Background: 14-3-3 proteins are highly conserved phosphoamino acid-binding proteins that may play a role in tumorigenesis.
Results: The oncogenic activity of 14-3-3γ requires a small variable domain in the N terminus.
Conclusion: The biological activity of 14-3-3 proteins involves protein motifs outside of the phosphoamino acid binding domain.
Significance: Understanding how structure determines 14-3-3 function is crucial to understanding how they function in tumorigenesis.
Keywords: Bioinformatics, Flow Cytometry, Oncogene, Protein Structure, Transformation, 14–3-3
Abstract
Members of the 14-3-3 superfamily regulate numerous cellular functions by binding phosphoproteins. The seven human isoforms (and the myriad of other eukaryotic 14-3-3 proteins) are highly conserved in amino acid sequence and secondary structure, yet there is abundant evidence that the various isoforms manifest disparate as well as common functions. Several of the human 14-3-3 isoforms are dysregulated in certain cancers and thus have been implicated in oncogenesis; experimentally, 14-3-3γ behaves as an oncogene, whereas 14-3-3σ acts as a tumor suppressor. In this study, we sought to localize these opposing phenotypes to specific regions of the two isoforms and then to individual amino acids therein. Using a bioinformatics approach, six variable regions (VRI–VRVI) were identified. Using this information, two sets of constructs were created in which N-terminal portions (including either VRI–IV or only VRI and VRII) of 14-3-3γ and 14-3-3σ were swapped; NIH3T3 cells overexpressing the four chimeric proteins were tested for transformation activity (focus formation, growth in soft agar) and activation of PI3K and MAPK signaling. We found that the specific phenotypes of 14-3-3γ are associated with the N-terminal 40 amino acids (VRI and VRII); in like fashion, VRI and VRII of 14-3-3σ dictated its tumor suppressor function. Using individual amino acid substitutions within the 14-3-3γ VRII, we identified two residues required for and two contributing to the γ-specific phenotypes. Our observations suggest that isoform-specific phenotypes are dictated by a relatively few amino acids within variable regions.
Introduction
Members of the 14-3-3 protein superfamily are ubiquitous and highly conserved in eukaryotes and serve as regulatory scaffolds, which bind phosphothreonine/phosphoserine proteins (1). Seven different isoforms (β, γ, ϵ, σ, ζ, τ, η) have been identified in humans, and nearly all eukaryotes have at least two isoforms (1). The 14-3-3 proteins lack catalytic domains but exert their regulatory activities by binding with a vast array of phosphoproteins, which often bear the binding motifs RSX(pS/pT)XP (mode-1) and RXXX(pS/pT)XP (mode-2), where X denotes any amino acid, pS denotes phospho-serine, and pT denotes phospho-threonine (2). Studies aimed at identification of the processes that 14-3-3 proteins are involved in have identified a large number of potential client proteins, suggesting that they regulate most cellular processes. Indeed they have been implicated in regulating metabolic pathways, redox regulation, transcription, RNA processing, protein synthesis, protein folding and degradation, cell cycle, cytoskeletal organization, and cellular trafficking (3).
The human 14-3-3 isoforms share nearly 80% similarity of their amino acid sequence, within which there are five highly conserved blocks, as defined by Wang and Shakes (4). The crystal structures of all of the 14-3-3 proteins have been resolved, and all have the same structural organization. Moreover, the 14-3-3 residues that are key for binding phosphoamino acids and that form the phosphoamino acid binding motif correspond to Lys49, Arg56, Arg127, and Tyr128 of human 14-3-3ζ, and these are completely conserved in all known 14-3-3 proteins (2).
Despite the common binding motif present on client proteins and the common phosphoamino acid binding domain of all of the family of 14-3-3 proteins, each of the family members appears to interact preferentially with specific intracellular signaling proteins. For example, PKCθ and Cbl interact preferentially with 14-3-3τ in T-cells (5, 6), IGF1 receptor and IRS1 interact preferentially with 14-3-3ϵ (7), the apoptosis inhibitor A20 interacts preferentially with 14-3-3β (8), and the glucocorticoid receptor interacts preferentially with 14-3-3η (9). Consistent with this is the observation that there is only a 15% overlap in the proteins that interact with 14-3-3ϵ and that also interact with 14-3-3ζ, -γ, and -σ (10). Hence, the cadre of proteins that is bound by each of the 14-3-3 isoforms is distinctive, suggesting that they have different biological activities and that the specificity of their interactions with cellular proteins is unique to each isoform. The 14-3-3 proteins even show specificity in their interactions with each other. For example, 14-3-3σ forms homodimers exclusively, whereas the other family members are more promiscuous, forming heterodimers with most of the other family members. The specificity of 14-3-3/target protein interactions does not appear to result from different affinities for the phosphopeptide binding motifs but rather appears to arise from contacts made on the variable surface of 14-3-3 outside the binding cleft. The highly conserved phosphopeptide motifs are the minimal sequence criteria required for binding and additional structural constraints then dictate whether the target actually binds (11).
There is compelling evidence that individual 14-3-3 proteins have distinct biological functions (3). In Drosophila, which express both 14-3-3ζ and 14-3-3ϵ, deletion of 14-3-3ζ results in embryonic lethality despite having normal amounts of 14-3-3ϵ (12). Hence, ϵ cannot compensate for loss of ζ, suggesting that ζ has distinct functions not mediated by ϵ. More recently, we have demonstrated an oncogenic phenotype of overexpressed 14-3-3γ protein versus the tumor suppressor phenotype of overexpressed 14-3-3σ (13). These observations imply that the dissimilar amino acids, about 20% of the sequence, determine the functions of the various isoforms.
Mounting evidence supports the notion that 14-3-3 proteins play an important role in human tumorigenesis. Changes in expression of various 14-3-3 isoforms have been associated with tumor suppressor or oncogenic properties, and differential expression of one or more isoforms has been documented in various malignancies; for example, 14-3-3ζ is up-regulated in hepatocellular carcinoma (14) and breast cancer (15) and has been suggested as a target for cancer treatment. Previously, we reported that 14-3-3γ is up-regulated in certain lung tumors and, when overexpressed in NIH3T3 cells, induces transformation activity: an oncogenic phenotype (13). In contrast, overexpression of the 14-3-3σ isoform (which is expressed exclusively in cells of epithelial origin, hence its name, stratifin or SFN (16)) inhibited transformation induced by H-Ras overexpression, thus acting as a tumor suppressor (13). Notably, SFN is a downstream target of p53 (17). Consonant with its possible role as a tumor suppressor, 14-3-3σ has been reported to be down-regulated in carcinomas of breast (18), cervical (19), and ovarian (20) origins, as well as transitional cell carcinoma of the bladder (21). However, other groups have found it to be up-regulated in carcinomas of the prostate (22); pancreas (23, 24); the stomach, colon, and rectum (25); scirrhous tumors of the stomach (26); and lung tumors (27).
Here, we report experiments in which we sought, using a combination of bioinformatics and cell biology, to determine whether the antithetical phenotypes of 14-3-3γ and 14-3-3σ could be localized to one or more domains and perhaps even to specific amino acids of these two highly similar proteins. Using bioinformatics, we identified in both isoforms six variable regions; then, chimeric protein were constructed in which N-terminal portions were swapped between σ and γ. In addition, specific amino acids within the γ sequence were individually mutated, changing each one to the cognate σ residue. Using the various constructs, we determined that the opposing phenotypes of overexpressed σ and γ reside within their N-terminal 40 amino acids (variable regions I and II); more specifically, four amino acids within VRII,2 which is close to but not within the phosphopeptide binding site, of 14-3-3γ are required for or contribute to its transformation activity. A second phenotype of overexpressed 14-3-3γ, the induction of polyploidy, was also mapped to VRII but to a larger subset of amino acids within it. These results suggest that discrete functions of various 14-3-3 isoforms may be linked to a limited number of identifiable amino acids residing within the variable regions.
EXPERIMENTAL PROCEDURES
Cell Culture
NIH3T3 cells (obtained from ATCC) and mouse embryonic fibroblast cells (provided by Thomas Doetschman, BIO5 Institute, University of Arizona) were grown in DMEM medium containing 10% fetal calf serum and supplemented with antibiotics. Plasmids were transfected into cells using the FuGENE 6.0 transfection reagent (Promega) according to the manufacturer's protocol. Stable clonal cell lines were established after G418 selection.
Plasmid Constructs for Chimeric and Mutated 14-3-3 Proteins
The plasmids pCMV-FLAG-14-3-3γ and pCMV-FLAG-14-3-3σ were described previously (13). The chimeric protein plasmid constructs were produced by a two-step PCR method, as described by Higuchi et al. (28) and modified by Wurch et al. (29), using primers based upon human 14-3-3γ and 14-3-3σ cDNA sequences and high fidelity Taq DNA polymerase (AccuPrime Pfx DNA polymerase, Invitrogen). Briefly, the initial PCR product served as a “megaprimer” for the second PCR reaction; the primers for the second set of reactions incorporated a BamHI site at the 5′ termini and an EcoRI site at the 3′ termini of the 14-3-3 cDNA sequences. The final PCR products were ligated into the pGEM-T Easy vector (Promega). After confirmatory sequencing, the plasmids were digested with BamHI and EcoRI, and the released inserts were ligated into a pCMV-FLAG plasmid (Stratagene). Inserts were also ligated into the pGEX-2T vector (GE Healthcare) for the production of recombinant GST fusion proteins. Details of the templates and primers used to generate the different chimeras are provided in supplemental Table 1.
The point mutations A26F, N29G, V30A, T31V, L33K, N34G, P36E, and N39C were produced by PCR synthesis using the QuikChange II site-directed mutagenesis kit (Stratagene) and were verified by sequencing. The primers for these reactions are listed in supplemental Table 2.
Focus Formation Assays
NIH3T3 cells were cultured in 6-well dishes and transfected with the appropriate 14-3-3 plasmid singly or in combination with one of the following plasmids: pT24-C3 (activated H-Ras inserted into the pBR322 plasmid (kindly provided by Dr. Barbacid, National Institutes of Health), pCMV-AKT, or pCDNA3.1-ERK2. Cells were maintained in DMEM containing 6% FCS (Gibco), which was refreshed every 2–3 days. After staining with crystal violet, the number of foci per plate was determined using a colony counter (Oxford Optronix Colony Counter). Focus formation assays were performed in duplicate in three independent experiments.
PI3K Assays
After transient transfections of NIH3T3 cells, cell-free lysates were prepared and assayed for PI3K activity using the method described by Higaki et al. (30). Anti-p110 antibody (Santa Cruz Biotechnology) was used to immunoprecipitate the p110 catalytic subunits of PI3K from the total lysate. The immunoprecipitates were washed once with cold phosphate-buffered saline (PBS) and twice with 0.5 mol/liter LiCl, 0.1 mol/liter Tris (pH 7.4). The kinase reactions contained 20 mm Tris-HCl, pH 7.6, 75 mm NaCl, 10 mm MgCl2, 2.5 mm EGTA, 0.2 mg/ml phosphatidylinositol, 0.3 mg of phosphatidylserine, 20 mm ATP, 10 μCi of [γ-32P]ATP, and washed immunoprecipitates; the reactions were incubated at room temperature for 10 min, then terminated with 100 μl of 1 n HCl. Phospholipids were extracted twice, first with 200 μl of CHCl3:MeOH (1:1) and then with 160 μl of 1 n HCl:MeOH (1:1), after which the organic phase was dried and resuspended in 50 μl of CHCl3:MeOH (1:1). Phosphorylated products were separated by thin-layer chromatography, identified by autoradiography, and quantified using a PhosphorImager (Amersham Biosciences).
MAPK Activity Assays
ERK2 activity was assayed as described elsewhere (31). The transfected NIH3T3 cells were starved overnight, washed with PBS, and lysed with radioimmunoprecipitation assay buffer. 200 μg of precleared lysates were immunoprecipitated with 5 μg of ERK2 antibody (Santa Cruz Biotechnology) and incubated at 4 °C for 4 h with gentle rocking followed by an additional 1-h incubation with protein A/G (Santa Cruz Biotechnology). The pellets were then washed and resuspended with 50 μl of kinase buffer (Cell Signaling) supplemented with 200 mm ATP and 2 μg of recombinant Elk-1 protein (Cell Signaling) and incubated at room temperature for 45 min. The reaction was stopped by adding lysis buffer, and the proteins were separated by SDS-10% PAGE followed by immunoblotting with rabbit antiphospho-Elk-1 antibody (Cell Signaling); the blot was stripped and probed with anti-ERK2 (rabbit polyclonal antibody, Santa Cruz Biotechnology) to confirm equal loading.
GST-14-3-3 Recombinant Proteins and in Vitro Kinase Assays
Lysates from actively growing NIH3T3 cells were immunoprecipitated with anti-p110 antibody, as in the PI3K assay. In vitro PI3K assays were then performed as described above, except that the 30-μl reactions also included 0, 250, or 500 ng of recombinant GST fusions of intact or chimeric 14-3-3 proteins, or GST alone. After incubating the reactions for 40 min at 37 °C with constant shaking, the products were separated by thin layer chromatography and analyzed by autoradiography. Phosphatidylinositol 3-kinase activity was quantified using a PhosphorImager.
Immunoblotting
Whole-cell lysates for Western blotting were obtained by scraping NIH3T3 cells from 6- or 10-cm culture dishes into radioimmunoprecipitation assay buffer. The protein content of the lysate was measured using the Bio-Rad protein assay. After SDS-PAGE was performed, the separated proteins were transferred to a nitrocellulose membrane (Millipore), blocked for at least 1 h with 5% fat-free milk in TBST buffer (which contained 0.1% Tween 20), and probed overnight at 4 °C with primary antibody. After washing the membrane three times with TBST buffer, it was incubated with the appropriate horseradish peroxidase-coupled secondary antibodies. An ECL kit (SuperSignal, Pierce) was used to detect HRP activity. All Western blots were performed at least twice; the images shown are from individual, representative experiments.
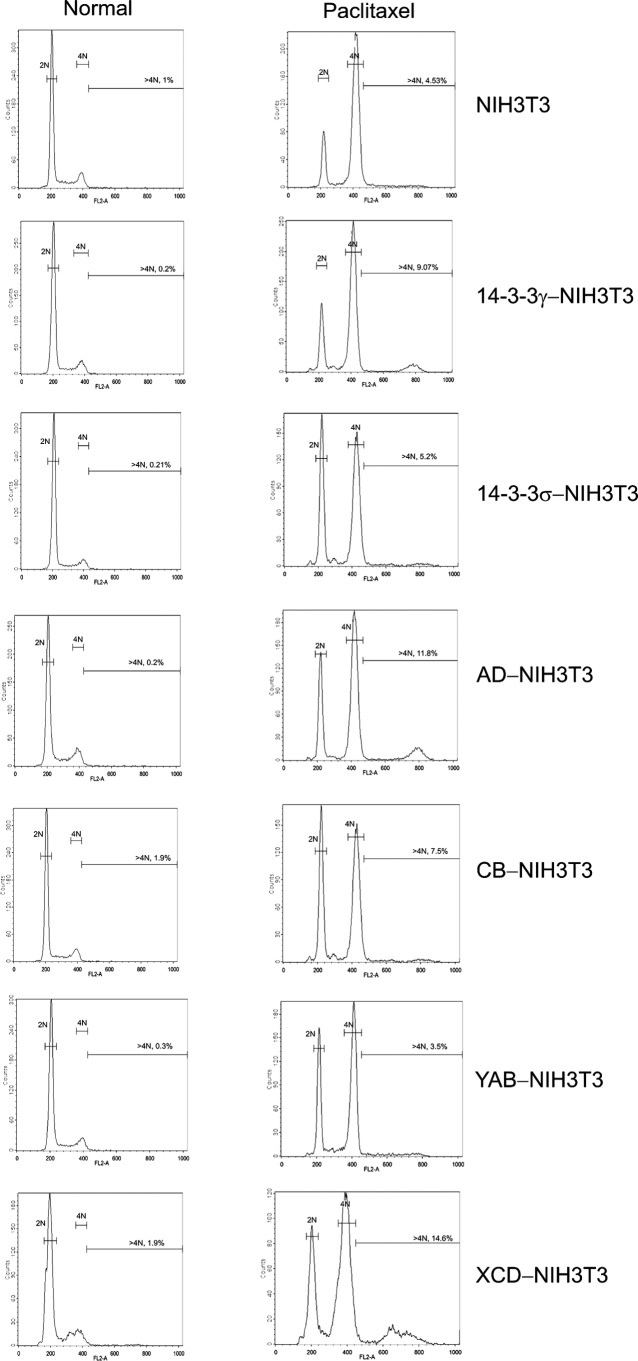
Detection of Polyploidy by Flow Cytometry
Intact or chimeric 14-3-3 proteins were expressed in NIH3T3 cells, and stable clones were selected using G418; protein expression was confirmed by Western blotting with anti-FLAG antibody. Asynchronous cultures were treated with paclitaxel or nocodazole for 16 h and analyzed for DNA content by flow cytometry.
Amino Acid Sequence Analysis of 14-3-3 Proteins
We utilized PSI-BLAST (National Center for Biotechnology Information (NCBI), blast.ncbi.nlm.nih.gov/Blast.cgi) to develop a broadly based consensus sequence for the 14-3-3 superfamily. The PSI-BLAST algorithm was initiated with the 14-3-3γ (YWHAG) amino acid sequence, retrieving 500 or 1000 sequences per iteration from the complete nonredundant reference protein sequence database. Multiple iterations were performed, continuing until the list of 500 proteins was stabilized (i.e. no more than one or two proteins appearing or disappearing from the entire list in successive iterations); typically, six or seven iterations were required. The first sequence of the ultimate list was adopted as representing consensus.
Alignments of paired protein sequences were performed with EMBOSS Needle (European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI)). Multiple sequences were aligned with MUSCLE (Multiple Sequence Alignment Comparison by Log-Expectation), using the CLUSTALW format (EMBL-EBI).
Statistical Analysis
Data are expressed as means ± S.D. Statistical significance was determined by Student's t test. Results in which it was determined that p < 0.05 were accepted as statistically significant.
RESULTS
Identifying Variable Regions and Designing Chimeric Proteins of 14-3-3σ and 14-3-3γ
Despite the striking similarities of amino acid sequence and structure of the many members of the eukaryotic 14-3-3 family of proteins, the multiple isoforms found in nearly all eukaryotic species, including the seven human isoforms, often manifest disparate functions. In a previous study (13), the 14-3-3σ (SFN) and 14-3-3γ (YWHAG) isoforms were found to have opposing phenotypes; we sought to take advantage of the stark phenotypic contrast to identify certain regions and, ultimately, specific amino acid disparities of these two isoforms that might contribute to their antithetical phenotypes.
The amino acid sequences of 14-3-3σ and 14-3-3γ share 63.3% identity and 79.3% similarity (EMBOSS Needle pairwise sequence alignment); the remaining ∼52 dissimilar residues assuredly determine the individualistic functions of each protein. We sought to further cull this list of 52 disparate residues by taking advantage of the enormous number of 14-3-3 sequences available in databases such as GenBankTM. The PSI-BLAST algorithm is conventionally employed to mine databases for proteins that share similar sequence patterns and are therefore likely to belong to the same family as the query protein. In light of the striking sequence similarities of 14-3-3 family members, we reasoned that the algorithm might be used in converse fashion, that is, to identify variant residues. After multiple iterations with PSI-BLAST (see “Experimental Procedures”), we identified a broadly based consensus sequence (accession number XP_002196990.1, Taeniopygia guttata, the zebra finch) representative of the many thousands of 14-3-3 family members. Then, using the MUSCLE algorithm for multiple sequence alignments, we compared the consensus sequence and the sequences of 14-3-3σ and 14-3-3γ. We reasoned that ascendant amino acid residues of the σ and γ isoforms would likely differ not only from each other but also from the consensus sequence.
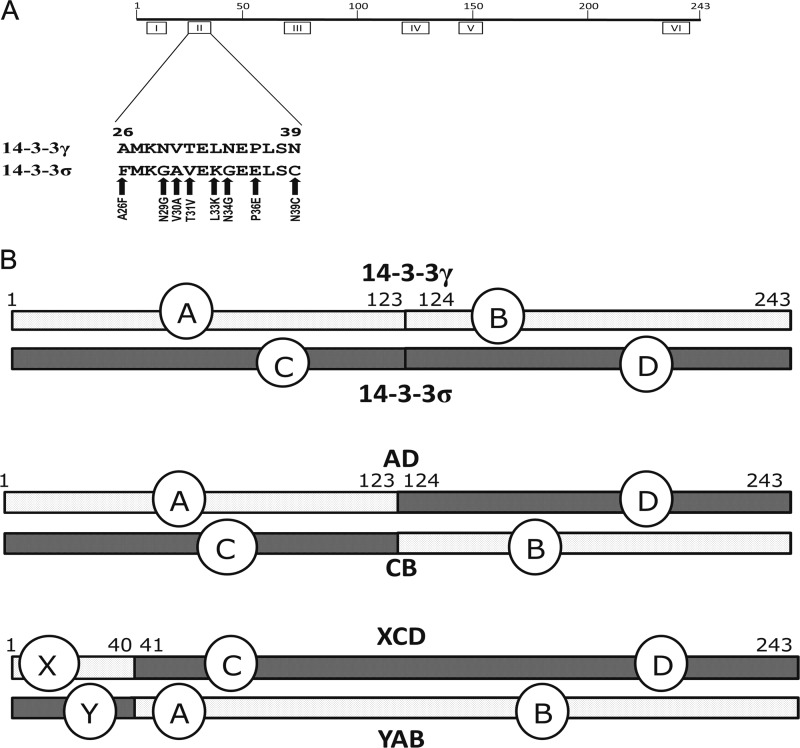
Using this approach, we identified amino acids that fulfilled two criteria. First, at a given position, the 14-3-3σ and 14-3-3γ residues must be different from each other, and second, each must also differ from the cognate residue in the consensus sequence. This analysis withdrew an additional ∼13% of the residues from further consideration; in sum, we identified 44 and 46 (∼15% of each sequence) “highly variable” residues in 14-3-3γ and 14-3-3σ, respectively. Importantly, when we mapped the variable residues to the two linear amino acid sequences, we found that they were clustered in six regions separated from one another by longer stretches (∼20–70 residues) of identical or nearly identical sequence. We labeled the clusters variable regions I–VI (Fig. 1A).
FIGURE 1.
Construction of the chimeric 14-3-3 proteins and location of the variable regions. A, a diagram of the linear amino acid sequence of a consensus 14-3-3 protein is depicted. Numbers above the line mark the amino acid numbers. Boxes below the line indicate the position of the variable regions as defined by amino acid sequence comparisons. Each variable region is identified with a roman numeral. The 14 amino acid sequence of VRII from both the γ (top line) and the σ (bottom line) 14-3-3 proteins is also shown. Vertical arrows mark amino acid positions where substitutions were made in the γ VRII. Each arrow is named with the amino acid substitution and the numerical position of the amino acid. B, the parent 14-3-3γ and 14-3-3σ proteins along with four chimeric proteins constructed from them are depicted in linear fashion. Segments of the protein are labeled with letters, and numbers indicate the coordinates of amino acids at the beginning and end of each segment in the parent molecule. The two parent proteins are at the top of the diagram. The 14-3-3γ parent molecule and segments derived from it are depicted in light gray, and the 14-3-3σ parent molecule and segments derived from it are depicted in dark gray. Protein segments were given a letter designation (seen near the middle of each diagram) and were used to name the chimeric molecule (e.g. AD contained amino acids 1–123 from 14-3-3γ and amino acids 124–243 from 14-3-3σ). Each chimera contained the FLAG tag appended to its N terminus (not shown).
Based upon the variable regions identified by the bioinformatics analysis, we devised an experimental approach using two sets of chimeric proteins in which portions of 14-3-3γ and 14-3-3σ were swapped (Fig. 1B). In the first set, approximately half of the 14-3-3σ and 14-3-3γ cDNA sequences, i.e. VRI–VRIV and VRV–VRVI, were switched, creating proteins AD and CB (Fig. 1B). Subsequently, as experimental observations drew our attention to the N-terminal variable regions, we created chimeric proteins XCD and YAD, in which an N-terminal fragment of ∼40 amino acids, encompassing VRI and VRII, was paired with the remainder of the sequence (including VRIII–VRVI) of the other isoform. Thus, in all of the chimeric proteins, VRI and VRII from σ or from γ remained coupled with each other within the swapped segments.
Variable Regions I and/or II of 14-3-3γ Transform NIH3T3 Cells
As discussed earlier, overexpressed 14-3-3γ induces transformation, whereas 14-3-3σ does not (13). Consequently, we sought to localize this γ phenotype to one or more variable regions. To this end, plasmids encoding chimeric proteins (in pCMV-based vectors) were transfected into NIH3T3 cells; because the pCMV vector also includes the neomycin resistance gene, we were able to develop stable cell lines by selection with the antibiotic G418. 14 days after plating, the cultures were scored for focus formation. Transfected H-Ras, ERK-2, and AKT were used as positive controls for transformation activity.
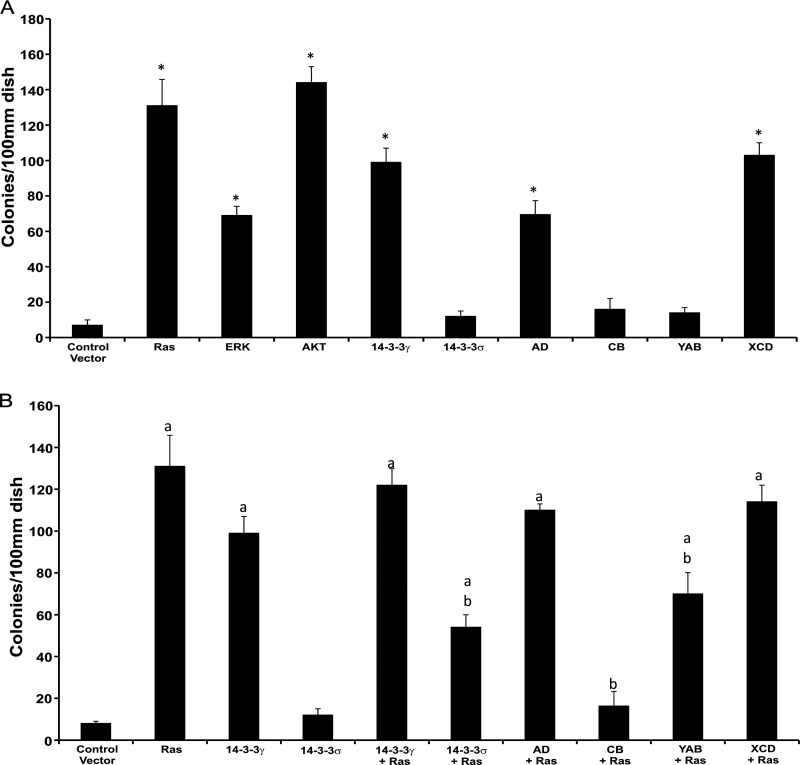
NIH3T3 cells bearing overexpressed intact 14-3-3γ or the chimeric proteins AD and XCD (both of these chimeric proteins include VRI and VRII of γ origin (Fig. 1, A and B)) formed numerous foci of transformed cells, whereas no foci were detected with cells transformed with intact 14-3-3σ, with the chimeric proteins CB and YAB, or with vector alone (Fig. 2A). The foci induced by the AD and XCD chimeric proteins exhibited morphologic features characteristic of transformed cells and were, in fact, indistinguishable from those resulting from transfection with intact 14-3-3γ. The transformed cell lines were likewise similar in their ability to grow as anchorage-independent colonies in soft agar (data not shown). In contrast, the colonies obtained by G418 selection after transfection with 14-3-3σ or the chimeric proteins CB and YAB appeared morphologically normal and failed to grow in soft agar. We therefore conclude that the transformation activity of 14-3-3γ likely resides within VRI and/or VRII.
FIGURE 2.
Focus formation and suppression of focus formation by chimeric 14-3-3 proteins. A, NIH3T3 cells were transfected with plasmids expressing 14-3-3γ, 14-3-3σ, or one of several chimeric proteins (10 μg each) in 6-well plates. Cells transfected with plasmids expressing H-Ras, ERK, or AKT served as positive controls. After 48 h, the cells were transferred into 10-cm dishes (1:5 ratio) and grown for 14 days in 5% calf serum, fixed, and then stained as described under “Experimental Procedures.” The colonies were counted using a colony counter, and experiments were repeated three times. The y axis shows the mean number of colonies per 100-mm dish. An asterisk above a bar indicates a significant difference when compared with cells transfected with the empty vector (p < 0.05). B, NIH3T3 cells were co-transfected with either 14-3-3γ or 14-3-3σ or one of the chimeric proteins and H-Ras, AKT, or ERK1. The number of foci per 100-mm-diameter dish was determined. Bars depict the mean number of colonies per 100-mm-diameter dish from three different experiments. Error bars indicate S.D. Significant differences when compared with cells transfected with an empty vector are indicated by a lowercase a (p < 0.05). Significant differences when compared with cells transfected with an activated H-Ras are indicated by a lowercase b (p > 0.05).
Previously, we had noted that 14-3-3σ not only lacks the capacity to transform but indeed suppresses transformation induced by the oncogene Ras (13). Co-transfection of the 14-3-3σ, CB, or YAB plasmids with the Ras plasmid significantly reduced transformation activity in comparison with that recorded with the Ras plasmid alone (Fig. 2B), whereas co-transfection of 14-3-3γ or the chimeric proteins AD and XCD with Ras had no effect upon Ras activity (Fig. 2B). Together, these data are consistent with the notion that VRI and/or VRII confer transformation activity upon 14-3-3γ and that the tumor suppressor activity of 14-3-3σ also resides within the same variable regions of its protein sequence.
Chimeric Proteins Encompassing VRI and VRII from 14-3-3γ Activate PI3K and MAPK Signaling
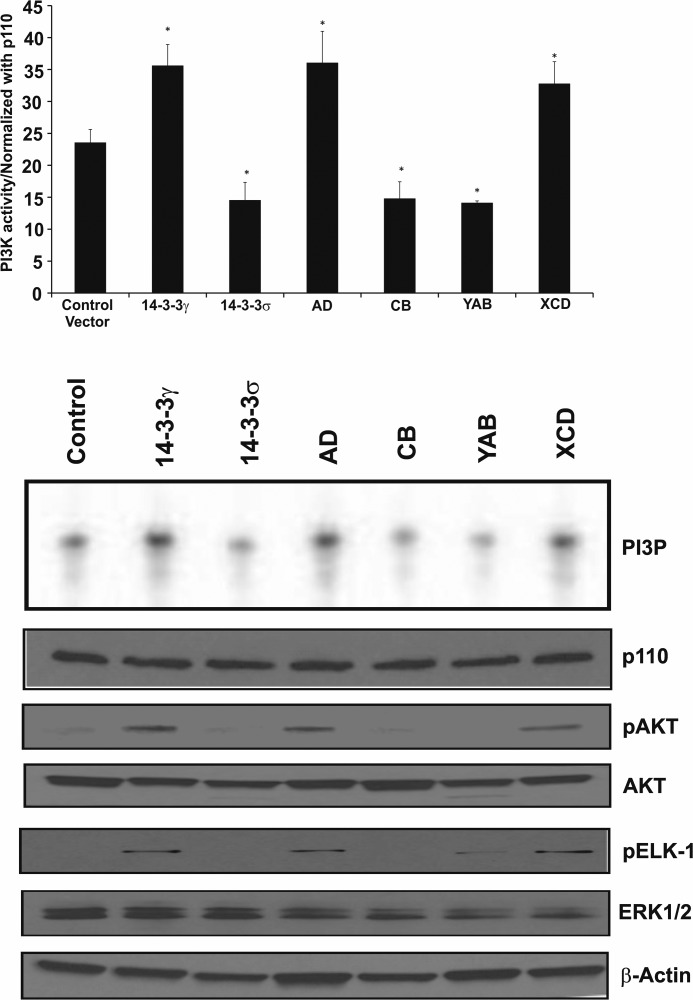
Activation of p85/p110 PI3K is an important mechanism for the transformation of NIH3T3 cells. Previously, we reported that overexpressed 14-3-3γ and 14-3-3σ exhibit contrary effects upon p85/p110 PI3K; 14-3-3γ activates p85/p110 PI3K, and 14-3-3σ suppresses p85/p110 PI3K (13). Consequently, we wondered whether these antithetical effects of the γ and σ isoforms also reside within VRI and VRII; we predicted that chimeric proteins in which VRI and VRII were taken from the γ sequence would activate p85/p110 PI3K, whereas chimeric proteins in which VRI and VRII originated from the σ sequence would suppress p85/p110 PI3K. To measure activated PI3K, extracts obtained from NIH3T3 transfectants were immunoprecipitated with an anti-p110 and assayed for their ability to phosphorylate α-phosphatidylinositol (see “Experimental Procedures”). PI3K activation was substantially increased when 14-3-3γ or the AD and XCD proteins were overexpressed (Fig. 3), whereas constitutive PI3K activity was significantly less in cells overexpressing 14-3-3σ or the chimeric proteins CB and YAB than in parental cells (Fig. 3), indicating that VRI and/or VRII of 14-3-3γ are required for PI3K activation in NIH3T3 cells. Moreover, when FLAG-tagged 14-3-3 proteins and chimeric proteins were immunoprecipitated with anti-FLAG antibody, PI3K was detected in the resulting complexes (data not shown), suggesting that 14-3-3 proteins physically interact with PI3K.
FIGURE 3.
PI3K and MAPK are activated by 14-3-3γ proteins containing variable regions I and II. NIH3T3 cells were transfected with pCMV (empty vector), pCMV-14-3-3γ, pCMV-14-3-3σ, pCMV-AD, pCMV-CB, pCMV-YAB, and pCMV-XCD. 48 h later, the cells were harvested, extracts were prepared, and p110 was immunoprecipitated. PI3K activity was determined by testing for phosphorylation of phosphoinositol in an in vitro assay, and the products were fractionated using thin layer chromatography (phosphatidylinositol 3-phosphate (PI3P)). Aliquots were also tested using immunoblotting for quantity of p110 protein (p110), and the values were used to normalize PI3K activity. The graph shows the average PI3K activity from three experiments ± S.D. An asterisk signifies significant differences when compared with cells that were transfected with the empty vector (*, p < 0.05). Aliquots of the lysates were also analyzed for the expression of phospho-AKTSer473 (pAKT). ERK1/2 activity was assayed by detecting phosphorylated Elk-1 (pELK-1) in an in vitro Elk-1 kinase assay that detected phosphorylated ELK-1. Immunodetection of total p110, AKT, ERK1/2, and β-actin protein served as loading controls.
AKT is a known downstream mediator of phosphatidylinositol 3′-kinase activation (32). Consequently, it was not surprising that elevated levels of phospho-AKT (Ser473) were detected in 14-3-3γ-, AD-, and XCD-transfected cells but not in 14-3-3σ-, CB-, and YAB-expressing cells (Fig. 3), findings consistent with the observed PI3K enzyme activities in the same cell lines. Thus, VRI and/or VRII of 14-3-3γ are required for the activation of both PI3K and its downstream effector, AKT.
The MAP kinase signaling cascade is also modulated by 14-3-3 proteins. Binding of 14-3-3 proteins to Raf-1 kinase activates it; activated Raf-1 kinase in turn leads to increased MAP kinase signaling, one target of which is the nuclear protein, Elk-1. Because 14-3-3γ, but not 14-3-3σ, has been shown to activate the MAP kinase pathway, we postulated that VRI and/or VRII of the 14-3-3γ isoform is required for MAPK activation, just as for PI3K enzyme activity. To test this prediction, NIH3T3 cells were transfected with expression vectors of intact 14-3-3 proteins and of the four chimeric proteins. The cells were serum-starved for 48 h and then harvested; extracts were immunoprecipitated with anti-ERK2 antibody, and the activity of the precipitated Erk2 was assayed using recombinant Elk-1 protein as substrate. Consistent with our prediction, cells transfected with intact 14-3-3γ or the AD and XCD chimeric proteins showed increased Erk2 kinase activity. Erk2 activity was undetectable in cells transfected with 14-3-3σ or the CB chimeric protein; however, barely detectable Erk2 kinase activity was induced by the YAB chimeric protein (Fig. 3). Thus, just as the transforming activity of 14-3-3γ requires variable region I and/or variable region II, so does its ability to activate the PI3K and the MAPK signal transduction pathways.
Recombinant Chimeric Proteins Modulate PI3K Activity in Vitro
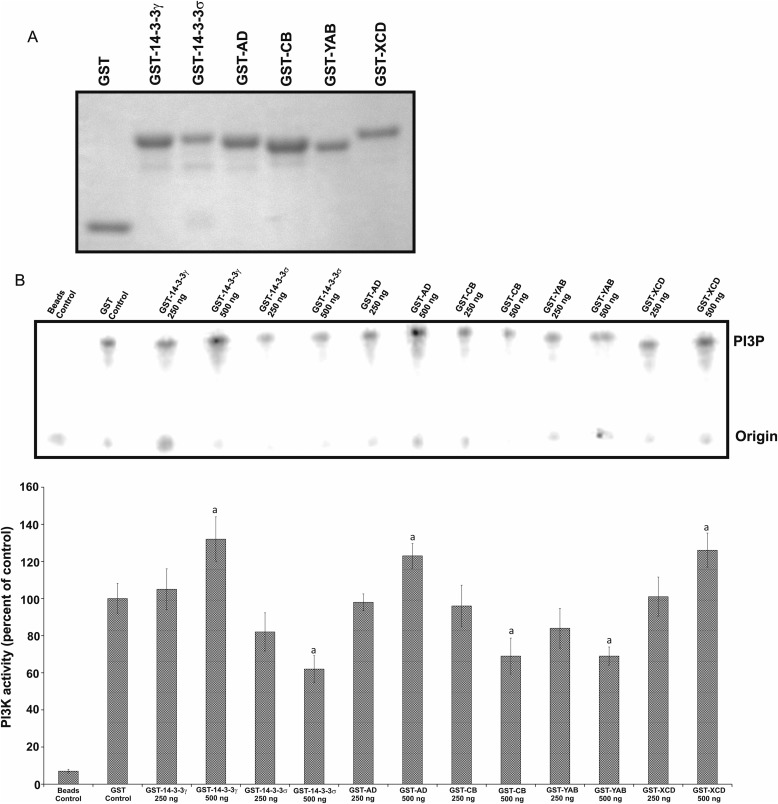
Although the in vivo data presented above clearly implicate VRI and VRII of 14-3-3γ in the activation of PI3K, we sought to directly test the functionality of that activation in vitro, using recombinant chimeric 14-3-3 proteins in an assay of PI3K catalytic activity. The GST chimeric proteins (Fig. 4A) were expressed in BL.23 bacteria and purified (see “Experimental Procedures”). Then, equal quantities (either 250 ng or 500 ng in a reaction volume of 30 μl) of the recombinant proteins were added to immunoprecipitated PI3K obtained from extracts of quiescent NIH3T3 cells. Recombinant GST-14-3-3γ, GST-AD, and GST-XCD (at the greater concentration) significantly (p < 0.05) increased catalytic activity of the PI3K enzyme when compared with GST alone (Fig. 4B). Interestingly, the inclusion of recombinant GST-14-3-3σ, GST-CB, or GST-YAB significantly (p < 0.05) decreased PI3K catalytic activity in vitro when compared with control reactions in which only GST was added (Fig. 4B). These data support the notion that direct interactions with PI3K of 14-3-3γ VRI and/or VRII versus 14-3-3σ VRI and/or VRII differentially modulate the catalytic activity of the enzyme.
FIGURE 4.
Variable region II of 14-3-3γ enhances PI3K activity in vitro. A, GST fusion proteins were created from the parental 14-3-3γ, 14-3-3σ, and other chimera cDNA by inserting into the pGEX-2T expression vector. The recombinant proteins were expressed in and purified from BL.23 bacteria. Purified proteins were fractionated by SDS-PAGE. B, the catalytic subunit of PI3K (p110) was immunoprecipitated from NIH3T3 cell lysates using anti-p110 antibody. In vitro kinase assays using the immunopurified p110 were performed in the presence of increasing quantities of purified GST-14-3-3 recombinant proteins (250 and 500 ng), and the quantity of radiolabeled phosphatidyl inositol was determined through fractionation using thin layer chromatography. A representative figure is shown (phosphatidylinositol 3-phosphate (PI3P)). In the graph, bars depict the mean ± S.D. of at least three independent experiments. Significant differences when compared with GST controls are marked with a lowercase a (p < 0.05).
Chimeric Proteins That Include VRI and VRII Induce Polyploidization of NIH3T3 cells
Previously, we reported an additional phenotype of overexpression of 14-3-3γ, the induction of polyploidy (33). In the human lung cancer cell line H322, overexpressing 14-3-3γ results in abnormal DNA replication and polyploidization. Cells become resistant to microtubule inhibitors and appear to reenter the cell cycle without executing mitosis (33), suggesting that elevated levels of 14-3-3γ enable cells to bypass the mitotic checkpoint. We wondered whether the polyploidy phenotype of γ overexpression is also associated with VRI and/or VRII. Asynchronous cultures of NIH3T3 cells stably overexpressing intact or chimeric 14-3-3 proteins were treated with paclitaxel or nocodazole for 16 h and analyzed for DNA content by flow cytometry. As in our previous study (13), 14-3-3γ overexpression in combination with the microtubule inhibitors paclitaxel (Fig. 5) or nocodazole (data not shown) resulted in modest but consistently detectable polyploidy, whereas 14-3-3σ overexpression did not. Similarly, overexpression of the chimeric proteins AD and XCD in combination with microtubule inhibitors likewise resulted in the generation of cells exhibiting ≥4N DNA content; overexpressing 14-3-3σ or the chimeric constructs CB and YAB did not produce polyploidy. These data indicate that a second phenotype of overexpressed 14-3-3γ, the induction of polyploidy, is also associated with VRI and/or VRII. The relatively weak polyploidy phenotypes of 14-3-3γ and the chimeric proteins AD and XCD, as well as the lack of corroborating biochemical activities for the induction of the polyploidy phenotype, similar to PI3K and MAPK signaling for transformation activity, led us to abandon efforts to localize this phenotype with greater precision.
FIGURE 5.
Variable regions I and II promote >4N DNA content in conjunction with paclitaxel treatment. Asynchronous cultures of NIH3T3 cells overexpressing 14-3-3γ or 14-3-3σ or one of the chimeric proteins were either left untreated or treated with paclitaxel (5 ng/ml). Untransfected NIH3T3 cells treated similarly were used as controls. 16 h later, the cells were harvested, fixed in 70% ethanol, and resuspended in a PBS-propidium iodide (50 μg/ml; Sigma)-RNase A (70 μg/ml; Sigma) solution and analyzed by flow cytometry. Peaks of diploid (2N), tetraploid (4N), and polyploid (>4N) are marked by horizontal bars on each of the histograms.
Functional Characterization of Individual Amino Acid Residues within VRII of 14-3-3γ
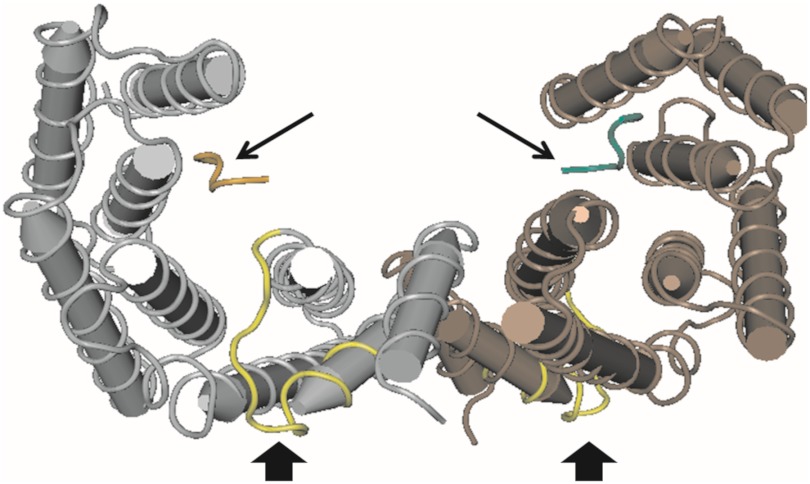
The observations described so far clearly implicate VRI and/or VRII in a specific phenotype of overexpressed 14-3-3γ not seen with overexpressed 14-3-3σ, namely the ability to transform, manifested by focus formation and anchorage-independent growth and substantiated by activation of PI3K and MAPK signaling. However, experiments with the chimeric proteins AD and XCD do not perforce distinguish the contribution(s) of VRI versus VRII to the γ-specific phenotype. Mapping VRI and VRII to images of the solved crystal structure of 14-3-3γ revealed that VRII, but not VRI, lies in proximity to the phosphopeptide binding pocket (Fig. 6) thus positioning VRII to influence client protein selection and hence function. It follows from this reasoning that within VRII, specific amino acid residues must be required for the phenotypes associated with the N terminus of 14-3-3γ. To test this notion, single amino acids within VRII of 14-3-3γ were switched to the cognate residues of 14-3-3σ by creating point mutations in the γ sequence; eight mutations were individually created: A26F, N29G, V30A, T31V, L33K, N34G, P36E, and N39C (Fig. 1A).
FIGURE 6.
The three-dimensional diagram of a 14-3-3γ homodimer generated using CND3 (NCBI) from the crystal structure (Molecular Modeling Database (MMDB) ID: 35668; Protein Data Bank (PDB) ID: 2B05). The phosphopeptide bound to each monomer is indicated by a small arrow. VRII is colored yellow and is indicated by thick black arrows. Note the proximity of variable region II to the phosphopeptide binding site.
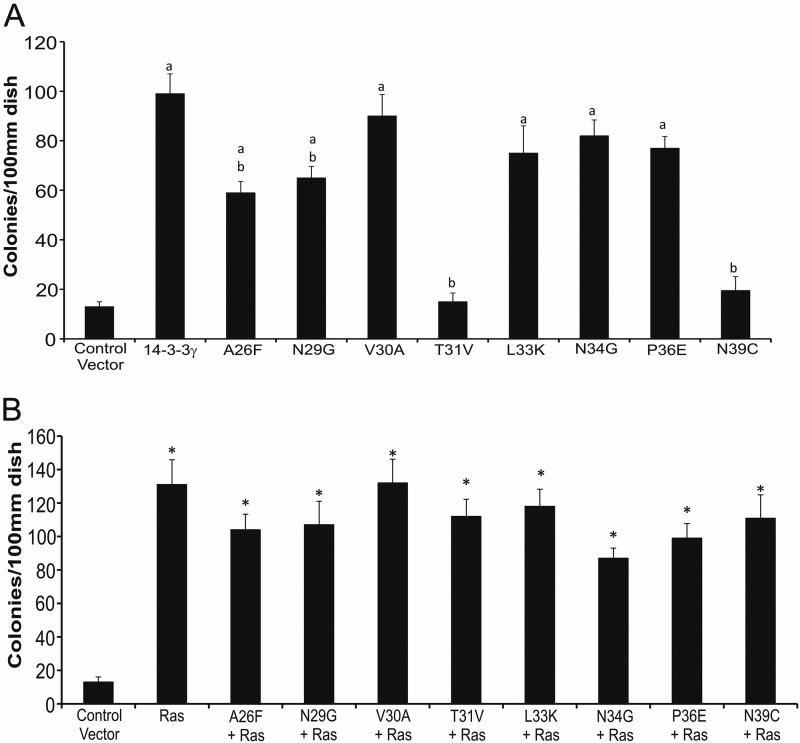
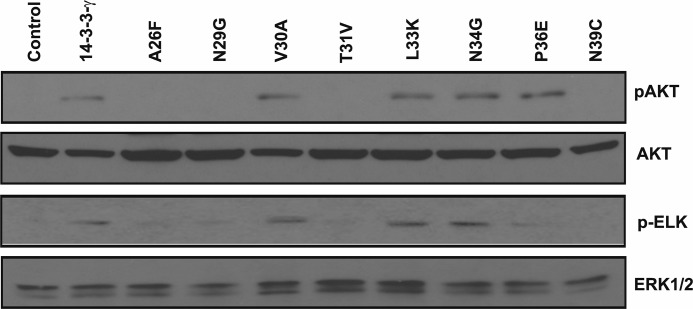
We first examined the ability of each of the 14-3-3γ amino acid changes to induce transformation in NIH3T3 cells (Fig. 7A). The mutations T31V and N39C each resulted in complete abolition of transformation induction ascertained by focus formation, whereas two other mutations, A26F and N29G, decreased focus formation by almost 40%; the other four mutants (V30A, L33K, N34G, and P36E) exhibited transformation ability comparable with wild type 14-3-3γ. Importantly, although each mutation represented a replacement of the 14-3-3γ residue with the cognate 14-3-3σ residue, only one of the mutants (N34G) exhibited tumor suppressor function after co-transfection with the H-Ras oncogene (Fig. 7B), a property of the intact σ isoform. Transfection with A26F, N29G, T31V, and N39C point mutation plasmids in NIH3T3 cells also led to decreased levels of phospho-AKT (Ser473) when compared with wild type 14-3-3γ. Similarly, when the extracts were analyzed for MAP kinase activity, as measured by the ability of ERK2 to phosphorylate the substrate Elk-1, the mutants A26F, N29G, T31V, and N39C manifested demonstrably less ERK2 kinase activity when compared with cells overexpressing 14-3-3γ (Fig. 8).
FIGURE 7.
Focus formation assay with 14-3-3γ point mutants. A, NIH3T3 cells were plated onto 10-cm plates and then transfected either with an empty vector (vector control) or with a plasmid expressing either 14-3-3γ or one of the 14-3-3γ VRII point mutants. Focus forming capacity was quantified as described in Fig. 2. Bars depict the averages from three experiments ± S.D. A lowercase a indicates a significant difference when compared with cells transfected with the control vector (p < 0.05). A lowercase b indicates a significant difference when compared with cells transfected with the 14-3-3γ parent molecule (p < 0.05). B, NIH3T3 cells were transfected with the control vector, H-Ras, 14-3-3γ, or 14-3-3σ only or were co-transfected with H-Ras plus one of the 14-3-3γ VRII point mutants. The bars depict the average from three independent experiments + S.D. An asterisk indicates there is a significant difference when compared with cells that are transfected with the empty vector (p < 0.05).
FIGURE 8.
Activation of AKT and MAPK by 14-3-3γ VRII point mutants. NIH3T3 cells were transiently transfected with the control vector (Control), a plasmid expressing 14-3-3γ, or one of the eight 14-3-3γ VRII point mutants. After 48 h, whole cell lysates were prepared, and phosphorylated AKT (pAKT) was detected by immunoblotting. An aliquot of the cell lysate was also tested for ERK1/2 kinase activity using an in vitro assay followed by immunodetection of phospho-Elk (p-ELK). Total AKT (AKT) and total ERK1/2 (ERK1/2) protein were detected by immunoblotting and served as loading controls.
We conclude from these observations that two amino acids (Thr31 and Asn39) residing in VRII of the 14-3-3γ sequence are necessary for its transformative capacity and related phenotypes and that two others (Ala26 and Asn29) contribute to these activities. However, only one of the single amino acid swaps tested (N34G) was sufficient to confer the tumor suppressor function of 14-3-3σ upon the γ isoform.
DISCUSSION
Previously, we reported that in NIH3T3 cells, the overexpression of 14-3-3γ induces transformation, i.e. it behaves as an oncogene, whereas the overexpression of 14-3-3σ suppresses transformation induced by H-Ras, qualifying it as a tumor suppressor (13). In this study, we identify a specific domain and within it certain amino acids, which are required for the antithetical phenotypes of 14-3-3γ and 14-3-3σ; both the oncogenic and the tumor suppressor functions are localized to the N-terminal 40 amino acids. Additionally, within variable region II of 14-3-3γ, a 13-amino acid span within the N-terminal 40 amino acids, the transformation ability of 14-3-3γ requires two specific residues; two other amino acids in VRII contribute to this phenotype. Interestingly, one of the amino acids in the σ VRII could confer a modest level of tumor suppressor activity when transferred from σ to γ (N34G). Importantly, swapping the γ and σ VRIIs did not alter the oligomerizing properties of either the γ or the σ parent proteins (data not shown). Hence, we propose that a very few residues within VRII of 14-3-3σ are required for determining the phenotype of the protein and that this phenotype results from the interaction with a specific subset of cellular proteins.
The quite remarkable sequence similarity not only of the seven human isoforms but also among the myriad eukaryotic members of the 14-3-3 superfamily, as well as their propensity to heterodimerize, has rendered difficult the assignment of discrete functions to individual 14-3-3 proteins. No less complex is the task of identifying specific domains or individual amino acids of importance within their sequences. Undoubtedly, the multiple species-specific isoforms found in nearly all eukaryotic species subserve certain communal functions; conversely, evidence suggests that each of the isoforms also executes individualistic functions. The latter conclusion is supported in principle by studies of the evolution of duplicated genes or genomes (34). More directly, analyses of genetic and protein interactions of 14-3-3 proteins in a variety of species indicate that different isoforms engage a variety of client phosphoproteins and thus are expected to support varying functions. In budding yeast (Saccharomyces cerevisiae), for example, the two 14-3-3 homologs, Bmh1 and Bmh2, share physical and genetic interactions with at least 51 proteins; individually, however, Bmh1 participates in interactions with an additional ∼234 proteins, and Bmh2 engages ∼110 other proteins. Thus, both lines of evidence support the search for individualistic phenotypes of the seven human isoforms.
Because of the profound sequence similarities of 14-3-3σ and 14-3-3γ, applying conventional approaches for exploring structure/function relationships, such as random mutagenesis, was considered by us and rejected as being, at the very least, inefficient. Instead, we utilized a bioinformatics approach to narrow our search for relevant domains and amino acid residues. The vast number of 14-3-3 protein sequences available in the GenBank and similar databases allowed us to deduce a consensus sequence using PSI-BLAST (see “Experimental Procedures”). We reasoned that ascendant residues, that is, amino acids crucial to a particular individualistic function, would differ not only between the isoforms in question but also from the consensus sequence. When the human isoforms were aligned with this consensus sequence, distinct variable regions were identifiable; moreover, even within the variable region, not all residues differed from each other or from the cognate consensus residues, thus further focusing our search for critical amino acids. The utility of this approach is supported by our analysis of specific amino acids within the 14-3-3γ VRII. The two residues, Thr31 and Asn39, essential to the 14-3-3 transformation phenotype and the two amino acids contributing to the phenotype, Ala26 and Asn29, all differed from the cognate amino acids of the 14-3-3σ sequence and of the consensus sequence. Conversely, of the four mutations tested that had no effect upon the phenotype of the γ isoform, two shared identity with the consensus sequence (although they differed between the σ and γ isoforms).
Based on the identification of variable regions, two sets of chimeric proteins were constructed; the first set bisected the σ/γ isoforms, swapping the N-terminal halves. Analyses of transformation, PI3K, and MAP kinase activities and induction of polyploidy indicated the N-terminal portion in the 14-3-3γ specific phenotype. Experiments with the second set of chimeric proteins, in which only the N-terminal 40 amino acids (VRI and VRII) were swapped, further refined the search. Then, based on the published crystal structures of 14-3-3γ, we deduced that VRII was more likely than VRI to influence client protein selection because of the proximity of the former to the phosphopeptide binding site. Finally, mutations of specific amino acid residues within VRII identified two that were obligatory and two that contributed to the transformation ability of 14-3-3γ.
Three points deserve emphasis regarding our observations with chimeric and mutated 14-3-3σ and -γ. First, the identification of a domain, VRII, and certain amino acids within this domain that are critical to the transformation phenotype of 14-3-3γ does not preclude contributions from other domains or residues, for example, by favoring or abolishing certain heterodimeric partner choices.
Secondly, in all of our experiments, swaps of domains or amino acids in the γ sequence were with the cognate σ domains or residues. Consequently, we assumed that the chimeric or mutated proteins would maintain structural features and phosphopeptide binding functions (which might not be the case with random mutagenesis) but perhaps not client protein selectivity. It was therefore not surprising that the σ phenotype, suppression of H-Ras transformation, also resided within its N-terminal regions VRI and VRII; chimeric proteins bearing the σ N terminus also suppressed H-Ras transformation. Interestingly, we found that conversion to the suppressor phenotype could be detected when one individual amino acid in the γ VRII was switched to the σ residue (N34G). Although we have not yet tested the hypothesis, we predict that single amino acid changes of VRII within the σ isoform context will identify residues necessary for or contributing to its tumor suppressor function.
Thirdly, with regard to the transformation phenotype of 14-3-3γ, each N-terminal swap produced consistent, parallel findings with each of the cellular and molecular measures tested: focus formation and anchorage-independent growth; PI3K and MAPK signaling; and the induction of polyploidy. However, the subset of VRII amino acids required for induction of polyploidy was somewhat broader than the pattern associated with the other phenotypes, suggesting that in turn, client protein selection may differ. Although an abundance of literature links the first two categories to each other, the induction of polyploidy is not easily linked at a molecular level with either transformation or PI3K or MAPK signaling. With regard to oncogenesis, however, the generation of polyploidy is of considerable interest.
This work was supported, in whole or in part, by National Institutes of Health Grants CA107510, R56CA107510, and CA023074 (to J. D. M.).

This article contains supplemental Tables 1 and 2.
- VR
- variable region
- SFN
- stratifin.
REFERENCES
- 1. Mhawech P. (2005) 14-3-3 proteins–an update. Cell Res. 15, 228–236 [DOI] [PubMed] [Google Scholar]
- 2. Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 3. Dougherty M. K., Morrison D. K. (2004) Unlocking the code of 14-3-3. J. Cell Sci. 117, 1875–1884 [DOI] [PubMed] [Google Scholar]
- 4. Wang W., Shakes D. C. (1996) Molecular evolution of the 14-3-3 protein family. J. Mol. Evol. 43, 384–398 [DOI] [PubMed] [Google Scholar]
- 5. Meller N., Liu Y. C., Collins T. L., Bonnefoy-Bérard N., Baier G., Isakov N., Altman A. (1996) Direct interaction between protein kinase Cθ (PKCθ) and 14-3-3τ in T cells: 14-3-3 overexpression results in inhibition of PKCθ translocation and function. Mol. Cell. Biol. 16, 5782–5791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y. C., Liu Y., Elly C., Yoshida H., Lipkowitz S., Altman A. (1997) Serine phosphorylation of Cbl induced by phorbol ester enhances its association with 14-3-3 proteins in T cells via a novel serine-rich 14-3-3-binding motif. J. Biol. Chem. 272, 9979–9985 [DOI] [PubMed] [Google Scholar]
- 7. Craparo A., Freund R., Gustafson T. A. (1997) 14-3-3 (ϵ) interacts with the insulin-like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. J. Biol. Chem. 272, 11663–11669 [DOI] [PubMed] [Google Scholar]
- 8. Vincenz C., Dixit V. M. (1996) 14-3-3 proteins associate with A20 in an isoform-specific manner and function both as chaperone and adapter molecules. J. Biol. Chem. 271, 20029–20034 [DOI] [PubMed] [Google Scholar]
- 9. Wakui H., Wright A. P., Gustafsson J., Zilliacus J. (1997) Interaction of the ligand-activated glucocorticoid receptor with the 14-3-3η protein. J. Biol. Chem. 272, 8153–8156 [DOI] [PubMed] [Google Scholar]
- 10. Ballif B. A., Cao Z., Schwartz D., Carraway K. L., 3rd, Gygi S. P. (2006) Identification of 14-3-3ϵ substrates from embryonic murine brain. J. Proteome Res. 5, 2372–2379 [DOI] [PubMed] [Google Scholar]
- 11. Wilker E. W., Grant R. A., Artim S. C., Yaffe M. B. (2005) A structural basis for 14-3-3σ functional specificity. J. Biol. Chem. 280, 18891–18898 [DOI] [PubMed] [Google Scholar]
- 12. Skoulakis E. M., Davis R. L. (1996) Olfactory learning deficits in mutants for leonardo, a Drosophila gene encoding a 14-3-3 protein. Neuron 17, 931–944 [DOI] [PubMed] [Google Scholar]
- 13. Radhakrishnan V. M., Martinez J. D. (2010) 14-3-3γ induces oncogenic transformation by stimulating MAP kinase and PI3K signaling. PLoS One 5, e11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi J. E., Hur W., Jung C. K., Piao L. S., Lyoo K., Hong S. W., Kim S. W., Yoon H. Y., Yoon S. K. (2011) Silencing of 14-3-3ζ over-expression in hepatocellular carcinoma inhibits tumor growth and enhances chemosensitivity to cis-diammined dichloridoplatium. Cancer Lett. 303, 99–107 [DOI] [PubMed] [Google Scholar]
- 15. Neal C. L., Yao J., Yang W., Zhou X., Nguyen N. T., Lu J., Danes C. G., Guo H., Lan K. H., Ensor J., Hittelman W., Hung M. C., Yu D. (2009) 14-3-3ζ overexpression defines high risk for breast cancer recurrence and promotes cancer cell survival. Cancer Res. 69, 3425–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leffers H., Madsen P., Rasmussen H. H., Honoré B., Andersen A. H., Walbum E., Vandekerckhove J., Celis J. E. (1993) Molecular cloning and expression of the transformation sensitive epithelial marker stratifin. A member of a protein family that has been involved in the protein kinase C signalling pathway. J. Mol. Biol. 231, 982–998 [DOI] [PubMed] [Google Scholar]
- 17. Hermeking H., Lengauer C., Polyak K., He T. C., Zhang L., Thiagalingam S., Kinzler K. W., Vogelstein B. (1997) 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol Cell 1, 3–11 [DOI] [PubMed] [Google Scholar]
- 18. Moreira J. M., Ohlsson G., Rank F. E., Celis J. E. (2005) Down-regulation of the tumor suppressor protein 14-3-3σ is a sporadic event in cancer of the breast. Mol. Cell. Proteomics 4, 555–569 [DOI] [PubMed] [Google Scholar]
- 19. Sano T., Shimooka H., Weixa P., Segawa A., Jian Z., Motegi A., Nakayama H., Oyama T., Nakajima T. (2004) Immunohistochemical expression of 14-3-3σ protein in various histological subtypes of uterine cervical cancers. Pathol. Int. 54, 743–750 [DOI] [PubMed] [Google Scholar]
- 20. Akahira J., Sugihashi Y., Suzuki T., Ito K., Niikura H., Moriya T., Nitta M., Okamura H., Inoue S., Sasano H., Okamura K., Yaegashi N. (2004) Decreased expression of 14-3-3σ is associated with advanced disease in human epithelial ovarian cancer: its correlation with aberrant DNA methylation. Clin. Cancer Res. 10, 2687–2693 [DOI] [PubMed] [Google Scholar]
- 21. Moreira J. M., Gromov P., Celis J. E. (2004) Expression of the tumor suppressor protein 14-3-3σ is down-regulated in invasive transitional cell carcinomas of the urinary bladder undergoing epithelial-to-mesenchymal transition. Mol. Cell. Proteomics 3, 410–419 [DOI] [PubMed] [Google Scholar]
- 22. Ito Y., Miyoshi E., Uda E., Yoshida H., Uruno T., Takamura Y., Miya A., Kobayashi K., Matsuzuka F., Matsuura N., Kakudo K., Kuma K., Miyauchi A. (2003) 14-3-3σ possibly plays a constitutive role in papillary carcinoma, but not in follicular tumor of the thyroid. Cancer Lett. 200, 161–166 [DOI] [PubMed] [Google Scholar]
- 23. Rodriguez J. A., Li M., Yao Q., Chen C., Fisher W. E. (2005) Gene overexpression in pancreatic adenocarcinoma: diagnostic and therapeutic implications. World J. Surg. 29, 297–305 [DOI] [PubMed] [Google Scholar]
- 24. Okada T., Masuda N., Fukai Y., Shimura T., Nishida Y., Hosouchi Y., Kashiwabara K., Nakajima T., Kuwano H. (2006) Immunohistochemical expression of 14-3-3σ protein in intraductal papillary-mucinous tumor and invasive ductal carcinoma of the pancreas. Anticancer Res. 26, 3105–3110 [PubMed] [Google Scholar]
- 25. Tanaka K., Hatada T., Kobayashi M., Mohri Y., Tonouchi H., Miki C., Nobori T., Kusunoki M. (2004) The clinical implication of 14-3-3σ expression in primary gastrointestinal malignancy. Int. J. Oncol. 25, 1591–1597 [PubMed] [Google Scholar]
- 26. Kuramitsu Y., Baron B., Yoshino S., Zhang X., Tanaka T., Yashiro M., Hirakawa K., Oka M., Nakamura K. (2010) Proteomic differential display analysis shows up-regulation of 14-3-3σ protein in human scirrhous-type gastric carcinoma cells. Anticancer Res. 30, 4459–4465 [PubMed] [Google Scholar]
- 27. Radhakrishnan V. M., Jensen T. J., Cui H., Futscher B. W., Martinez J. D. (2011) Hypomethylation of the 14-3-3σ promoter leads to increased expression in non-small cell lung cancer. Genes Chromosomes Cancer 50, 830–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Higuchi R., Krummel B., Saiki R. K. (1988) Phosphatidylinositol 3-kinase-independent signal transduction pathway for platelet-derived growth factor-induced chemotaxis. Nucleic Acids Res. 16, 7351–73673045756 [Google Scholar]
- 29. Wurch T., Colpaert F. C., Pauwels P. J. (1998) Chimeric receptor analysis of the ketanserin binding site in the human 5-hydroxytryptamine1D receptor: importance of the second extracellular loop and fifth transmembrane domain in antagonist binding. Mol. Pharmacol. 54, 1088–1096 [DOI] [PubMed] [Google Scholar]
- 30. Higaki M., Sakaue H., Ogawa W., Kasuga M., Shimokado K. (1996) Phosphatidylinositol 3-kinase-independent signal transduction pathway for platelet-derived growth factor-induced chemotaxis. J. Biol. Chem. 271, 29342–29346 [DOI] [PubMed] [Google Scholar]
- 31. Ballarè E., Persani L., Lania A. G., Filopanti M., Giammona E., Corbetta S., Mantovani S., Arosio M., Beck-Peccoz P., Faglia G., Spada A. (2001) Mutation of somatostatin receptor type 5 in an acromegalic patient resistant to somatostatin analog treatment. J. Clin. Endocrinol. Metab. 86, 3809–3814 [DOI] [PubMed] [Google Scholar]
- 32. Cantley L. C. (2002) The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 33. Qi W., Liu X., Chen W., Li Q., Martinez J. D. (2007) Overexpression of 14-3-3γ causes polyploidization in H322 lung cancer cells. Mol. Carcinog 46, 847–856 [DOI] [PubMed] [Google Scholar]
- 34. Fernández A., Tzeng Y. H., Hsu S. B. (2011) Subfunctionalization reduces the fitness cost of gene duplication in humans by buffering dosage imbalances. BMC Genomics 12, 604. [DOI] [PMC free article] [PubMed] [Google Scholar]