Background: Transglycosylases are important enzymes in bacterial glycogen metabolism.
Results: The tertiary structure and function of a novel α-transglucosylase have been defined.
Conclusion: In addition to previously known activities, glycoside hydrolase family 31 (GH31) contains a group of enzymes with 1,4-α-glucan 4-α-glucosyltransferase activity.
Significance: This gives new insight into bacterial glycogen utilization and will inform future bioinformatics analyses of (meta)genomes.
Keywords: Carbohydrate, Carbohydrate Metabolism, Enzyme Catalysis, Enzyme Kinetics, Glycogen, GH31, Acarbose, Starch, Transglucosylase, Transglycosylase
Abstract
The metabolism of the storage polysaccharides glycogen and starch is of vital importance to organisms from all domains of life. In bacteria, utilization of these α-glucans requires the concerted action of a variety of enzymes, including glycoside hydrolases, glycoside phosphorylases, and transglycosylases. In particular, transglycosylases from glycoside hydrolase family 13 (GH13) and GH77 play well established roles in α-glucan side chain (de)branching, regulation of oligo- and polysaccharide chain length, and formation of cyclic dextrans. Here, we present the biochemical and tertiary structural characterization of a new type of bacterial 1,4-α-glucan 4-α-glucosyltransferase from GH31. Distinct from 1,4-α-glucan 6-α-glucosyltransferases (EC 2.4.1.24) and 4-α-glucanotransferases (EC 2.4.1.25), this enzyme strictly transferred one glucosyl residue from α(1→4)-glucans in disproportionation reactions. Substrate hydrolysis was undetectable for a series of malto-oligosaccharides except maltose for which transglycosylation nonetheless dominated across a range of substrate concentrations. Crystallographic analysis of the enzyme in free, acarbose-complexed, and trapped 5-fluoro-β-glucosyl-enzyme intermediate forms revealed extended substrate interactions across one negative and up to three positive subsites, thus providing structural rationalization for the unique, single monosaccharide transferase activity of the enzyme.
Introduction
Glycogen is a highly branched, mixed linkage α(1→4)/α(1→6)-glucan polymer that serves as a readily accessible, osmotically neutral, cellular energy reserve in all domains of life (1–3). Glycogen is structurally related to amylopectin, which together with the linear polysaccharide amylose (α(1→4)-glucan) comprises the plant storage reserve starch (4). Prokaryotic and eukaryotic glycogen biosynthesis and degradation are a complex, highly conserved, and tightly controlled process involving a myriad of enzymes and regulatory factors (2, 3). In bacteria such as Escherichia coli, glycogen is synthesized from ADP-glucose by the combined action of glycogen synthase, which builds linear α(1→4)-glucan chains, and glycogen branching enzyme, which catalyzes chain rearrangement via α(1→4)-to-α(1→6) transglycosylation, thereby yielding polydisperse molecules with molar masses of up to 107–108 Da (2, 3). In turn, catabolism under carbon-limited conditions occurs via the sequential action of glycogen phosphorylase and debranching enzyme to yield glucose 1-phosphate.
In bacteria, glycogen metabolism is closely linked to the metabolism of storage maltodextrins that involves the buildup and rearrangement of linear α(1→4)-gluco-oligosaccharides by transglycosylation (5). This process has been well described in E. coli in which the amylomaltase MalQ, a member of glycoside hydrolase family 77 (GH774; Ref. 6), catalyzes the transfer of a 4-α-glucanosyl fragment from the non-reducing end of malto-oligosaccharide donor substrates and possibly the disaccharide maltose to malto-oligosaccharide acceptors (4-α-glucanotransferase activity; EC 2.4.1.25) (5, 7, 8). The bacterial amylomaltases are structurally and functionally related to the plant disproportionating enzymes (“D-enzymes”) of GH77, which transfer maltosyl and longer 4-α-glucanosyl units from maltotriose and higher congeners (9). Likewise, certain thermophilic bacterial 4-α-glucanotransferases of GH13 catalyze the disproportionation of maltotriose (10, 11), maltotetraose (12, 13), and longer 4-α-glucan chains.
Indeed, transglycosylation reactions leading to the rearrangement of α-glucans are widespread among bacteria. Glycogen branching and debranching aside, diverse enzymes with 4-α-glucanotransferase activity (α(1→4)-glucan:α(1→4)-glucan transferase activity) can produce a range of linear and cyclic maltodextrin products via freely reversible disproportionation and cyclization reactions, respectively (9, 14). The production of six-, seven-, and eight-membered α(1→4)-linked cyclodextrins by the cyclodextrin glucanotransferases (EC 2.4.1.19) of GH13 represents an especially important process in industrial starch valorization (15). Analogous α(1→6)-linked cycloisomalto-oligosaccharides are the main products of some GH66 enzymes (16–18). Likewise, certain bacterial members of GH77 catalyze the production of large cyclic α-glucans (degrees of polymerization ≥22) through intramolecular transglycosylation (9). The GH31 enzymes CtsY and CtsZ from Arthrobacter and Sporosarcina species generate commercially interesting cycloalternan tetrasaccharides (cyclo[→6)-α-d-Glcp-(1→3)-α-d-Glcp-(1→6)-α-d-Glcp-(1→3)-α-d-Glcp-(1→]) from α(1→4)-glucans via a remarkable three-step, coupled reaction involving α(1→4)-to-α(1→6) transglycosylation, intermolecular isomaltosyl transfer, and cyclization (19, 20). In addition to these predominant transglycosylases, a number of retaining α-glucoside hydrolases with degrees of transglycosylation ability have also been identified (Refs. 21–23 and references therein).
The Gram-negative soil saprophyte Cellvibrio japonicus is best known for its ability to efficiently utilize a plethora of plant cell wall polysaccharides as energy sources (24). Additionally, the genome sequence of this organism has revealed a large number of predicted α-glucan-active enzymes. In total, the C. japonicus genome encodes 22 enzymes from GH13, GH15, GH31, GH57, and GH77 (25) that may be predicted to act on starch and/or glycogen. However, none of these have been biochemically or structurally characterized (6, 25).
GH31 in particular is one of the major α-glucosidase-containing glycoside hydrolase families. This family is functionally diverse; it also contains α-xylosidases and α-glucan lyases in addition to the aforementioned CtsY and CtsZ α-transglycosylases. A phylogenetic analysis has recently been presented that partially delineates these activities in clades, although sequence-based functional prediction is not absolute (26). The generally exo-acting GH31 enzymes, which are members of Clan GH-D together with GH27 and GH36, have been suggested to share a common ancestor with members of clan GH-H, which comprises generally endo-acting α-glucan-active enzymes of GH13, GH70, and GH77 (27).
Building upon our interest in the postgenomic characterization of GH31 enzymes from C. japonicus (26), we present here a detailed structural enzymology study of CjAgd31B, whose coding sequence resides within a gene cluster encoding predicted α-glucan-active enzymes and sugar transporters. Biochemical analysis revealed that CjAgd31B is a predominant transglucosylase with strict α(1→4) linkage specificity, which represents a previously undiscovered activity in GH31. Crystallography of the enzyme in free, acarbose-complexed, and trapped 5-fluoro-β-d-glucopyranosyl-enzyme intermediate forms has highlighted the structural basis for the strict transfer of a single glucosyl residue and preference for maltotriose and longer substrates. Taken together, the data suggest a biological role for CjAgd31B in glycogen or maltodextrin metabolism that may be complementary to that predicted for the GH77 homologue CjMal77Q.
EXPERIMENTAL PROCEDURES
Curve fitting and processing of kinetics data were performed using Origin 8 software (OriginLab). p-Nitrophenyl (pNP) α-glycosides, sucrose, d-maltose, and starch from corn were purchased from Sigma. Malto-oligosaccharides (maltotriose to maltohexaose), isomaltose, melibiose, and acarbose were purchased from Carbosynth. α-Glucosyl fluoride and 5-fluoro-α-d-glucopyranosyl fluoride were kind gifts from Professor Stephen Withers (Department of Chemistry, University of British Columbia, Canada). Ultrapure water was used in all experiments and refers to water purified on a Milli-Q system (Millipore) with a resistivity (ρ) >18.2 megaohms·cm.
Cloning of CjAgd31B
The open reading frame encoding CjAgd31B (GenBank accession number ACE84782.1) was amplified by PCR from genomic DNA of C. japonicus Ueda107 using Phusion polymerase (Finnzymes) and the following primers (Thermo Fischer Scientific): 5′-CACCATGAATCCGGTCAAACG-3′ and 5′-ATGCAACCTGAGGTTAAGCGCTTC-3′ with the forward primer incorporating the CACC overhang (underlined) needed for TOPO cloning and excluding the predicted signal peptide (cleavage site between amino acid residues 24 and 25). The PCR product was cloned into the pENTR/SD/D-TOPO entry vector (Invitrogen) and recombined into the pET-DEST42 destination vector (Invitrogen) as described previously (26).
Gene Expression and Protein Purification
Plasmids harboring the CjAgd31B gene were transformed into E. coli BL21(DE3) by electroporation, the gene was expressed and the resulting protein was purified by immobilized metal affinity chromatography following an established protocol (26). Analysis by SDS-PAGE showed the protein to be electrophoretically pure. LC electrospray ionization MS was used for protein molar mass determination as described previously (28). For crystallization studies, the protein was further purified by size exclusion chromatography and ion exchange chromatography. The eluted protein solution was concentrated to 5 ml by a Vivaspin 20 concentrator (Sartorius Stedim Biotech) and loaded onto a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) equilibrated with 20 mm Tris (pH 8.0), 300 mm sodium chloride. The eluted protein solution was dialyzed into 20 mm Tris (pH 8.0) at 4 °C for 16 h. The dialyzed protein solution was loaded onto a Resource Q column (GE Healthcare) equilibrated with 20 mm Tris (pH 8.0) and eluted with a linear gradient of 20 mm Tris (pH 8.0), 400 mm sodium chloride. Two major peaks of CjAgd31B were obtained, and the peak eluted in lower salt concentration was collected and used for protein crystallization. Protein concentrations were determined from A280 values of suitably diluted samples using an extinction coefficient of 139,245 m−1·cm−1 as calculated by the ProtParam tool on the ExPASy server (29).
Thin Layer Chromatography (TLC)
TLC was performed using normal phase silica on aluminum plates eluted with acetonitrile-water (2:1). Analytes were visualized by immersion in 8% H2SO4 in ethanol followed by charring.
High Performance Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD)
Oligo- and monosaccharides were analyzed on a Dionex ICS-3000 HPLC system operated by Chromelion software version 6.80 (Dionex) using a Dionex Carbopac PA200 column. Solvent A was water, solvent B was 1 m sodium hydroxide, and solvent C was 1 m sodium acetate.
Gradient A
Conditions used were 0–5 min, 10% B, 2% C; 5–12 min, 10% B and a linear gradient from 2 to 30% C; 12–12.1 min, 50% B, 50% C; 12.1–13 min, an exponential gradient of B and C back to the initial conditions; 13–17 min, initial conditions.
Gradient B
Conditions used were 0–4 min, 10% B, 5% C; 4–8 min, 10% B and a linear gradient from 5 to 25% C; 8–8.1 min, 50% B, 50% C; 8.1–9 min, an exponential gradient of B and C back to the initial conditions; 9–13 min, initial conditions.
Gradient C
Conditions used were 0 to 4 min, 10% B, 6% C; 4 to 17 min, 10% B and a linear gradient from 6–25% C; 17 to 17.1 min, 50% B, 50% C; 17.1 to 18 min, an exponential gradient of B and C back to initial conditions; 18 to 22 min, initial conditions.
Gradient D
Conditions used were 0–4 min, 10% B, 6% C; 4–10 min, 10% B and a linear gradient from 5 to 25% C; 10–10.1 min, 50% B, 50% C; 10.1–11 min, an exponential gradient of B and C back to the initial conditions; 11–15 min, initial conditions.
Gradient E
Conditions used were 0–4 min, 10% B, 6% C; 4–15 min, 10% B and a linear gradient from 5 to 25% C; 15–15.1 min, 50% B, 50% C; 15.1–16 min, an exponential gradient of B and C back to the initial conditions; 16–20 min, initial conditions.
pH-Rate Profile
Measurements of the pH dependence of CjAgd31B catalysis were carried out using maltose as substrate in an HPAEC-PAD-based transglycosylation assay described below. The buffers used (50 mm) were sodium citrate (pH 3–6.5), sodium phosphate (pH 6.5–8), glycylglycine (pH 8–9), and glycine (pH 9–10) (supplemental Fig. S1).
Enzyme Assays
Activity on pNP-glycosides was analyzed by a stopped assay as described previously (26) using an enzyme concentration of 6.5 μm. The transglycosylation activity of CjAgd31B on various oligosaccharides was performed in 100-μl reactions at 25 °C in 50 mm citrate buffer (pH 6). For initial rate saturation kinetics experiments, CjAgd31B was added to a final concentration of 1.6 μm for maltose and 270 pm for maltotriose, maltotetraose, and maltopentaose. Reactions typically proceeded for 10 min and were stopped by addition of 4 μl of 5 m sodium hydroxide. HPAEC-PAD was used for product analysis using Gradients A, B, D, and E for reactions on maltose, maltotriose, maltotetraose, and maltopentaose, respectively. Commercial malto-oligosaccharides (maltose to maltohexaose) were used as standards.
To test different acceptors, starch was used as a glucosyl donor. Starch from corn was dissolved to 1% (w/v) in water followed by dialysis in deionized water using a 5-kDa-cutoff membrane to remove monosaccharides and small oligosaccharides. 50-μl reactions containing 0.4% starch as donor, 1 mm acceptor (glucose or isomaltose), and 2 μm enzyme were incubated at 25 °C for 10 min and terminated by addition of 2 μl of 5 m sodium hydroxide. Products were analyzed by HPAEC-PAD using Gradient C.
IC50 Measurements
The inhibition of CjAgd31B by acarbose (0–1050 μm) was determined by using maltotriose (100 μm) as a substrate in reactions as described above. Product formation was analyzed by HPAEC-PAD (Gradient B).
Crystallization and Data Collection
CjAgd31B was stored in 5 mm Bistris propane (pH 8.5) and concentrated to 7 mg/ml by using a Vivaspin 20 concentrator. In initial crystal screens using Crystal Screen HT, Index HT, SaltRx HT (Hampton Research), and modified Newcastle Screen prepared at the York Structural Biology Laboratory, small single crystals were obtained in several conditions. Well diffracting crystals were obtained after 3–4 days in 1.8 m ammonium sulfate, 0.1 m HEPES (pH 7.0), 2% PEG 400 at 20 °C by the sitting drop vapor diffusion method. The structure was solved using experimental phasing with an iodine derivative. This was prepared by placing ∼1 μl of a 0.25 g/ml potassium iodide solution into a 2-μl crystallization droplet to allow slow diffusion of iodine into the crystal. Crystallization droplets with iodine solution were left at 20 °C for 16 h prior to freezing and data collection. For the complex structures, crystals were soaked in 1.8 m ammonium sulfate, 0.1 m HEPES (pH 7.0), 2% PEG 400 with either 5 mm 5-fluoro-α-d-glucopyranosyl fluoride or 5 mm acarbose for 1 h at 20 °C.
All crystals were cryoprotected by 2.0 m lithium sulfate, 0.1 m HEPES (7.0), 2% PEG 400. The x-ray data for the free enzyme and iodine-soaked crystals were collected at 100 K on an ADSC Q315 charge-coupled device detector on beamline I02 of the Diamond Light Source. X-ray data for the complex with acarbose and the 5FβGlc-enzyme were collected at 100 K on an ADSC Q315 charge-coupled device detector at BL-ID14-4 and BL-ID29 at the European Synchrotron Radiation Facility, respectively. The details of the data collections are listed in Table 1. All data were processed using iMOSFLM (30) and programs from the CCP4 suite (31) unless otherwise stated. The statistics of the data processing and structure refinement are listed in Table 1.
TABLE 1.
X-ray data collection and refinement statistics
OXL, oxalate; EDO, 1,2-ethanediol; PEG, 2-(2-hydroxyethoxy)ethanol; PGE, 2-[2-(2-hydroxyethoxy)ethoxy]ethanol; PG4, 2-[2-[2-(2-hydroxyethoxy)ethoxy]ethoxy]ethanol; r.m.s.d., root mean square; 5FβGlc, 5-fluoro-β-glucosyl; ESRF, European Synchrotron Radiation Facility; SO4, sulfate.
| Name (Protein Data Bank code) |
|||
|---|---|---|---|
| Free enzyme (4b9y) | 5FβGlc (4ba0) | Acarbose (4b9z) | |
| Data collection | |||
| Beamline | Diamond I02 | ESRF ID29 | ESRF14-4 |
| Space group | P622 | P622 | P622 |
| Cell dimensions | |||
| a, b, c (Å) | 197.3, 197.3, 103.0 | 197.2, 197.2, 103.1 | 196.9, 196.9, 102.8 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) (outer shell) | 49.85–1.90 (2.00–1.90) | 44.48–1.85 (1.95–1.85) | 51.38–2.0 (2.11–2.00) |
| Rmerge | 0.10 (0.52) | 0.10 (0.52) | 0.11 (0.46) |
| I/σI | 21.5 (6.0) | 19.8 (5.9) | 22.2 (7.4) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 22.0 | 20.8 | 21.5 |
| Refinement | |||
| Resolution (Å) | 49.3–1.90 | 44.5–1.85 | 49.7–2.00 |
| No. reflections | 92,667 | 100,216 | 79,078 |
| Rwork/Rfree | 0.16/0.19 | 0.17/0.19 | 0.16/0.19 |
| No. atoms | |||
| Protein | 6,273 | 6,270 | 6,251 |
| Ligand | 93 | 80 | 142 |
| Water | 647 | 557 | 444 |
| B-factors (Å2) | |||
| Protein (TLS refinement) | 27.9 | 31.2 | 29.3 |
| Water | 33.8 | 36.8 | 32.4 |
| Ligands | SO4, 80.8; PG4, 44.8; OXL, 42.7; EDO, 48.2 | 5FβGlc, 21.2; SO4, 71.3; PGE, 47.7; Arg, 48.2; PEG, 34.4; EDO, 53.8 | Acarbose, 40.2; SO4, 55.3; OXL, 55.1; PEG, 37.5; EDO, 53.1 |
| r.m.s.d. | |||
| Bond lengths (Å) | 0.007 | 0.006 | 0.007 |
| Bond angles (°) | 1.1 | 1.1 | 1.1 |
Experimental phasing was performed by single wavelength anomalous diffraction methods at a wavelength of 1.8 Å. Heavy atom substructure solution and initial phasing was performed on the 2.9-Å resolution iodine-derivatized crystal data with autoSHARP (32) followed by phase extension with the 1.9-Å free enzyme data set using DM (33). The 1.9-Å data were used as a starting point for automatic model building using ARP/wARP (34) (Table 2). Structure refinement, including TLS refinement of molecular motions, was performed using PHENIX (35) interspersed with manual rebuilding using COOT (36). Complex structures were solved by molecular replacement using MOLREP (37) with the free enzyme structure as a search model and refined as above.
TABLE 2.
Iodine single wavelength anomalous diffraction (SAD) phasing statistics
FOM, figure of merit.
| Data collection | |
|---|---|
| Beamline | Diamond I02 |
| Wavelength (Å) | 1.80 |
| Space group | P622 |
| Cell dimensions | |
| a, b, c (Å) | 197.7, 197.7, 102.6 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) (outer shell) | 71.17–2.9 (3.06–2.90) |
| Total no. of measured reflections | 1,127,119 |
| No. of unique reflections | 26,692 |
| Rmerge | 0.16 (0.74) |
| I/σI | 24.9 (6.7) |
| Anomalous completeness (%) | 100.0 (100.0) |
| Anomalous redundancy | 22.6 (22.5) |
| FOM SAD/density modification | 0.17/0.84 |
RESULTS
Bioinformatics Analyses
CjAgd31B is found among putative receptors, transporters, and α-glucan-active enzymes in the genome of C. japonicus (supplemental Fig. S2). Notably, these genomic neighbors include a predicted α-amylase, cyclomaltodextrin glucanotransferase, 6-phospho-β-glucosidase, and a glucokinase. CjAgd31B has a predicted secretion signal peptide and is thus likely to be localized in the periplasm or to be secreted extracellularly. In addition to CjAgd31B, the genome of C. japonicus encodes two other GH31 members, the putative α-glucosidase CjAgd31A and the biochemically and structurally characterized α-xylosidase CjXyl31A (26). Although all are members of GH31, CjAgd31B has a low sequence similarity to both CjAgd31A and CjXyl31A with amino acid identities of 28 and 27% and similarities of 43 and 45%, respectively. From our recent phylogenetic analysis (26), the biochemically characterized member of GH31 most similar to CjAgd31B is YihQ from E. coli (sequence identity, 28%; similarity, 44%). E. coli YihQ has been annotated as an α-glucosidase based on a weak ability to hydrolyze the artificial substrate α-glucosyl fluoride, although the enzyme was impotent toward a range of other α-glucosides (38).
Gene Expression
A gene construct encoding CjAgd31B with a C-terminal hexahistidine tag and lacking the predicted native signal peptide was expressed in E. coli BL21(DE3) cells. The protein product was purified by immobilized metal affinity chromatography for kinetics analyses and additionally by size exclusion chromatography and ion exchange for crystallization studies; purity in both cases was confirmed by SDS-PAGE (data not shown). The molar mass of CjAgd31B, corresponding to the C-terminal His-tagged enzyme starting at Asn-25 (the natural site of signal peptide cleavage), was verified by LC electrospray ionization MS (expected, 94,478.8 Da; observed, 94,478.1 Da; supplemental Fig. S3). The overall yield was typically around 100 mg/liter of culture broth.
Transglycosylation Activity on Malto-oligosaccharides
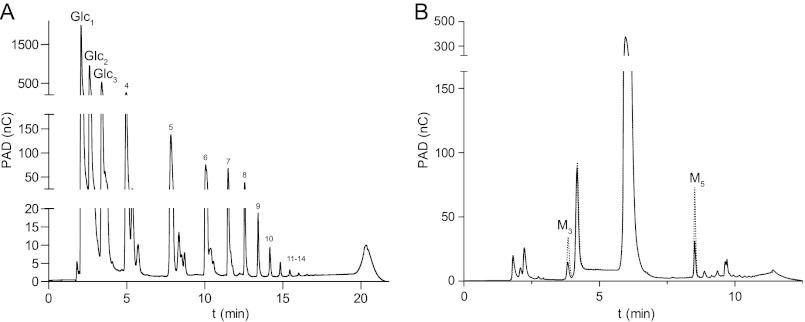
Based on membership in GH31, the substrate specificity of CjAgd31B was initially tested using pNP-α-glucoside and pNP-α-xyloside; the enzyme showed no apparent liberation of the aglycone from either of these substrates after extended incubation (1 mm substrate and 6.5 μm enzyme and up to 4-h incubation). The enzyme also displayed no detectable activity on sucrose, melibiose, isomaltose, or α-glucosyl fluoride (a substrate for E. coli YihQ (38)). On maltose (Glcpα(1→4)Glc), however, formation of both glucose and longer oligosaccharides could be observed using TLC (data not shown). The transglycosylation potential of CjAgd31B was confirmed by HPAEC-PAD following incubation of 6.5 μm enzyme with 10 mm maltose for 30 min, which led to a buildup of malto-oligosaccharides. Products with a degree of polymerization of up to 14 glucose residues could be detected (Fig. 1A).
FIGURE 1.
HPAEC-PAD chromatograms of reaction products of malto-oligosaccharides, catalyzed by CjAgd31B. A, incubation with maltose (10 mm; 6.5 mm enzyme; 30 min). Shoulder peaks are assumed to be transglycosylation products arising from contaminants in the commercial maltose preparation. B, incubation with maltotetraose (0.5 mm; 10-min reaction). Solid line, commercial maltotetraose sample prior to addition of enzyme; dotted line, enzyme reaction indicating production of maltotriose (M3) and maltopentaose (M5) by disproportionation. Commercial malto-oligosaccharides were used as standards to determine retention times. nC, nanocoulomb.
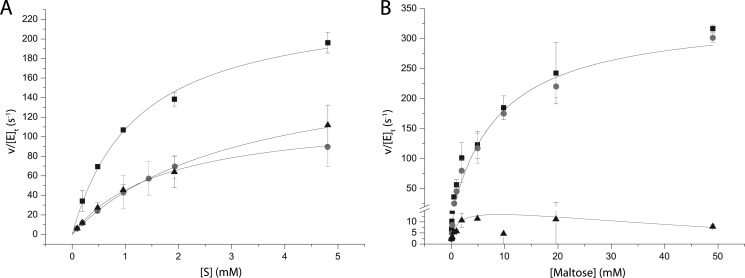
To further analyze the catalytic properties of CjAgd31B, HPAEC-PAD was used to measure product formation from reactions on malto-oligosaccharides. Under conditions of low substrate conversion (<10% of substrate consumed), the enzyme was shown to transfer a single glucose moiety from a donor to an acceptor molecule. Incubation of the enzyme with linear malto-oligosaccharides (maltotriose to maltopentaose) exclusively yielded Glcn − 1 and Glcn + 1 products via transglucosylation (Fig. 1B). On maltotriose, maltotetraose, and maltopentaose, the production of glucose, which would indicate competing substrate hydrolysis, was not observed under initial rate conditions. Thus, apparent Michaelis-Menten kinetics parameters for these substrates could be directly determined from plots of v0/[E]t versus [Glcn] (Fig. 2A) where the rate of transglycosylation product formation is given by Equation 1 (in this case, the rate of Glcn − 1 formation can also be used). The best substrate for the enzyme was maltotriose with a (kcat/Km)app value of 196 s−1·mm−1, whereas maltotetraose and maltopentaose displayed comparable (kcat/Km)app values of 72 and 58 s−1·mm−1, respectively (Table 3).
 |
Hydrolysis could, however, be detected when maltose was used as a substrate. Here, the production of Glc was measurably higher than the 1:1 stoichiometric ratio of Glc to Glc3 expected for disproportionation (Glc2 → Glc3 + Glc). In this case, the velocity of the transglycosylation reaction was directly measured according to Equation 1 by quantifying the maltotriose produced. Determination of the hydrolytic rate required subtraction of the amount of glucose co-produced by disproportionation from that arising from hydrolysis, including accounting for the stoichiometry of glucose release by hydrolysis (2 mol of Glc/mol of maltose) (Equation 2). From the specific transglycosylation activity on 0.5 mm maltose, the pH profile was found to be broad with an optimum of pH 6.5 (supplemental Fig. S1).
 |
Fig. 2B shows plots of v0/[E]t versus [Glc2] for both the hydrolytic and transglycosylation reactions, which clearly indicate the predominance of transglycosylation over a wide range of substrate concentrations. The (kcat/Km)app value determined for the transglycosylation reaction was 39 s−1·mm−1 (Table 3). Although the (kcat)app value for maltose transglycosylation is actually higher than that of maltotriose, the lower (kcat/Km)app value on the disaccharide is due to a high (Km)app value. This high value may reflect the observation of up to four enzyme subsites (−1 to +3; nomenclature according to Ref. 39) in the crystal structure (see below) of which only two would be occupied by maltose acting as a donor substrate.
FIGURE 2.
A, initial rate kinetics curves fitted to the Michaelis-Menten equation for maltotriose (black squares), maltotetraose (gray circles), and maltopentaose (black triangles). B, initial rate kinetics curves fitted to the Michaelis-Menten equation for maltose. Black squares represent formed glucose, gray circles represent maltotriose, and black triangles represent the rate of hydrolysis. Error bars represent two standard deviations from the mean value.
TABLE 3.
Transglycosylating activity of CjAgd31B on various substrates
ΔΔG‡ was calculated using the following formula: ΔΔG‡ = RTln(kcat/Km[maltotriose]/kcat/Km[maltooligo]). No activity was detected on pNP-α-Glc, pNP-α-Xyl, α-glucosyl fluoride, isomaltose, sucrose, or melibiose.
| Substrate | (kcat)app | (Km)app | (kcat/Km)app | ΔΔG‡ |
|---|---|---|---|---|
| s−1 | mm | s−1· mm−1 | kJ mol−1 | |
| Maltose | 341 ± 12 | 8.8 ± 0.87 | 38.8 | 4.17 |
| Maltotriose | 239 ± 8.3 | 1.2 ± 0.13 | 195.5 | |
| Maltotetraose | 123 ± 9.0 | 1.7 ± 0.29 | 72.3 | 2.57 |
| Maltopentaose | 181 ± 17 | 3.1 ± 0.55 | 57.8 | 3.14 |
Across the range of maltose concentrations examined (50 μm to 50 mm), the rate of hydrolysis was consistently low (Fig. 2B). At 50 μm maltose, the rate of hydrolysis was equal to the rate of transglycosylation (v0/[E]t = 2 s−1) after which the hydrolytic rate increased to a maximum of 10 s−1 at 2 mm substrate. Nonetheless, the hydrolytic rate was only 13% of the transglycosylation rate at this maximum. The rate of hydrolysis steadily decreased to ∼8 s−1 at 50 mm maltose as the transglycosylation rate continued to increase, resulting in a relative hydrolysis rate of <2% at this concentration.
Starch and Isomaltose as an Alternate Donor/Acceptor Pair
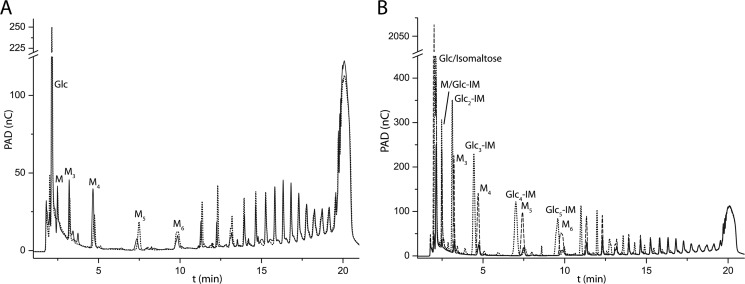
Starch was also tested as a donor substrate with high degree of polymerization with glucose and isomaltose as alternate acceptor substrates. Despite dialysis to reduce the content of short malto-oligosaccharides, the commercial starch sample contained a minor amount of α(1→4)-glucans with a degree of polymerization of 2 and higher. When CjAgd31B was incubated with starch alone, the amounts of shorter malto-oligosaccharides (maltose to maltotetraose) still present in the substrate mixture decreased significantly, whereas longer malto-oligosaccharides (maltopentaose to maltododecaose) increased in keeping with previous observations on purified malto-oligosaccharides during kinetics analyses (Fig. 3A). In contrast, when the reaction was supplemented with 1 mm glucose, a large increase of malto-oligosaccharides (maltose to maltododecaose) could be observed (Fig. 3B). Here, glucose (which as a monosaccharide cannot act as a glycosyl donor) acted as viable acceptor substrate in the breakdown of the glycosyl-enzyme intermediate formed via initial attack of starch chain ends. This reaction is the reverse of glycosyl-enzyme formation when maltose acts as a donor substrate, consistent with the principle of microscopic reversibility. The large increase in medium length malto-oligosaccharides compared with the glucose-free reaction both demonstrates how the non-reducing ends present in starch are utilized as glucose donors and suggests that glucose moieties are mainly “shuffled” between the non-reducing ends of the starch molecules when no additional acceptor is included in the reactions.
FIGURE 3.
HPAEC-PAD chromatograms of donor/acceptor specificity experiments using starch, isomaltose (IM), and glucose. A, starch blank (0.4%, w/v; solid line) and following incubation with CjAgd31B (dotted line) showing a reduction of short malto-oligosaccharides (M to M5) and an increased amount of medium length malto-oligosaccharides (M7 to M12). B, starch (0.4%) following incubation with CjAgd31B with no additional acceptor substrate added (solid line), with added isomaltose (1 mm; dashed line), and with added glucose (1 mm; dotted line). With both alternate acceptor substrates, there is a clear increase in the amount of short to medium length oligosaccharides formed compared with starch alone. nC, nanocoulomb.
The disaccharide isomaltose (Glcpα(1→6)-Glc) on its own was not a substrate for CjAgd31B, indicating that the enzyme was unable to utilize α(1→6)-linked glucosides as donor substrates. Glycogen and starch both contain α(1→6)-linked branch points, which prompted us to test whether such branches might be extended by CjAgd31B using isomaltose as a model. Indeed, the enzyme transferred glucosyl moieties from starch to isomaltose (1 mm) as indicated by the appearance of an alternate series of peaks on the chromatogram, each slightly preceding the corresponding all-α-linked congener, which suggests the formation of Glcn-α(1→4)-Glc-α(1→6)-Glc saccharides (Fig. 3B). As such, these data indicated that the +2 subsite of the active site is not strictly specific for α(1→4)-linked sugars but will accommodate α(1→6)-linked isomers.
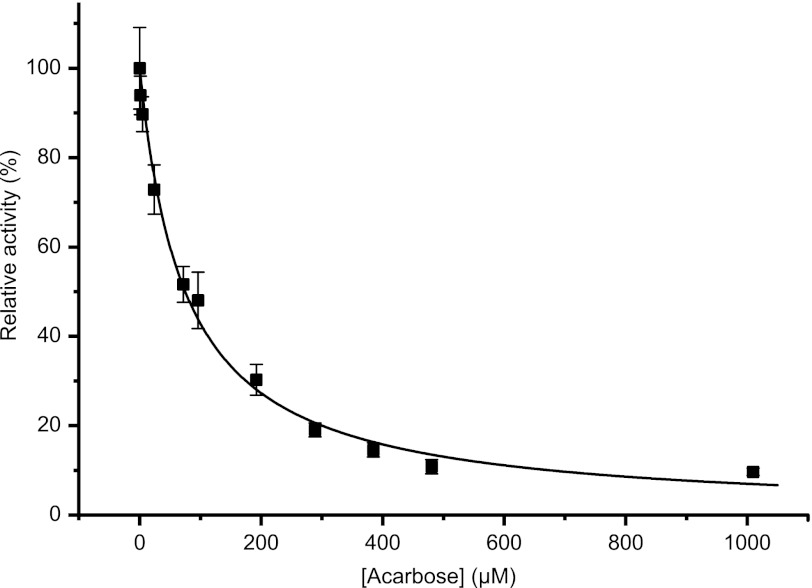
Inhibition by Acarbose
The inhibitory effect of the pseudotetrasaccharide acarbose, a common α-glucanase and α-glucosidase inhibitor, was assayed using maltotriose as the substrate at a fixed concentration of 100 μm. The IC50 value was determined to be 75.1 ± 3.4 μm by plotting the relative activity versus the concentration of acarbose and fitting Equation 3 by non-linear regression (Fig. 4). With reactions performed at a substrate concentration of 100 μm, which is much lower than the apparent Km value (1.2 mm), the IC50 value is approximately equal to the Ki value. The Ki may be more accurately calculated using Equation 4 (40), which yielded 81.4 μm.
 |
 |
The calculated Ki value of CjAgd31B is similar to Ki values for acarbose with other GH31 enzymes such as the human maltase-glucoamylase N-terminal subunit (62 μm; Ref.41) and sucrase N-terminal subunit (14 μm; Ref. 42), whereas it is significantly higher than the Ki values for the human maltase-glucoamylase C-terminal subunit (1.72 μm; Ref. 43), the Gracilariopsis lemaneiformis α-glucan lyase (0.02 μm; Ref. 44), and the Thermoplasma acidophilum AglA α-glucosidase (2.99 μm; Ref. 23).
FIGURE 4.
Inhibition of CjAgd31B by acarbose. Maltotriose was used as substrate (100 μm; 10-min reaction), and the inhibitory effect of acarbose on transglycosylation (formation of maltotetraose) was calculated by non-linear regression, fitting Equation 3 to the data. nC, nanocoulomb. Error bars represent two standard deviations from the mean value.
Tertiary Structures of CjAgd31B
Three tertiary structures of CjAgd31B were obtained at 1.9-, 2.0-, and 1.85-Å resolution, respectively: the free enzyme, a non-covalent complex with the inhibitor acarbose, and a trapped 5-fluoro-β-d-glucopyranosyl-enzyme intermediate. All crystals contained one molecule in the asymmetric unit. The CjAgd31B structures consisted of a typical GH31 fold comprising four domains with two insertions (Fig. 5A): the N-terminal domain (N-terminal; residues 35–240), the catalytic (β/α)8 domain (residues 241–586) with insertion domain 1 (Insert 1; residues 345–384) and insertion domain 2 (Insert 2; residues 415–435), the C-terminal proximal domain (C-proximal; residues 587–667), and the C-terminal distal domain (C-distal; residues 668–817). In all structures, the electron density map of the 10 N-terminal residues from 25 to 34 and C-terminal residues from 818 to the end (859), including the V5 epitope and His tag provided by the expression vector, were disordered. The free and acarbose structures had a disordered region from 137 to 140, and the 5-fluoro-β-glucosyl-enzyme structure had a disordered region from 139 to 140. The visible secondary structures of the four domains, including the insertion domains, in CjAgd31B were well conserved with the human sucrase-isomaltase (Protein Data Bank code 3lpp) of GH31 with a root mean square deviation of 1.9–2.4 Å.
FIGURE 5.
The three-dimensional structure and ligand binding site of CjAgd31B. A, three-dimensional structure of CjAgd31B as a protein schematic. The N-terminal β-sandwich domain is colored red; the central catalytic (β/α)8 domain is shown in blue with Inserts 1 and 2 colored purple and pink, respectively; the C-terminal proximal β-sandwich is orange; and the C-terminal distal β-sandwich domain is green. B, observed electron density for the acarbose complex of CjAgd31B (map is at 1 σ in divergent stereo). The nucleophile Asp-412, acid-base Asp-480, and hydrophobic “clamp” residues (discussed in text) are shown. This figure was drawn with CCP4MG (66).
The free enzyme structure reveals a water-lined pocket where the conserved catalytic aspartic acid residues (Asp-412 and Asp-480) are located (45, 46). The pocket was 318 Å3 as calculated by the Pocket-Finder server (47) with a depth of ∼11 Å. The active site pocket was composed of residues Tyr-179, Phe-271, Asp-299, Leu-300, Ile-307, Met-311, Ile-341, Glu-343, Tyr-376, Phe-377, Trp-410, Asp-412, Leu-413, Glu-417, Arg-463, Trp-477, Asp-480, Asp-509, Phe-513, Arg-538, His-540, and Gln-542 (supplemental Fig. S4). To define enzyme-substrate interactions, two complexes, with the inhibitory tetrasaccharide acarbose (see above) and the covalent 5-fluoro-β-d-glucopyranosyl-enzyme intermediate, were obtained.
Structure of the Acarbose Complex
The CjAgd31B complex with acarbose revealed clear, unambiguous density for the inhibitor in the −1 to +3 subsites (Fig. 5B; subsite nomenclature according to Ref. 39). In the −1 subsite, the enzyme-derived nucleophile, Asp-412, is indeed poised for nucleophilic attack, lying 3.2 Å “above” the pseudo-anomeric carbon of acarbose and with a nucleophile-C1-NH angle of 164.1°. The catalytic acid, Asp-480, lies 2.5 Å from the “interglycosidic” nitrogen of acarbose as expected. The hydrophobic residues Leu-300, Ile-341, Trp-410, Trp-477, Phe-271, and Phe-513 all lie within a 4-Å distance from the −1 subsite pseudosugar of acarbose (supplemental Fig. S4).
The +1 subsite contains the 6-deoxyglucosyl moiety of acarbose. Hydrogen bonds are made to Arg-463, Glu-417, and a water molecule, and enzyme-substrate distances suggest van der Waals contacts to Phe-377. In most other solved structures of GH31, the hydrogen bonds provided here by Glu-417 (Insert 2) are made instead by an aspartate residue from a loop in the N-terminal domain (42, 48–50); the other exception to this is found in CjXyl31A in which a PA14 domain insert in the N-terminal domain extends the active site (26, 42, 48–50).
The substrate-interacting residues in the −1 and +1 subsites of the CjAgd31B structure are essentially homologous to those in the human maltase-glucoamylase and the Ro-αG1 α-glucosidase from Ruminococcus obeum with the exception of Phe-271, Leu-300, Phe-377, and Leu-413, which instead are Trp, Ile, Trp, and Met, respectively, in the maltase-glucoamylase and Ro-αG1 structures (supplemental Fig. S5 and Refs. 48 and 49). These side chains all make van der Waals contacts to acarbose as well as the 5-fluoro-β-glucosyl residue (see below). It is possible that Phe-271 of CjAgd31B, corresponding to Trp-169 and Tyr-299 in Ro-αG1and maltase-glucoamylase, respectively, contributes to substrate specificity as wild type Ro-αG1 prefers isomaltose to maltose as a substrate, whereas the W169Y mutant inverts this preference (49).
Notable features of the +2 and +3 subsites are the tyrosine clamp of Tyr-179 (part of a long loop extending from the N-terminal domain) and Tyr-376 of Insert 1, which together form van der Waals contacts to the internal glucose moiety (third ring) of acarbose (cf. Fig. 5B and supplemental Fig. S4). A similar hydrophobic clamp has not been found in other GH31 structures apart from the PA14-mediated protein-sugar interaction in CjXyl31A (26).
Structure of the Trapped Covalent 5-Fluoro-β-glucosyl-enzyme Intermediate
To assess factors that may lead to the strict transglycosylation activity of CjAgd31B, a near mimic of the covalent glycosyl-enzyme intermediate was accessed using a classic “Withers” reagent, 5-fluoro-α-d-glucopyranosyl fluoride. The electron density map clearly reveals the trapped 5-fluoro-β-d-glucopyranosyl-enzyme observed in 1S3 skew boat conformation (conformational aspects of catalysis are reviewed in Ref. 51) formed via covalent linkage to the Oδ2 atom of Asp-412 with 1.34-Å distance (Fig. 6A). The majority of interactions of this sugar are the same as previously observed for the −1 subsite sugar of the acarbose pseudotetrasaccharide (supplemental Fig. S4), but in addition, two water molecules (Fig. 6A) bind to Oδ2 atom of Asp-480, the acid-base residue. These water molecules form a hydrogen bond network with Asp-480, Arg-463, Glu-417, and a third water molecule binding to Gln-542. Although transglycosylation is always kinetically favored over hydrolysis (52), what is unusual about CjAgd31B and indeed other transglycosylases is how they overcome the thermodynamically favored hydrolysis reaction in 55 m water as discussed below.
FIGURE 6.
The covalent intermediate of CjAgd31B. A, observed electron density for the trapped covalent 5-fluoro-β-glucosyl-enzyme intermediate of CjAgd31B (map is contoured at 1 σ in divergent stereo). The nucleophile Asp-412, acid-base Asp-480, and a pair of solvent water (Wat) molecules are shown. B, overlay of the −1 and +1 subsites of the covalent intermediate complex (pink; waters are shown as red spheres) with the acarbose (yellow) complex (Fig. 5B). Of particular note is that neither (nearby) water of the intermediate complex is in an appropriate position for nucleophilic attack of the intermediate by hydrolysis, implying that hydrogen bonding is optimized to prevent hydrolysis and facilitate transglycosylation. This figure was drawn with CCP4MG (66).
DISCUSSION
Through a combination of enzymological and structural analysis, we have revealed that CjAgd31B from the soil saprophyte C. japonicus possesses the ability to exclusively transfer single glucosyl units from α(1→4)-glucans to the non-reducing terminal 4-OH of glucose and α(1→4)- and α(1→6)-linked glucosyl residues; weak hydrolysis activity is only observed on the disaccharide maltose. As outlined in the Introduction and discussed below, this type of transglycosylase has not previously been described in GH31 nor any other CAZyme family to our knowledge.
GH31 enzymes utilize a double displacement mechanism involving a covalent glycosyl-enzyme intermediate, which, as was the case here for CjAgd31B, can be trapped and directly observed using kinetic probes derived from fluorosugars (46). In the natural reactions catalyzed by GH31 members, the glycosyl-enzyme is most commonly decomposed by water, yielding substrate hydrolysis. However, this intermediate can also be intercepted by saccharide acceptor substrates to generate transglycosylation products with varying efficiencies in a substrate- and enzyme-dependent manner (53, 54). Indeed, several members of GH31 have been shown to possess transglycosylation ability, although yields are typically low due to dominating hydrolytic reactions (21–23, 55, 56). In this context, the strict transglycosylating activity of CjAgd31B on malto-oligosaccharide substrates with degree of polymerization ≥3 is particularly noteworthy.
CjAgd31B is also distinct from homologues of bacterial ctsY and ctsZ gene products, which are the only other predominant transglycosylases to have been identified in GH31 thus far. Working in concert, CtsY and CtsZ generate cycloalternan tetrasaccharides from α(1–4)-glucans via a three-step reaction (19, 20). In the first step, CtsZ acts as an α(1→4)-to-α(1→6) transglucosylase to generate isomaltosyl moieties at the end of α(1→4)-glucan chains. Our HPAEC-PAD data indicate that in contrast CjAgd31B effects α(1→4)-to-α(1→4) transglycosylation and moreover cannot address (1→6) linkages; isomaltose is not a donor substrate. In the second and third steps of cycloalternan tetrasaccharide synthesis, CtsY catalyzes an intermolecular isomaltosyl transfer to yield a α-d-Glcp-(1→6)-α-d-Glcp-(1→3)-α-d-Glcp-(1→6)-α-d-Glcp-(1→4)-α-glucan structure followed by intramolecular cyclization to yield cyclo[→6)-α-d-Glcp-(1→3)-α-d-Glcp-(1→6)-α-d-Glcp-(1→3)-α-d-Glcp-(1→]. These two reactions are clearly distinct from that catalyzed by CjAgd31B. Unfortunately, the three-dimensional structures of both CtsZ and CtsY are currently unknown, which precludes comparison with CjAgd31B to understand the structural basis for these divergent transglycosylation activities in GH31.
The reactions catalyzed by CjAgd31B bear some similarity to, but are again distinct from, those catalyzed by 4-α-glucanotransferases (EC 4.2.1.25) of GH13 and GH77. GH13 encompasses a huge diversity of α-glucan-hydrolyzing and -transglycosylating enzymes (57) of which the Thermotoga spp. 4-α-glucanotransferases are perhaps the most relevant to the present study (12, 13, 58). Thermotoga maritima and Thermotoga neapolitana 4-α-glucanotransferases catalyze disproportionation reactions of malto-oligosaccharides, utilizing maltotetraose as the smallest donor substrate (12, 13). The structure of the T. maritima enzyme has revealed that the active site is an open cleft comprising at least 5 subsites (−2 to +3), which provides clear rationalization for the ability of the enzyme to randomly transfer longer α-glucan chains. In contrast, the T. maritima maltosyltransferase, also of GH13, strictly transfers maltosyl (Glc2) units from maltotriose and longer malto-oligosaccharides to the 4-position of α-glucan acceptor substrates (58) due to the presence of a unique protein motif that blocks the active site cleft (59).
GH77 enzymes are structurally related to GH13 enzymes in clan GH-H and thus generally possess open cleft-shaped active sites (60–62) that confer specificity for longer glucan donor substrates (9). For example, the eukaryotic Solanum tuberosum (potato) starch disproportionating (“D”) enzyme has such an extended active site (Protein Data Bank code 1x1n), transfers long α-glucan chains, and does not use maltose as a donor substrate (14, 63, 64). The disaccharide is also not a substrate for the Thermus aquaticus amylomaltase of GH77 that is distinguished by its propensity to form large cyclic α-glucans from long α(1→4)-glucan donors (14, 60). In the context of bacterial malto-oligosaccharide metabolism, the E. coli GH77 amylomaltase MalQ appears to favor the transfer of longer α-glucan chains, although there appears to be some debate whether this enzyme can utilize maltose as a donor, thereby transferring a single glucosyl residue to longer congeners.
CjAgd31B thus occupies a unique catalytic place as a 4-α-glucosyltransferase among the broader spectrum of 4-α-glucanotransferases. As a member of GH31, CjAgd31B belongs to clan GH-D, which is also composed of GH27 and GH36. Clan GH-D and GH-H members are built on a common triose isomerase (β/α)8 barrel scaffold and may share a distant evolutionary relationship (27). However, in contrast to the clefted clan GH-H members, clan GH-D members are typified by shallow, pocket-shaped active sites comprising only one negative subsite accommodating a monosaccharide residue, the glycosidic bond of which undergoes catalysis. CjAgd31B likewise presents an active site pocket as revealed by acarbose and 5-fluoro-glycosyl-enzyme complex structures, allowing speculation regarding its strict glucosyl transfer capacity.
The inherent challenge for transglycosylases is overcoming the thermodynamic preference for water as a nucleophile versus saccharide acceptor substrates. It has long been established by Withers et al. (52) through analysis of the reactivation of trapped intermediates that transglycosylation is kinetically favored over hydrolysis. Crystallographic analysis of the trapped covalent glycosyl-enzyme intermediate of CjAgd31B suggests that the hydrogen-bonding scheme does not place a water molecule with appropriate geometry or interaction with the catalytic acid-base residue Asp-480 to facilitate hydrolysis of the intermediate. Instead, two water molecules (one of them disordered) appear to lie on either side of the position expected of a catalytically competent nucleophile. However, the interglycosidic nitrogen of acarbose does interact with the acid-base (and with its O3 and C5 groups, binding in positions corresponding to the observed waters of the trapped intermediate).
This solvent hydrogen-bonding arrangement suggests that the CjAgd31B active site has evolved to avoid deprotonation and activation of water, whereas optimization of hydrogen bonds to O6 and O3 of acceptor glucosides in the +1 subsite allows a favorable placement of the O4 atom for deprotonation and concerted electrophilic migration of C1 of the β-glucosyl-enzyme. In this process, Glu-417 and Arg-463 play particularly important roles in binding the O3 of the +1 sugar and legislating against a water molecule positioned to enable hydrolysis. A caveat is that the trapped intermediate observed is that of a 5-fluoroglycoside, so it is possible that the observed solvent network is perturbed by the unnatural 5-fluoro substituent. However, the solvent network of the free enzyme structure is similar to that of the trapped intermediate complex especially in context of the O6- and O3-mimicking water molecules, where there is no suitably poised “nucleophilic” water molecule bound to Asp-480.
It is currently unclear what role(s) CjAgd31B might play in the biology of C. japonicus. Interestingly, the gene encoding CjAgd31B is located among a cluster of genes predicted to encode α-glucan-active enzymes (α-amylase, cyclomaltodextrin glucanotransferase, 6-phospho-β-glucosidase, and glucokinase) and transporter proteins (TonB-dependent receptors and ATP-binding cassette transporters; supplemental Fig. S2). Whether these genes are co-regulated or comprise an operon is currently not known. This genomic association together with the observation that CjAgd31B is encoded with a native secretion signal peptide hints toward a role in glycogen or starch metabolism in the periplasm. Indeed, CjAgd31B could possibly have a function similar to the GH77 amylomaltase MalQ from E. coli, which creates longer α-glucan chains from shorter malto-oligosaccharides as substrates for maltodextrin phosphorylases; these phosphorylases require maltopentaose as a minimal substrate to generate glucose 1-phosphate for further metabolism (5, 65, 66). C. japonicus, however, does possess a predicted GH77 homologue (CjMal77Q, CJA_1882; Ref. 25), which is located elsewhere in the genome and in proximity to other predicted glycogen/starch-active enzymes. This would suggest that CjAgd31B and CjMal77Q most likely have independent or perhaps complementary functions.
Another possible clue to the physiological function of CjAgd31B can be gleaned from analysis of potential GH31 orthologs. The biochemically characterized GH31 member closest to CjAgd31B is the α-glucosidase YihQ from E. coli K12 MG1655 (38). Biological data on E. coli YihQ are currently lacking; however, a reverse genetics analysis of a YihQ orthologue in Salmonella enterica serovar Enteriditis indicates that ΔyihQ mutants are deficient in capsular polysaccharide formation (67). Notably, the LPS of this organism consists of a repeating core glycan comprising tyvelose, l-rhamnose, galactose, and mannose that is appended with extended α(1→4)-glucan chains. It is therefore tempting to speculate that YihQ and by extension CjAgd31B may act as a transglucosylase to extend or restructure these chains. In this context, it is interesting to note that E. coli YihQ has previously been designated as an α-glucosidase based on a weak activity on α-glucosyl fluoride but no other α-glucosides (38). A reassessment of YihQ activity both in vitro and in vivo in light of the transglycosylation capacity of GH31 enzymes demonstrated in the present study may well be warranted. In conclusion, the detailed enzyme structure-function analysis of Agd31B from the model soil bacterium C. japonicus presented here that has defined a previously unknown α-transglucosylase activity in GH31 will inform future functional genomics studies in bacteria and other microorganisms.
Acknowledgments
We are grateful to Prof. Harry J. Gilbert (Newcastle University) for insightful discussions on C. japonicus and early access to genomic sequence data. We thank Prof. Stephen G. Withers (University of British Columbia) for gifts of α-glucosyl fluoride and 5-fluoro-α-d-glucopyranosyl fluoride and Dr. Gustav Sundqvist (KTH Glycoscience) for assistance with protein mass spectrometry. The staffs of the Diamond Light Source and European Synchrotron Radiation Facility are thanked for provision of data collection facilities.
Work in Stockholm was supported by the Mizutani Foundation for Glycoscience and The Swedish Research Council Formas (via the European Union WoodWisdom-Net ERA-NET project “FiberSurf: New biotechnological tools for wood fiber surface modification and analysis” and via “CarboMat-the KTH Advanced Carbohydrate Materials Centre”), and work in York was supported by Biotechnology and Biological Sciences Research Council Grant BB/I014802/1.

This article contains supplemental Figs. S1–S5.
The atomic coordinates and structure factors (codes 4b9y, 4b9z, and 4ba0) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GH
- glycoside hydrolase family
- HPAEC-PAD
- high performance anion exchange chromatography with pulsed amperometric detection
- Cj
- C. japonicus
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- pNP
- p-nitrophenyl
- Ro
- R. obeum.
REFERENCES
- 1. Elbein A. D. (2009) in Microbial Glycobiology: Structures, Relevance and Applications (Moran A., Holst O., Brennan P. J., von Itzstein M., eds) pp. 185–201, Academic Press, London [Google Scholar]
- 2. Wilson W. A., Roach P. J., Montero M., Baroja-Fernández E., Muñoz F. J., Eydallin G., Viale A. M., Pozueta-Romero J. (2010) Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol. Rev. 34, 952–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roach P. J., Depaoli-Roach A. A., Hurley T. D., Tagliabracci V. S. (2012) Glycogen and its metabolism: some new developments and old themes. Biochem. J. 441, 763–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ball S., Colleoni C., Cenci U., Raj J. N., Tirtiaux C. (2011) The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis. J. Exp. Bot. 62, 1775–1801 [DOI] [PubMed] [Google Scholar]
- 5. Boos W., Shuman H. (1998) Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62, 204–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Palmer T. N., Ryman B. E., Whelan W. J. (1976) Action pattern of amylomaltase from Escherichia coli. Eur. J. Biochem. 69, 105–115 [DOI] [PubMed] [Google Scholar]
- 8. Dippel R., Boos W. (2005) The maltodextrin system of Escherichia coli: metabolism and transport. J. Bacteriol. 187, 8322–8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takaha T., Smith S. M. (1999) The functions of 4-α-glucanotransferases and their use for the production of cyclic glucans. Biotechnol. Genet. Eng. Rev. 16, 257–280 [DOI] [PubMed] [Google Scholar]
- 10. Roujeinikova A., Raasch C., Burke J., Baker P. J., Liebl W., Rice D. W. (2001) The crystal structure of Thermotoga maritima maltosyltransferase and its implications for the molecular basis of the novel transfer specificity. J. Mol. Biol. 312, 119–131 [DOI] [PubMed] [Google Scholar]
- 11. Roujeinikova A., Raasch C., Sedelnikova S., Liebl W., Rice D. W. (2002) Crystal structure of Thermotoga maritima 4-α-glucanotransferase and its acarbose complex: implications for substrate specificity and catalysis. J. Mol. Biol. 321, 149–162 [DOI] [PubMed] [Google Scholar]
- 12. Berezina O. V., Zverlov V. V., Lunina N. A., Chekanovskaya L. A., Dubinina E. N., Liebl W., Velikodvorskaya G. A. (1999) Gene and properties of thermostable 4-α-glucanotransferase of Thermotoga neapolitana. Mol. Biol. 33, 801–806 [PubMed] [Google Scholar]
- 13. Liebl W., Feil R., Gabelsberger J., Kellermann J., Schleifer K. H. (1992) Purification and characterization of a novel thermostable 4-α-glucanotransferase of Thermotoga maritima cloned in Escherichia coli. Eur. J. Biochem. 207, 81–88 [DOI] [PubMed] [Google Scholar]
- 14. Terada Y., Fujii K., Takaha T., Okada S. (1999) Thermus aquaticus ATCC 33923 amylomaltase gene cloning and expression and enzyme characterization: production of cycloamylose. Appl. Environ. Microbiol. 65, 910–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leemhuis H., Kelly R. M., Dijkhuizen L. (2010) Engineering of cyclodextrin glucanotransferases and the impact for biotechnological applications. Appl. Microbiol. Biotechnol. 85, 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim Y. M., Kiso Y., Muraki T., Kang M. S., Nakai H., Saburi W., Lang W., Kang H. K., Okuyama M., Mori H., Suzuki R., Funane K., Suzuki N., Momma M., Fujimoto Z., Oguma T., Kobayashi M., Kim D., Kimura A. (2012) Novel dextranase catalyzing cycloisomaltooligosaccharide formation and identification of catalytic amino acids and their functions using chemical rescue approach. J. Biol. Chem. 287, 19927–19935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim Y. M., Yamamoto E., Kang M. S., Nakai H., Saburi W., Okuyama M., Mori H., Funane K., Momma M., Fujimoto Z., Kobayashi M., Kim D., Kimura A. (2012) Bacteroides thetaiotaomicron VPI-5482 glycoside hydrolase family 66 homolog catalyzes dextranolytic and cyclization reactions. FEBS J. 279, 3185–3191 [DOI] [PubMed] [Google Scholar]
- 18. Suzuki R., Terasawa K., Kimura K., Fujimoto Z., Momma M., Kobayashi M., Kimura A., Funane K. (2012) Biochemical characterization of a novel cycloisomaltooligosaccharide glucanotransferase from Paenibacillus sp. 598K. Biochim. Biophys. Acta 1824, 919–924 [DOI] [PubMed] [Google Scholar]
- 19. Mukai K., Maruta K., Satouchi K., Kubota M., Fukuda S., Kurimoto M., Tsujisaka Y. (2004) Cyclic tetrasaccharide-synthesizing enzymes from Arthrobacter globiformis A19. Biosci. Biotechnol. Biochem. 68, 2529–2540 [DOI] [PubMed] [Google Scholar]
- 20. Kim Y. K., Kitaoka M., Hayashi K., Kim C. H., Côté G. L. (2003) A synergistic reaction mechanism of a cycloalternan-forming enzyme and a d-glucosyltransferase for the production of cycloalternan in Bacillus sp NRRL B-21195. Carbohydr. Res. 338, 2213–2220 [DOI] [PubMed] [Google Scholar]
- 21. Wang Y. H., Jiang Y., Duan Z. Y., Shao W. L., Li H. Z. (2009) Expression and characterization of an α-glucosidase from Thermoanaerobacter ethanolicus JW200 with potential for industrial application. Biologia 64, 1053–1057 [Google Scholar]
- 22. Zhou C., Xue Y., Zhang Y., Zeng Y., Ma Y. (2009) Recombinant expression and characterization of Thermoanaerobacter tengcongensis thermostable α-glucosidase with regioselectivity for high-yield isomaltooligosaccharides synthesis. J. Microbiol. Biotechnol. 19, 1547–1556 [DOI] [PubMed] [Google Scholar]
- 23. Seo S. H., Choi K. H., Hwang S., Kim J., Park C. S., Rho J. R., Cha J. (2011) Characterization of the catalytic and kinetics properties of a thermostable Thermoplasma acidophilum α-glucosidase and its transglucosylation reaction with arbutin. J. Mol. Catal. B Enzym. 72, 305–312 [Google Scholar]
- 24. Hazlewood G. P., Gilbert H. J. (1998) Structure and function analysis of Pseudomonas plant cell wall hydrolases. Prog. Nucleic Acid Res. Mol. Biol. 61, 211–241 [DOI] [PubMed] [Google Scholar]
- 25. DeBoy R. T., Mongodin E. F., Fouts D. E., Tailford L. E., Khouri H., Emerson J. B., Mohamoud Y., Watkins K., Henrissat B., Gilbert H. J., Nelson K. E. (2008) Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J. Bacteriol. 190, 5455–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larsbrink J., Izumi A., Ibatullin F. M., Nakhai A., Gilbert H. J., Davies G. J., Brumer H. (2011) Structural and enzymatic characterization of a glycoside hydrolase family 31 α-xylosidase from Cellvibrio japonicus involved in xyloglucan saccharification. Biochem. J. 436, 567–580 [DOI] [PubMed] [Google Scholar]
- 27. Janecek S., Svensson B., MacGregor E. A. (2007) A remote but significant sequence homology between glycoside hydrolase clan GH-H and family GH31. FEBS Lett. 581, 1261–1268 [DOI] [PubMed] [Google Scholar]
- 28. Sundqvist G., Stenvall M., Berglund H., Ottosson J., Brumer H. (2007) A general, robust method for the quality control of intact proteins using LC-ESI-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 852, 188–194 [DOI] [PubMed] [Google Scholar]
- 29. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M., ed.) pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 30. Leslie A. G. (1999) Integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 55, 1696–1702 [DOI] [PubMed] [Google Scholar]
- 31. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 33. Cowtan K. D. (1994) ‘dm’: an automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newslett. Protein Crystallogr. 31, 34–38 [Google Scholar]
- 34. Perrakis A., Morris R., Lamzin V. S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 35. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37. Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 38. Okuyama M., Mori H., Chiba S., Kimura A. (2004) Overexpression and characterization of two unknown proteins, YicI and YihQ, originated from Escherichia coli. Protein Expr. Purif. 37, 170–179 [DOI] [PubMed] [Google Scholar]
- 39. Davies G. J., Wilson K. S., Henrissat B. (1997) Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 321, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burlingham B. T., Widlanski T. S. (2003) An intuitive look at the relationship of Ki and IC50: a more general use for the Dixon plot. J. Chem. Educ. 80, 214–218 [Google Scholar]
- 41. Rossi E. J., Sim L., Kuntz D. A., Hahn D., Johnston B. D., Ghavami A., Szczepina M. G., Kumar N. S., Sterchi E. E., Nichols B. L., Pinto B. M., Rose D. R. (2006) Inhibition of recombinant human maltase glucoamylase by salacinol and derivatives. FEBS J. 273, 2673–2683 [DOI] [PubMed] [Google Scholar]
- 42. Sim L., Willemsma C., Mohan S., Naim H. Y., Pinto B. M., Rose D. R. (2010) Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains. J. Biol. Chem. 285, 17763–17770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ren L., Cao X., Geng P., Bai F., Bai G. (2011) Study of the inhibition of two human maltase-glucoamylases catalytic domains by different α-glucosidase inhibitors. Carbohydr. Res. 346, 2688–2692 [DOI] [PubMed] [Google Scholar]
- 44. Lee S. S., Yu S., Withers S. G. (2003) Detailed dissection of a new mechanism for glycoside cleavage: α-1,4-glucan lyase. Biochemistry 42, 13081–13090 [DOI] [PubMed] [Google Scholar]
- 45. Lee S. S., He S., Withers S. G. (2001) Identification of the catalytic nucleophile of the family 31 α-glucosidase from Aspergillus niger via trapping of a 5-fluoroglycosyl-enzyme intermediate. Biochem. J. 359, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lovering A. L., Lee S. S., Kim Y. W., Withers S. G., Strynadka N. C. (2005) Mechanistic and structural analysis of a family 31 α-glycosidase and its glycosyl-enzyme intermediate. J. Biol. Chem. 280, 2105–2115 [DOI] [PubMed] [Google Scholar]
- 47. Hendlich M., Rippmann F., Barnickel G. (1997) LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J. Mol. Graph. Model. 15, 359–363, 389 [DOI] [PubMed] [Google Scholar]
- 48. Sim L., Quezada-Calvillo R., Sterchi E. E., Nichols B. L., Rose D. R. (2008) Human intestinal maltase-glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J. Mol. Biol. 375, 782–792 [DOI] [PubMed] [Google Scholar]
- 49. Tan K., Tesar C., Wilton R., Keigher L., Babnigg G., Joachimiak A. (2010) Novel α-glucosidase from human gut microbiome: substrate specificities and their switch. FASEB J. 24, 3939–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ernst H. A., Lo Leggio L., Willemoës M., Leonard G., Blum P., Larsen S. (2006) Structure of the Sulfolobus solfataricus α-glucosidase: implications for domain conservation and substrate recognition in GH31. J. Mol. Biol. 358, 1106–1124 [DOI] [PubMed] [Google Scholar]
- 51. Davies G. J., Planas A., Rovira C. (2012) Conformational analyses of the reaction coordinate of glycosidases. Acc. Chem. Res. 45, 308–316 [DOI] [PubMed] [Google Scholar]
- 52. Withers S. G., Warren R. A. J., Street I. P., Rupitz K., Kempton J. B., Aebersold R. (1990) Unequivocal demonstration of the involvement of a glutamate residue as a nucleophile in the mechanism of a retaining glycosidase. J. Am. Chem. Soc. 112, 5887–5889 [Google Scholar]
- 53. Sinnott M. L. (1990) Catalytic mechanisms of enzymatic glycosyl transfer. Chem. Rev. 90, 1171–1202 [Google Scholar]
- 54. Stick R., Williams S. (2009) Carbohydrates: the Essential Molecules of Life, pp. 262–263 and 273–276, Elsevier, Oxford [Google Scholar]
- 55. Kato N., Suyama S., Shirokane M., Kato M., Kobayashi T., Tsukagoshi N. (2002) Novel α-glucosidase from Aspergillus nidulans with strong transglycosylation activity. Appl. Environ. Microbiol. 68, 1250–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamamoto T., Unno T., Watanabe Y., Yamamoto M., Okuyama M., Mori H., Chiba S., Kimura A. (2004) Purification and characterization of Acremonium implicatum α-glucosidase having regioselectivity for α-1,3-glucosidic linkage. Biochim. Biophys. Acta 1700, 189–198 [DOI] [PubMed] [Google Scholar]
- 57. Stam M. R., Danchin E. G., Rancurel C., Coutinho P. M., Henrissat B. (2006) Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein. Eng. Des. Sel. 19, 555–562 [DOI] [PubMed] [Google Scholar]
- 58. Humphry D. R., Black G. W., Cummings S. P. (2003) Reclassification of ‘Pseudomonas fluorescens subsp. cellulosa’ NCIMB 10462 (Ueda et al. (1952) as Cellvibrio japonicus sp. nov., and revival of Cellvibrio vulgaris sp. nov., nom. rev., and Cellvibrio fulvus sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 53, 393–400 [DOI] [PubMed] [Google Scholar]
- 59. Meissner H., Liebl W. (1998) Thermotoga maritima maltosyltransferase, a novel type of maltodextrin glycosyltransferase acting on starch and malto-oligosaccharides. Eur. J. Biochem. 258, 1050–1058 [DOI] [PubMed] [Google Scholar]
- 60. Przylas I., Tomoo K., Terada Y., Takaha T., Fujii K., Saenger W., Sträter N. (2000) Crystal structure of amylomaltase from Thermus aquaticus, a glycosyltransferase catalysing the production of large cyclic glucans. J. Mol. Biol. 296, 873–886 [DOI] [PubMed] [Google Scholar]
- 61. Jung J. H., Jung T. Y., Seo D. H., Yoon S. M., Choi H. C., Park B. C., Park C. S., Woo E. J. (2011) Structural and functional analysis of substrate recognition by the 250s loop in amylomaltase from Thermus brockianus. Proteins 79, 633–644 [DOI] [PubMed] [Google Scholar]
- 62. Barends T. R., Bultema J. B., Kaper T., van der Maarel M. J., Dijkhuizen L., Dijkstra B. W. (2007) Three-way stabilization of the covalent intermediate in amylomaltase, an α-amylase-like transglycosylase. J. Biol. Chem. 282, 17242–17249 [DOI] [PubMed] [Google Scholar]
- 63. Jones G., Whelan W. J. (1969) Action pattern of D-enzyme, a transmaltodextrinylase from potato. Carbohydr. Res. 9, 483 [Google Scholar]
- 64. Takaha T., Yanase M., Okada S., Smith S. M. (1993) Disproportionating enzyme (4-α-glucanotransferase; EC 2.4.1.25) of potato. Purification, molecular-cloning, and potential role in starch metabolism. J. Biol. Chem. 268, 1391–1396 [PubMed] [Google Scholar]
- 65. Schwartz M., Hofnung M. (1967) Maltodextrin phosphorylase of Escherichia coli. Eur. J. Biochem. 2, 132–145 [DOI] [PubMed] [Google Scholar]
- 66. Becker S., Palm D., Schinzel R. (1994) Dissecting differential binding in the forward and reverse reaction of Escherichia coli maltodextrin phosphorylase using 2-deoxyglucosyl substrates. J. Biol. Chem. 269, 2485–2490 [PubMed] [Google Scholar]
- 67. Gibson D. L., White A. P., Snyder S. D., Martin S., Heiss C., Azadi P., Surette M., Kay W. W. (2006) Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 188, 7722–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]