Abstract
Translocation renal cell carcinoma is a newly recognized subtype of renal cell carcinoma (RCC) with chromosomal translocations involving TFE3 (Xp11.2) or, less frequently, TFEB (6p21). Xp11 translocation RCC was originally described as a pediatric neoplasm representing 20–40% of pediatric RCCs with a much lower frequency in the adult population. TFEB translocation RCC is very rare, with approximately 10 cases reported in the literature. Here, we describe the clinicopathological features of adult translocation RCC from a single institution. Utilizing tissue microarray (TMA), immunohistochemistry, cytogenetic examination, and FISH, we identified 6 (~5%) cases of TFE3 translocation RCC and 1 (<1%) case of TFEB translocation RCC in 121 consecutive adult renal cell carcinoma cases between 2001 and 2009. Our results suggest that weak TFE3 staining of a significant proportion of RCC cases may due to expression of the full length TFE3 protein rather than the chimeric fusion protein resulting from chromosomal translocation.
Keywords: translocation renal cell carcinoma, TFE3, TFEB
INTRODUCTION
Renal cell carcinomas (RCCs) are a heterogeneous group of tumors that account for around 90% of all adult renal malignancies. The most common subtypes are clear cell (60–75%), papillary (10–15%), chromophobe (5%) and collecting duct carcinoma, each with unique associated features at the molecular and genetic levels. Recent progress in the understanding of molecular alterations defining kidney tumorigenesis has led to the development of a new array of ancillary studies, providing a better approach to the sub-classification of these tumors(28, 29).
Xp11 translocation renal cell carcinomas have been recently recognized as a distinct subset of renal carcinomas associated with a number of gene rearrangements involving the TFE3 gene on chromosome Xp11.2. The gene rearrangements result in the overexpression of several fusion proteins which contain the C-terminal portion of TFE3, a member of the Microphtalmia-associated transcriptional factor family (MITF). At least 5 known fusion gene partners including ASPL on 17q25(2, 24), PRCC on 1q21(32), PSF on 1q34(17), NonO on Xq12(17), and CLTC on 17q23(10) have been reported to date. Furthermore, t(X;3)(p11;q23)(11, 34) and t(X;19)(p11.2;q13.1) (13) have also been reported without a defined gene partner. A closely related but rare variant of RCC harboring the t(6;11)(p21; q12)(8, 19) translocation involves the transcription factor EB(TFEB), also a member of the MITF family. The Alpha–TFEB fusion in TFEB translocation RCC results in dysregulated expression of the full-length TFEB protein (8, 19). It has been demonstrated that nuclear labeling for TFE3 and TFEB is a sensitive and specific diagnostic marker for tumors with TFE3 (9) and TFEB (8) translocation RCC respectively.
MiTF, TFE3, TFEB and TFEC belong to the same microphthalmia transcription factor (MiTF) family (30). Members of the MiTF family form homodimers and heterodimers in all combinations to bind similar or identical DNA sequences of similar downstream target genes (30). Because TFE3 and TFEB translocation RCCs share clinical, histopathological, immunohistochemical, and molecular features, Argani and Ladanyi proposed to regroup these neoplasms under the category of “MiTF/TFE translocation carcinoma family”(6).
The frequency of adult TFE3 translocation carcinoma has been reported to be about 1% (14) to 1.6% (25) of all renal tumors; however, its actual incidence remains largely underestimated. In the present study, we examined all adult RCCs diagnosed at our institution between 2001 and 2009 using tissue microarray (TMA), immunohistochemistry (IHC), cytogenetics, and fluorescence in situ hybridization (FISH), a combined tissue based and molecular diagnostic algorithmic approach that has never been done before. Six cases of TFE3 translocation RCCs and one case of TFEB translocation RCCs were identified and their clinical and histopathological features are reviewed here. Additionally, RCCs presenting with weak nuclear TFE3 staining were investigated in-depth.
MATERIALS AND METHODS
Tissue Samples and Tissue Microarray Construction
After Institutional Review Board approval (protocol# 0120080157), a total 121 consecutive cases (from 2001–2009) with a diagnosis of renal cell carcinoma were obtained from files of the Pathology Department of the New Jersey Medical School, University Hospital, Newark. In all cases, tissues were fixed in neutral buffered formalin and embedded in a paraffin block as routine surgical pathology procedure.
For tissue microarray construction, three cores (1.0mm in diameter) of each tumor from patient paraffin blocks were paired with one core from non-neoplastic kidney taken from donor paraffin blocks. The final tissue microarrays consisted of up to 280 tissue cores per slide with a 1.5mm space between each core. After construction, 4μm sections were cut and H&E staining was performed on the initial slide to verify the histology.
IHC
After initial deparaffinization, endogenous peroxidase activity was blocked with 0.3% H2O2. Deparaffinized sections were microwaved in 10 mmol/l citrate buffer (pH 6.0). The slides were then incubated for 1 h at room temperature using goat polyclonal anti-human TFE3 (1:300; Santa Cruz sc-5958) or TFEB(1:400; Santa Cruz sc-11004) followed by biotinylated rabbit anti-goat IgG (Vector Laboratories, Burlingame, CA, USA) for 30 min, and finally ABComplex (Vector Laboratories, Burlingame, CA, USA)(3). The bound complex was visualized with 0.125% amino-ethyl carbazole (AEC, Sigma, St Louis, MO, USA) and 0.003% (v/v) H2O2. The sections were then counterstained in Mayer’s hematoxylin. For negative controls, the primary antibody was replaced with PBS.
TFE3 Gene Rearrangement Assessment by FISH
A dual-color, break-apart FISH assay for TFE3 gene rearrangement was performed on the formalin-fixed paraffin-embedded tissue microarray slides as described earlier. The normal result is a co-localization of red and green signals, whereas a TFE3 rearrangement results in a split signal.
Cytogenetic analysis
A fresh sample of tumor was collected in RPMI tissue culture medium and was sent directly to the cytogenetics laboratory. The tumor tissue was carefully sliced into small pieces and further treated with collagenase (3 mg/mL Hanks Balanced Salt Solution, Roche/Boehringer, IN) for 2 hours to yield the highest possible concentration of cells. These cells were cultured in RPMI 1640 solution for 3–5 days on a coverslip dish (MatTek Corp., Ashland, MA). After adding 10 uL of colchicine (10 mg/mL Hanks Balanced Salt Solution) to each chamber for 45 minutes, the cells were harvested. The slides were prepared using standard in situ techniques. The chromosomes on the slides were G-banded after trypsin pretreatment. The aberrations were designated according to the International System for Human Cytogenetic Nomenclature (1995).
RT-PCR
RNA extraction was performed on a frozen tissue using a standard organic extraction method (Trizol; Life Technologies, Inc., Friendsworth, TX). PSF-TFE3 hybrid transcripts were detected using the PSF forward primer 5′-TGGTGGTGGCATAGGTTATG-3′ and TFE3 reverse primer 5′-CGTTTGATGTTGGGCAGCTC-3′. NonO (p54nrb)-TFE3 hybrid transcripts were detected using the NonO (p54nrb) forward primer 5′-GAGAAACTAGACACAGCAAC-3′ and the TFE3 reverse primer 5′-CTTTCTTCTGCCGTTCCTTC-3′. PRCC-TFE3 hybrid transcripts were detected using the PRCC forward primer 5′-CCAAGCCAAAGAAGAGGA-3′ and the TFE3 reverse primer 5′-AGTGTGGTG-GACAGGTACTG-3′. CLTC-TFE3 hybrid transcripts were detected using the CLTC forward primer 5′-AGTCGCGTTGTTGGAAAGTATTGTG-3′ and a TFE3 reverse primer 5′-CAAAAGGGCCTTTGCCTCGGTC-3′. ASPL-TFE3 fusion transcript were detected using a forward primer from ASPL 5′-AAAGAAGTCCAAGTCGGGCCA-3′ and a TFE3 exon 4 reverse primer 5′-CGTTTGATGTTGGGCAGCTCA-3′. Normal TFE3 transcripts were detected using the forward primers 5′-CCCGCAAGTGC-CCAGCCACTG-3′ (exon 3) and reverse primer 5′-CAGTTCCTTGATCCTGTCG-3′ (exon4). β-actin transcripts were detected using the forward primer 5′-ATCACCATTGGCAATGAGCG-3′ and reverse primer 5′-TTGAAGGTAGTTTCGTGGAT-3′
RESULTS
We collected 121 consecutive adult renal cell carcinoma cases from 2001–2009. The first TMA contained tumor and non-neoplastic adjacent tissue from all 121 cases. TFE3 IHC was applied to the first TMA. There were 6 cases with strong and moderate TFE3 staining and 5 cases with weak TFE3 staining. The second TMA, constructed after the first, included all TFE3 positive (6 strong/moderate and 5 weak) cases and 5 TFE3 negative cases. A dual color break-apart FISH assay which detects chromosomal rearrangements at the Xp11.2 region was applied to the 2nd TMA. The 6 cases (Table 1) with strong/moderate TFE3 staining were confirmed as TFE3 translocation RCC by FISH. The cases #1 to #4 have been reported previously (34). The 5 cases with weak TFE3 staining and 5 cases with negative TFE3 staining did not show chromosomal rearrangement at the Xp11.2 region by FISH. We concluded that the 6 cases with strong/moderate TFE3 staining were TFE3 translocation RCC. Clinical and pathological features of these cases are reported below. Among the rest 115 RCC cases, around 73% cases were clear cell RCC; 20% of cases were papillary RCC; and the remaining 7% cases were consistent with chromophobe and unclassified RCC. This reflects that our RCC cases collection is a representative cross section of that of the general population.
Table 1.
Clinical information of TFE3 translocation RCCs
Clinical pathological information of Xp11 translocation RCC in this study. Cases #1 to #4 have been reported previously (34).
| Age | sex | Location | Size (cm) | Initial Presentation | Metastases | Follow-up (month) | Status | Cytogenetic or RT- PCR Result | |
|---|---|---|---|---|---|---|---|---|---|
| 1 RCC | 38 | F | L Kidney | 14 | Asymptomatic, Pick up by CT due to gastric by-pass work up | Abdomen Omentum | 21 | Alive | +7, +12, +16, +20 ASPL-TFE3 Type I |
| 2 RCC | 65 | F | R Kidney | 9.5 | Asymptomatic | Peri-aortic LN | 19 | Alive | Cytogenetic N/A ASPL-TFE3 Type I |
| 3 RCC | 42 | F | R Kidney | 11.5 | Pick up by CT due to incarcerated ventral hernia | No | 42 | Alive | +7, t(X; 1) (p11:p34) |
| 4 RCC | 45 | F | R kidney | 7.2 | Pathological fracture | Bone, Liver | 5 | Dead | der(X)t(X; 1) (p11:p34) PSF-TFE3 |
| 5 RCC | 30 | F | R Kidney | 25 | Hematuria | Peri-aortic LN | 47 | Alive | Numerical Abberation |
| 6 RCC | 48 | F | Left Kidney | 8 | Asymptomatic | Retro- peritoneum | 42 | Alive | Numerical Abberation |
Clinical and histopathological features of TFE3 Translocation RCC
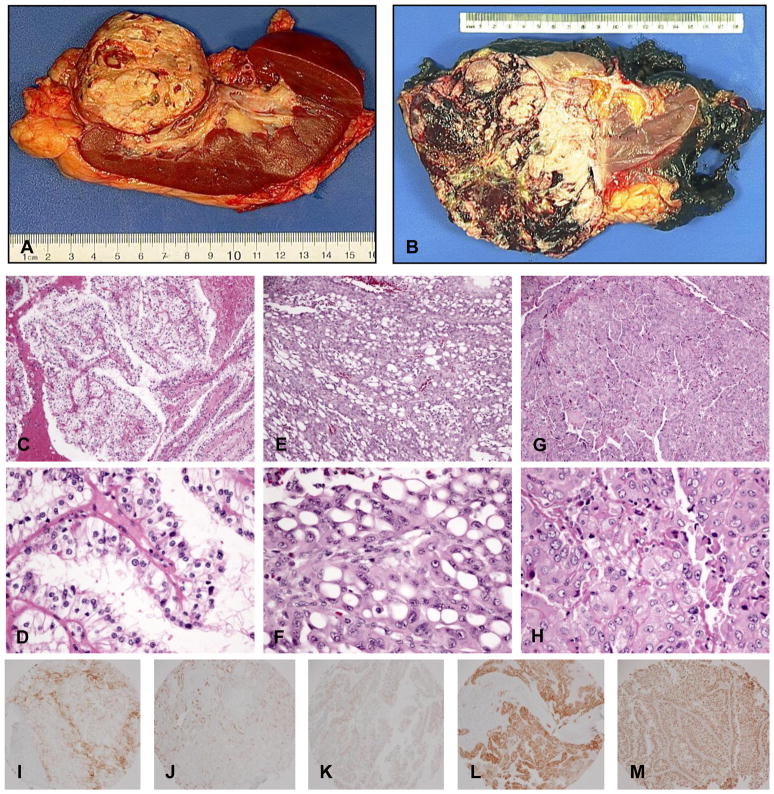
Patients with TFE3 translocation RCC were all female with a mean age of 43 years (range: 30 to 65). None of the patients had been previously exposed to cytotoxic chemotherapy. The most common presenting symptom was metastatic lesion(s), however, the majority of patients were asymptomatic. Five of the six patients had distant and/or lymph node metastasis either synchronously or metachronously. One patient died 5 months after diagnosis. The remaining patients were alive at follow up which ranged from 21 to 47 months. The tumors ranged in size from 7.2 cm to 25 cm (Table 1). Grossly, the tumors were not distinguishable from other RCC subtypes. They contained tan-yellow necrotic areas, hemorrhage, and cystic degeneration (Fig 1A&B).
Figure 1.
Gross pictures, H&Es and IHC of Xp11 translocation RCC. (A), (B) Gross pictures from case #4 and #1, respectively. (C), (D) H&E from case #3 showed typical clear cells with papillary architecture. (E), (F) H&E from case #1 showed polygonal cells with large cytoplasmic vacuoles and eccentric nuclei (signet ring-like) with a microcystic growth pattern. (G), (H) H&E from case #5 showed a solid/syncytial growth pattern and grade 4 nuclear atypia. IHC (I) for CD10, (J) for AE1/3, (K) for Melan-A, (L) for HMB 45, and (M) for TFE3.
On microscopic examination, several of the TFE3 translocation RCC cases had similar morphologies. The tumors were composed of large polygonal cells with discrete borders, granular eosinophilic to clear cytoplasm, vesicular nuclear chromatin, and prominent nucleoli. Cells were arranged in nests and papillary/pseudopapillary structures (Fig 1C&D). Pigment and hyaline nodules were not seen. Few psammoma bodies were present. Although the tumors resembled clear cell and papillary RCC in that they contained clear cytoplasm and papillary architecture, they did not contain the typical delicate and uniform small vessels typical of clear cell RCC. In addition, collections of foamy histiocytes are rare in Xp11 translocation RCC compared to papillary RCC. Other histological patterns can be seen. The specimen from patient #1 showed polygonal cells with large cytoplasmic vacuoles and eccentric nuclei (signet ring-like) with a microcystic growth pattern (Fig 1E&F). The specimen from patient #5 showed a solid/syncytial growth pattern and grade 4 nuclear atypia (Fig 1G&H).
Xp11 translocation RCC demonstrates a different immunoprofile compared to other RCC subtypes. Although there was slight variation of immunostaining from case to case in our 6 Xp11 translocation RCCs, most cases showed weak and focal staining for renal marker CD10 (Fig 1I) and epithelial marker AE1/3 (Fig 1J) and EMA. The melanocytic markers Melan-A and HMB-45(Fig 1K&L) were positive in 3 cases. Xp11 translocation RCC showed variable moderate/weak to negative vimentin staining, in comparison to the strong and diffuse staining typically seen in clear cell RCC. All cases were strongly positive to moderately positive for TFE3 (Fig 1M).
Biological meaning of weak TFE3 nuclear staining
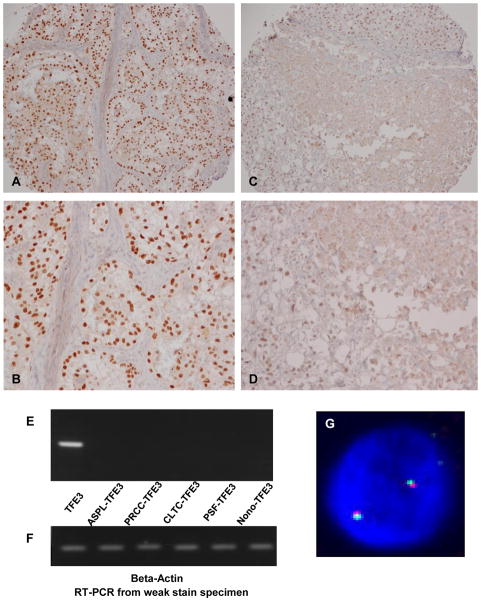
During our initial screening of TFE3 translocation RCC by TMA and IHC, we found that a small but significant portion of cases (5 cases, approximately 4%) showed weak nuclear staining for TFE3 (Fig 2 C&D). Using a FISH assay which we previously developed (34), we did not find any evidence of chromosomal translocations involving the Xp11 region in any of the 5 cases (Fig 2G). We also performed RT-PCR on one of the cases in which fresh frozen tissue was available. We used primers for wild-type (non-translocation) and all 5 known translocation types (ASPL-TFE3, PRCC-TFE3, PSF-TFE3 CLTC-TFE3 and Nono-TFE3). Only wild-type (non-translocation) TFE3 transcript was detected (Fig 2E). Beta-actin loading controls were detected in each reaction (Fig 2F). Taken together, the FISH and RT-PCR results strongly suggest that weak nuclear staining appears to be due to the expression of the full length TFE3 protein, rather than a chimeric fusion protein resulting from translocation. The similar results were also been observed by other group (3).
Figure 2.
Biological meaning of weak TFE3 nuclear staining. (A), (B) an example of Strong TFE3 nuclear staining at low and high power, respectively. (C), (D) an example of weak nuclear TFE3 staining at low and high power, respectively. (E) RT-PCR results from a weak nuclear TFE3 staining case using primers for wild-type (non-translocation) and all 5 known translocation types. Only wild-type (non-translocation) TFE3 transcript was detected. (F) RT-PCR Beta-actin control results from the same sample. (G) Example of FISH results from weak TFE3 nuclear staining cases. There was no evidence of TFE3 gene rearrangement in these cases.
Clinical and histopathological features of TFEB translocation RCC
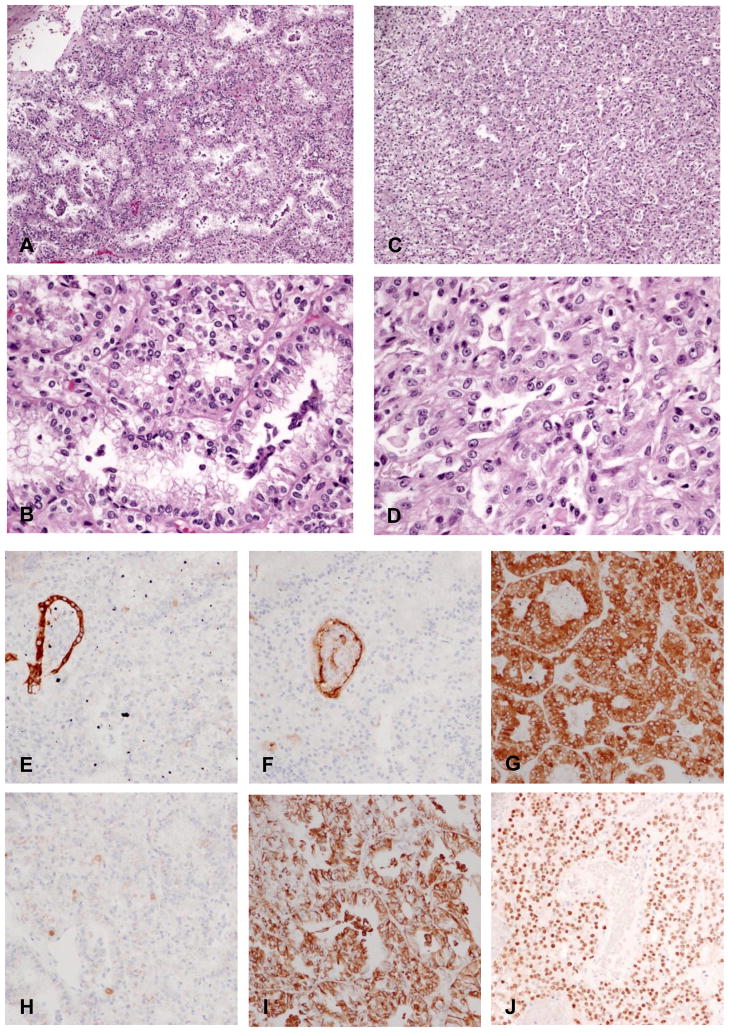
In our records, there was one case of renal cell carcinoma with t(6,11). The patient is an African-American male with a renal neoplasm diagnosed incidentally on CT scan after presenting with abdominal trauma (age 17). He was lost to follow up and returned three years later (age 20) complaining of abdominal pain. A left total nephrectomy was performed. The tumor measured 19 cm in greatest dimension. Grossly, the tumor was tan to yellow-brown with diffuse hemorrhage and necrosis. Degenerative cystic areas were seen in the central portion of tumor.
Microscopic examination of viable tumor showed solid sheets of medium-sized, monomorphic, polygonal epithelioid cells with multiple areas of tubular and papillary architecture (Fig 3A&C). The cells contained clear to granular eosinophilic cytoplasm, well-defined cell borders, and small, round nuclei with occasional small nucleoli (Fig 3B&D). Mitoses were rare and no psammoma bodies or hyaline nodules were present. The tumor was extensively sampled. Clusters of smaller epithelioid cells, a previously described phenomenon in this tumor (6), were not identified. The tumor also showed a unique immunoprofile consisting of absent staining for pan-cytokeratin (AE1/3)(Fig 3E) and CD10 (Fig 3F), strong diffuse strong positivity for Melan-A and focal positivity for HMB45 (Fig 3H). The TFEB immunohistochemical stain was heterogeneous and ranged from moderate to strong nuclear staining at periphery of the tissue (Fig 3J) to weak or completely negative nuclear staining. This pattern may be due to fixation artifact affecting antibody binding, an occurrence which has been reported previously (6). We also applied the TFEB immunohistochemical stain to the first TMA which contained 121 cases of RCC. Positive TFEB staining was not identified. Although the possibility of a false negative arises due to the nature of the TMA and TFEB antibodies, we believe that this possibility is unlikely due to the rare nature of TFEB translocation RCC.
Figure 3.
H&E and IHC of TFEB translocation RCC. (A)& (B), showing tubular and papillary architecture.. (C)&(D), showing solid growth pattern. IHC (E) for AE1/3, (F) for CD10, Please note that trapped tubule serve as internal positive control and tumor cells were completely negative for these 2 staining. (G) for Melan-A, (H) for HMB45, (I) for Vimentin and (J) for TFEB.
DISCUSSION
Renal cell carcinomas (RCCs) encompass a spectrum of tumors with unique clinical and histopathological features as well as characteristic cytogenetic aberrations (28, 29). Cytogenetic analysis has provided a valuable tool in the diagnosis of the various histological subtypes and has elucidated how their distinctive molecular profiles correlate with different morphological patterns. Pediatric renal cell carcinomas, though uncommon, usually show distinctive chromosomal abnormalities most often involving translocations involving TFE3 and TFEB, members of the MiTF family of transcription factors (12, 15).
While Xp11 translocation RCC is typically considered a pediatric malignancy, its frequency in the adult population remains underestimated (14). Several reasons could explain this occurrence. One possibility lies in the fact the translocation RCCs usually overlap morphologically or mimic other RCCs (primarily clear cell and papillary RCC). In contrast to pediatric neoplasms, fresh tissue collection for cytogenetics and molecular analysis is not routinely performed in adult translocation RCC making further diagnostic testing difficult.
Previous cytogenetic and immunohistochemistry-based studies attempting to estimate the frequency of Xp11 translocation RCC have revealed an incidence of around 1% (14) and 1.6 % (25) respectively among all RCCs examined in these series. Taking the aforementioned into consideration along with the fact that RCC is more commonly encountered in the adult population, it is possible that the frequency of Xp11 translocation RCCs in adults may outnumber cases in the pediatric group (31). Interestingly, our study revealed a frequency close to 5%, likely because routine fresh tissue collection for cytogenetic testing and cryopreservation of tumor samples is a common practice at our institution allowing us to perform in depth ancillary studies including cytogenetic analysis, RT-PCR and FISH. This emphasizes that the combination of histopathology along with molecular studies increases the diagnostic accuracy of these tumors. Our case series is relatively small (121 cases). A more accurate frequency of Xp11 translocation RCC needs to be confirmed by larger and multi-institutional studies. If our results are representative of the general population, there will be approximately 2500 new TFE3 translocation RCC cases each year in US, considering NCI estimates of 58,240 new kidney tumor cases in the US in 2011.
Although prior exposure to chemotherapy remains the only known risk factor for the development of Xp11 translocation RCC (7), none of the patients in our study had a history of chemotherapy exposure. Female predominance, as described in the literature (11, 14), was seen in our study. All TFE3 translocation RCC cases were present in females with a mean age of 43, contrasting with the much younger mean age (~15 years less) of all 121 RCC patients examined.
Grossly, Xp11 translocation RCCs are large tumors resembling conventional clear cell RCC with a variegated appearance including tan-yellow areas admixed with hemorrhagic and necrotic tissue foci. The mean tumor size in our series was 12.5 cm, almost double of that reported in other series (11, 14). This may be due to the fact that our hospital serves a population of predominantly low socioeconomic status in which patients tend to seek medical attention at more advanced stages of disease due to limited access to health care.
As described by Argani and Ladanyi, TFE3 gene fusions translate histologically to a distinct morphological pattern consisting mainly of mixed papillary and nested cells with granular eosinophilic cytoplasm (1, 6). The majority of our cases were consistent with this histological picture. Intriguingly, we also found some unique histological features that have not been previously described in the literature. These include signet ring-like cells in a microcystic arrangement (Fig 1E&F) with high grade cytologic atypia and non-clear cells laid in solid/syncytial architectural patterns (Fig. 1G&H). Similar morphology of high grade cytologic atypia and non-clear cells have been previously reported in Xp11 RCC (9). Abundant psammoma bodies and hyaline nodules were not seen in our cases. These tumors did not contain the typical delicate and uniform small vessels common in clear cell RCC or collections of foamy histiocytes common in papillary RCC. Xp11 translocation RCC should also be distinguished from clear cell papillary renal cell carcinoma (23), a newly described entity. Most clear cell papillary RCCs contain both clear cell and papillary features in a similar style to Xp11 translocation RCCs. A cardinal feature to differentiate these tumors is based on the location of the nuclei towards either the middle or upper pole of the cell (23). This reverse polarity is a feature not seen in Xp11 translocation RCC.
Xp11 translocation RCCs are known to variably express epithelial immunohistochemical markers and are consistently positive for renal cell carcinoma marker antigens and CD10 (5, 11, 14). Still, nuclear immunoreactivity to TFE3 protein remains the most sensitive, specific, and distinct immunohistochemical marker for these neoplasms (5, 11, 14). For a comprehensive immunohistochemical profile of these tumors, please see a recent publication (5). The TFE3 protein antibody recognizes the C-terminal portion of TFE3. The C-terminal portion is retained in the gene fusion (9). Its over-expression in TFE3 translocation RCC is believed to be due to the up-regulation of a chimeric TFE3 protein resulting from a swapping to a stronger promoter (9). In our study, 6 cases expressed moderate to strong nuclear staining while 5 cases exhibited weak TFE3 staining. Further investigation of the latter cases by FISH and/or RT-PCR failed to reveal any evidence of TFE3 rearrangement suggesting that the weak nuclear TFE3 staining may be the result of an up-regulation of the full-length TFE3 protein, translating into a weak expression(3). This up-regulation is in contrast to the chimeric fusion proteins resulting from Xp11.2 gene rearrangements. These results re-confirm that weak nuclear staining should not be interpreted as true positive staining in these tumors and that TFE3 IHC is a sensitive marker for Xp11 translocation RCC (9). Additionally, in all six Xp11 translocation RCC cases determined by FISH and/or cytogenetics, RT-PCR showed strong to moderate nuclear TFE3 staining. These findings also showed that with careful evaluation, TFE3 IHC can be a specific marker for these tumors as well (9).
The diagnosis of Xp11 translocation RCC requires the integration of clinical information, histopathologic features, TFE3 IHC stain, cytogenetic, and molecular studies. Any of the following scenarios should prompt work up to further investigate the possibility of an Xp11 translocation RCC: young/middle age patient (<45) with metastatic RCC; typical clear cell with papillary histopathological features along with minimal immunohistochemical reactivity to cytokeratins and EMA. TFE3 IHC is a sensitive and relatively specific marker for Xp11 translocation RCC that could be used as a screening method, followed by FISH assay which should be reserved as a helpful ancillary technique in those cases with equivocal TFE3 IHC staining. To date, the diagnosis of Xp11 translocation RCC remains largely as an academic exercise due to the lack of a specific therapy for these neoplasms. However, awareness among pathologist is pivotal in order to arrive to an accurate diagnosis.
Xp11 translocations have been implicated in several tumors other than RCC. Alveolar soft part sarcoma (ASPS) with the ASPL-TFE3 gene fusion is one example (26). Interestingly, Xp11 translocation RCC with identical gene rearrangements have shown similar histopathological patterns to ASPS (6). Perivascular epithelioid cell tumors (PEComas) have also shown immunoreactivity for TFE3. Folpe, et al showed TFE3 positivity in 5 of 17 PEComas (21). Two cases of PEComa with a PSF-TFE3 gene fusion demonstrated by FISH and RT-PCR have also been reported (16, 33). More recently, Argani reported a subset of lesions currently classified as PEComas which harbor TFE3 gene fusions (3). He also reported a distinctive type of renal cancer with overlapping features of PEComa, Xp11 translocation carcinoma, and melanoma (4).
In the present series we also report on TFEB t(6;11)(p21;q12) translocation RCC, another rare subtype of RCC. These carcinomas are characterized by fusion of the intronless Alpha gene (11q12) with the first intron of the TFEB transcription factor gene (6p21)(8, 19). TFEB translocation RCCs share similar histopathological features with TFE3 translocation RCCs, but at the same time reveal distinct clinicopathologic features (6). Microscopically, these tumors display a dimorphic pattern composed of large polygonal epithelioid cells with clear to eosinophilic cytoplasm in a nested pattern co-mingled with a population of smaller epitheliod-like cells typically clustered around hyaline nodules (6). In our case, however, this dimorphic pattern was not present. Instead, the tumor was composed of medium sized polygonal cells with small round nuclei distributed in solid sheets with multiple foci of papillary and tubular architectural patterns, a feature that has been observed in other TFEB translocation RCCs confirmed by cytogenetic studies (Argani, personal communication). Immunoprofiling on this case revealed the distinctive features of this tumor subtype including entirely absent staining for pan-cytokeratins AE1/3 and kidney tubule marker CD10 and a diffuse strong immunoreacivity with HMB-45. TFEB staining showed moderate to strong nuclear staining which was more prominent at the periphery of the tumor, perhaps due to better antigen preservation from more complete fixation at the periphery of the tissue (6).
MITF, an MIT family protein which also includes TFE3, TFEB and TFEC, has also been implicated in various cancers. MiTF amplification (22) and somatic mutations (18) have been reported in human melanoma. Another correlation of MITF with tumorigenesis has been made for clear cell sarcoma (CCS) (20), a rare sarcoma that expresses melanocytic markers and harbors the EWS–ATF1 chromosomal translocation. The resulting fusion protein occupies the MITF promoter and induces its expression, mediating CCS survival. CCS cells can not survive in the absence of MITF, although their survival can be rescued by either MITF or the closely related family members TFE3 or TFEB. Reciprocally, UOK109, an Xp11 translocation RCC cells that harbors the NONO-TFE3 translocation, could not survive upon TFE3 knockdown by small interference RNA (siRNA) but were rescued by expression of MITF(20). These elegant experiments demonstrated functional oncogenic overlap among MITF, TFE3 and TFEB. Based on oncogenic gene fusion–translocation events, dysregulation of MITF-related family members, and genetic evidence of functional redundancy among several of these factors, Fisher et al suggested a re-evaluation of the classification of these tumors (27). They coined the term “MiT family cancers” which includes a subset of melanoma, TFE3 and TFEB translocation RCC, alveolar soft part sarcoma (ASPS), and clear cell sarcoma (CCS) (27).
This observation is of crucial importance, as recent therapeutic strategies focusing on targeting common molecular pathways that contribute to critical steps in malignant transformation have proven successful. One such example is with the tyrosine kinase inhibitor Imatinib, which targets BCR-ABL and c-KIT and significantly improves the prognosis of chronic myeloid leukemia (CML) and gastrointestinal stromal tumors (GIST) respectively. Even though MiT family cancers differ in many clinical aspects, these cancers share common or closely related oncogenic factors. It is reasonable to predict that targeted therapies for the MITF family proteins will be effective for all Mit family cancers including TFE3 translocation RCC.
In conclusion, this study reports on the unique clinicopathologic features of adult RTCs using TMA, immunohistochemistry, cytogenetics and FISH. In addition to histological diagnosis, these adjunctive techniques could potentially be used in an algorithmic combination to strengthen diagnostic accuracy and greatly increase the chances of therapeutic success. We found that these tumors are not uncommon in light of a single institution experience. Despite the significant advances and recent progress in the understanding of the molecular basis of translocation-associated renal cell carcinomas, many aspects of these tumors remain to be uncovered.
Acknowledgments
The authors thank Dr. Argani for his helpful discussion on TFEB translocation RCC. The authors also thank Frank Tian, Dana Settembre, and Taozhi Lin for their technical support. This study is partly supported by the NJMS pathology department resident research fund.
Footnotes
Disclosures: The authors have no conflicts of interest or funding to disclose. The order of authors is base on the length of the time involved in this project, in descending order.
References
- 1.Argani P, Antonescu CR, Couturier J, et al. PRCC-TFE3 renal carcinomas: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2;q21) Am J Surg Pathol. 2002;26:1553–1566. doi: 10.1097/00000478-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Argani P, Antonescu CR, Illei PB, et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–192. doi: 10.1016/S0002-9440(10)61684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argani P, Aulmann S, Illei PB, et al. A distinctive subset of PEComas harbors TFE3 gene fusions. Am J Surg Pathol. 2010;34:1395–1406. doi: 10.1097/PAS.0b013e3181f17ac0. [DOI] [PubMed] [Google Scholar]
- 4.Argani P, Aulmann S, Karanjawala Z, et al. Melanotic Xp11 translocation renal cancers: a distinctive neoplasm with overlapping features of PEComa, carcinoma, and melanoma. Am J Surg Pathol. 2009;33:609–619. doi: 10.1097/PAS.0b013e31818fbdff. [DOI] [PubMed] [Google Scholar]
- 5.Argani P, Hicks J, De Marzo AM, et al. Xp11 translocation renal cell carcinoma (RCC): extended immunohistochemical profile emphasizing novel RCC markers. Am J Surg Pathol. 2010;34:1295–1303. doi: 10.1097/PAS.0b013e3181e8ce5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argani P, Ladanyi M. Translocation carcinomas of the kidney. Clin Lab Med. 2005;25:363–378. doi: 10.1016/j.cll.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Argani P, Lae M, Ballard ET, et al. Translocation carcinomas of the kidney after chemotherapy in childhood. J Clin Oncol. 2006;24:1529–1534. doi: 10.1200/JCO.2005.04.4693. [DOI] [PubMed] [Google Scholar]
- 8.Argani P, Lae M, Hutchinson B, et al. Renal carcinomas with the t(6;11)(p21;q12): clinicopathologic features and demonstration of the specific alpha-TFEB gene fusion by immunohistochemistry, RT-PCR, and DNA PCR. Am J Surg Pathol. 2005;29:230–240. doi: 10.1097/01.pas.0000146007.54092.37. [DOI] [PubMed] [Google Scholar]
- 9.Argani P, Lal P, Hutchinson B, et al. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am J Surg Pathol. 2003;27:750–761. doi: 10.1097/00000478-200306000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Argani P, Lui MY, Couturier J, et al. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23) Oncogene. 2003;22:5374–5378. doi: 10.1038/sj.onc.1206686. [DOI] [PubMed] [Google Scholar]
- 11.Argani P, Olgac S, Tickoo SK, et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol. 2007;31:1149–1160. doi: 10.1097/PAS.0b013e318031ffff. [DOI] [PubMed] [Google Scholar]
- 12.Armah HB, Parwani AV. Xp11.2 translocation renal cell carcinoma. Arch Pathol Lab Med. 2010;134:124–129. doi: 10.5858/2008-0391-RSR.1. [DOI] [PubMed] [Google Scholar]
- 13.Armah HB, Parwani AV, Surti U, et al. Xp11.2 translocation renal cell carcinoma occurring during pregnancy with a novel translocation involving chromosome 19: a case report with review of the literature. Diagn Pathol. 2009;4:15. doi: 10.1186/1746-1596-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camparo P, Vasiliu V, Molinie V, et al. Renal translocation carcinomas: clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of the literature. Am J Surg Pathol. 2008;32:656–670. doi: 10.1097/PAS.0b013e3181609914. [DOI] [PubMed] [Google Scholar]
- 15.Carcao MD, Taylor GP, Greenberg ML, et al. Renal-cell carcinoma in children: a different disorder from its adult counterpart? Med Pediatr Oncol. 1998;31:153–158. doi: 10.1002/(sici)1096-911x(199809)31:3<153::aid-mpo5>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 16.Chang IW, Huang HY, Sung MT. Melanotic Xp11 translocation renal cancer: a case with PSF-TFE3 gene fusion and up-regulation of melanogenetic transcripts. Am J Surg Pathol. 2009;33:1894–1901. doi: 10.1097/PAS.0b013e3181ba7a5f. [DOI] [PubMed] [Google Scholar]
- 17.Clark J, Lu YJ, Sidhar SK, et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–2239. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 18.Cronin JC, Wunderlich J, Loftus SK, et al. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 2009;22:435–444. doi: 10.1111/j.1755-148X.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis IJ, Hsi BL, Arroyo JD, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A. 2003;100:6051–6056. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis IJ, Kim JJ, Ozsolak F, et al. Oncogenic MITF dysregulation in clear cell sarcoma: defining the MiT family of human cancers. Cancer Cell. 2006;9:473–484. doi: 10.1016/j.ccr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Folpe AL, Mentzel T, Lehr HA, et al. Perivascular epithelioid cell neoplasms of soft tissue and gynecologic origin: a clinicopathologic study of 26 cases and review of the literature. Am J Surg Pathol. 2005;29:1558–1575. doi: 10.1097/01.pas.0000173232.22117.37. [DOI] [PubMed] [Google Scholar]
- 22.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 23.Gobbo S, Eble JN, Maclennan GT, et al. Renal cell carcinomas with papillary architecture and clear cell components: the utility of immunohistochemical and cytogenetical analyses in differential diagnosis. Am J Surg Pathol. 2008;32:1780–1786. doi: 10.1097/PAS.0b013e31818649ed. [DOI] [PubMed] [Google Scholar]
- 24.Heimann P, El Housni H, Ogur G, et al. Fusion of a novel gene, RCC17, to the TFE3 gene in t(X;17)(p11.2;q25.3)-bearing papillary renal cell carcinomas. Cancer Res. 2001;61:4130–4135. [PubMed] [Google Scholar]
- 25.Komai Y, Fujiwara M, Fujii Y, et al. Adult Xp11 translocation renal cell carcinoma diagnosed by cytogenetics and immunohistochemistry. Clin Cancer Res. 2009;15:1170–1176. doi: 10.1158/1078-0432.CCR-08-1183. [DOI] [PubMed] [Google Scholar]
- 26.Ladanyi M, Lui MY, Antonescu CR, et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57. doi: 10.1038/sj.onc.1204074. [DOI] [PubMed] [Google Scholar]
- 27.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Linehan WM, Bratslavsky G, Pinto PA, et al. Molecular diagnosis and therapy of kidney cancer. Annu Rev Med. 2010;61:329–343. doi: 10.1146/annurev.med.042808.171650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linehan WM, Zbar B. Focus on kidney cancer. Cancer Cell. 2004;6:223–228. doi: 10.1016/j.ccr.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Rehli M, Den Elzen N, Cassady AI, et al. Cloning and characterization of the murine genes for bHLH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics. 1999;56:111–120. doi: 10.1006/geno.1998.5588. [DOI] [PubMed] [Google Scholar]
- 31.Ross H, Argani P. Xp11 translocation renal cell carcinoma. Pathology. 2010;42:369–373. doi: 10.3109/00313021003767348. [DOI] [PubMed] [Google Scholar]
- 32.Shipley JM, Birdsall S, Clark J, et al. Mapping the X chromosome breakpoint in two papillary renal cell carcinoma cell lines with a t(X;1)(p11.2;q21.2) and the first report of a female case. Cytogenet Cell Genet. 1995;71:280–284. doi: 10.1159/000134127. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka M, Kato K, Gomi K, et al. Perivascular epithelioid cell tumor with SFPQ/PSF-TFE3 gene fusion in a patient with advanced neuroblastoma. Am J Surg Pathol. 2009;33:1416–1420. doi: 10.1097/PAS.0b013e3181a9cd6c. [DOI] [PubMed] [Google Scholar]
- 34.Zhong M, De Angelo P, Osborne L, et al. Dual-color, break-apart FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of Xp11 translocation renal cell carcinoma and alveolar soft part sarcoma. Am J Surg Pathol. 2010;34:757–766. doi: 10.1097/PAS.0b013e3181dd577e. [DOI] [PMC free article] [PubMed] [Google Scholar]