Background: Pih1 is an unstable protein and forms an R2TP complex with Rvb1, Rvb2, and Tah1.
Results: Pih1 contains two intrinsically disordered regions that mediate different protein-protein interactions within R2TP complex.
Conclusion: Pih1 contains an N-terminal Rvb1/Rvb2-binding domain and a C-terminal regulatory domain.
Significance: The study provides important insights into the mechanism of intrinsically disordered proteins in protein complex formation.
Keywords: Molecular Chaperone, Protein Assembly, Protein Degradation, Protein Domains, Protein Folding, Protein Stability, Protein Turnover, rRNA Processing, Yeast Genetics
Abstract
Pih1 is an unstable protein and a subunit of the R2TP complex that, in yeast Saccharomyces cerevisiae, also contains the helicases Rvb1, Rvb2, and the Hsp90 cofactor Tah1. Pih1 and the R2TP complex are required for the box C/D small nucleolar ribonucleoprotein (snoRNP) assembly and ribosomal RNA processing. Purified Pih1 tends to aggregate in vitro. Molecular chaperone Hsp90 and its cochaperone Tah1 are required for the stability of Pih1 in vivo. We had shown earlier that the C terminus of Pih1 destabilizes the protein and that the C terminus of Tah1 binds to the Pih1 C terminus to form a stable complex. Here, we analyzed the secondary structure of the Pih1 C terminus and identified two intrinsically disordered regions and five hydrophobic clusters. Site-directed mutagenesis indicated that one predicted intrinsically disordered region IDR2 is involved in Tah1 binding, and that the C terminus of Pih1 contains multiple destabilization or degron elements. Additionally, the Pih1 N-terminal domain, Pih11–230, was found to be able to complement the physiological role of full-length Pih1 at 37 °C. Pih11–230 as well as a shorter Pih1 N-terminal fragment Pih11–195 is able to bind Rvb1/Rvb2 heterocomplex. However, the sequence between the two disordered regions in Pih1 significantly enhances the Pih1 N-terminal domain binding to Rvb1/Rvb2. Based on these data, a model of protein-protein interactions within the R2TP complex is proposed.
Introduction
Pih1,4 protein interacting with Hsp90, is a highly conserved protein in eukaryotes. It was initially identified to interact with Hsp90 in our yeast two hybrid screen for Hsp90 interactors and has been shown to form a complex with Rvb1 and Rvb2 helicases as well as a tetratricopeptide repeat (TPR)-containing protein Tah1 (1); the complex has been named the R2TP complex. In yeast Saccharomyces cerevisiae, Pih1 consists of 344 amino acids and contains what is now known as the Pih1 domain (residues Ile32–Gly166) (2). Tah1 is the smallest known protein (111 amino acids) that contains a functional TPR domain, which is composed of only two TPR motifs and a long helix that is sufficient to bind the C terminus of Hsp90 (3, 4). Rvb1 and Rvb2 are two highly conserved AAA+ family helicases that form a heterohexamer (5). They are also part of many critical complexes such as SWR-C and Ino80 inside the cell (5, 6).
Pih1, also known as Nop17, is mainly localized in the nucleolus under normal growth conditions (7), suggesting a major role in pre-rRNA processing (3). The R2TP complex originally identified in baking yeast (1) is also conserved in human cells (8). The complex has been shown to be required for the proper assembly of box C/D snoRNPs (3, 9), for the assembly of RNA polymerase II complex (10), and for the stability of the phosphatidylinositol 3-kinase-related kinases (PIKKs) (11). Furthermore, both human Pih1 (PIH1D1) and Tah1 (RPAP3) in the R2TP complex have been shown to regulate doxorubicin-induced apoptosis (12, 13). The human Pih1 is also shown to be associated with histone H4 and is involved in the recovery from nutrient starvation (14). In addition to S. cerevisiae and human cells, Pih1 homologues have been identified in other animals such as mouse, Drosophila, and other fungi species such as Torulaspora delbrueckii, Ashbya gossypii, and Candida albicans. However, it seems that there is no close Pih1 homologue in higher plant Arabidopsis or fission yeast Schizosaccharomyces pombe.
Despite many important roles for R2TP complex in vivo (8), neither Pih1 nor Tah1 is essential for yeast cell growth. However, deletion of Pih1 in yeast cells causes slight temperature sensitivity in certain genetic backgrounds (7). We have previously shown that in the R2TP complex, Pih1 bridges the interaction between Rvb1/Rvb2 heterohexamer complex and Tah1, and that the association of Pih1 with Rvb1/Rvb2 is independent of Tah1 (3). Additionally, Pih1 protein is very unstable when expressed and purified from Escherichia coli and is prone to degradation in vivo in yeast when Tah1 is depleted (3). Therefore, it is likely that one role of Pih1 and Tah1 is to regulate the essential Rvb1/Rvb2 complex activity that is required for a variety of cellular processes (6). We proposed that, Hsp90 with its cochaperone Tah1 stabilizes Pih1 and promotes the formation of Rvb1-Rvb2-Tah1-Pih1 (R2TP) complex (3). However, the mechanism of action of Pih1 and Tah1 in regulating Rvb1/Rvb2 function in vivo is still unclear.
Pih1 C-terminal 145 amino acid fragment Pih1199–344 is able to bind Tah1 in a 1:1 stoichiometry as shown by size exclusion chromatography and mass spectrometry (15). Our recent study showed that even the last 115 amino acid fragment Pih1231–344 is sufficient to bind Tah1 and that the binding involves the Tah1 C terminus (4). Interestingly, the recently solved NMR structure of Tah1 revealed only five helices from the Tah1 N terminus, which are enough for binding the MEEVD motif at the Hsp90 C terminus, and a very short Tah1 C-terminal disordered region (4). It is unknown how the Tah1 C terminus binds and stabilizes Pih1 with the aid of Hsp90 in vivo. To investigate Pih1 stability and the effect of Tah1 on Pih1 stability, we constructed Pih1 truncation and point mutants and analyzed their roles in vivo. We found that the Pih1 N-terminal fragments Pih11–230 and Pih11–195 are able to bind the Rvb1/Rvb2 complex. But a small fragment beyond Pih11–230 significantly enhances the binding of Pih1 to Rvb1/Rvb2. Additionally, we show that the C-terminal fragment of Pih1 contains two intrinsically disordered regions and multiple destabilization factors, which likely form degron elements (16) to confer proteolysis activity and are protected by Tah1 under normal conditions. Based on this study, a model of how Tah1 binds and controls the exposure of Pih1 degron and thus controls the R2TP complex integrity and function is proposed.
EXPERIMENTAL PROCEDURES
Bioinformatics Analysis
The potential proline, glutamic acid, serine, and threonine enriched PEST motif that is involved in rapid in vivo proteolysis (17) in Pih1 sequence was identified by ePESTfinder. The hydrophobic amino acid clusters were predicted by hydropathy plot using Kyte-Doolittle hydropathy index (18). Amino acids that have an average hydrophobicity score greater than 1.6 with a window size of 5 amino acids were considered as real hydrophobic amino acid clusters. The intrinsically disordered regions within Pih1 were analyzed using IUPred (19) with the prediction of long disordered sequences and using a meta-predictor PONDR-FIT (20). The scores of disordered tendency can be between 0 and 1. The continuous sequence fragment with all scores above 0.5 was considered as a disordered region.
Plasmid Construction
The yeast expression plasmids for Pih1 and Pih1 truncation mutants Pih11–230, Pih11–284 were constructed by PCR amplification of the genes from yeast genome and insertion into the expression plasmid p416GPD that contains a constitutive glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter (21). GFP-Pih1282–344 fusion protein construct in p416ADH vector was previously described (4). The GFP-Pih1282–344 fusion protein used in this study was constructed by cloning the GFP-Pih1282–344 cassette into SmaI/HindIII sites of p415GPD vector that contains a LEU2 selection marker gene (21). GFP fusion construct GFP-Pih1231–344 was constructed based on GFP-Pih1282–344 with Pih1282–344 replaced by Pih1231–344. GFP-Pih1282–344HP2M, GFP-Pih1282–344HP3M, GFP-Pih1282–344HP4M, and GFP-Pih1282–344HP5M are similar to GFP-Pih1282–344 except that the four predicted hydrophobic amino acid clusters 307ILYIN311, 314LSIPL318, 322IVVNA326, and 340LYIYI344 were mutated to DDRSN, RSDPL, DRSNA, and LDRDR, respectively, using Stratagene QuickChangeTM Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA). Similarly, GFP-Pih1231–344HP1M was constructed from GFP-Pih1231–344 with the hydrophobic cluster 281LRILI285 mutated to LRDRS. All constructs were confirmed by sequencing.
Yeast Strains
To generate C-terminal 3xFLAG tagged Pih1, Pih11–248, Pih11–230, or Pih11–195 strains, yeast S. cerevisiae strain (W303 background, MATa, leu2–3,112 trp1–1 can1–100 ura3–1 ade2–1 his3–11,15) was used, and PCR-based gene tagging procedures (22) were used with the plasmid pFA6a-3FLAGkanMX6 as template (3). The 3×FLAG-tagged Pih1 or Pih1 C-terminal truncation mutant complexes were purified using anti-FLAG antibody resin (A2220, Sigma-Aldrich) according to established procedure as previously described (23).
Yeast Dilution Assay
Pih1 and Pih1 truncation mutants in p415GPD were transformed into pih1Δ cells (S288C genetic background, MATa leu2 his3 lys2 can1::STE2pr-Sp_his5 pih1Δ::NAT). The transformed cells were grown in synthetic dropout medium without supplement of leucine (SD-Leu). The overnight cultures were diluted to the optical absorbance density of 2.0 measured at 600 nm. Then the cultures were 10-fold diluted and 10 μl of each culture was inoculated onto SD-Leu selection medium plates using replica pin tool (V&P Scientific, Inc), and cells were grown for 36 h.
Circular Dichroism Measurements
CD spectra were obtained using a Jasco J-810 spectropolarimeter equipped with a Peltier temperature control device. Each spectrum was collected by averaging the signal at every 1 nm for 1 s. The protein concentration was 0.2 mg/ml in 10 mm NaH2PO4, pH 7.5. A 1-mm path length cuvette was used, and the temperature was set at 25 °C.
Size Exclusion Chromatography
His6-tagged Pih11–195, Pih11–230, or Pih11–248 was expressed in p11 vector and purified according to Jimenez et al. (4). His6-tagged Rvb1 and Rvb2 were expressed and purified as described previously (3). Then the proteins were cleaved by tobacco etch virus protease and the untagged Rvb1 and Rvb2 proteins were further purified by passing through Ni-NTA column to remove cleaved His6 tag. The concentrations of purified proteins were determined using the Bradford assay (24). Size exclusion chromatography was performed using a calibrated Superdex 200 10/30 column attached to an ÄKTA FPLC system (GE Healthcare). The column was equilibrated with buffer containing 25 mm Tris-HCl, pH 7.5, 100 mm KCl, 10% glycerol, and 1 mm DTT. To test the interaction between Pih11–230 and Rvb1/Rvb2 complex, equal molar concentrations of Rvb1 and Rvb2 were mixed and a total of 100 μg of Rvb1/Rvb2 complex was mixed with 250 μg of Pih11–230 and loaded onto the Superdex 200 column. Molecular mass standards used were purchased from Sigma or Bio-Rad: thyroglobulin (669 kDa), apoferritin (443 kDa), α-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), ovalbumin (44 kDa), carbonic anhydrase (29 kDa), myoglobin (17 kDa), cytochrome c (12.4 kDa), aprotinin (6.5 kDa), and vitamin B12 (1.4 kDa).
In Vivo Analysis of Protein Stability
To test the stability of Pih1, Pih1 truncation mutants or GFP fusion proteins in vivo, plasmids expressing these constructs were transformed into yeast S. cerevisiae strain in W303 or in S288C background. Cells were grown to mid-log phase and then cycloheximide was added to the culture at a final concentration of 50 μg/ml to inhibit translation. Equal volumes of cell cultures were then withdrawn at different time points and lysed. Proteins were separated on 12% SDS-PAGE gels followed by immunoblotting using anti-GFP antibody (Sigma G1544), anti-Pih1 antibody (3), or anti-Hsp82 antibody (1).
In Vitro Pulldown Assays
His6-tagged Tah1 protein was expressed and purified as previously described (3). To test interactions between Tah1 and different GFP fusion proteins, yeast cells expressing the GFP fusion proteins were lysed and incubated with 100 μg of His6-tagged Tah1 and 50 μl of Ni-NTA resin for 1 h. The Ni-NTA resin was washed four times using a buffer containing 25 mm Tris-HCl, 150 mm NaCl, 25 mm imidazole, 10% glycerol, and 0.5 mm DTT, and the bound proteins in the Ni-NTA resin were resolved on 12% SDS-PAGE and immunoblotted with anti-GFP antibody.
RESULTS
The C-Terminal Fragment of Pih1 Contains Intrinsically Disordered Regions and a PEST Motif
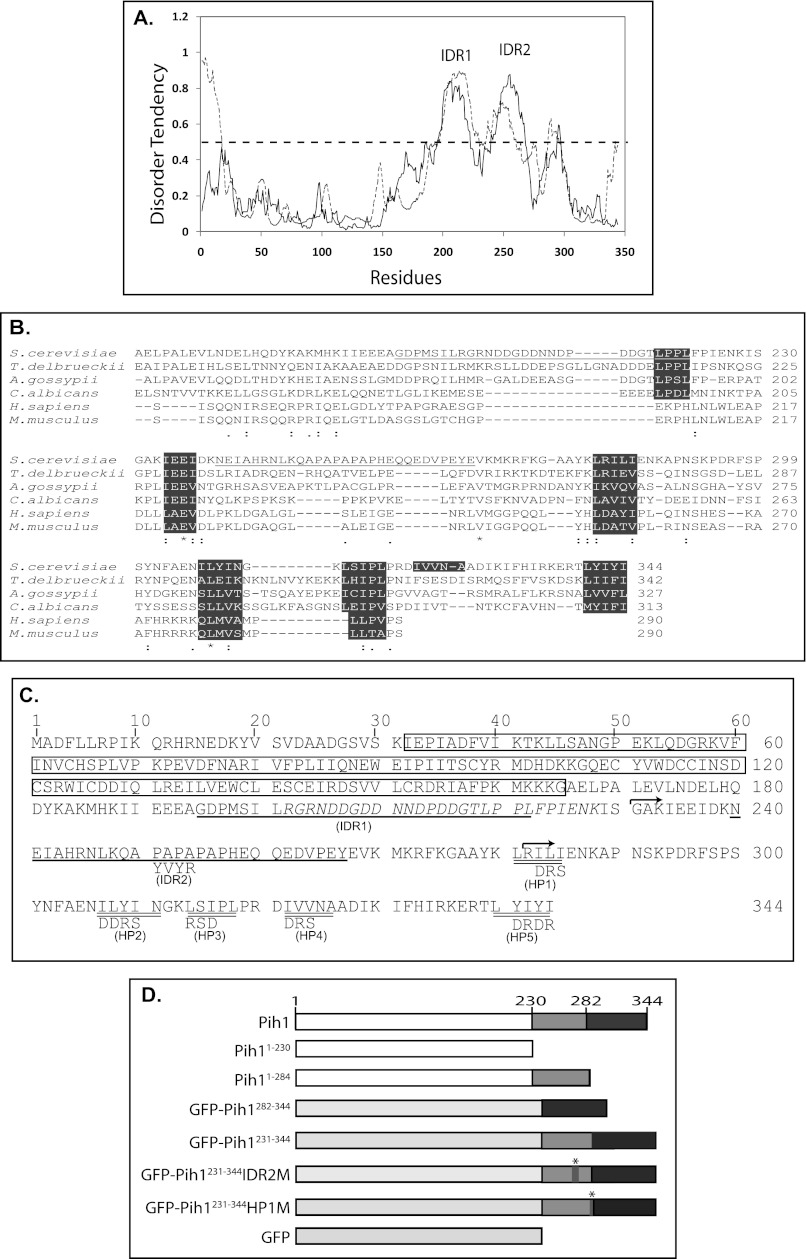
Heat shock protein Hsp90 and its cochaperone Tah1 work together to stabilize Pih1 in yeast cells. We observed that Pih1 is degraded rapidly in vivo if its binding partner Tah1 is depleted (3). To further understand the role of Pih1 in the formation of the R2TP complex and its regulatory role in the R2TP complex integrity, we first performed bioinformatics analysis on the Pih1 primary sequence. A search for intrinsically disordered regions in Pih1 shows that Pih1 C terminus consistently contains two long intrinsically disordered regions as predicted using both IUPred (19) and PONDR-FIT (20) programs (Fig. 1A). Since the two intrinsically disordered regions predicted by IUPred are well separated (spanning amino acids Gly195–Leu222 and Asn240–Tyr267), they are named specifically as IDR1 and IDR2 in this study (underlined in Fig. 1, B and C). Intrinsically disordered sequences are usually involved in mediating protein-protein interactions (25, 26) and they might become structured when associated with binding partners. Therefore, the two intrinsically disordered regions may be important for interaction with Rvb1/Rvb2 complex and/or Tah1 in forming the R2TP complex (3). Because Pih1 orthologues have also been identified in other species (8), to examine if the identified disordered regions are conserved, we analyzed the Pih1 orthologues from human, mouse and three other yeast species. Interestingly, all analyzed proteins show higher disorder tendency in a region corresponding to the IDR1 region in Pih1 (data not shown), although their primary sequences do not show high similarity until the very end of the IDR1 region, where there is a highly conserved LPXL motif in four analyzed yeast species (Fig. 1B). The second disordered region IDR2 is not well conserved, but it follows a conserved region ΦXEΦ, where Φ represents a hydrophobic amino acid (Fig. 1B). IDR2 also contains a proline-rich APAPAPAP cluster. It may form a unique structure for the Pih1 C terminus since it does not appear in any other Pih1 homologues. Previous in vitro size exclusion chromatography assays have shown that Pih1231–344, which only contains the second intrinsically disordered region, is sufficient to bind Tah1 (4), suggesting that IDR1 is not involved in binding Tah1.
FIGURE 1.
Bioinformatics analysis of Pih1. A, disorder tendency of Pih1 protein. The disorder tendency was analyzed using IUPred (19) (solid line), and a meta predictor PONDR-FIT (20) (dashed line). The two major peaks as predicated by IUPred representing the intrinsically disordered regions IDR1 and IDR2 are labeled. B, Pih1 and its five orthologues from T. delbrueckii (XP_003681764.1), A. gossypii (NP_982480.1), C. albicans (XP_719405.1), H. sapiens (NP_060386.1), and M. musculus (AAH68254.1) were aligned using CLUSTALW program. Only sequences corresponding to Pih1 C-terminal fragment are shown. Identical amino acids are indicated by *, IDR2 in Pih1 are underlined. The conserved LPXL and ΦXEΦ motifs, and the five hydrophobic amino acid clusters are highlighted. C, Pih1 primary amino acid sequence and predicted features. The predicted PEST sequence (Arg204–Lys228) is shown in italics. The predicted intrinsically long disordered regions IDR1 and IDR2 as shown in A are singly underlined, and hydrophobic amino acid clusters (HP1 to HP5) at the Pih1 C terminus are doubly underlined. The start amino acids of the two Pih1 C-terminal fragments Pih1231–344 and Pih1282–344 that were used to construct GFP fusion proteins are indicated by arrows. The mutant amino acids are shown under WT amino acids. The predicted Pih1 domain from the conserved domain database (2) is highlighted in the rectangle box. D, schematic diagrams of Pih1 and Pih1 truncation mutants Pih11–230 and Pih11–284 as well as the GFP fusion proteins used in this study. GFP-Pih1231–344 represents the GFP fusion protein in which Pih1231–344 is fused to the C terminus of GFP. GFP-Pih1282–344 represents the GFP fusion protein in which Pih1282–344 is fused to the C terminus of GFP. GFP-Pih1231–344IDR2M and GFP-Pih1231–344HP1M are similar to GFP-Pih1231–344 except that the predicted IDR2 and HP1 were mutated as shown in B. The * in GFP-Pih1231–344IDR2M and GFP-Pih1231–344HP1M represent the positions of the mutation.
Pih1 is also predicted to contain a proline (P)-, glutamic acid (E)-, serine (S)-, and threonine (T)-rich PEST (17) motif from Arg204 to Lys228. The PEST motif partially overlaps with the first intrinsically disordered region IDR1 (Fig. 1C). A PEST motif is defined as hydrophilic stretches of at least 12 amino acids long with a high local concentration of critical amino acids such as those that are negatively charged. PEST sequence has been well recognized as a destabilization factor for many short-lived proteins such as fos, ornithine decarboxylase, and IκBα (27) and it has been hypothesized that the PEST sequence serves as a degradation targeting signal and causes rapid protein degradation in vivo (17). It is likely that the identified PEST motif plays a role in the regulation of Pih1 turnover in vivo. Interestingly, the mouse Pih1 orthologue is also predicted to contain a potential PEST motif, but at the N terminus (data not shown). Moreover, it is noticed that Pih1 C-terminal fragment contains five hydrophobic amino acid clusters, designated as HP1, HP2, HP3, HP4, and HP5 respectively (Fig. 1, B and C), which may form hydrophobic patches (28) in the Pih1 C terminus. Alignment with other Pih1 homologues shows that hydrophobic region HP1 HP2, HP3 are conserved between the yeast and mammalian homologues and the C-terminal HP5 is highly conserved in all four analyzed yeast species (Fig. 1B).
Pih1 C-terminal Domain Is Dispensable for Pih1 in Vivo Function
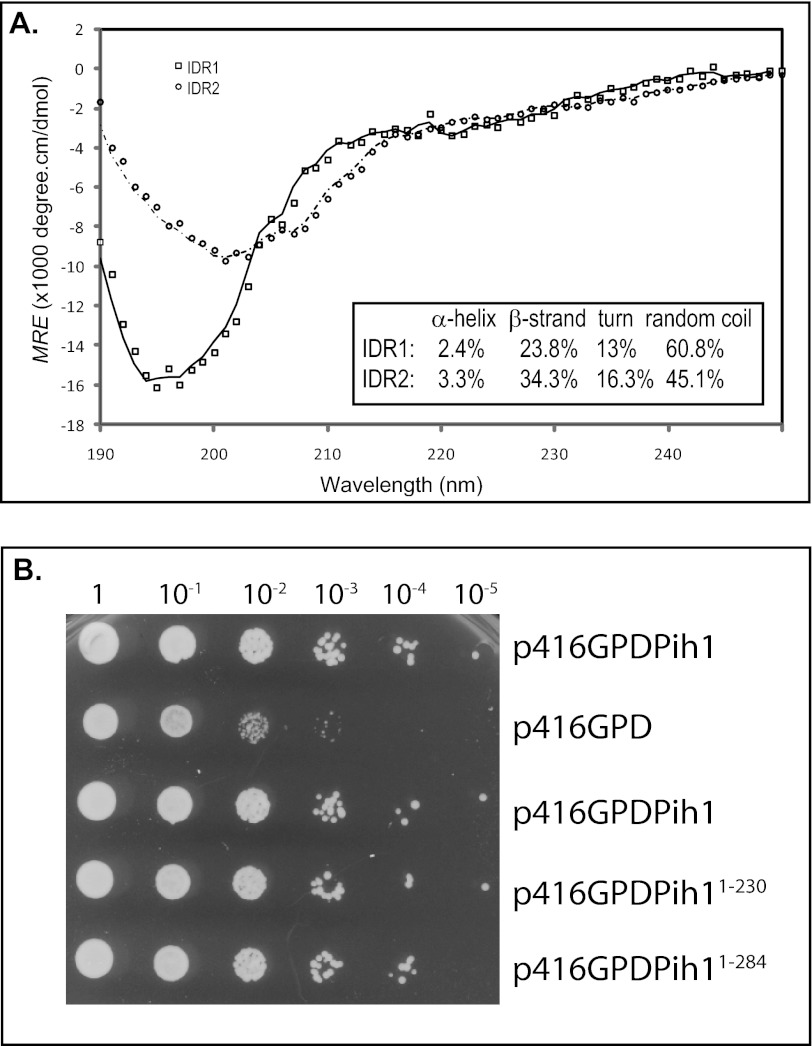
To analyze the role of the two predicted intrinsically disordered regions, we first tried to experimentally analyze the possible secondary structure of the two disordered regions. We cloned the two IDR regions, expressed and purified the two His6-tagged disordered fragments from E. coli. Circular dichroism spectra of the two isolated fragments were measured and analyzed using CONTINLL program (29), and it was shown that IDR1 and IDR2 contain almost no α-helices, but as high as 61 and 45% disordered regions respectively (Fig. 2A), suggesting that IDR1 and IDR2 are indeed disordered. Additionally, we made Pih1 truncation mutants that contain either one (Pih11–230) or two (Pih11–284) intrinsically disordered regions (Fig. 1D) and introduced these Pih1 truncation mutants into yeast cells that have the endogenous PIH1 gene deleted. It was shown that pih1Δ mutant displays a slightly temperature sensitive phenotype, particularly for yeast cells in S288C background (7). When grown at 22 °C for 36 h, pih1Δ does not show any significant growth defect (data not shown). However, when grown at 37 °C, pih1Δ cells show a clear growth defect as shown in Fig. 2B. Interestingly, similar to the full-length Pih1, both Pih11–230 and Pih11–284 are able to complement endogenous Pih1 at 37 °C. This suggests that the Pih1 C-terminal fragment Pih1231–344 is not essential for Pih1 function under these growth conditions, and that the intrinsically disordered region Asn240–Tyr267, the IDR2 fragment, is not essential for Pih1 in vivo function under the tested conditions.
FIGURE 2.
Circular dichroism spectra and in vivo function of the two disordered regions. A, far-ultraviolet CD spectra at 25 °C of His6-tagged IDR1 (solid line) and His6-tagged IDR2 (dashed line). The CD signal is expressed as the mean residue ellipticity (MRE). The calculated percentages of α-helix, β-strand, and random coil are shown in the inset. B, full-length Pih1 and Pih1 truncation mutants were expressed from GPD promoter (indicated as p416GPDPih1, p416GPDPih11–230, and p416GPDPih11–284 respectively) in yeast cells that have endogenous PIH1 gene deleted. The yeast cells carrying Pih1 expression plasmids or the empty vector (p416GPD) were grown on medium without supplement of uracil for 36 h at 37 °C. The optical density of the starting cultures and 10× dilutions are indicated on the top.
The Pih1 C-terminal Fragment Pih1231–344 Contains Degron Elements That Are Normally Blocked by Tah1
To understand the function of Pih1 C-terminal fragment Pih1231–344 in vivo, we constructed a green fluorescent protein (GFP) fusion protein in which Pih1231–344 was fused to the GFP C terminus (Fig. 1D) and GFP-Pih1282–344 (4) construct was used as a control. Additionally, to determine the role of IDR2 in mediating Tah1 binding, we disrupted the APAPAPAP cluster in IDR2 and mutated it to APYVYRAP (Fig. 1C). As a result, the mutant sequence, designated as Pih1231–344IDR2M (Fig. 1D) does not contain significantly long intrinsically disordered regions as predicted using IUPred (19). Analysis of the mutated sequence by a secondary structure prediction program GOR IV (30) indicated that a short extended strand is likely generated in the original disordered AP cluster region (data not shown). Moreover, we mutated the hydrophobic amino acid cluster HP1 from LRILI to LRDRS (Fig. 1C), which is not fully included in the GFP-Pih1282–344, resulting in a construct GFP-Pih1231–344HP1M (Fig. 1D), and tested if the first hydrophobic amino acid cluster is particularly involved in Pih1 C-terminal stability and Tah1 binding.
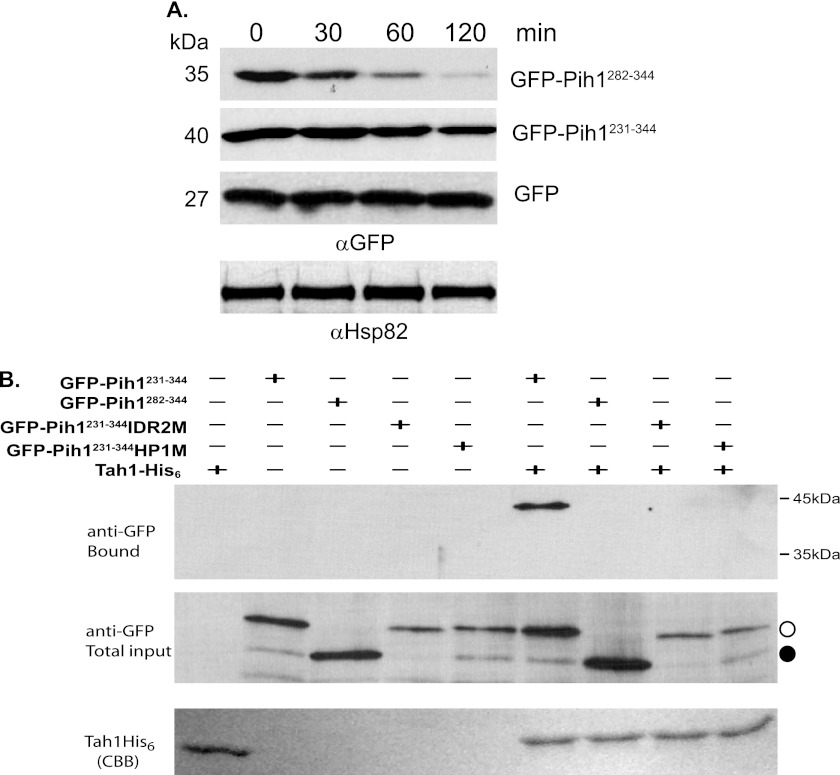
As reported in our previous study (4), the Pih1 C-terminal 63-amino acid sequence, Pih1282–344 is able to induce rapid GFP degradation as also shown on the top panel of Fig. 3A. Interestingly, the longer Pih1 C-terminal fragment Pih1231–344, which contains IDR2, resulted in a slower degradation rate of the fused GFP (second panel from the top in Fig. 3A). Since Hsp90 cochaperone Tah1 binds to Pih1231–344 (4), it is likely that, although fused to GFP, Pih1231–344 fragment can still bind Tah1 and that the GFP-Pih1231–344 is protected from degradation by the Hsp90/Tah1 chaperone system. To test this possibility, we performed an in vitro pulldown assay to test if Tah1 is able to bind GFP-Pih1231–344 using His6-tagged Tah1 and yeast lysates expressing GFP-Pih1231–344, GFP-Pih1282–344, GFP-Pih1231–344 IDR2M, or GFP-Pih1231–344HP1M. As shown in Fig. 3B, Tah1 is indeed able to pulldown GFP-Pih1231–344, but not GFP-Pih1282–344, which contains only a short Pih1 C terminus. Interestingly, the in vitro pulldown assay indicates that Tah1 does not bind GFP-Pih1231–344 if either IDR2 or the hydrophobic amino acid cluster HP1 is mutated (Fig. 3B). This suggests that both IDR2 and HP1 are involved in Tah1 binding.
FIGURE 3.
Degradation of GFP Pih1 fusion proteins in vivo. A, degradation of GFP fusion proteins in yeast strain W303 after inhibition of protein translation using cycloheximide at 30 °C. Immunoblotting using anti-Hsp82 was used as an internal control. B, in vitro pulldown assays using His6-tagged Tah1 purified as bait from yeast cell lysates that express GFP-Pih1231–344, GFP-Pih1282–344, GFP-Pih1231–344IDR2M, or GFP-Pih1231–344HP1M. The top and middle panels are immunoblotting using anti-GFP antibody for the bound and total input fractions. The bottom panel shows Tah1 proteins bound to Ni-NTA. The open circle indicates the position of GFP-Pih1231–344 and its mutants. The closed circle indicates GFP-Pih1282–344.
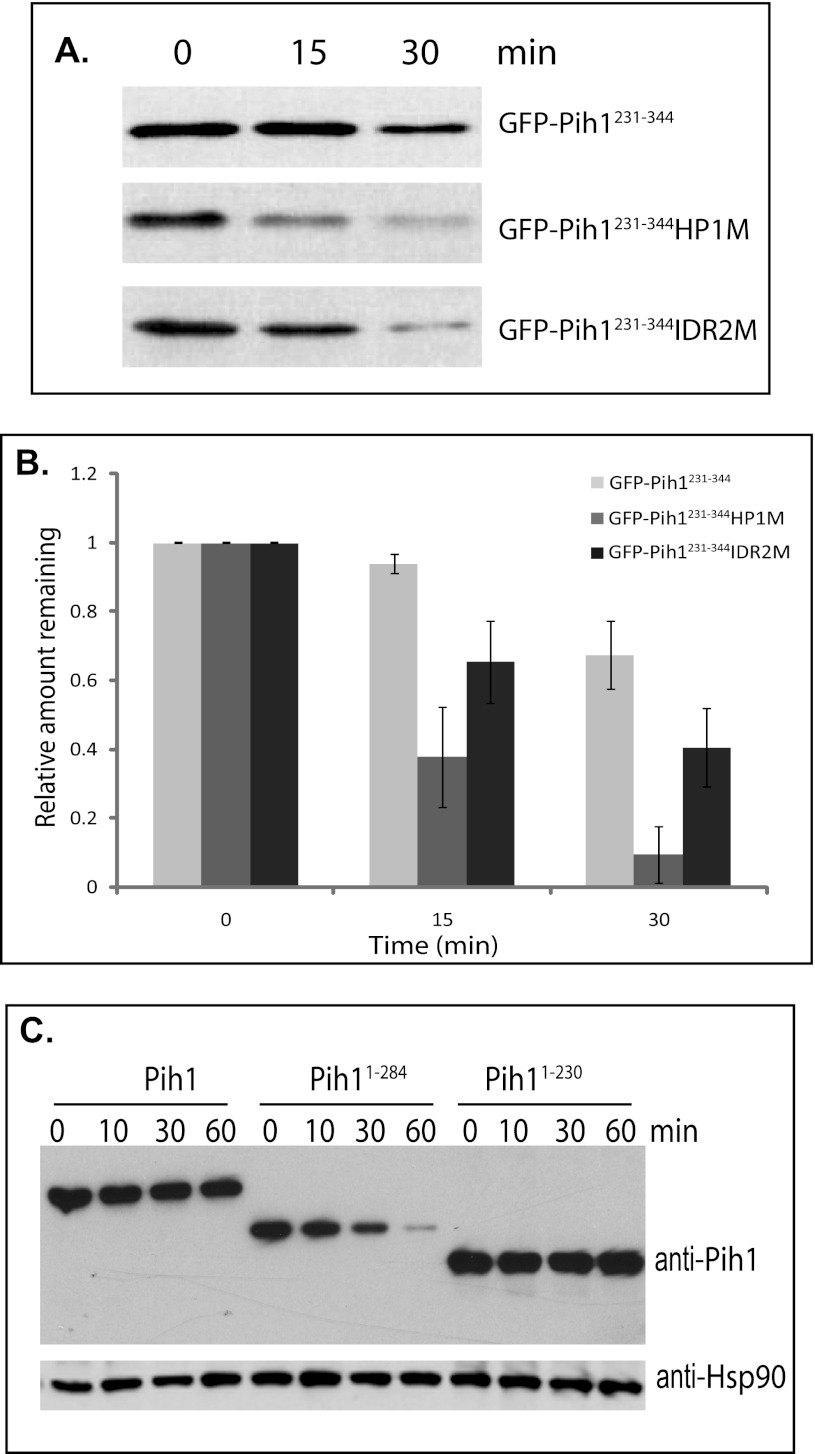
To examine the mechanism by which the Pih1 C terminus mediates Pih1 degradation, we tested if the two mutated Pih1231–344 fragments, GFP-Pih1231–344HP1M and GFP-Pih1231–344IDR2M, which have the hydrophobic amino acid cluster HP1 and the second predicted disordered region mutated, respectively, are able to cause faster GFP degradation since they are no longer able to bind Tah1. As shown in Fig. 4A, the mutations do cause rapid GFP protein degradation. Quantitative analyses indicated that mutation in the first hydrophobic region HP1 causes significantly faster GFP degradation than that in IDR2 region (Fig. 4B). This implies that, although both IDR2 and HP1 regions are required for Tah1 binding, amino acids surrounding HP1 region may play a more important role for Pih1 to bind Tah1.
FIGURE 4.
Degradation of GFP Pih1231–344 fusion proteins and Pih1 truncation mutants in vivo. A, immunoblotting using anti-GFP antibody of GFP-Pih1231–344 fusion protein and its mutants expressed in yeast cells after inhibition of protein translation. B, quantitative analyses of GFP fusion protein degradation. The immunoblot was scanned and analyzed using Quantity One™ software (Bio-Rad) and the relative signal remaining was calculated by using the signal at 0 min as 100%. The averages and standard errors shown were from at least three independent degradation assays within 30 min following inhibition of protein translation with cycloheximide. C, immunoblotting of Pih1 and Pih1 truncation mutants Pih11–284 and Pih11–230 expressed in yeast cells which have their endogenous PIH1 genes deleted after inhibition of protein translation using cycloheximide. Immunoblotting using anti-Hsp82 was performed to detect Hsp90 expression and used as an internal control.
Interestingly, the sequence surrounding HP1 region shows a relatively low score in the disorder tendency profile (Fig. 1A). This suggests that the region surrounding HP1 may be important for a stable structure formation in the Pih1 C terminus. We have also shown that the Pih1 C-terminal truncation mutant Pih11–284 is able to complement the full-length of Pih1 in vivo as well as the shorter Pih1 C-terminal truncation mutant Pih11–230 (Fig. 2B). Pih11–284 ends at Leu284, which is actually in the middle of the HP1 (Fig. 1C). To examine how endogenous Pih1 is complemented by its C-terminal truncation mutants, the expression and degradation of Pih11–284 and Pih11–230 in vivo were tested in yeast cells that have the endogenous PIH1 gene deleted. As shown in Fig. 4C, in the presence of cycloheximide, the full-length Pih1 is not significantly degraded. This is reasonable since the yeast cells used in this assay contain both Tah1 and Hsp90 that stabilize Pih1 in vivo. Surprisingly, Pih11–230, which contains the first intrinsically disordered region and the PEST motif (Fig. 1C), is not degraded significantly either. However, Pih11–284 is significantly degraded within 60 min. These data suggest that the intrinsically disordered region IDR2 is involved in Pih1 instability in vivo and truncation mutant Pih11–284 exposes hydrophobic regions. Taken together with the mutation analysis around HP1 region (Fig. 4A), it is likely that HP1 region is involved in a stable structure formation and that disruption of this region by site directed mutagenesis or C-terminal truncation at Leu284 exposes degradation signals, thus causing fast degradation of both GFP-Pih1231–344HP1M and Pih11–284.
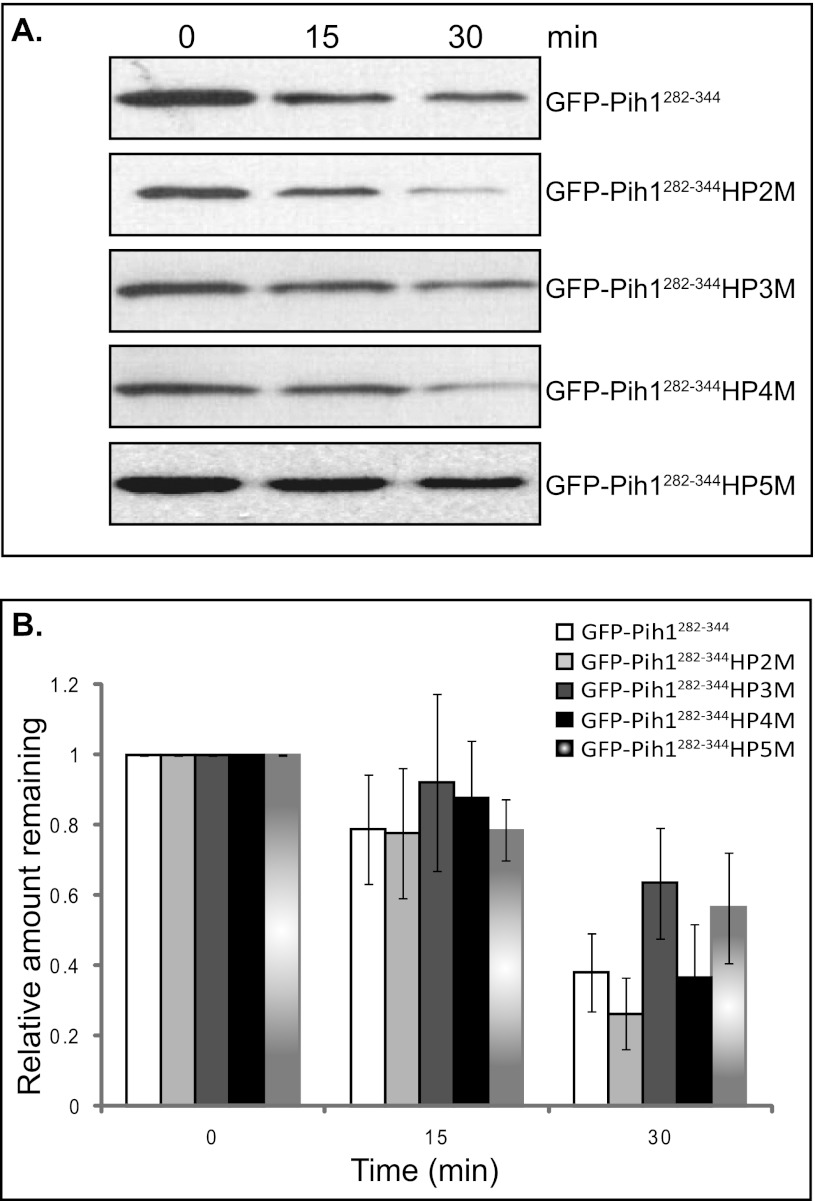
To further elucidate the role of hydrophobic amino acid clusters in the Pih1 C terminus, we mutated the other four C-terminal hydrophobic amino acid clusters, namely HP2 HP3, HP4, and HP5 to charged amino acids to lower their hydrophobicities (Fig. 1C), and then compared their abilities to cause GFP degradation in vivo. After inhibition of protein translation with cycloheximide, the degradation of GFP fusion proteins was followed for 30 min. As shown in Fig. 5A and the corresponding quantitative analyses in Fig. 5B, mutations in HP2 and HP4 regions do not significantly change the stability of GFP fusion proteins, while mutations in HP3 and HP5 seem to slow down the degradation of GFP fusion proteins. These results suggest that the hydrophobic regions have different roles in forming the Pih1 C terminus structure and in contributing to the degradation signal in Pih1 C terminus. Hydrophobic regions HP3 and HP5 seem to have more important roles than the other two in exposing degradation signals.
FIGURE 5.
Degradation of GFP-Pih1282–344 fusion protein and its mutants in vivo. A, immunoblotting of GFP-Pih1282–344 fusion protein and its mutants expressed in yeast cells after inhibition of protein translation. B, quantitative analyses of GFP fusion protein degradation. The immunoblot was scanned and analyzed using Quantity One™ software (Bio-Rad) and the relative signal remaining was calculated by using the signal at 0 min as 100%. The averages and standard errors shown were from three independent degradation assays within 30 min following inhibition of protein translation with cycloheximide.
Intrinsically Disordered Region IDR1 and Surrounding Region Are Essential for Pih1 Association with Rvb1/Rvb2 Heterocomplex
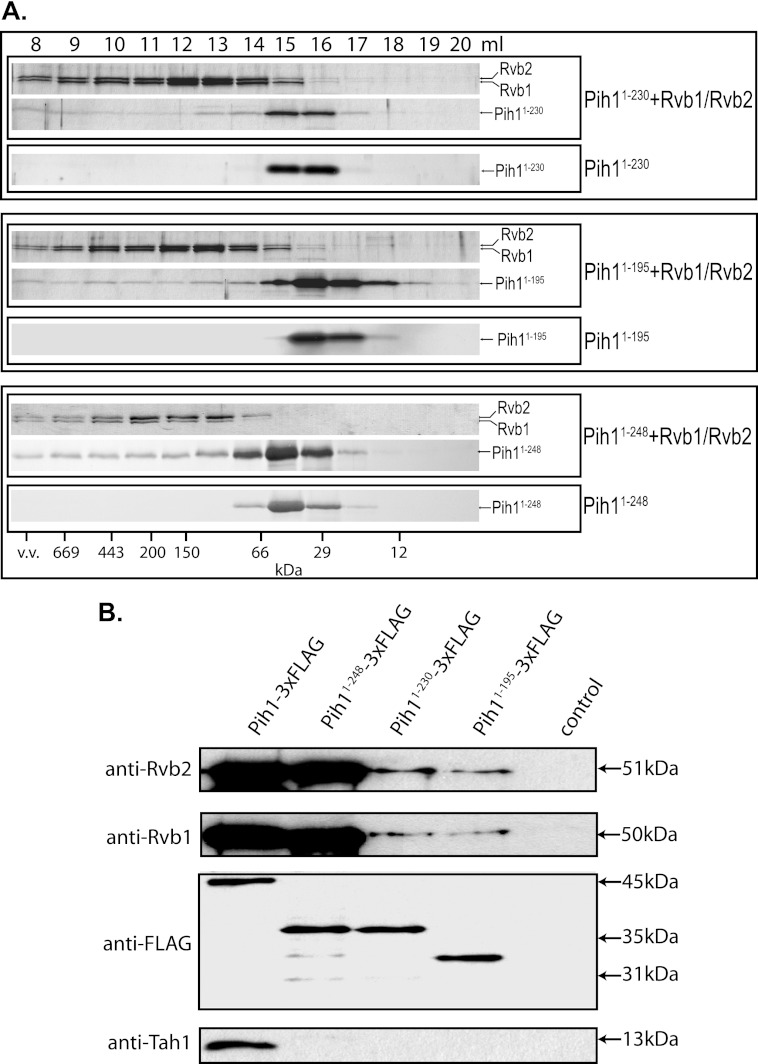
Based on the aforementioned studies (Fig. 4C) and our previous work (4), Pih11–230 seems to form a stable fold and is very stable when expressed in yeast cells. However, it is predicted that Pih11–230 contains an intrinsically disordered region IDR1 and a putative PEST motif (Fig. 1C), which are generally thought to cause protein degradation. It is likely that, similar to IDR2 that is required to bind Tah1, the region around IDR1 and the PEST motif mediates protein-protein interaction and is protected by other proteins in vivo. Since Pih1 forms an R2TP complex with Tah1 and the two DNA helicases Rvb1 and Rvb2 (3), we hypothesize that IDR1 is essential in binding Rvb1/Rvb2 heterocomplex in vivo. To test this hypothesis, we first determined if purified Pih11–230, which is lacking the long C-terminal fragment Pih1231–344, is able to bind Rvb1/Rvb2 complex using size exclusion chromatography. As shown on the top panel in Fig. 6A, Pih11–230 was co-eluted with preformed Rvb1/Rvb2 complex right after the void volume when using the Superdex 200 size exclusion column, while Pih11–230 alone was only observed in fractions corresponding to monomers. This indicates that Pih11–230 is able to interact with Rvb1/Rvb2 complex directly. It should be noted that in the size exclusion chromatography assay, 20 μm Pih11–230 was mixed with 6 μm Rvb1/Rvb2 heterocomplex and that there were excess amount of Pih11–230 in the analyzed mixtures. Therefore, the majority of Pih1 truncation mutants was eluted as a single peak and relatively only a small fraction of the Pih11–230 was bound by and co-eluted with Rvb1/Rvb2. The interaction between Rvb1/Rvb2 complex with Pih11–230 also supports the in vivo complementation results as shown in Fig. 2B. Surprisingly, Pih11–195, a further Pih1 C terminus truncation mutant that is lacking the IDR1 and the PEST motif, and expressed from a D196Stop mutation, however, is still able to bind Rvb1/Rvb2 complex as shown in the middle panel of Fig. 6A.
FIGURE 6.
Protein interactions between Rvb1/Rvb2 and Pih1. A, size exclusion chromatography of Pih11–230, Pih11–195, Pih11–248, Rvb1/Rvb2 complex, Pih11–230 + Rvb1/Rvb2 complex, Pih11–195 + Rvb1/Rvb2, or Pih11–248 + Rvb1/Rvb2 complex (20 μm respective Pih1 truncation mutant alone or mixed with 6 μm Rvb1/Rvb2 complex was applied). Samples were fractionated on a size exclusion column Superdex 200. Sample fractions were run on 15% SDS-PAGE and stained by Coomassie Brilliant Blue. B, immunoprecipitatin of Pih1 or Pih1 C-terminal truncation mutant complexes. 3×FLAG tagged Pih1, Pih11–248, Pih11–230, or Pih11–195 was expressed from the endogenous chromosomal PIH1 promoter and purified using anti-FLAG antibody resin. The purified samples were run on 15% SDS-PAGE, transferred to nitrocellulose membrane and immunoblotted using anti-Rvb2, anti-Rvb1, anti-FLAG, or anti-Tah1 antibodies. Control lane represents sample purified using wild type yeast lysate that does not contain any FLAG-tagged proteins.
Pih1 C-terminal truncation mutant Pih11–230 was chosen for analysis previously (3) because of the differences of predicted secondary structure before and after the S230 (data not shown). Actually, by examining Pih1 homologues from fungi and mammalian cells, E236 in Pih1 is highly conserved before the second intrinsically disordered region IDR2 (Fig. 1B). We therefore made a shorter Pih1 C terminus truncation mutant Pih11–248, which contains 18 more amino acids compared with Pih11–230 and retains the region surrounding the conserved amino acid E236. Similar to Pih11–230, Pih11–248 is able to bind Rvb1/Rvb2 heterocomplex (bottom panel in Fig. 6A). However, the difference of binding affinity to Rvb1/Rvb2 does not seem to be very significant as revealed by size exclusion chromatography. Since it is usually technically difficult to quantitatively analyze protein-protein interaction via size exclusion chromatography, to better analyze the binding affinity of Pih1 C-terminal truncation mutants to Rvb1/Rvb2, we constructed three different 3xFLAG tagged Pih1 C-terminal truncation mutants from the chromosomal copy of Pih1. The endogenous Pih1 or Pih1 truncation mutant complexes were purified using anti-FLAG antibody resin. As shown in Fig. 6B, immunoblotting of Rvb1 and Rvb2 proteins indicated that, although all four Pih1 proteins could pulldown Rvb1/Rvb2 complex, the full-length Pih1 and Pih11–248 immunoprecipitated much more Rvb1/Rvb2 complex and there is no significant difference between Pih1 and Pih11–248 in binding Rvb1/Rvb2. The immunoprecipitation results confirm the results of size exclusion chromatography shown in Fig. 6A and additionally indicate that, the region surrounding the conserved E236 in Pih1 is important in enhancing the Rvb1/Rvb2 binding, but not for Tah1 binding as immunoblotting did not detect any Tah1 protein in the Pih11–248 protein complex (bottom panel in Fig. 6B).
DISCUSSION
Proteins that contain intrinsically disordered regions are very common inside the cells and they often participate in many vital biological processes by mediating protein-protein interactions (26, 31). Pih1 is predicted to contain two major intrinsically disordered regions, IDR1 and IDR2 that surround Gly195–Leu227 and Asn240–Tyr267, respectively (Fig. 1, A and C). Based on the size exclusion chromatography results (Fig. 6A) and in vitro pulldown assays (Fig. 6B), it is reasonable to propose that, intrinsically disordered region IDR1 and the following conserved region are involved in association with Rvb1/Rvb2 complex, and that IDR2 is required for binding Tah1. A protein interaction model within the R2TP complex along Pih1 protein sequence is proposed and shown in Fig. 7. In this model, Rvb1/Rvb2 complex is interacting with Pih1 N terminus that includes IDR1 and the following region up to Lys248. Tah1 C terminus is interacting with Pih1 C terminus surrounding amino acid Leu284 and overlapping with the intrinsically disordered region IDR2, from Asn240–Tyr267, thus preventing the exposure of Pih1 C-terminal degrons. However, whether there is a distinct boundary between the Rvb1/Rvb2 interaction domain and the Tah1 interaction domain is still not clear since Pih11–248, which binds Rvb1/Rvb2 well, overlaps with the fragment Pih1231–344, which binds Tah1 in our study. Based on the prediction of the intrinsically disordered regions in Pih1 (Fig. 1A) and sequence alignment (Fig. 1B), it is also likely that the region that contains the conserved ΦXE236Φ between the two IDRs forms a distinct domain/motif that is required for Rvb1/Rvb2 binding. Further mutagenesis analyses within this region and/or crystallization of the Rvb1/Rvb2/Pih1 complex are required to elucidate the exact role of the ΦXE236Φ motif. Nevertheless, Pih11–248 is able to bind Rvb1/Rvb2, but not Tah1 (Fig. 6B), suggesting that sequences beyond Lys248 in Pih1 C terminus is more important for Tah1 binding.
FIGURE 7.
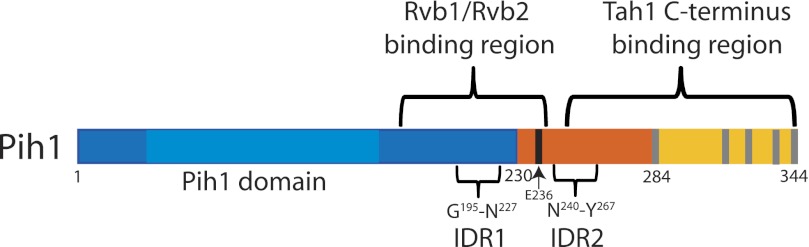
The diagram of Pih1 regions and motifs that are involved in R2TP complex formation. Pih1 is represented by a rectangle from amino acid 1 to amino acid 344. The Pih1 fragments used in this study were color coded as Pih11–230 (blue), Pih1231–284 (orange), and Pih1285–344 (yellow). It should be noted that fusion protein GFP-Pih1282–344 used in this study contains Pih1 fragment Pih1282–344 that starts two amino acids before Leu284. The two predicted intrinsically disordered regions IDR1 (Gly195–Leu227) and IDR2 (Asn240–Tyr267) are also highlighted. The five hydrophobic amino acids clusters in the Pih1 C terminus are indicated as gray bars, and the E236 is indicated as black bar. The overall sequence fragments required for Rvb1/Rvb2 and Tah1 bindings are indicated on the top of the Pih1.
We analyzed the role of the Pih1 C-terminal fragment in triggering protein degradation and determined the essential elements in the Pih1 C terminus that are required for Tah1 binding. Intrinsically disordered region IDR2 (particularly the unique APAPAP amino acid cluster) and the hydrophobic amino acid clusters HP1 are essential for Tah1 binding and when the Pih1 C-terminal fragment is not able to bind Tah1, it induces much faster degradation of GFP reporter protein as indicated in Fig. 3. Therefore, we propose that one major role of Tah1 is to block the exposure of the Pih1 degron elements, which are mainly conferred by IDR2 and hydrophobic amino acid clusters HP1, HP3, and HP5.
However, it should be noted that, although we propose that Tah1 binds directly to the Pih1 C-terminal intrinsically disordered region and blocks the degradation signals of Pih1 in the R2TP complex, we cannot rule out the role of Pih1 C-terminal fragment in binding other proteins in yeast cells. It has been reported that Pih1 is also able to interact with Rrp43 and Nop58 (7), Rsa1 (9), while human Pih1 homologue PIH1D1 interacts with TEL2 (11) and histone H4 (14). It is plausible to hypothesize that Pih1 in the R2TP complex may transiently interact with its target protein or protein complexes when R2TP is performing its regulatory roles in box C/D snoRNP biogenesis, RNA polymerase II assembly or axonemal dynein assembly (8).
Additionally, when Pih1 and Pih1 C-terminal truncation mutants were expressed in yeast cells that had their endogenous Pih1 depleted, we observed that the full-length Pih1 was very stable for over the examined 60 min (Fig. 4C). Because the full-length Pih1 was expressed from a constitutively high expression GPD promoter (Fig. 2B), it would be suspicious if assuming that all overexpressed Pih1 had been bound and protected by Tah1 as there might be limited endogenous Tah1. As discussed above, the other proteins such as Rrp43 and Nop58 (7) may potentially bind and slow down the degradation of Pih1. On the other hand, overexpression of Pih1 may transiently increase Tah1 steady state protein level in vivo by forming tight Pih1/Tah1 complex. Nevertheless, the change of Tah1 expression level and the other potential effects exerted by Pih1 overexpression remain for future investigation.
We showed in this study and our previous work (4) that Pih1 C-terminal fragment can cause efficient protein degradation in vivo. We have also previously shown that by fusing to a temperature sensitive proteasome-dependent degron, Pih1 can be degraded at elevated temperature (3). These studies suggest that in vivo degradation of Pih1 might be through the ubiquitin-proteasome system. Indeed, a previous proteomics study aimed at identifying all ubiquitylated proteins in yeast cells identified Pih1 as a potential ubiquitylated protein (32). It is possible that the Pih1 C-terminal fragment Pih1231–344 contains elements that can be efficiently recognized by certain E3 ligase complexes. Based on our mutagenesis analyses of Pih1282–344 (Fig. 5), hydrophobic amino acid clusters HP3 and HP5 may play a more important role in forming a recognition signal, if there is such one. Additionally, since Pih1 is predicted to contain two obvious intrinsically disordered regions IDR1 and IDR2, degradation of Pih1 in vivo may be through other pathways such as the ubiquitin-independent proteasomal degradation. It has been well documented that intrinsically disordered fragments within a protein often cause protein instability and target protein degradation by the proteasome (33, 34). Intrinsically disordered proteins can be easily degraded by 20S proteasome in a ubiquitin-independent mechanism, and this property has been practically used to define intrinsically disordered proteins (35). It is likely that Pih1, particularly without the protection of Tah1, can be degraded by 20S proteasome and 26S proteasome in vivo in both ubiquitin-dependent and ubiquitin-independent pathways as degradation by dual modes has been observed for proteins such as the oncoprotein p53 and cyclin-dependent kinase inhibitor p21CIP1 (36).
Acknowledgments
We thank Victoria Feiyang Liu for help with the construction of GFP fusion protein constructs and Jennifer Huen for help in the purification of Rvb1 and Rvb2 proteins.
This work was supported by a University of Toronto Connaught New Stuff Matching Fund (487199) and NSERC Discovery Grant 371789-2009 (to R. Z.) and by Grant MOP-93778 from the Canadian Institutes of Health Research (to W. A. H.).
- Pih1
- protein interacting with Hsp90
- TPR
- tetratricopeptide repeat
- R2TP
- Rvb1-Rvb2-Tah1-Pih1 complex
- IDR
- intrinsically disordered region
- snoRNP
- small nucleolar ribonucleoprotein
- GPD
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1. Zhao R., Davey M., Hsu Y. C., Kaplanek P., Tong A., Parsons A. B., Krogan N., Cagney G., Mai D., Greenblatt J., Boone C., Emili A., Houry W. A. (2005) Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120, 715–727 [DOI] [PubMed] [Google Scholar]
- 2. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D., Zhang N., Zheng C., Bryant S. H. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., Costanzo M., Brost R. L., Boone C., Hughes T. R., Yip C. M., Houry W. A. (2008) Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180, 563–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jiménez B., Ugwu F., Zhao R., Ortí L., Makhnevych T., Pineda-Lucena A., Houry W. A. (2012) The structure of the minimal tetratricopeptide repeat domain protein Tah1 reveals the mechanism of its interaction with Pih1 and Hsp90. J. Biol. Chem. 287, 5698–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gribun A., Cheung K. L., Huen J., Ortega J., Houry W. A. (2008) Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J. Mol. Biol. 376, 1320–1333 [DOI] [PubMed] [Google Scholar]
- 6. Huen J., Kakihara Y., Ugwu F., Cheung K. L., Ortega J., Houry W. A. (2010) Rvb1-Rvb2: essential ATP-dependent helicases for critical complexes. Biochem. Cell Biol. 88, 29–40 [DOI] [PubMed] [Google Scholar]
- 7. Gonzales F. A., Zanchin N. I., Luz J. S., Oliveira C. C. (2005) Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J. Mol. Biol. 346, 437–455 [DOI] [PubMed] [Google Scholar]
- 8. Kakihara Y., Houry W. A. The R2TP complex: discovery and functions. Biochim. Biophys. Acta 1823, 101–107 [DOI] [PubMed] [Google Scholar]
- 9. Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jády B. E., Rothé B., Pescia C., Robert M. C., Kiss T., Bardoni B., Krol A., Branlant C., Allmang C., Bertrand E., Charpentier B. (2008) The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 180, 579–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boulon S., Pradet-Balade B., Verheggen C., Molle D., Boireau S., Georgieva M., Azzag K., Robert M. C., Ahmad Y., Neel H., Lamond A. I., Bertrand E. (2010) HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell 39, 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horejsí Z., Takai H., Adelman C. A., Collis S. J., Flynn H., Maslen S., Skehel J. M., de Lange T., Boulton S. J. (2010) CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol. Cell 39, 839–850 [DOI] [PubMed] [Google Scholar]
- 12. Ni L., Saeki M., Xu L., Nakahara H., Saijo M., Tanaka K., Kamisaki Y. (2009) RPAP3 interacts with Reptin to regulate UV-induced phosphorylation of H2AX and DNA damage. J. Cell. Biochem. 106, 920–928 [DOI] [PubMed] [Google Scholar]
- 13. Inoue M., Saeki M., Egusa H., Niwa H., Kamisaki Y. (2010) PIH1D1, a subunit of R2TP complex, inhibits doxorubicin-induced apoptosis. Biochem. Biophys. Res. Commun. 403, 340–344 [DOI] [PubMed] [Google Scholar]
- 14. Zhai N., Zhao Z. L., Cheng M. B., Di Y. W., Yan H. X., Cao C. Y., Dai H., Zhang Y., Shen Y. F. Human PIH1 associates with histone H4 to mediate the glucose-dependent enhancement of pre-rRNA synthesis. J. Mol. Cell. Biol. [DOI] [PubMed] [Google Scholar]
- 15. Eckert K., Saliou J. M., Monlezun L., Vigouroux A., Atmane N., Caillat C., Quevillon-Chéruel S., Madiona K., Nicaise M., Lazereg S., Van Dorsselaer A., Sanglier-Cianférani S., Meyer P., Moréra S. (2010) The Pih1-Tah1 cochaperone complex inhibits Hsp90 molecular chaperone ATPase activity. J. Biol. Chem. 285, 31304–31312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Varshavsky A. (1991) Naming a targeting signal. Cell 64, 13–15 [DOI] [PubMed] [Google Scholar]
- 17. Rogers S., Wells R., Rechsteiner M. (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234, 364–368 [DOI] [PubMed] [Google Scholar]
- 18. Kyte J., Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 19. Dosztányi Z., Csizmok V., Tompa P., Simon I. (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 [DOI] [PubMed] [Google Scholar]
- 20. Xue B., Dunbrack R. L., Williams R. W., Dunker A. K., Uversky V. N. (2010) PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 1804, 996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mumberg D., Müller R., Funk M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 22. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 23. Shen X. (2004) Preparation and analysis of the INO80 complex. Methods Enzymol. 377, 401–412 [DOI] [PubMed] [Google Scholar]
- 24. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 25. Tompa P. (2002) Intrinsically unstructured proteins. Trends Biochem. Sci. 27, 527–533 [DOI] [PubMed] [Google Scholar]
- 26. Meszaros B., Simon I., Dosztanyi Z. The expanding view of protein-protein interactions: complexes involving intrinsically disordered proteins. Phys. Biol. 8:035003, 2011 [DOI] [PubMed] [Google Scholar]
- 27. Rechsteiner M., Rogers S. W. (1996) PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21, 267–271 [PubMed] [Google Scholar]
- 28. Lijnzaad P., Argos P. (1997) Hydrophobic patches on protein subunit interfaces: characteristics and prediction. Proteins 28, 333–343 [PubMed] [Google Scholar]
- 29. Provencher S. W., Glöckner J. (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 30. Garnier J., Gibrat J. F., Robson B. (1996) GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266, 540–553 [DOI] [PubMed] [Google Scholar]
- 31. Dyson H. J., Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 32. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 33. Baugh J. M., Viktorova E. G., Pilipenko E. V. (2009) Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 386, 814–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsvetkov P., Myers N., Moscovitz O., Sharon M., Prilusky J., Shaul Y. (2012) Thermo-resistant intrinsically disordered proteins are efficient 20S proteasome substrates. Mol. Biosyst. 8, 368–373 [DOI] [PubMed] [Google Scholar]
- 35. Tsvetkov P., Asher G., Paz A., Reuven N., Sussman J. L., Silman I., Shaul Y. (2008) Operational definition of intrinsically unstructured protein sequences based on susceptibility to the 20S proteasome. Proteins 70, 1357–1366 [DOI] [PubMed] [Google Scholar]
- 36. Jariel-Encontre I., Bossis G., Piechaczyk M. (2008) Ubiquitin-independent degradation of proteins by the proteasome. Biochim. Biophys. Acta 1786, 153–177 [DOI] [PubMed] [Google Scholar]