Background: Synergistic interplay of cellulases is key for efficiency of cellulose hydrolysis.
Results: In situ observation of individual and synergistic action of endo- and exo-cellulases on a polymorphic cellulose substrate reveals specificity of individual enzyme components for crystalline or amorphous regions.
Conclusion: Cellulase synergism is governed by mesoscopic morphological characteristics of the cellulose substrate.
Significance: Advanced knowledge basis for rational optimization of cellulose saccharification.
Keywords: Atomic Force Microscopy, Biotechnology, Carbohydrate-binding Protein, Carbohydrate Processing, Cellulase, Enzyme Mechanisms, Cellobiohydrolase, Cellulose Polymorphism, Endoglucanase, Synergism
Abstract
Cellulose is the most abundant biopolymer and a major reservoir of fixed carbon on earth. Comprehension of the elusive mechanism of its enzymatic degradation represents a fundamental problem at the interface of biology, biotechnology, and materials science. The interdependence of cellulose disintegration and hydrolysis and the synergistic interplay among cellulases is yet poorly understood. Here we report evidence from in situ atomic force microscopy (AFM) that delineates degradation of a polymorphic cellulose substrate as a dynamic cycle of alternating exposure and removal of crystalline fibers. Direct observation shows that chain-end-cleaving cellobiohydrolases (CBH I, CBH II) and an internally chain-cleaving endoglucanase (EG), the major components of cellulase systems, take on distinct roles: EG and CBH II make the cellulose surface accessible for CBH I by removing amorphous-unordered substrate areas, thus exposing otherwise embedded crystalline-ordered nanofibrils of the cellulose. Subsequently, these fibrils are degraded efficiently by CBH I, thereby uncovering new amorphous areas. Without prior action of EG and CBH II, CBH I was poorly active on the cellulosic substrate. This leads to the conclusion that synergism among cellulases is morphology-dependent and governed by the cooperativity between enzymes degrading amorphous regions and those targeting primarily crystalline regions. The surface-disrupting activity of cellulases therefore strongly depends on mesoscopic structural features of the substrate: size and packing of crystalline fibers are key determinants of the overall efficiency of cellulose degradation.
Introduction
Lignocellulosic plant biomass is generally considered as the most promising renewable feedstock for sustainable bioproduction of transportation fuels and commodity chemicals. A bioeconomy built on lignocellulose utilization offers compelling advantages: reduced dependence on crude oil, decreased carbon dioxide emission, productive use of agricultural, forestal, and municipal waste, and elimination of “food versus fuel” concerns which arise from the use of an edible raw material (1–4). The critical bottleneck in the development of bioconversion applications, however, remains the production of soluble sugars from cellulose (5). Chemically, cellulose, which constitutes the main polysaccharide component of plant biomass, is a linear β-(1,4)-linked d-glucose homopolymer (6–9). The complex structural organization of cellulose imparts it a pronounced resistance to chemical and enzymatic degradation into soluble hydrolysis products (6, 8): depending on its source, cellulose consists of varying amounts of highly ordered (i.e. crystalline) regions and such of lower order, referred to as amorphous cellulose, and additionally, it can be intertwined with hemicellulose and lignin (9, 10). Cellulases are highly proficient catalysts for the hydrolysis of β-glycosidic linkages in isolated, soluble short-chain fragments of the cellulose. By contrast, hydrolysis rates of the natural insoluble substrate often decrease dramatically at later stages of hydrolysis (11–14). Therefore, many attempts have been made to enhance the intrinsic efficiency of cellulases (6, 15) and various types of cellulose pretreatment have been developed with the aim of maximizing substrate accessibility and reactivity toward enzymatic hydrolysis (6, 16). The actual source of this limitation, however, be it the enzymes, the substrate, or both, is a fundamentally unsolved puzzle (14). Identifying the limiting factors by achieving a deeper mechanistic understanding of enzymatic cellulose degradation is pivotal for advancing biomass-to-fuel process development (5–7).
Comprehension of cellulase activity on the cellulose surface has turned out to be remarkably difficult, essentially because of two main complexities: firstly, natural cellulase systems are a multi-component mixture of at least two basic types of enzyme activity: chain-end-cleaving cellobiohydrolase (CBH)4 and internally chain-cleaving endoglucanase (EG). CBH enzymes are additionally distinguished according to their preference for hydrolysis of the cellulose chain from either the reducing (CBH I) or the non-reducing end (CBH II) (12, 17–19). Another highly distinctive feature of the action of CBH and EG is their synergy in the hydrolysis of insoluble cellulose: a combination of the three enzymes is much more active in terms of soluble sugar release than it would be expected from the sum of the individual activities (12, 17, 18, 20–22). Although the molecular structure and function of the enzymes are known (23–25) and synergism has been extensively studied (12, 17, 20, 22, 26–28), the distinct roles of the single enzymes in cellulose degradation are unclear. Secondly, cellulose is polymorphic at several levels of its structural organization, which leads to a spatial variation in crystallinity and hence to resistance toward hydrolysis (6, 29). With the molecular determinants of the hydrolytic chain cleavage being well understood, the factors affecting the hydrolysis rate most are presumably associated with substrate morphology (28, 30, 31). Mechanistic questions of cellulase synergy will therefore only become tractable when adequate visualization on a nano- and mesoscale is applied to a suitable cellulosic substrate. The cellulose surface exhibits a large amount of meso- and microscopic heterogeneity, thus complicating visualization of surface degradation events at the requisite nanometer scale. Morphological characteristics of the substrate will moreover change with conversion (10, 17). One way of dealing with cellulose polymorphism is by using a highly crystalline model substrate thus reducing the structural complexity. This has proven to be particularly successful in the application of AFM in order to study the mode of action of CBH I, which is able to bind to and degrade crystalline cellulose (12, 32). Recent studies visualized single CBH I molecules moving unidirectionally along accessible lanes on the surface of crystalline cellulose, which is consistent with the proposed processivity of cellulose chain degradation by this enzyme (28, 33). The strictly crystalline Valonia cellulose employed in these studies is a suitable model substrate for CBH I, but does not allow direct observation of CBH II (34). EG, which hardly releases soluble sugars from crystalline cellulose (17, 32), is likely to present a similar problem. Considering the key importance of CBH II and EG for full cellulase activity on real-life cellulose substrates, it is pivotal to expand mesoscale visualization to a polymorphic (i.e. mixed amorphous-crystalline) cellulose preparation.
EXPERIMENTAL PROCEDURES
Cellulase Preparation and Characterization
All materials were purchased of the highest purity available from Carl Roth (Karlsruhe, Germany) unless stated otherwise. The complete cellulase system of Trichoderma reesei mutant SVG 17 was produced and stored as previously described (7). Pure preparations of β-glucosidase (BGL) from Aspergillus niger and CBH I and EG, both from T. longibrachiatum, were purchased from Megazyme (Dublin, Ireland). T. longibrachiatum is closely related to T. reesei and its CBH I and EG are isofunctional and structurally analogous to the respective T. reesei enzymes (35, 36). Recombinant T. reesei CBH II, obtained by heterologous expression in Pichia pastoris, was a gift from Anton Glieder (Institute of Molecular Biotechnology, Graz University of Technology). The absence of relevant cross-contaminations by other cellulolytic activities in these preparations was verified by employing the well-established carboxymethyl cellulose (CMC) and 4-methylumbelliferyl-β-d-cellobioside (MUF-cellobioside) assays (30, 37, 38). Reducing sugars were measured using the 3,5-dinitrosalicylic acid (DNS) assay (39).
Generally, a 50 mm sodium citrate buffer (pH 5.0) was used for all experiments in this study unless stated otherwise. Using the filter paper unit (FPU) assay as recommended by the IUPAC (39), the activity of the complete cellulase system was determined as 2.1 FPU/mg. We measured the protein concentration of the complete cellulase system according to Bradford (40), employing Roti®-Quant and Roti®-Nanoquant assays (both from Carl Roth, Karlsruhe, Germany) standardized with BSA. The concentration of pure enzyme preparations was determined via their intrinsic UV absorbance at a wavelength of 280 nm using a Nanodrop 1000 Spectrometer (Thermo Fisher Scientific Inc., Walham, MA). The respective molar extinction coefficients were calculated using ProtParam (41): ϵCBHI = 88250 m−1cm−1; ϵCBHII = 97665 m−1cm−1; ϵEG = 74940 m−1cm−1.
Polymorphic Cellulose and Specimen Preparation
The cellulosic substrate employed in this study was prepared and characterized as previously described (7). The protocol involves dissolution of the microcrystalline cellulose Avicel (Sigma-Aldrich) in ionic liquid. We used 1-butyl-3-methylimidazolium chloride (BMIMCl, Sigma-Aldrich). The resulting transparent gel was subjected to a fractional dehydration with ethanol (30% to absolute). This procedure removed the residual ionic liquid, leaving a pure and polymorphic cellulosic substrate (7).
To generate a stationary substrate for AFM studies, we embedded the substrate in epoxy resin without employing elevated temperatures or pressure. The absence of epoxy material diffusion into the substrate was confirmed previously (7). To prepare a nano-flat specimen surface, the epoxy-embedded substrate was first pre-cut with an Ultracut UCT ultramicrotome (Leica Microsystems, Wetzlar, Germany), then, the substrate was allowed to swell in buffer for one hour. The wet specimen was finally cut again to receive a nano-flat surface with a root-mean-squared (RMS) roughness of 10–15 nm.
Enzymatic Hydrolysis Studies
All hydrolysis experiments were conducted in triplicate at 50 °C, pH 5.0, in buffer. The cellulosic substrate concentration was 7.2 mg/ml in a total volume of 350 μl. Enzyme loadings were the same as in the respective AFM experiments: 3.6 mg/gsubstrate (complete system), 36 mg/gsubstrate (CBH I and EG) and 1.3 g/gsubstrate (CBH II). Sample tubes were centrifuged for 1 min at 10,000 rpm at defined breakpoints. The supernatant was heated to 95 °C for 7 min to terminate cellulase activity, and then again centrifuged for 1 min at 10,000 rpm. The reducing sugars were determined in the resulting supernatant by means of the DNS assay (30).
For the hydrolysis experiment probing cellulase synergism, the cellulosic substrate concentration was 1 mg/ml in a total volume of 1 ml of buffer. To prevent inhibition by accumulating cellobiose, 1 unit of BGL was added. The setup was designed to resemble the AFM synergism experiment at 20 °C: in the beginning, solely EG (9 mg/gsubstrate) was present. CBH II (320 mg/gsubstrate) was added after 160 min and CBH I (9 mg/gsubstrate) after 280 min total reaction time. After 350 min, the reaction was stopped. Samples were drawn accordingly, treated as described above and the amount of reducing sugars was determined.
In Situ AFM Observation, Data Collection, and Analyses
For the in situ AFM measurements at 20 °C in buffer a commercial Dimension 3100 AFM equipped with a Hybrid scan head, a liquid cell tip holder, and Nanoscope IVa controller (Bruker AXS, Santa Barbara) was used. Imaging was performed with a TR400PSA cantilever (Olympus Probes, Tokyo, Japan) in TappingMode®. A laboratory-built liquid cell featuring an injection valve was used for specimen mounting. Scan rates, set points, and drive amplitudes were chosen accordingly to guarantee stable scanning with the lowest energy dissipation possible. During image recording we gave careful consideration to avoiding tip related artifacts by permanently evaluating side wall angles and morphological features of the surface.
Initially, AFM reference images of the specimen surface were recorded in buffer (3 ml) and areas for continuous in situ observation were selected. Suitable areas featured one larger, characteristic crystalline region, which served as a marker. Then, the respective enzyme solution was added, and the dynamics of the enzymatic degradation were recorded continuously. The protein loadings used were 3.6 mg/gsubstrate for the complete cellulase system, 36 mg/gsubstrate for EG and CBH I, and 1.3 g/gsubstrate for CBH II. These loadings were chosen after preliminary studies showed a good time-dependent resolution of activity for the settings of our AFM experiments. For the hydrolysis experiment probing cellulase synergism, solely EG (9 mg/gsubstrate) was present in the beginning. CBH II (320 mg/gsubstrate) was added after 160 min and CBH I (9 mg/gsubstrate) after 280 min total reaction time. After 350 min, the reaction was stopped. 1 unit of BGL was present in the reaction solution to prevent inhibition by accumulating cellobiose.
Image processing and detailed analysis was performed with NanoScope Analysis 1.20 (Build R1Sr3.64571, Veeco Instruments Inc.) and Gwyddion (Gwyddion 2.25). Data analysis of depth evolution and degradation velocity was done in OriginLab 8.5 (OriginLab Corp, Northampton).
We analyzed the vertical degradation quantitatively by evaluating multiple points on the surface and tracking their change over time using large crystallites as a reference. Careful post-processing of the images allowed for measuring the depth of a distinct point in reference to a crystallite on each of 18 images which were taken in distinct time intervals. Evaluation of the height difference per time allowed to calculate the degradation velocity at different positions on the substrate. Evidence for two distinct velocities was found through statistical evaluation of velocities and double peak fitting.
RESULTS AND DISCUSSION
A Mesoscopic View on Synergistic Degradation of Polymorphic Cellulose
Here we present a study on the cellulolytic degradation of a representative mixed amorphous-crystalline cellulose (7). We prepared the substrate by incomplete dissolution of Avicel in an ionic liquid and subsequent removal of the solvent. The supramolecular structure of the resulting polymorphic cellulose exhibits an amorphous cellulose matrix in which larger crystallites (100 nm-10 μm) and smaller fibrils (width: ≤15 nm; length: 100–300 nm) are embedded randomly as schematically shown in supplemental Fig. S1. Overall, 30% of the substrate consists of crystallites and small crystalline fibrils. The primarily amorphous character of the cellulose preparation was reported previously (7). The surface roughness in liquid environments was lower than 10 nm, as determined by using AFM in a liquid cell. As previously confirmed, the respective crystalline phases are predominantly made of cellulose I (7), which is the main allomorph of natural cellulose in higher plants (42). This substrate is well suited for drawing conclusions to natural and pretreated biomass cellulose because it features amorphous as well as crystalline regions of allomorph I. Furthermore, it fulfills the requirements for a comprehensive analysis of cellulase synergy by AFM: it is sufficiently flat and it features crystalline and amorphous regions which make a suitable substrate for the diverse cellulases.
A Complete Cellulase System Preferentially Degrades the Amorphous Regions of the Polymorphic Substrate to Bare Crystalline Regions
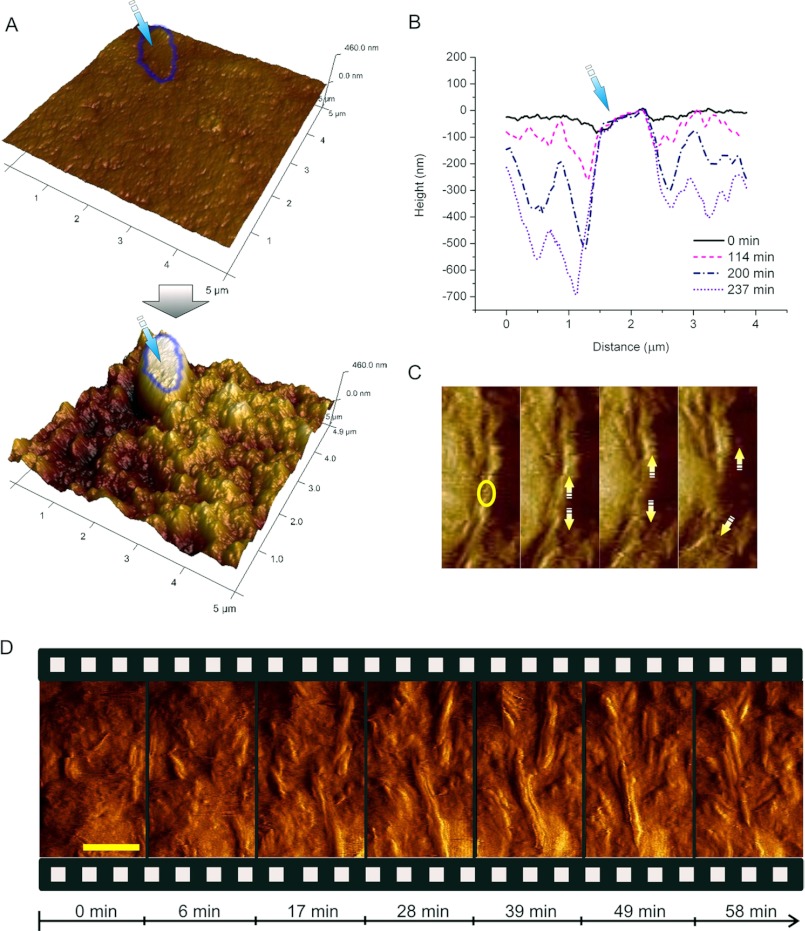
First of all, we aimed at visualizing the mesoscopic structural consequences of exposure of the polymorphic substrate to the synergistic degradation by a complete cellulase system in situ. We used a complete cellulase preparation from the cellulolytic fungus Trichoderma reesei, which contains CBH I (60%), CBH II (20%), and EG (12%) as its major enzyme components (17, 19, 33). Enzymatic attack resulted in a considerable volume degradation of the polymorphic substrate in dependence of the incubation time (Fig. 1, A and B). After 4 h, the height differed up to 700 nm compared with the start of the reaction (Fig. 1B). Eventually, the cellulose surface was completely rugged with the large crystallite marker remaining (Fig. 1A). This implies that the bulk of the amorphous material had been degraded (Fig. 1A). In situ AFM imaging shows that homogeneous substrate areas, corresponding to amorphous cellulose, are degraded to bare crystalline nanofibrils and crystallite surfaces with a defined and highly ordered structure (crystalline cellulose), comparable to AFM images of crystalline model substrates and fibrils (28, 43). Generally, we observed that large crystallites were degraded extremely slowly. Therefore, they were used as markers for the scan area and to follow height changes over time. This is reflected in the graph in Fig. 1B where the surface evolution is illustrated by superimposition of time-resolved section profiles: while the material at the side of the crystallites is removed and leads to the formation of cavities, the crystallite itself remains unchanged.
FIGURE 1.
In situ observation of the synergistic degradation of polymorphic cellulose by the complete cellulase system of T. reesei. A, substrate surface at the beginning of (t = 0 min) and after incubation (t = 237 min) with the complete cellulase system. A typical large crystallite, which was used as a marker for height change during degradation, is indicated. The substrate around it is degraded, while the crystallite itself remains virtually unaltered (see also supplemental Video S2). B, section profiles of the area shown in B reflect the volumetric degradation with time. Relative to the marker, the substrate around it was degraded up to 700 nm in 237 min. C, in situ observation of a defect (circle) being introduced into a fibril. Subsequently, the fibril is attacked and degraded from both new ends of the generated defect (arrows). D, snapshots along the time course of cellulose degradation (see also supplemental Video S1) show initial degradation of amorphous regions (17 min). Previously buried crystalline fibrils appear (28 min) and are quickly degraded in a processive manner starting at their ends as well as at previously introduced defects (49 min). The scale bar represents 100 nm.
Interestingly, in time-resolved AFM sequences we observed that small cellulose fibrils (width: ≤15 nm) appeared and were degraded in an alternating manner (Fig. 1D; supplemental Videos S1 and S2). At first, these fibrils were polished from covering amorphous material. Subsequently, they were degraded rapidly. This leads to the conclusion that cellulases remove amorphous material surrounding the crystalline fibrils first, and then degrade the fibrils to uncover subjacent amorphous material again. Fibrils are degraded in various ways: primarily by thinning starting from the side walls, but also by shortening starting from fibril tips and introduction of defects in the middle of the fibrils (Fig. 1, C and D; supplemental Videos S1 and S2). As soon as a new defect was generated (Fig. 1C, circle in the left panel), degradation proceeded rapidly from the two new ends thus produced (Fig. 1C, arrows). Eventually, the fibril was degraded quickly (Fig. 1C; supplemental Video S1). However, in contrast to the smaller crystalline fibrils (width: ≤15 nm; length: 100–300 nm), the large crystallites (100 nm-10 μm) remained unchanged. This is probably due to a majority of the cellulase binding sites being buried within the crystal, which would impede enzymatic attack. The thin fibrils, by contrast, exhibit easily accessible cellulase binding sites at their ends and therefore are quickly attacked as soon as they are bared from the amorphous matrix.
We analyzed vertical degradation quantitatively by evaluating multiple points on the surface and tracking their change over time. Strikingly, we thus found evidence for two distinct degradation velocities (supplemental Fig. S2). This corroborates what we observed in the AFM images: once bared, small crystalline fibrils are degraded significantly faster (3.8 ± 0.2 nm/min) than the residual amorphous matrix (0.7 ± 0.2 nm/min). Taking into account that large crystallites are degraded infinitely slower, this leads to the practically important conclusion that the retarding effect of cellulose crystallinity on the reaction rate is strongly dependent on crystal size and shape and the accessibility of binding sites at crystals.
CBH II and EG Efficiently Remove Amorphous Cellulose and Polish Crystalline Regions for CBH I Attack
Consequently, the next question was how each of the major cellulase enzymes contributes to the mesoscopic destruction of the cellulose as observed with the complete system. Therefore, we dissected the synergistic action of the complete system by separate in situ imaging and analysis of its major individual activities: CBH I, CBH II, and EG. In contrast to the complete cellulase system, neither of the single enzymes caused the massive volumetric degradation as observed in the complete cellulase system. With each of the individual cellulases, the surface roughness was only modestly increased and no volume loss was observed, despite the high loadings used: the amount of CBH I, CBH II, and EG used corresponded to 17 times, 1800 times, and 83 times their respective relative abundance in the experiment with the complete system. These loadings were chosen with respect to a suitable time-dependent resolution of their activity in our AFM experiments.
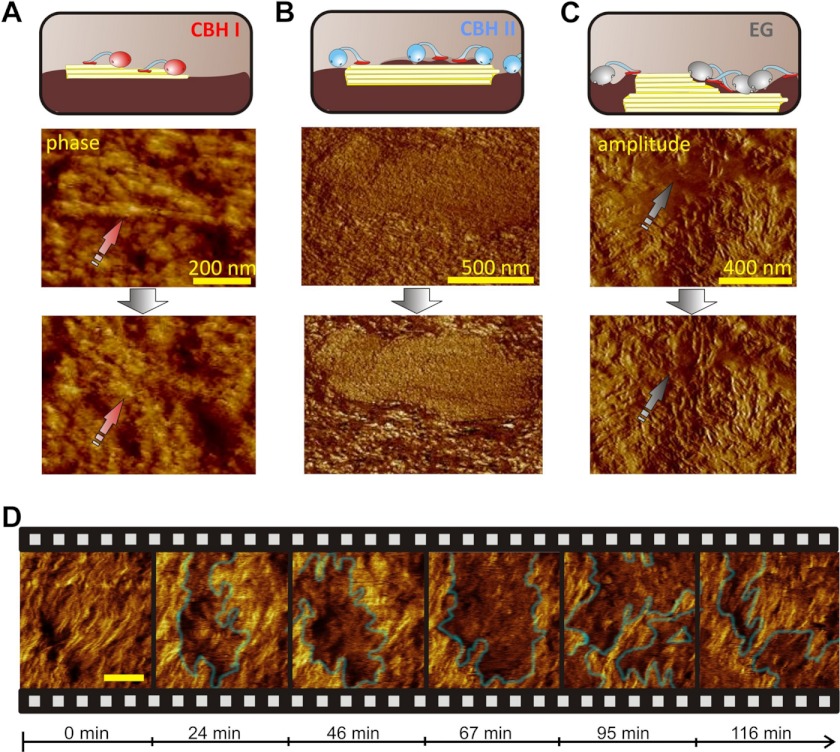
Each of the enzymes produced a remarkably distinct pattern of cellulose surface disruption (Fig. 2, A–C). We observed that EG, which has been shown to preferably attack amorphous cellulose (31, 43, 44), efficiently degraded amorphous regions of the specimen. Thus, EG action caused gradual exposure of well-defined crystalline nanofibrils (“polishing”), which were previously covered by amorphous material. The crystalline fibrils themselves were not attacked by EG. This is exemplified by selected AFM images (Fig. 2C), where an amorphous spot (Fig. 2C, arrow) is polished by EG and thus cellulose of higher order is bared: in the second image, the previously blurry surface is highly defined. Interestingly, treatment with CBH II caused a similar polishing effect in combination with clustering of the enzymes. Generally, CBH II enzymes are known to be essential in deconstruction of crystalline cellulose (12, 31, 45). However, it has been proposed from indirect biochemical data that CBH II also degrades amorphous cellulose (45). We here show direct and conclusive evidence on the specificity of this enzyme for amorphous areas. In the AFM image shown in Fig. 2B, a large crystallite is at first buried beneath amorphous material. CBH II polishes it from amorphous material and the result is a defined crystallite surface visible in the second image. Additionally, we observed the appearance of CBH II “clusters” using AFM phase imaging, which allows a clear distinction between materials with different characteristics (e.g. cellulose and enzymes). In the AFM phase images these clusters are visible as darker cloud-like areas (Fig. 2D; supplemental Video S3). Initially, they bound to areas that did not exhibit a defined surface structure. Then, the clusters removed the amorphous cellulose to leave behind a more defined (i.e. crystalline) area (Fig. 2B; supplemental Video S3). However, in contrast to EG, and despite its observed distinct preference for amorphous cellulose, CBH II was bi-specific and also showed minor degradation of crystalline cellulose, observable as slight shortening of the fibrils. Accompanying biochemical studies of the substrate saccharification showed that EG and CBH II reached almost 60% conversion (Fig. 3). This is in good agreement with the relative abundance of amorphous cellulose in our specimen (7).
FIGURE 2.
Dissecting synergism: in situ observation of single enzymes. A, CBH I degrades small fibrils (phase image). B, CBH II polishes the surface of a large crystallite by removing amorphous cellulose (phase image). Right: (C), an example of EG polishing which leads to a highly defined fibrillar surface (amplitude image). D, “clustering” effect of CBH II in a phase image sequence (see also supplemental Video S3). The scale bar represents 100 nm.
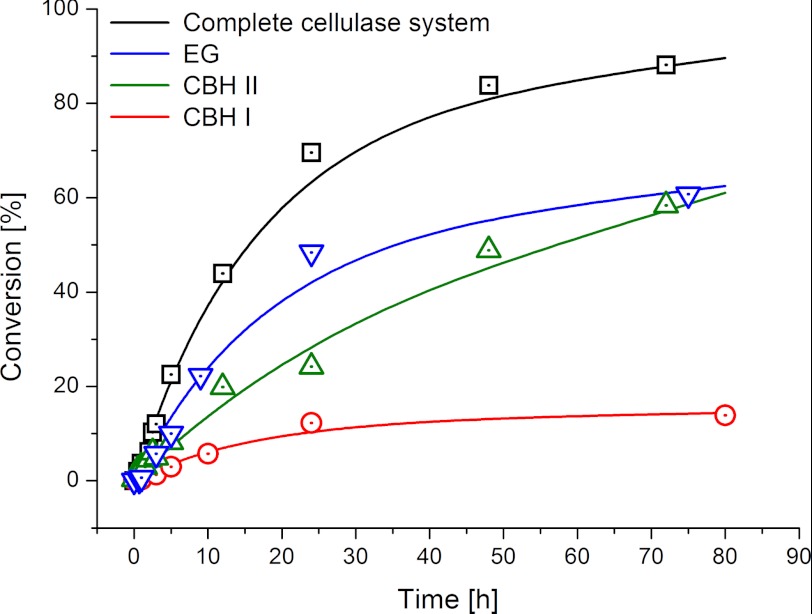
FIGURE 3.
Conversion versus time profiles of the complete cellulase system and the single cellulases used in this study. The highest degree of conversion is achieved by the synergistic action of the complete cellulase system (3.6 mg/gsubstrate). When solely CBH I (36 mg/gsubstrate) is present, the hydrolysis rate declines at a pronouncedly lower degree of conversion. CBH II (1.3 g/gsubstrate) and EG (36 mg/gsubstrate) show a pronounced preference for the amorphous regions, which is reflected in a higher overall conversion of the primarily amorphous substrate as compared with CBH I.
Notably, EG and CBH I behaved as monospecific cellulases, showing complementary substrate preference for amorphous and crystalline cellulose, respectively. The surface roughness stayed constant during CBH I treatment (RMS roughness: 10.6 ± 0.5 nm). The AFM images in Fig. 2A depict the selective degradation of a small crystalline fibril (arrow). CBH I was exclusively active toward such small crystalline nanofibrils (≤ 10 nm width) on the surface (Fig. 2A), but did neither attack the surrounding amorphous material nor fibrils coated with amorphous material. Accordingly, sugar release by CBH I became stalled at low conversion (≤ 14%), this supposedly is the moment when the limited amount of accessible crystalline fibrils on the surface has been degraded (Fig. 3). This implies that, in order to degrade crystalline cellulose, CBH I requires helpers, which remove the amorphous layers beforehand. Overall, the absence of synergism renders single cellulases incapable of rapid and complete three-dimensional degradation of polymorphic cellulose. Accordingly, biochemical studies showed that synergistic degradation of the polymorphic substrate by the complete cellulase system resulted in 90% conversion (Fig. 3).
A Combination of EG, CBH II, and CBH I Enables Reconstruction of Synergism as Observed in the Complete System
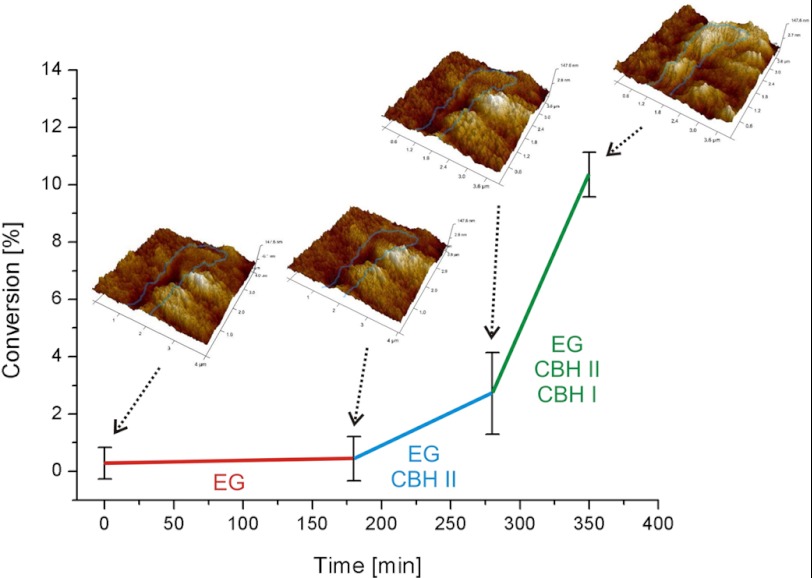
While we observed considerable volumetric degradation with the complete cellulase system, this was not the case with the dissected single activities although we used them at significantly higher loadings compared with their abundance in the complete system. In the single enzyme experiments, CBH I only degraded small crystalline fibrils, which were exposed on the surface. The main effect of CBH II and EG was to polish crystalline regions by removing amorphous material covering those. In that way, CBH II and EG would prepare crystalline regions for CBH I attack. Accordingly, a mixture of solely EG and CBH II should not result in a significant rate acceleration or increase in volumetric degradation. Only when CBH I is added to these two, complete synergism would be re-established and boost the rate. In order to test this hypothesis, we added EG, CBH II, and CBH I in a controlled manner (Fig. 4). We also provided a sufficient amount of BGL to prevent inhibition by cellobiose (14, 17, 30). As expected, the pre-incubation of the substrate with EG led to minor hydrolysis through removal of some amorphous material. Subsequently, we added CBH II, which aided in the removal of amorphous cellulose, and besides it slowly started degrading the small crystalline fibrils, which had been exposed by foregoing EG action. In contrast to the single enzyme experiments, we now also observed a beginning volume loss and an increase in surface roughness from 12 nm to 16 nm RMS roughness. Over time, large crystalline areas, which were not attacked, became prevalent on the surface due to removal of the amorphous matrix. The rate of sugar release was enhanced 4-fold as a consequence of the synergy between EG and CBH II (Fig. 4). Finally, we added CBH I which immediately caused a massive degradation of both amorphous and crystalline areas of the cellulose. Large crystallites were unaltered, and small crystalline fibrils were degraded instantly upon being bared. RMS roughness and total volume loss increased significantly over time. Accordingly, we noted a 6-fold increase in the hydrolysis rate after addition of CBH I (Fig. 4). The presence of CBH I enhances the degradation of small fibrils in particular. As a consequence, we observed alternating appearance and disappearance of fibrils on the cellulose surface, as it was the case with the complete cellulase system (supplemental Video S1). Globally, the performance of the three-component enzyme mixture on the cellulose surface was identical to that of the complete cellulase system: large cellulose crystals were degraded more slowly than surrounding areas featuring small fibrils embedded in amorphous material.
FIGURE 4.
Reconstructing synergism, substrate degradation with sequential addition of single cellulases. This biochemical study of synergism shows how synergism becomes effective after addition of CBH I, which boosts the hydrolysis rate on the polymorphic substrate. The AFM image insets depict changes of the substrate along the reaction timeline. Rapid synergistic degradation and consequential three-dimensional disruption of the substrate does not happen until all three cellulases (EG, CBH II, and CBH I) are present.
The Rate of Enzymatic Cellulose Degradation Is Dependent on Substrate Morphology
Our results show that the appearance of large crystallites hardly changed as they were attacked slowly, probably due to the fact that most of the cellulase binding sites were buried inside the crystal and hence not accessible to the enzymes. Exposed small crystalline fibrils, by contrast, were degraded faster even than amorphous regions through synergistic interaction of the cellulases (supplemental Fig. S2 and Video S1). This seemingly contradictory observation can be explained by size and shape of the crystallites in the respective substrates. There are various reasons for the slower degradation of big crystallites: the tight packing of the cellulose fibrils generally prevents interception and limits accessibility, cellulase binding sites might be buried inside the crystal, and traffic jams among surface-bound enzymes impede mobility and cause unproductive binding (22, 28).
Synergism among Cellulases Is Morphology-dependent and Dominated by Cooperativity between Enzymes Degrading Amorphous Cellulose and Those Degrading Crystalline Cellulose
We here demonstrated that synergism among cellulases is based upon the cooperation between those enzymes primarily attacking amorphous regions and those preferring crystalline areas: while EG and CBH II are utterly efficient in removing amorphous material, they show no (EG) or only marginal (CBH II) activity on crystalline cellulose (supplemental Fig. S3). Conversely, CBH I has a pronounced preference for regions of higher order, but is dependent on having these made accessible by EG and CBH II. A recent mechanistic study (30) found evidence that the rates of complexation of CBH and accessible chain ends are low but are significantly enhanced by presence of EG, which was reported to amplify CBH activity (30, 31). Moreover, the study authors assume a morphology-influenced synergy, meaning that other cellulases aid in removal of obstacles, which might impede CBH. This is in good agreement with several other in vitro and in silico studies (28, 30, 31, 46–48). According to the in situ visualization presented here, these obstacles are amorphous regions covering the substrate of CBH I, i.e. crystalline cellulose. We showed that CBH I efficiently degrades crystalline nanofibrils, which have easily accessible chain ends as compared with the large crystallites, where lots of chain ends might be buried within. Recently it was reported that amorphous regions of bacterial cellulose caused CBH I processivity to halt (31). In the polymorphic substrate used in our study, the majority of the nanofibrils are enclosed in the amorphous matrix of the substrate and hence inaccessible for CBH I. This is also reflected in the low activity of CBH I on the polymorphic substrate: when the surface-exposed nanofibrils are degraded, its activity stalls (Fig. 3). Accordingly, when the obstacles are removed by EG and CBH II, it is mainly CBH I that boosts the hydrolysis rate (Fig. 4).
Both CBH I and the bi-specific CBH II lack the ability to efficiently degrade crystalline cellulose unless other cellulases are present (Fig. 3; supplemental Fig. S2). Hence, cooperativity between an endo-exo system (EG and CBH II), which prepares the cellulose, and an exo-cellulase (CBH I) is pivotal, and this morphology-dependent synergy therefore constitutes the primary synergism. We showed that the three major cellulase components EG, CBH II, and CBH I are sufficient to reconstruct the synergism that can be observed for the complete cellulase system. Therefore, these three are the main cellulolytic enzymes of a system, which is ideally adapted for degradation of crystalline cellulosic substrates.
The degradation of crystalline regions once they are bared is furthermore strongly dependent on the size and shape of these areas. Aspect ratio, packing, and accessibility of free chain ends are responsible for the rapid degradation of small crystalline fibrils as compared with the cumbersome attack on the large crystallites (Figs. 1 and 2). It is conceivable, that the nanofibrils have easily accessible chain ends and allow for rapid processive degradation as shown in Fig. 1 and supplemental Video S1. This relatively rapid degradation of small crystalline fibrils explains the puzzling observations made in a number of other studies, which showed that crystallinity did not increase substantially with enzymatic conversion in some cellulose substrates while it did in others (12, 21).
Dependent on its source and possible pretreatment, biomass cellulose contains amorphous and crystalline regions to a variable extent (10). The polymorphic substrate we used is typical for a substrate with amorphous and crystalline regions of varying size. Our study visualizes that morphology is the main determinant of cellulase synergism. The finding that synergy among cellulases is dependent on substrate morphology furthermore explains why synergism between cellulases was observed on some substrates but not on others (49). Hence, enzyme mixtures for efficient saccharification of biomass need to be adapted for the morphology of their respective substrate.
Acknowledgments
We thank Karin Longus for producing the complete cellulase system; Gregor Trimmel for characterization of the substrate; Anton Glieder for the kind gift of CBH II; Ferdinand Hofer for discussions; and Dominika Stiger for commenting on the manuscript.
This work was supported by the Austrian Science Fund FWF (Grant P 24156-B21, to B. N.).

This article contains supplemental Figs. S1–S3 and Videos S1–S3.
- CBH
- cellobiohydrolase
- AFM
- atomic force microscopy
- BGL
- β-glucosidase
- BMIMCl
- 1-butyl-3-methylimidazolium chloride
- CMC
- carboxymethyl cellulose
- DNS
- 3,5-dinitrosalicylic acid
- EG
- endoglucanase
- FPU
- filter paper unit
- MUF-cellobioside
- methylumbelliferyl-β-d-cellobioside
- MS
- mass spectrometry
- RMS
- root-mean-squared.
REFERENCES
- 1. Farrell A. E., Plevin R. J., Turner B. T., Jones A. D., O'Hare M., Kammen D. M. (2006) Ethanol can contribute to energy and environmental goals. Science 311, 506–508 [DOI] [PubMed] [Google Scholar]
- 2. Graham-Rowe D. (2011) Agriculture: Beyond food versus fuel. Nature 474, S6–8 [DOI] [PubMed] [Google Scholar]
- 3. Hill J., Polasky S., Nelson E., Tilman D., Huo H., Ludwig L., Neumann J., Zheng H., Bonta D. (2009) Climate change and health costs of air emissions from biofuels and gasoline. Proc. Natl. Acad. Sci. U.S.A. 106, 2077–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmer M. R., Vogel K. P., Mitchell R. B., Perrin R. K. (2008) Net energy of cellulosic ethanol from switchgrass. Proc. Natl. Acad. Sci. U.S.A. 105, 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lynd L. R., Laser M. S., Bransby D., Dale B. E., Davison B., Hamilton R., Himmel M., Keller M., McMillan J. D., Sheehan J., Wyman C. E. (2008) How biotech can transform biofuels. Nat. Biotechnol. 26, 169–172 [DOI] [PubMed] [Google Scholar]
- 6. Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., Foust T. D. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 [DOI] [PubMed] [Google Scholar]
- 7. Bubner P., Dohr J., Plank H., Mayrhofer C., Nidetzky B. (2012) Cellulases dig deep: In situ observation of the mesoscopic structural dynamics of enzymatic cellulose degradation. J. Biol. Chem. 287, 2759–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chundawat S. P., Bellesia G., Uppugundla N., da Costa Sousa L., Gao D., Cheh A. M., Agarwal U. P., Bianchetti C. M., Phillips G. N., Jr., Langan P., Balan V., Gnanakaran S., Dale B. E. (2011) Restructuring crystalline cellulose hydrogen bond network enhances its depolymerization rate. J. Am. Chem. Soc. 133, 11163–11174 [DOI] [PubMed] [Google Scholar]
- 9. Jarvis M. (2003) Chemistry: cellulose stacks up. Nature 426, 611–612 [DOI] [PubMed] [Google Scholar]
- 10. Klemm D., Heublein B., Fink H. P., Bohn A. (2005) Cellulose: fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Ed. Engl. 44, 3358–3393 [DOI] [PubMed] [Google Scholar]
- 11. Bansal P., Hall M., Realff M. J., Lee J. H., Bommarius A. S. (2009) Modeling cellulase kinetics on lignocellulosic substrates. Biotechnol. Adv. 27, 833–848 [DOI] [PubMed] [Google Scholar]
- 12. Lynd L. R., Weimer P. J., van Zyl W. H., Pretorius I. S. (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang B., Willies D. M., Wyman C. E. (2006) Changes in the enzymatic hydrolysis rate of Avicel cellulose with conversion. Biotechnol. Bioeng. 94, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 14. Levine S. E., Fox J. M., Blanch H. W., Clark D. S. (2010) A mechanistic model of the enzymatic hydrolysis of cellulose. Biotechnol. Bioeng. 107, 37–51 [DOI] [PubMed] [Google Scholar]
- 15. Becker D., Braet C., Brumer H., 3rd, Claeyssens M., Divne C., Fagerström B. R., Harris M., Jones T. A., Kleywegt G. J., Koivula A., Mahdi S., Piens K., Sinnott M. L., Ståhlberg J., Teeri T. T., Underwood M., Wohlfahrt G. (2001) Engineering of a glycosidase Family 7 cellobiohydrolase to more alkaline pH optimum: the pH behaviour of Trichoderma reesei Cel7A and its E223S/A224H/L225V/T226A/D262G mutant. Biochem. J. 356, 19–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blanch H. W., Simmons B. A., Klein-Marcuschamer D. (2011) Biomass deconstruction to sugars. Biotechnol. J. 6, 1086–1102 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y. H., Lynd L. R. (2004) Toward an aggregated understanding of enzymatic hydrolysis of cellulose: noncomplexed cellulase systems. Biotechnol. Bioeng. 88, 797–824 [DOI] [PubMed] [Google Scholar]
- 18. Boisset C., Fraschini C., Schülein M., Henrissat B., Chanzy H. (2000) Imaging the enzymatic digestion of bacterial cellulose ribbons reveals the endo character of the cellobiohydrolase Cel6A from Humicola insolens and its mode of synergy with cellobiohydrolase Cel7A. Appl. Environ. Microbiol. 66, 1444–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goyal A., Ghosh B., Eveleigh D. (1991) Characteristics of Fungal Cellulases. Biores. Technol. 36, 37–50 [Google Scholar]
- 20. Nidetzky B., Steiner W., Hayn M., Claeyssens M. (1994) Cellulose hydrolysis by the cellulases from Trichoderma reesei: a new model for synergistic interaction. Biochem. J. 298, 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bansal P., Vowell B. J., Hall M., Realff M. J., Lee J. H., Bommarius A. S. (2012) Elucidation of cellulose accessibility, hydrolysability and reactivity as the major limitations in the enzymatic hydrolysis of cellulose. Bioresour. Technol. 107, 243–250 [DOI] [PubMed] [Google Scholar]
- 22. Väljamäe P., Sild V., Nutt A., Pettersson G., Johansson G. (1999) Acid hydrolysis of bacterial cellulose reveals different modes of synergistic action between cellobiohydrolase I and endoglucanase I. Eur. J. Biochem./FEBS 266, 327–334 [DOI] [PubMed] [Google Scholar]
- 23. Divne C., Stahlberg J., Reinikainen T., Ruohonen L., Pettersson G., Knowles J. K., Teeri T. T., Jones T. A. (1994) The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science 265, 524–528 [DOI] [PubMed] [Google Scholar]
- 24. Koivula A., Reinikainen T., Ruohonen L., Valkeajarvi A., Claeyssens M., Teleman O., Kleywegt G. J., Szardenings M., Rouvinen J., Jones T. A., Teeri T. T. (1996) The active site of Trichoderma reesei cellobiohydrolase II: the role of tyrosine 169. Protein Eng. Des. Sel. 9, 691–699 [DOI] [PubMed] [Google Scholar]
- 25. Kleywegt G. J., Zou J. Y., Divne C., Davies G. J., Sinning I., Stâhlberg J., Reinikainen T., Srisodsuk M., Teeri T. T., Jones T. A. (1997) The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 Å resolution, and a comparison with related enzymes. J. Mol. Biol. 272, 383–397 [DOI] [PubMed] [Google Scholar]
- 26. Henrissat B., Driguez H., Viet C., Schülein M. (1985) Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Nat. Biotechnol. 3, 722–726 [Google Scholar]
- 27. Hu J., Arantes V., Saddler J. N. (2011) The enhancement of enzymatic hydrolysis of lignocellulosic substrates by the addition of accessory enzymes such as xylanase: is it an additive or synergistic effect? Biotechnol. Biofuels 4, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Igarashi K., Uchihashi T., Koivula A., Wada M., Kimura S., Okamoto T., Penttilä M., Ando T., Samejima M. (2011) Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333, 1279–1282 [DOI] [PubMed] [Google Scholar]
- 29. Sturcová A., His I., Apperley D. C., Sugiyama J., Jarvis M. C. (2004) Structural details of crystalline cellulose from higher plants. Biomacromolecules 5, 1333–1339 [DOI] [PubMed] [Google Scholar]
- 30. Fox J. M., Levine S. E., Clark D. S., Blanch H. W. (2012) Initial- and processive-cut products reveal cellobiohydrolase rate limitations and the role of companion enzymes. Biochemistry 51, 442–452 [DOI] [PubMed] [Google Scholar]
- 31. Jalak J., Kurašhin M., Teugjas H., Väljamäe P. (2012) Endo-exo synergism in cellulose hydrolysis revisited. J. Biol. Chem. 287, 28802–28815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Medve J., Karlsson J., Lee D., Tjerneld F. (1998) Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol. Bioeng. 59, 621–634 [PubMed] [Google Scholar]
- 33. Liu Y. S., Baker J. O., Zeng Y., Himmel M. E., Haas T., Ding S. Y. (2011) Cellobiohydrolase hydrolyzes crystalline cellulose on hydrophobic faces. J. Biol. Chem. 286, 11195–11201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Igarashi K., Koivula A., Wada M., Kimura S., Penttilä M., Samejima M. (2009) High speed atomic force microscopy visualizes processive movement of Trichoderma reesei cellobiohydrolase I on crystalline cellulose. J. Biol. Chem. 284, 36186–36190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kubicek C. P., Bolzlbauer U. M., Kovacs W., Mach R. L., Kuhls K., Lieckfeldt E., Borner T., Samuels G. J. (1996) Cellulase formation by species of Trichoderma sect. Longibrachiatum and of Hypocrea spp. with anamorphs referable to Trichoderma sect. Longibrachiatum. Fungal Genet. Biol. 20, 105–114 [DOI] [PubMed] [Google Scholar]
- 36. Druzhinina I. S., Komon-Zelazowska M., Ismaiel A., Jaklitsch W., Mullaw T., Samuels G. J., Kubicek C. P. (2012) Molecular phylogeny and species delimitation in the section Longibrachiatum of Trichoderma. Fungal Genet. Biol. 49, 358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marx M.-C., Wood M., Jarvis S. C. (2001) A microplate fluorimetric assay for the study of enzyme diversity in soils. Soil Biol. Biochem. 33, 1633–1640 [Google Scholar]
- 38. Josefsson P., Henriksson G., Wågberg L. (2008) The physical action of cellulases revealed by a quartz crystal microbalance study using ultrathin cellulose films and pure cellulases. Biomacromolecules 9, 249–254 [DOI] [PubMed] [Google Scholar]
- 39. Ghose T. K. (1987) Measurement of Cellulase Activities. Pure Appl. Chem. 59, 257–268 [Google Scholar]
- 40. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 41. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M., ed), Humana Press [Google Scholar]
- 42. Lee I., Evans B. R., Woodward J. (2000) The mechanism of cellulase action on cotton fibers: evidence from atomic force microscopy. Ultramicroscopy 82, 213–221 [DOI] [PubMed] [Google Scholar]
- 43. Wang J., Quirk A., Lipkowski J., Dutcher J. R., Hill C., Mark A., Clarke A. J. (2012) Real-Time Observation of the Swelling and Hydrolysis of a Single Crystalline Cellulose Fiber Catalyzed by Cellulase 7B from Trichoderma reesei. Langmuir 28, 9664–9672 [DOI] [PubMed] [Google Scholar]
- 44. Nieves R. A., Ellis R. P., Todd R. J., Johnson T. J., Grohmann K., Himmel M. E. (1991) Visualization of Trichoderma reesei cellobiohydrolase I and endoglucanase I on aspen cellulose by using monoclonal antibody-colloidal gold conjugates. Appl. Environ. Microbiol. 57, 3163–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koivula A., Kinnari T., Harjunpää V., Ruohonen L., Teleman A., Drakenberg T., Rouvinen J., Jones T. A., Teeri T. T. (1998) Tryptophan 272: an essential determinant of crystalline cellulose degradation by Trichoderma reesei cellobiohydrolase Cel6A. FEBS Lett. 429, 341–346 [DOI] [PubMed] [Google Scholar]
- 46. Zhou W., Xu Y., Schüttler H.-B. (2010) Cellulose Hydrolysis in Evolving Substrate Morphologies III: Time-Scale Analysis. Biotech. Bioeng. 107, 224–234 [DOI] [PubMed] [Google Scholar]
- 47. Cruys-Bagger N., Elmerdahl J., Praestgaard E., Tatsumi H., Spodsberg N., Borch K., Westh P. (2012) Pre-steady-state kinetics for hydrolysis of insoluble cellulose by cellobiohydrolase Cel7A. J. Biol. Chem. 287, 18451–18458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kurasin M., Väljamäe P. (2011) Processivity of cellobiohydrolases is limited by the substrate. J. Biol. Chem. 286, 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mansfield S. D., Mooney C., Saddler J. N. (1999) Substrate and enzyme characteristics that limit cellulose hydrolysis. Biotechnol. Prog. 15, 804–816 [DOI] [PubMed] [Google Scholar]