(1):238–46. doi: 10.1016/0042-6822(90)90399-c. [DOI] [PubMed] [Google Scholar]
- 56.Terhune SS, Milcarek C, Laimins LA. Regulation of human papillomavirus type 31 polyadenylation during the differentiation-dependent life cycle. J. Virol. 1999;73(9):7185–92. doi: 10.1128/jvi.73.9.7185-7192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terhune SS, Hubert WG, Thomas JT, Laimins LA. Early polyadenylation signals of human papillomavirus type 31 negatively regulates capsid gene expression. J. Virol. 2001;75(17):8147–57. doi: 10.1128/JVI.75.17.8147-8157.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rush M, Zhao X, Schwartz S. A splicing enhancer in the E4 coding region of human papillomavirus type 16 is required for early mRNA splicing and polyadenylation as well as inhibition of premature late gene expression. J. Virol. 2005;79(18):12002–15. doi: 10.1128/JVI.79.18.12002-12015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Öberg D, Fay J, Lambkin H, Schwartz S. A downstream polyadenylation element in human papillomavirus type 16 L2 encodes multiple GGG motifs and interacts with hnRNP H. J. Virol. 2005;79(14):9254–69. doi: 10.1128/JVI.79.14.9254-9269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao X, Oberg D, Rush M, et al. A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2 and polypyrimidine tract binding protein. J. Virol. 2005;79(7):4270–88. doi: 10.1128/JVI.79.7.4270-4288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14(5):802–13. doi: 10.1261/rna.876308. A good review of current knowledge of genome-wide splicing integration and control.
- 62.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 63.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417(1):15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 64.Martinez-Contreras R, Cloutier P, Shkreta L, et al. hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 2007;623:123–47. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 65.Eperon IC, Makarova OV, Mayeda A, et al. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 2000;20(2):8303–18. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barash Y, Calarco JA, Gao W, et al. Deciphering the splicing code. Nature. 2010;465(7294):53–9. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 67.Berget S. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270(6):2411–4. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 68.Zhao X, Fay J, Lambkin H, Schwartz S. Identification of a 17-nucleotide splicing enhancer in HPV-16 L1 that counteracts the effect of multiple hnRNP A1-binding splicing silencers. Virol. 2007;369:351–63. doi: 10.1016/j.virol.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 69.Mole S, McFarlane M, Chuen-Im T, et al. RNA splicing factors regulated by HPV16 during cervical tumour progression. J. Pathol. 2009;219:383–91. doi: 10.1002/path.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mole S, Milligan SG, Graham SV. Human papillomavirus type 16 E2 protein transcriptionally activates the promoter of a key cellular splicing factor, SF2/ASF. J. Virol. 2009;83(1):357–67. doi: 10.1128/JVI.01414-08. The first demonstration that HPV16 E2 regulates transcription of a cellular gene and the first example of a virus protein that controls expression of splicing proteins.
- 71.Stubenrauch F, Colbert AME, Laimins LA. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J. Virol. 1998;72(10):8115–23. doi: 10.1128/jvi.72.10.8115-8123.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sokolowski M, Furneaux H, Schwartz S. The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3′ untranslated region correlates with its affinity for the elav-like HuR protein. J. Virol. 1999;73(2):1080–91. doi: 10.1128/jvi.73.2.1080-1091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sokolowski M, Zhao C, Tan W, Schwartz S. AU-rich mRNA instability elements on human papillomavirus type 1 late mRNAs and c-fos mRNAs interact with the same cellular factors. Oncogene. 1997;15(19):2303–19. doi: 10.1038/sj.onc.1201415. [DOI] [PubMed] [Google Scholar]
- 74.Sokolowski M, Schwartz S. Heterogeneous nuclear ribonucleoprotein C binds exclusively to the functionally important UUUUU-motifs in the human papillomavirus type-1 AU-rich inhibitory element. Virus Res. 2001;73(2):163–75. doi: 10.1016/s0168-1702(00)00238-0. [DOI] [PubMed] [Google Scholar]
- 75.Tan W, Schwartz S. The rev protein of human immunodeficiency virus type 1 counteracts the effect of an AU-rich negative element in the human papillomavirus type 1 late 3′ untranslated region. J. Virol. 1995;69(5):2932–45. doi: 10.1128/jvi.69.5.2932-2945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao C, Tan W, Sokolowski M, Schwartz S. Identification of nuclear and cytoplasmic proteins that interact specifically with an AU-rich, cis-acting inhibitory sequence in the 3′ untranslated region of human papillomavirus type 1 late mRNAs. J. Virol. 1996;70(6):3659–67. doi: 10.1128/jvi.70.6.3659-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cumming SA, McPhillips MG, Veerapraditsin T, Milligan SG, Graham SV. Activity of the human papillomavirus type 16 late negative regulatory element is partly due to four weak consensus 5′ splice sites that bind a U1 snRNP-like complex. J. Virol. 2003;77(9):5167–77. doi: 10.1128/JVI.77.9.5167-5177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cumming SA, Repellin CE, McPhillips M, et al. The human papillomavirus type 31 late 3′ untranslated region contains a complex bipartite negative regulatory element. J. Virol. 2002;76(12):5993–6003. doi: 10.1128/JVI.76.12.5993-6003.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chuen-Im T, Zhang J, Milligan SG, McPhillips MG, Graham SV. The alternative splicing factor hnRNP A1 is up-regulated during virus-infected epithelial cell differentiation and binds the human papillomavirus type 16 late regulatory element. Virus Res. 2008;131:189–98. doi: 10.1016/j.virusres.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cumming SA, Chuen-Im T, Zhang J, Graham SV. The RNA stability regulator HuR regulates L1 protein expression in vivo in differentiating cervical epithelial cells. Virol. 2009;383:142–9. doi: 10.1016/j.virol.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koffa MD, Graham SV, Takagaki Y, Manley JL, Clements JB. The human papillomavirus type 16 negative regulatory element interacts with three proteins that act at different posttranscriptional levels. Proc. Natl. Acad. Sci. USA. 2000;97(9):4677–82. doi: 10.1073/pnas.070049097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dietrich-Goetz W, Kennedy IM, Levins B, Stanley MA, Clements JB. A cellular 65kDa protein recognizes the negative regulatory element of human papillomavirus late mRNA. Proc. Natl. Acad. Sci. USA. 1997;94(1):163–8. doi: 10.1073/pnas.94.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kennedy IM, Haddow JK, Clements JB. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J. Virol. 1991;65(4):2093–7. doi: 10.1128/jvi.65.4.2093-2097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brennan CM, Steitz JA. HuR and mRNA stability. Cell. Mol. Life Sci. 2001;58(2):266–77. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Collier B, Goobar L, Sokolowski M, Schwartz S. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogeneous ribonucleoprotein K and poly (rC)-binding proteins 1 and 2. J. Biol. Chem. 1998;273(35):22648–56. doi: 10.1074/jbc.273.35.22648. [DOI] [PubMed] [Google Scholar]
- 86.Sokolowski M, Tan W, Jellne M, Schwartz S. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J. Virol. 1998;72(2):1504–15. doi: 10.1128/jvi.72.2.1504-1515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wiklund L, Sokolowski M, Carlsson A, Rush M, Schwartz S. Inhibition of translation by UAUUUAU and UAUUUUUAU motifs of the AU-rich RNA instability element in the HPV-1 late 3′ untranslated region. J. Biol. Chem. 2002;277(43):40462–71. doi: 10.1074/jbc.M205929200. An interesting paper analysing translation regulation in HPV-1.
- 88.Newbury SF. Control of mRNA stability in eukaryotes. Biochem. Soc Trans. 2006;34(1):30–4. doi: 10.1042/BST20060030. [DOI] [PubMed] [Google Scholar]
- 89.Zhou J, Lui WJ, Peng SW, Sun XY, Frazer IH. Papillomavirus capsid protein expression levels depends on the match between codon usage and tRNA availability. J. Virol. 1999;73(6):4972–82. doi: 10.1128/jvi.73.6.4972-4982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao KN, Lui WJ, Frazer IH. Codon usage bias and A+T content variation in human papillomavirus genomes. Virus Res. 2003;98(2):95–104. doi: 10.1016/j.virusres.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 91.Gu W, Ding J, Wang X, et al. Generalized substitution of isoencoding codons shortens the duration of papillomavirus L1 protein expression in transiently gene-transfected keratinocytes due to cell differentiation. Nucleic Acids Res. 2007;35(14):4820–32. doi: 10.1093/nar/gkm496. A paper describing the impact of codon usage on HPV L1 protein expression.
- 92.Doorbar J. The papillomavirus life cycle. J. Clin. Virol. 2005;32S:S7–S15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 93.McPhillips MG, Veerapraditsin T, Cumming SA, et al. SF2/ASF binds the human papillomavirus type 16 late RNA control element and is regulated during epithelial differentiation. J. Virol. 2004;78(19):10598–605. doi: 10.1128/JVI.78.19.10598-10605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fay J, Kelehan P, Lambkin H, Schwartz S. Increased expression of cellular RNA-binding proteins in HPV-induced neoplasia and cervical cancer. J. Med. Virol. 2009;81(5):897–907. doi: 10.1002/jmv.21406. [DOI] [PubMed] [Google Scholar]
- 95.Bailer SM, Haas J. Connecting viral with cellular interactomes. Curr. Opin. Microbiol. 2009;12(4):453–9. doi: 10.1016/j.mib.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schmitt M, Dalstein V, Waterboer T, et al. Diagnosing cervical cancer and high grade precursors by HPV16 transcription patterns. Cancer Res. 2010;70(1):249–56. doi: 10.1158/0008-5472.CAN-09-2514. An up-to-the-minute study of HPV16 RNA detection and its potential use in diagnosis.
- 97.Green KL, Gaston K. Development of a topical protein therapeutic for human papillomavirus and associated cancers. BioDrugs. 2006;20(4):209–18. doi: 10.2165/00063030-200620040-00002. [DOI] [PubMed] [Google Scholar]
- 98.Tiemann K, Rossi JJ. RNAi-based therapeutics-current status, challenges and prospects. EMBO Mol. Med. 2009;1:142–51. doi: 10.1002/emmm.200900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.DiPaolo JA, Alvarez-Salas LM. Advances in the development of therapeutic nucleic acids against cervical cancer. Expert Opin. Biol. Ther. 2004;4(8):1251–64. doi: 10.1517/14712598.4.8.1251. An interesting review assessing the possible uses of therapeutic nucleic acids in anti-HPV therapy.
- 100.Doorbar J, Griffin H. Intrabody strategies for the treatment of human-papillomavirus-associated disease. Expert Opin. Biol. Ther. 2007;7(5):677–89. doi: 10.1517/14712598.7.5.677. [DOI] [PubMed] [Google Scholar]
- 101.Maitland NJ, Conway S, Wilkinson NS, et al. Expression patterns of the human papillomavirus type 16 transcription factor E2 in low- and high-grade cervical intraepithelial neoplasia. J. Pathol. 1998;186:275–80. doi: 10.1002/(SICI)1096-9896(1998110)186:3<275::AID-PATH159>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 102.Soret J, Bakkour N, Maire S, et al. Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc. Natl. Acad. Sci. USA. 2005;102(24):8764–9. doi: 10.1073/pnas.0409829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bakkour N, Lin Y-L, Maire S, et al. Small-molecule inhibition of HIV pre-mRNA splicing as a novel antiretroviral therapy to overcome drug resistance. PLosPath. 2007;3(10):e159. doi: 10.1371/journal.ppat.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Keriel A, Mahuteau-Betzer F, Jacquet C, et al. Protection against retrovirus pathogenesis by SR protein inhibitors. PLoS One. 2009;4(2):e4533. doi: 10.1371/journal.pone.0004533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meisner N-C, Hintersteiner M, Mueller K, et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat. Chem. Biol. 2007;3(8):508–15. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]
- 106.Jiang M, Rubbi CP, Milner J. Gel-based application of siRNA to human epithelial cancer cells induces RNAi-dependent apoptosis. Oligonucleotides. 2004;14:239–48. doi: 10.1089/oli.2004.14.239. [DOI] [PubMed] [Google Scholar]
- 107.Van Pachterbeke C, Bucella D, Rozenberg S, et al. Topical treatment of CIN2+ by cidofovir: results of a phase II, double-blind, prospective, placebo-controlled study. Gynecol. Oncol. 2010;115(1):69–74. doi: 10.1016/j.ygyno.2009.06.042. [DOI] [PubMed] [Google Scholar]