Background: FlaX is essential for archaella assembly in Sulfolobus acidocaldarius and its stability is dependent on FlaI-J.
Results: FlaX assembles into ring like oligomers and interacts with the motor ATPase FlaI.
Conclusion: FlaX oligomers depend on the presence of its C-terminal domain, which also interacts with FlaI.
Significance: The first structural analysis of an archaellum subunit, a rotating type IV pilus structure.
Keywords: Cell Motility, Microbiology, Protein assembly, Protein Secretion, Protein-Protein Interactions, Archaea, Membrane Biology, Type IV Pili
Abstract
Archaella are the archaeal motility structure, which are structurally similar to Gram-negative bacterial type IV pili but functionally resemble bacterial flagella. Structural and biochemical data of archaellum subunits are missing. FlaX, a conserved subunit in crenarchaeal archaella, formed high molecular weight complexes that adapted a ring-like structure with an approximate diameter of 30 nm. The C terminus of FlaX was not only involved in the oligomerization, but also essential for FlaX interaction with FlaI, the bifunctional ATPase that is involved in assembly and rotation of the archaellum. This study gives first insights in the assembly apparatus of archaella.
Introduction
As the ability to move toward favorable conditions is an important driving force for evolution most organisms have developed structures that enable them to translocate on surfaces or in liquid environments. Whereas in bacteria this function is fulfilled by flagella and in eukaryotes by cilia, archaea have developed a surface structure called the archaellum (1).
Archaella are widespread in archaea and have been shown to be involved in swimming motility (1, 2). Although archaella are functionally similar to bacterial flagella, they resemble structurally type IV pili from bacteria, as many of their subunits share a common origin with these (3, 4).
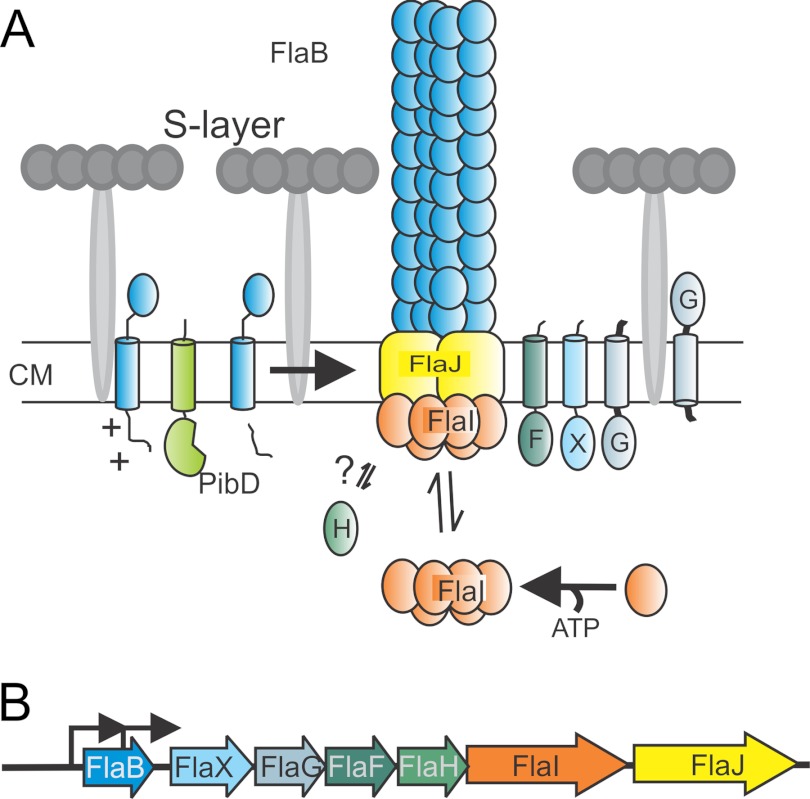
All archaella loci encode six conserved subunits, FlaB, FlaF, -G, -H, -I, and -J. FlaB, the archaellin, is the structural subunit and is like type IV pre-pilins processed by a type IV pre-pilin peptidase, PibD/FlaK (5, 6). Although FlaG and FlaF are monotopic membrane proteins of unknown function, FlaH, -I, and -J probably form the archaellum assembly platform. FlaH is a predicted ATP-binding protein, whereas FlaI, an ATPase (7), and FlaJ, a polytopic membrane protein, are homologous to PilB and PilC in bacterial type IV pili-like assembly systems, respectively (1–3) (Fig. 1, A and B). The three-dimensional reconstruction of the archaellum from different archaeal species has revealed that it has comparable symmetry and diameter to the type IV pilus in Gram-negative bacteria (8, 9). Furthermore, like type IV pili, archaella lack the central lumen (9) suggesting similar assembly mechanics by addition of the structural subunits at the bottom of the growing filament (2, 3, 10). In Halobacterium salinarum it has been shown that archaellum rotation is ATP dependent (11). Besides the core components the archaella loci contain unique accessory proteins that are different in the subphyla Crenarchaeota and Euryarchaeota.
FIGURE 1.
A, model of the subunits of the archaellum. Sulfolobus sp. have a single cell membrane that is surrounded with a proteinaceous layer called S-layer. So far it is known that PibD, a dedicated type IV prepilin signal peptidase, processes FlaB, the sole archaellin, prior to assembly. FlaI, the only characterized archaella subunit that forms an ATP-dependent hexamer, probably interacts with the sole polytopic membrane protein FlaJ. FlaH is a predicted ATP-binding protein but its role in archaella biosynthesis or rotation is unknown. The localization and function of FlaX, FlaF, and FlaG the other three conserved archaellum proteins in Sulfolobus sp. is not known. CM, cytoplasmic membrane. B, the operon structure of archaellum. Arrowhead showing the promoters involved in transcription mRNA of archaella-specific proteins.
In euryarchaeotes these are the FlaC, -D, and -E proteins, which were shown in H. salinarum to form a complex with proteins ORF2402F and ORF2404R that in turn were interacting with the Che signaling system (12). Consequently deletion mutants of these two proteins demonstrated a smooth swimming phenotype as switching was abolished. In contrast, crenarchaea like the Sulfolobales do not contain any of these chemotaxis associated genes in the archaella loci (1) nor are any che-genes present in their genome (4). The unique crenarchaeal archaellum component is encoded by flaX, a monotopic membrane protein, which shares a distant similarity with methyl accepting chemotaxis proteins (2). Previously, it was shown that FlaX is essential for archaellum assembly and that it is destabilized in the absence of FlaI, FlaH, and FlaJ (4) indicating that these proteins might form a complex within the archaellum assembly apparatus.
In the present study our biochemical and structural analysis shows that FlaX forms ring-shaped oligomers with a ∼30 nm diameter in solution, dependent on the presence of the C-terminal domain. Moreover, we established that the C terminus of FlaX interacts with FlaI, the archaellum motor ATPase. Our study provides first insights into the assembly pathway and interaction of the different archaellum subunits.
EXPERIMENTAL PROCEDURES
Bioinformatics Analysis
In silico analyses were carried out using available online tools as described in supplemental “Experimental Procedures”.
Strains, Growth Conditions and Used Plasmids
Sulfolobus acidocaldarius DSM639 was grown aerobically at 75 °C in Brock's basal salts medium adjusted to pH 3.5 with sulfuric acid and supplemented with 0.1% (w/v) tryptone (Roth) or NZ Amine AS (Sigma) and 0.2% (w/v) dextrin. The uracil auxotrophic S. acidocaldarius MW001 (13) and ΔflaXΔaapF (MW452, ΔflaX in S. acidocaldarius ΔaapF background) (4) strains were grown in basal Brock medium supplemented with 10 μg ml−1 of uracil. Other strains used are described in supplemental Table S1. Supplemental Table S2 and Table S3 describe primers used in this study. Detailed construction of plasmids is described in supplemental “Experimental Procedures”.
Expression of Recombinant Proteins in Escherichia coli
pET based overexpression constructs containing either the full-length flaX gene or different truncated variants of the flaX gene were transformed into E. coli BL21(DE3) RIL (codon plus) cells and grown in LB/ampicillin/chloramphenicol medium overnight at 37 °C as pre-culture for expression. 1 ml of pre-culture was used to inoculate 1 liter of Luria-Bertani medium supplemented with ampicillin (50 μg/ml) and chloramphenicol (34 μg/ml) and was grown at 37 °C until optical density (A600) of 0.4–0.5 was reached. About 0.25–0.5 mm isopropyl β-d-thiogalactopyranoside was added for induction of protein expression. The induction was carried out at 37 °C for 3–4 h. The cells were collected by centrifugation, frozen in liquid nitrogen, and stored at −80 °C. Expression of untagged FlaI was performed using the protocol described previously (7).
Processing of Cell Pellet and Purification of Recombinant Proteins
Cell pellets were thawed on ice and resuspended in lysis buffer containing 50 mm Tris-HCl, pH 8, 150 mm NaCl, 5% glycerol, 0.5% Triton X-100, 20 mm imidazole, 1 mm PMSF, and 1× Roche complete ULTRA protease inhibitor. Resuspended cells were sonicated using Sonoplus HD3100 sonicator (Bandelin Sonorex Biotechnique) with probe HD3100 until the lysate became translucent. Cell debris was removed using a centrifugation step with 10,000 × g at 4 °C for 25 min. The cell lysate was kept at 70 °C for 20 min to precipitate E. coli proteins and the heat-shocked lysate was cleared at 10,000 × g for 30 min. The clear supernatant was kept for further analysis. To obtain the membrane-associated FlaX and the truncated variants the cell lysate was further ultracentrifuged at 150,000 × g for 30 min at 4 °C in an OptimaTM MAX-XP ultracentrifuge (rotor MLA55; Beckman Coulter) and the membrane fraction was collected. The membrane proteins were solubilized by resuspending the membrane fraction in 50 mm Tris-HCl buffer, pH 8, 150 mm NaCl, 5% glycerol, 20 mm imidazole, 1× Roche complete ULTRA protease inhibitor, and 2% (w/v) n-dodecyl-β-maltoside (DDM)2 followed by an incubation at 50 °C for 1–2 h for proper solubilization of the membrane proteins (14). As S. acidocaldarius is a hyperthermophilic organism, a high temperature solubilization protocol was adopted for heterologously expressed proteins. The decreased membrane rigidity due to the high temperature treatment facilitates extraction by detergent of the membrane-associated protein (14). Immediately after a high temperature incubation the solubilized membrane fraction was centrifuged at 100,000 × g at 4 °C in a MAX-XP ultracentrifuge (rotor TLA110; Beckman Coulter) for 30 min to remove all insoluble particles. The soluble membrane fraction was diluted until the final concentration of DDM was decreased to 0.1% (w/v) and kept for further analysis.
To purify His6-tagged overexpressed protein Ni-NTA affinity chromatography was used. Either soluble lysate or the solubilized membrane fraction was loaded onto a Ni-NTA affinity column (native IMAC) on a Profinia purification system (Bio-Rad Laboratories). The protein was eluted using 50 mm HEPES, pH 7.2, 150 mm NaCl, 3% glycerol, 1× cocktail inhibitor, and 350 mm imidazole (0.05% (w/v) DDM was added in case of purifying from solublized membrane fraction) from the Ni-NTA affinity column. The eluted protein was desalted using 50 mm HEPES, pH 7.2, 150 mm NaCl, 3% (v/v) glycerol, 1× mixture inhibitor buffer (0.05% (w/v) DDM was added in case of solublized membrane fraction) with a native desalting column on Profinia. To ensure purity of the overexpressed protein, the His-purified protein was further loaded onto an anion exchange DEAE-Sepharose fast flow (GE Healthcare) column. The bound protein was eluted using 50 mm HEPES, pH 7.2, 250 mm NaCl, 3% (v/v) glycerol, 1× cocktail inhibitor buffer (0.05% DDM was added in case of solublized membrane fraction) from the anion exchange column. The purity of the protein was assessed on a Coomassie-stained SDS-PAGE and anti-His/anti-FlaX Western blot analyses (supplemental Fig. S2).
Far-UV CD Spectroscopy
CD spectra were recorded at room temperature with a Jasco J-810 spectropolarimeter. Experiments were conducted in 1-mm path length cells. Confirmation of the structural integrity of the constructed mutants was obtained by the acquisition of far-UV (190–280 nm) CD spectra. Far-UV spectra of the tested proteins are presented as the smoothed average of three accumulations. Secondary structure prediction analysis was performed with the supplied J-810 software for Windows, using the Yang statistical algorithm (15). All recorded CD spectra were baseline corrected by subtraction of the “buffer only” spectrum.
Yeast Two-hybrid Analysis
All vectors, yeast strains, and vector backbones were adopted from the BD MATCHMAKER Library Construction & Screening Kit (Clontech, Palo Alto, CA). The bacterial strain E. coli DH5α was used for the propagation of the plasmid constructs. The yeast strain Saccharomyces cerevisiae AH109 (MATa, trp1–901, leu2–3, 112, ura3–52, his3–200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ) was employed as hosts in the two-hybrid analysis. To construct bait and prey constructs on pGADT7 and pGBKT7 vector backbone, flaX and Saci_1176 genes were amplified from S. acidocaldarius DSM639 genomic DNA using p2152/p2133 and p2148/p2123, respectively.
Yeast transformation was performed using the protocol described in Ref. 16 with slight modification. The S. cerevisiae AH109 stain was grown on YAPD plates (supplemented with adenine and uracil; 1% (w/v) Bacto-yeast extract, 2% (w/v) Bacto-peptone, adenine hemisulfate 80 mg/liter; for plates 18 g liter−1 of Bacto Agar was added) at 30 °C. To prepare 500 μl of a one-step transformation buffer 300 μl of 70% PEG in 1× TE buffer was mixed with 100 μl of 1 m LiOAc, 2.5 μl of 14.4 m β-mercaptoethanol, and 97.5 μl of MQ water was added together and mixed thoroughly. One single colony was added to 50 μl of freshly prepared one-step transformation buffer and after thoroughly vortexing 2.5 μl of carrier DNA (Herrings testis DNA, Clontech) with a concentration of 10 mg/ml was added (carrier DNA was boiled for 10 min in a water bath and quickly chilled on ice before addition). The sample was mixed thoroughly and 1–2 μg of vector was added to the mixture and a heat shock was given at 45 °C for 30 min. The transformed cells were plated on the respective antibiotic selection plates.
The pGBKT7-derived vectors were transformed into AH109 and plated on SD (-Trp) plates and the pGADT7-derived vectors after AH109 transformation were plated on SD (-Leu) plates and both set of plates were incubated at 30 °C for 3–5 days. Upon positive transformation the growing colonies were transformed with complementary backbone vectors and plated onto DD (-Trp-Leu) plates and incubated at 30 °C for 3–5 days. After positive transformation of both vectors, the positive colonies were streaked onto QD (-Trp-Leu-His-Ade) plates. To check the positive interaction between proteins, a β-galactosidase assay was performed. Positive colonies on the QD medium were streaked onto another QD plate supplemented with α-X-gal where a blue colony indicated a positive interaction.
Cross-linking Experiment
The oligomerization of FlaXc was assessed using chemical cross-linking as described in Ref. 17 with slight modifications. Formaldehyde was used as chemical cross-linker. 37% formaldehyde was diluted to a final concentration of 5% (v/v) in 25 mm sodium phosphate, pH 7.5, 150 mm NaCl buffer to prepare the working stock. 0.5 mg/ml of protein was incubated with 0.1% (v/v) formaldehyde and the mixture was incubated at room temperature for 5 min. Immediately after the incubation the reaction was stopped using 125 mm glycine-NaOH, pH 10, buffer.
Finally, the cross-linked material was precipitated with trichloroacetic acid (TCA). 1 volume of 50% TCA stock was mixed with 2 volumes of cross-linked sample and incubated for 30 min on ice. Samples were centrifuged at 14,000 × g for 5 min. The supernatant was removed and the white fluffy pellet was washed three times with 200 μl of ice-cold 100% acetone for the complete removal of the TCA. The pellet was then dried in a heat block at 95 °C to remove acetone and resuspended in 25 mm sodium phosphate, pH 7.5, 150 mm NaCl buffer. The sample was diluted in 5× Laemmli buffer and the cross-linking was visualized on 11% SDS-PAGE with Coomassie Blue staining.
Refolding of FlaXc
Denaturation of FlaXc was performed by 30 min incubation at room temperature in 3 m GdnHCl. 0.2–0.3 mg/ml of FlaXc was incubated with 5 mm DTT (dithiothreitol) for 5 min at room temperature prior to GdnHCl treatment and then GdnHCl was added to a final concentration of 3 m and mixed thoroughly. To ensure denaturation the mixture was incubated at room temperature for 30 min. To refold FlaXc we used the rapid dilution method to reduce the concentration of GdnHCl. As l-arginine functions as a chemical chaperone during protein folding (18, 19), we included l-Arg in the refolding buffer. Immediately after the denaturation, the protein was diluted in 50 mm sodium phosphate buffer, pH 7.5, containing 150 mm NaCl, 10% glycerol, 0.05% DDM, 1× cocktail inhibitor, and 200 mm l-Arg until the final concentration of GdnHCl was reduced to 0.08–0.1 m and incubated on ice for 30–60 min for optimal refolding. After refolding, the protein was concentrated using Amicon filters until a 1–2 ml final sample volume was left. The concentrated refolded protein was then dialyzed overnight against the refolding buffer without l-arginine. The folding status of the refolded FlaXc was assessed using far UV CD spectroscopy (supplemental Fig. S3).
Cell-Lysate Interaction Analysis
In vitro binary complex formation through cell-lysate interaction analysis experiments was carried out essentially as described (20, 21) with minor modifications. In this experiment a “two cell” protocol was used in which His6-tagged FlaXc and untagged FlaI were expressed separately in E. coli BL21(DE3) RIL (codon plus) cells. Cells were grown in LB medium supplemented with ampicillin (50 μg/ml) and chloramphenicol (34 μg/ml) until an A600 of 0.6–0.8 has reached. The induction was carried out with an addition of 0.5–1 mm isopropyl β-d-thiogalactopyranoside followed by cultivation for 3–4 h at 37 °C for His6 FlaXc and overnight at 16 °C for untagged FlaI. Cell pellets were harvested by centrifugation and resuspended in 50 mm HEPES, pH 7.5, buffer containing 50 mm NaCl, 1× protease inhibitor cocktail. Resuspended cells were mixed together in 1:5 (v/v) for untagged FlaI and His6-tagged FlaXc, respectively. The mixed cell lysate was sonicated thoroughly on ice and centrifuged at 10,000 × g for 30 min at 4 °C, the supernatant was collected for further analyses. 1–2 ml of the soluble fraction was mixed with 100 μl of Ni-NTA resin (equilibrated with the lysis buffer) and incubated for 2–3 h at room temperature with gentle rotation to enhance binding. The unbound portion was collected using 2500 × g centrifugation for 5 min on a table top centrifuge. Beads were washed with 20 bead volume of 50 mm HEPES, pH 7.5, buffer containing 50 mm NaCl, 50 mm imidazole. Bound proteins were eluted using 2× bead volume of 50 mm HEPES, pH 7.5, buffer containing 150 mm NaCl, 350 mm imidazole. The control experiment was performed using the lysate containing untagged FlaI alone and passing it through the Ni-NTA resin. Interaction was visualized using SDS-PAGE and immunoblotted using anti-His, anti-FlaX, and anti-FlaI antibodies.
Co-expression and Pure Protein Interaction Analysis
His6-tagged FlaXc or its deletion/truncation variants were co-overproduced from pETDuet1 backbone plasmids with untagged FlaI in E. coli codon plus using ampicillin (50 μg/ml) and chloramphenicol (34 μg/ml) on Luria-Bertani media, and proteins were copurified by affinity chromatography on a Ni-NTA His select (Sigma) matrix. Cells were grown and induced at 16 °C overnight using 0.5 mm isopropyl β-d-thiogalactopyranoside. The cell-free lysate was prepared in 50 mm Tris-HCl, pH 8, 150 mm NaCl, 3% glycerol, 20 mm imidazole buffer by sonification and centrifugation at 10,000 × g and then filtered using 0.45-μm filter, in cases where the protein was going to the membrane, the membrane was collected and solubilized using the protocol described earlier to prepare the membrane lysate. The lysate was loaded onto a Ni-NTA gravity column preincubated with binding buffer (50 mm HEPES, pH 7.5, 150 mm NaCl). After collecting the unbound fraction the column bed was washed using 5–10 column volumes using 50 mm HEPES, pH 7.5, 150 mm NaCl, 50 mm imidazole buffer. Proteins were copurified using 50 mm HEPES, pH 7.5, 150 mm NaCl, 350 mm imidazole buffer and analyzed on SDS-PAGE and immunoblot against His tag and FlaI peptide antibody. To perform the binary complex formation using purified proteins, the proteins were mixed and incubated overnight and loaded onto Ni-NTA affinity resin, incubated and pulled down after a rigorous washing step.
In Trans Complementation and Swarming Assay
In trans complementation of MW452 (ΔflaXΔaapF) competent cells using flaX and its deletion/truncations and swarming assay of transformed strains were performed according to the protocol described previously (4).
Electron Microscopy and Image Analysis
Electron microscopy of S. acidocaldarius cells was performed using the protocol described previously (22).
FlaX complexes were negatively stained with 1% (w/v) uranyl acetate. Electron micrographs were collected using a Philips CM120 at 120 kV, at a magnification of ×44,000 on a Gatan 2kx2k CCD camera. For cryo-EM, samples were applied to glow-discharged Quantifoil grids (Quantifoil MicroTools) and vitrified by injection into liquid ethane using a Vitrobot plunge-freezing device (FEI). Samples were imaged in an FEI Polara at ×59,000 magnification (calibrated as ×61,400) at a defocus of 2–4 μm. Images were recorded on Kodak SO-163 film and scanned on a Zeiss Photoscan scanner (Intergraph) at a step size of 7 μm. Subsequently, adjacent pixels were averaged to yield a pixel size on the specimen of 3.42 Å. Particles were selected using the boxer module from EMAN (23). The CTF was determined using EMAN2 (24). A data set of 7,000 particles was processed with Imagic V (25). The images were subjected to reference-free alignment and multivariate statistical analysis. The images were classified and images assigned to the same class were averaged.
RESULTS
FlaX Oligomerizes
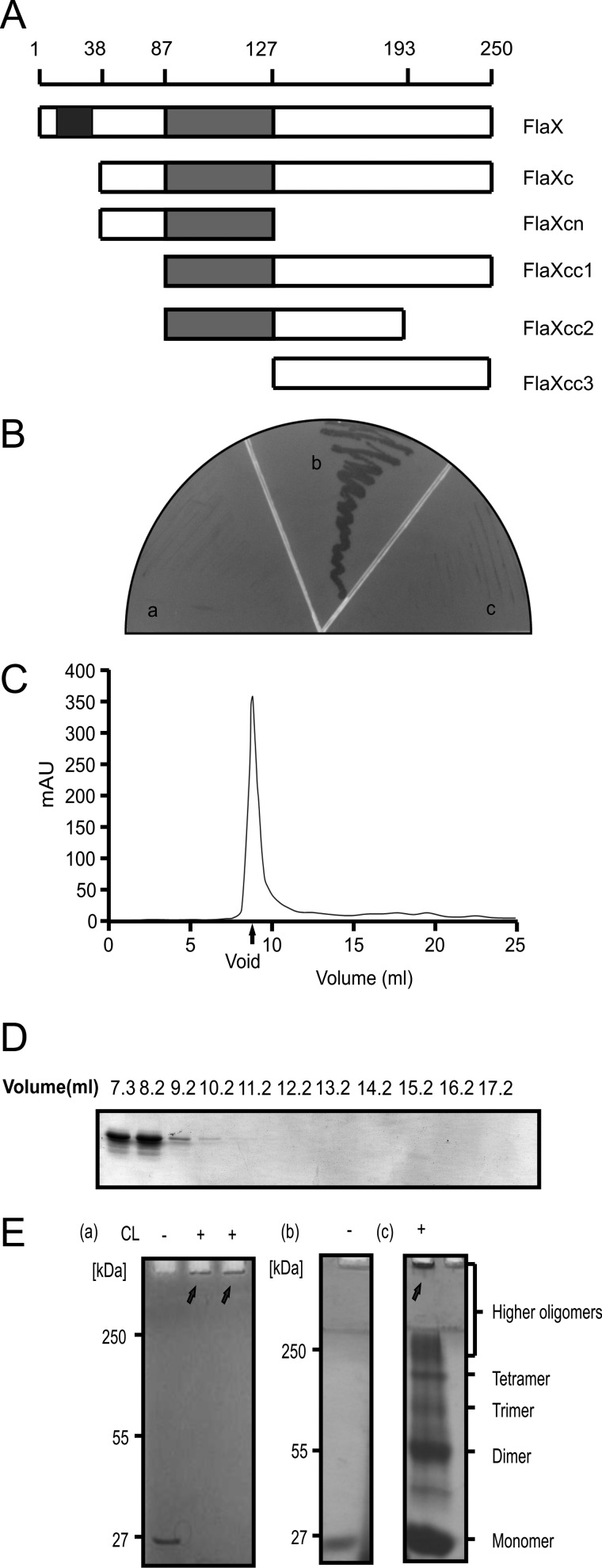
It has been described above that FlaX is one of the seven essential proteins that build the archaellum of the crenarchaeon S. acidocaldarius (Fig. 1, A and B) (4). It is a monotopic membrane protein of 28 kDa that is conserved in crenarchaeal archaella loci. Secondary structure predictions suggest that the N-terminal transmembrane domain is 22 amino acids long (amino acids 10–32) and that the soluble domain is rich in helices and contains a coiled-coil region (amino acids 87–127) predicted as a triple coil (26) (supplemental Fig. S1 and Fig. 2A).
FIGURE 2.
FlaX oligomerizes. A, schematic diagram of FlaX deletion/truncations. The predicted coiled-coil region is shown in gray and the membrane domain in black. B, yeast two-hybrid analysis to show interaction. Panel a shows the vector control (with FlaXc as bait and empty vector as prey), panel b reflects the interaction (where both bait and prey are FlaXc), and panel c is the protein control (FlaXc as bait and saci_1176 as prey). C, size exclusion chromatography (SEC analyis) of FlaX. D, SDS-PAGE profile of FlaXc SEC analysis. E, formaldehyde cross-linking of FlaXc. Panel a, chemical cross-linking of native FlaXc; panel b, refolded FlaXc; and panel c, chemical cross-linking of refolded FlaXc, where on SDS-PAGE different oligomeric forms are visible. CL, cross-linker, (+) with cross-linker and (−) without cross-linker. The arrows indicates formaldehyde cross-linked high oligomeric species.
As full-length FlaX-His6 expressed poorly in E. coli, a truncate version of FlaX was constructed missing the first 37 amino acids, called FlaXc-His6 (Fig. 2A), that could successfully be purified from E. coli (supplemental Fig. S2). To understand the influence of the different regions of FlaXc, we also constructed different truncates based on domain predictions from the bioinformatic analysis (Fig. 2A). All the different truncates of FlaX were expressed in E. coli and purified to homogeneity using Ni-NTA affinity chromatography (supplemental Fig. S3). Analysis by a anti-His Western blot of pure proteins revealed that most of the FlaX truncates can form SDS-resistant oligomers except for FlaXcc1 (supplemental Fig. S3 and Fig. 4). The SDS-resistant oligomeric pattern is, however, concentration and preparation dependent. All the higher molecular weight protein bands that appeared on the SDS-PAGE were identified as FlaX by using peptide mass fingerprinting and correct folding of FlaXc was determined by far-UV CD spectroscopy (supplemental Fig. S4).
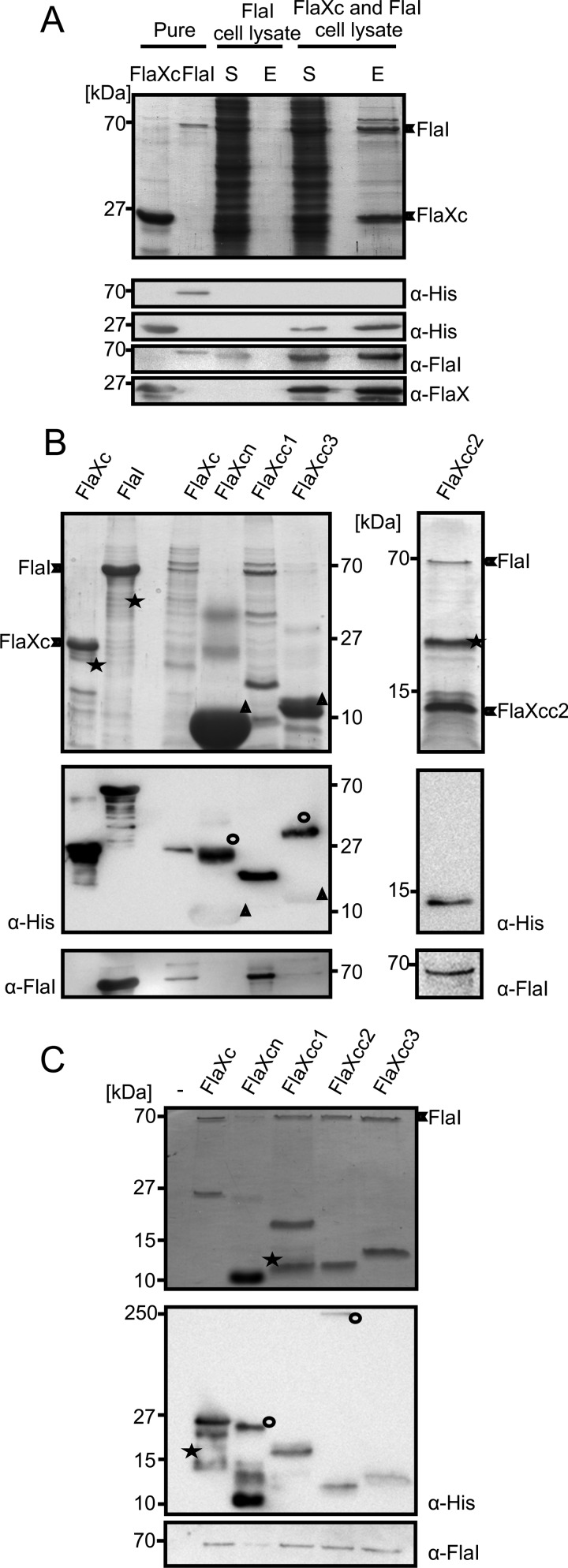
FIGURE 4.
FlaX and FlaI interact. A, cell lysates containing either untagged FlaI or FlaXc-His6 were mixed and FlaXc-His6 was isolated using Ni-NTA beads. Samples were analyzed on SDS-PAGE and immunoblotted with antibodies against the His6 tag, FlaI, and FlaX. Whereas untagged FlaI did not bind to the beads itself (“FlaI cell lysate”), it did co-elute with FlaXc-His6 (“FlaXc/FlaI cell lysate”). S, start material; E, elution fraction. His6-tagged FlaI was used in lane 2 to confirm the migration pattern of untagged FlaI. B, N-terminal His6-tagged FlaX truncates were co-expressed with untagged wt FlaI and Ni-NTA beads were used for pulldown analysis. The pulldown fraction was confirmed using SDS-PAGE (upper panels) and anti-His and anti-FlaI Western blot analysis (lower panels). ★, degraded protein. ○, higher oligomeric forms of the protein. Δ, monomer of the protein. FlaXcn and FlaXcc3 form SDS-resistant higher oligomers in a concentration-dependent manner and on Western blot analysis the higher oligomers provide more signal than the monomeric form, which is visible on the anti-His Western blot. The degradation product in the most right-hand panel reacted with the FlaI antibody. C, purified FlaX truncates were incubated with untagged FlaI as described in supplemental ”Experimental Procedures“ pulled down with Ni-NTA beads. Elution fractions were analyzed on SDS-PAGE (upper panel) and Western blot with either anti-His or anti-FlaI antibodies (lower panel). ★, degraded protein; ○, SDS-resistant oligomer.
The first indication that FlaXc indeed forms oligomers in vivo was obtained using yeast two-hybrid analysis with FlaXc as both bait and prey (Fig. 2B). As a negative control, the growth of S. cerevisiae cells expressing FlaXc as bait protein in combination with either empty vector or another protein as the prey was evaluated and no growth was detected (Fig. 2B). Analytical gel filtration of FlaXc showed that the protein eluted in the void volume of the Superdex 200 column, indicating that the oligomeric species had to be larger than 600 kDa (Fig. 2, C and D). The in vitro oligomerization behavior of FlaXc was further tested employing cross-linking in the presence of 0.1% formaldehyde. All the cross-linked species of FlaXc remained trapped at the stacking gel/separating gel interface on SDS-PAGE indicating the formation of a very high molecular weight oligomeric species (Fig. 2E). As it has been shown that the refolding of native protein can significantly enhance the probability to identify different intermediate oligomeric species, GdnHCl was used to denature FlaXc and subsequent refolding was achieved by using a rapid dilution method in the presence of l-arginine. The cross-linking experiment with refolded FlaXc indicated the presence of dimeric, trimeric, tetrameric, and several higher-oligomeric species (Fig. 2E).
To exclude the fact that SDS-resistant oligomerization patterns of most of the FlaX truncates are an artifact as mentioned by Wu and Filutowicz (27) we have performed SEC analysis of FlaX truncates. The elution patterns demonstrated that most of the FlaX truncates formed high molecular weight oligomers (supplemental Fig. S5). FlaXcn was the smallest truncate eluted at a molecular mass around 150 kDa, suggesting formation of oligomer under native conditions. On the other hand FlaXcc2 has a broad elution profile, whereas all other truncates eluted in the void volume including FlaXcc1 and FlaXcc3, suggesting the formation of high oligomeric complex.
FlaX Forms a Ring
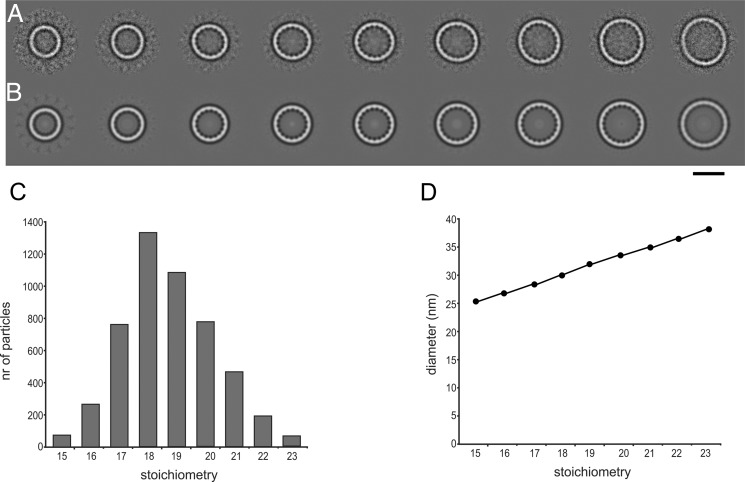
To study the oligomeric state of FlaX directly, we performed electron microscopy (EM) on the different truncates. The N-terminal truncate FlaXcn did not show any large structures, and FlaXcc2 formed large globular particles. FlaXc, FlaXcc1, and FlaXcc3 formed rings of various sizes with a diameter of 26–38 nm. As these three truncates have the C-terminal domain in common, we conclude that the ring-forming domain is in the C-terminal 57 amino acids, which are predicted to contain three α-helices (supplemental Fig. S1). A cryo-EM data set of 7,000 particles was collected of FlaXc, the largest construct. Class averages of cryo-EM images display a rotational symmetry of 15–23 (Fig. 3, A and B), 18 and 19 being most common with 23 and 21%, respectively (Fig. 3C). The difference in diameter of the rings was consistent with the symmetry, each additional unit increasing the diameter with 1.5 nm (Fig. 3D). The intersubunit spacing is constant at 5.3 nm. The rings have a ∼4 nm wide rim and thin spokes pointing into the ring interior.
FIGURE 3.
Cryo-EM of FlaXc. A, class averages after multireference alignment in order of increasing size. B same as A with rotational symmetry applied from 15 (left) to 23 (right). C, the number of particles per class follows a Gaussian distribution with a maximum at 18–19. D, the diameter of the rings increases linearly. The scale bar represents 25 nm in A and B.
FlaXc Interacts with the ATPase FlaI
So far no information was available on possible interactions between the 7 subunits of the archaellum assembly machinery. Because FlaX is a monotopic membrane protein, it was important to determine whether the FlaX ring is localized in the cytoplasm or the pseudo-periplasm. Recently, we observed that FlaX was destabilized in ΔflaH, ΔflaI, ΔflaJ mutants of S. acidocaldarius suggesting that FlaX, FlaH, FlaI, and FlaJ possibly interact (4). Moreover, the soluble motor ATPase FlaI has recently been shown to localize to the membrane, possibly via an interaction of the archaellum membrane components (7). We therefore hypothesized that FlaX might interact with FlaI during the assembly of archaella. To that end cell lysates of cells expressing either FlaXc-His6 or untagged FlaI were mixed and incubated with Ni-NTA beads to purify FlaXc-His6 and associated protein(s). Immunoblot analysis showed that under the experimental conditions, untagged FlaI eluted with FlaXc indicating an interaction between FlaXc and FlaI (Fig. 4A), whereas untagged FlaI did not bind to the Ni-NTA beads (Fig. 4A). The same result was obtained when purified untagged FlaI and FlaXc-His6 were mixed and a pulldown assay using Ni-NTA beads was performed (Fig. 4C). Taken together these data suggest a strong interaction between FlaX and FlaI and thereby localization of the FlaX ring at the cytoplasmic side of the membrane.
FlaXc Interacts with FlaI Using the C-terminal Region
To determine which part of FlaX is responsible for the interaction with FlaI, pulldown assays were performed with purified untagged FlaI and the different His6-tagged FlaX truncates (Fig. 4C), coexpression and pulldown experiments were also performed in parallel with pure protein interaction analysis (Fig. 4B). Although FlaI co-purified with FlaXc, FlaXcc1, FlaXcc2, and FlaXcc3, no interaction was evident for FlaXcn in which C-terminal amino acids 128–250 are missing (Fig. 4, B and C). These results indicate that the interaction with FlaI is mediated by the C-terminal half of FlaX.
Full-length FlaX Is Required for the Method Termed as in Trans Complementation
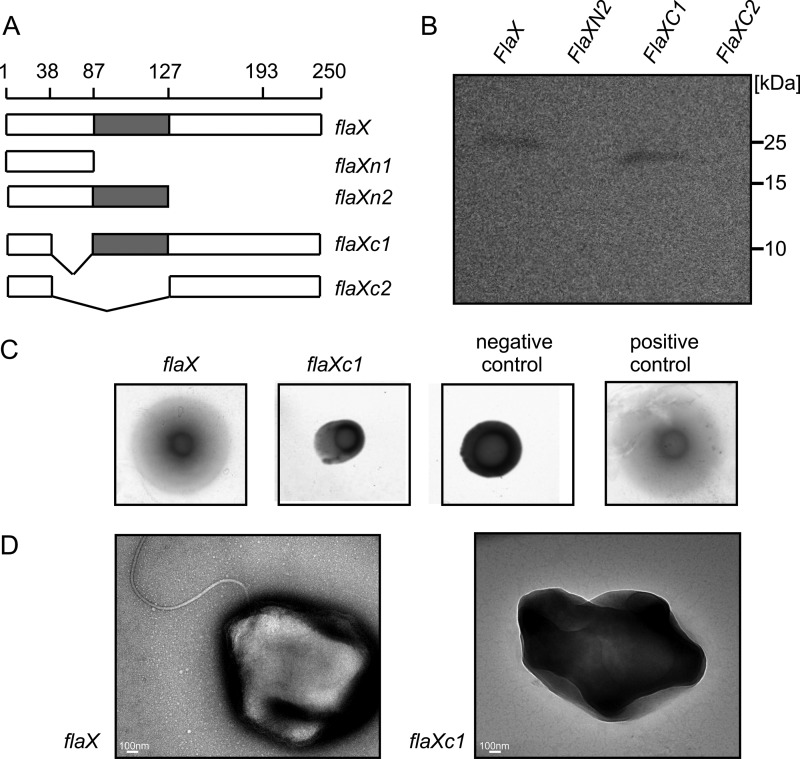
To test whether full-length FlaX is indispensible for the assembly of archaella, we introduced full-length as well as different truncated variants of the flaX gene (Fig. 5A) in a multicopy plasmid and transformed these into the ΔaapFΔflaX S. acidocaldarius mutant. The ΔaapF background strain was used as the deletion of the aap pili of which aapF is an essential component, leads to a hypermotile phenotype due to hyperarchaellation (4, 28). In the complementation strains only FlaX and FlaXC1 were stably expressed (Fig. 5B). Soft-gelrite motility assay and electron microscopy of these two strains revealed that only the full-length flaX gene product could restore the wild type phenotype (Fig. 5, C and D). This indicates that FlaX acts as a major core subunit in the archaella assembly.
FIGURE 5.
In trans complementation of ΔaapFΔflaX (MW452) strain. A, schematic representation of different flaX truncates that were tested for their ability to complement a ΔflaX strain. B, expression levels of the truncates in S. acidocaldarius. C, soft gelrite motility assay of complemented strains. As control experiment MW351 with pSVA1614 was used as positive control and MW456 with pSVA352 (a E to A mutant of the Walker B box in FlaI on pSVA1450 backbone (S. Reindl, A. Ghosh et. al., unpublished data) was used as negative control. D, transmission electron microscopic images of flaX and flaXC1 expressing MW452 strains, respectively.
DISCUSSION
The soluble domain of FlaX forms ∼30 nm diameter oligomeric rings of >600 kDa and the three C-terminal α-helices are responsible for ring formation. In addition, we showed that the C-terminal half of FlaXc interacts with the ATPase FlaI.
The archaellum is a unique motility structure as it resembles a simple type IV pilus that is able to rotate. A major unsolved question is how the force for filament rotation is obtained, as none of the archaellum proteins show homology to bacterial flagellar proteins. Bacterial flagella rotate on multiprotein ring complexes, like the C and MS ring (29, 30). The C ring is composed of multiple copies of FliN, FliM, and FliG (31, 32). The average diameter of the bacterial C ring, which is localized just beneath the inner membrane (6, 33), is around 45 nm (34) and has variable stoichiometry, centered on 34 (34, 35), and it was also shown that the C ring components are interacting with the type III machineries (36). Given the structural similarities and interaction with the motor ATPase FlaI, we propose that the FlaX ring might be important as a static part in the archaellum motor for force generation (Fig. 6). Previously, we showed that FlaX is destabilized also in the ΔflaJ and ΔflaB deletion strains (4), which further suggests that the membrane domain of FlaX is very mobile and may be stabilized upon interacting with other proteins and therefore might play a crucial role in the recruitment of processed FlaB into the archaellum filament.
FIGURE 6.
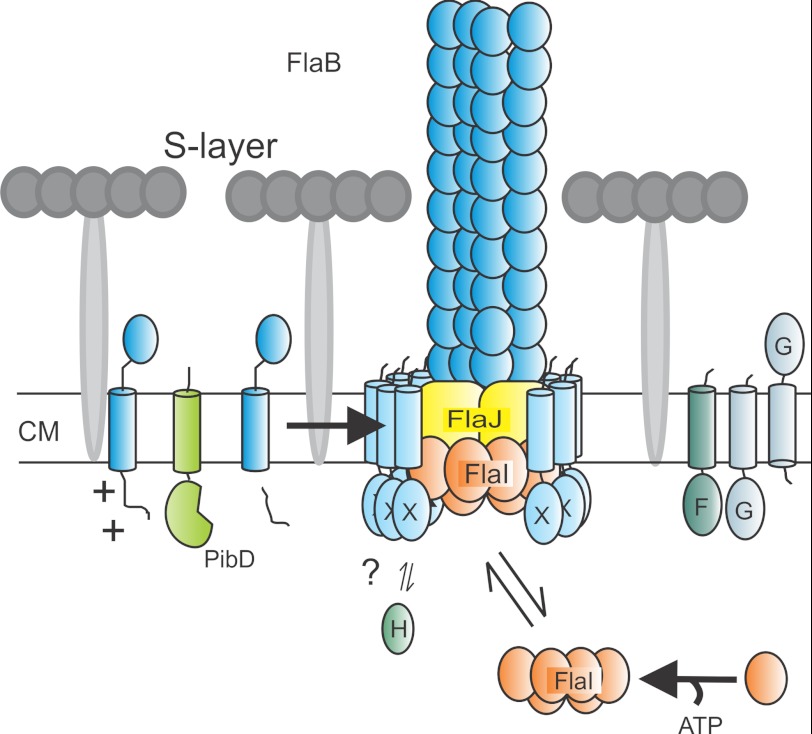
Current model of archaellum assembly. Taken the results of this manuscript into account we can now adapt the model of the assembled archaellum. We propose that the FlaX ring is positioned in such a way that its soluble domain is in contact with FlaI and that the membrane domain is placed all around the possible trimer of FlaJ. CM, cytoplasmic membrane.
With recent advancements in the understanding of the archaella assembly using biochemical approaches we suggest that in the crenarchaeal archaella model system FlaX is the central protein that is indeed needed as a priming subunit to successfully assemble the archaella. This unique nanomachine is very simplistic when compared with bacterial flagella and it will be exciting to learn in the future whether FlaX plays a role in torque generation.

This article contains supplemental ”Experimental Procedures,“ Tables S1-S3, and Figs. S1–S5.
- DDM
- n-dodecyl-β-maltoside
- Ni-NTA
- nickel-nitrilotriacetic acid
- GdnHCl
- guanidine hydrochloride.
REFERENCES
- 1. Jarrell K. F., Albers S. V. (2012) The archaellum. An old motility structure with a new name. Trends Microbiol. 20, 307–312 [DOI] [PubMed] [Google Scholar]
- 2. Ghosh A., Albers S. V. (2011) Assembly and function of the archaeal flagellum. Biochem. Soc. Trans. 39, 64–69 [DOI] [PubMed] [Google Scholar]
- 3. Pohlschroder M., Ghosh A., Tripepi M., Albers S. V. (2011) Archaeal type IV pilus-like structures. Evolutionarily conserved prokaryotic surface organelles. Curr. Opin. Microbiol. 14, 357–363 [DOI] [PubMed] [Google Scholar]
- 4. Lassak K., Neiner T., Ghosh A., Klingl A., Wirth R., Albers S. V. (2012) Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 83, 110–124 [DOI] [PubMed] [Google Scholar]
- 5. Albers S. V., Szabó Z., Driessen A. J. (2003) Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 185, 3918–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sosinsky G. E., Francis N. R., Stallmeyer M. J., DeRosier D. J. (1992) Substructure of the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 223, 171–184 [DOI] [PubMed] [Google Scholar]
- 7. Ghosh A., Hartung S., van der Does C., Tainer J. A., Albers S. V. (2011) Archaeal flagellar ATPase motor shows ATP-dependent hexameric assembly and activity stimulation by specific lipid binding. Biochem. J. 437, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen-Krausz S., Trachtenberg S. (2008) The flagellar filament structure of the extreme acidothermophile Sulfolobus shibatae B12 suggests that archaeabacterial flagella have a unique and common symmetry and design. J. Mol. Biol. 375, 1113–1124 [DOI] [PubMed] [Google Scholar]
- 9. Trachtenberg S., Cohen-Krausz S. (2006) The archaeabacterial flagellar filament. A bacterial propeller with a pilus-like structure. J. Mol. Microbiol. Biotechnol. 11, 208–220 [DOI] [PubMed] [Google Scholar]
- 10. Bardy S. L., Ng S. Y., Jarrell K. F. (2004) Recent advances in the structure and assembly of the archaeal flagellum. J. Mol. Microbiol. Biotechnol. 7, 41–51 [DOI] [PubMed] [Google Scholar]
- 11. Streif S., Staudinger W. F., Marwan W., Oesterhelt D. (2008) Flagellar rotation in the archaeon Halobacterium salinarum depends on ATP. J. Mol. Biol. 384, 1–8 [DOI] [PubMed] [Google Scholar]
- 12. Schlesner M., Miller A., Streif S., Staudinger W. F., Müller J., Scheffer B., Siedler F., Oesterhelt D. (2009) Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol. 9, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wagner M., van Wolferen M., Wagner A., Lassak K., Meyer B. H., Reimann J., Albers S. V. (2012) Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front. Microbiol. 3, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peng G., Fritzsch G., Zickermann V., Schägger H., Mentele R., Lottspeich F., Bostina M., Radermacher M., Huber R., Stetter K. O., Michel H. (2003) Isolation, characterization and electron microscopic single particle analysis of the NADH:ubiquinone oxidoreductase (complex I) from the hyperthermophilic eubacterium Aquifex aeolicus. Biochemistry 42, 3032–3039 [DOI] [PubMed] [Google Scholar]
- 15. Yang J. T., Wu C. S., Martinez H. M. (1986) Calculation of protein conformation from circular dichroism. Methods Enzymol. 130, 208–269 [DOI] [PubMed] [Google Scholar]
- 16. Gietz R. D., Schiestl R. H. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 [DOI] [PubMed] [Google Scholar]
- 17. Pecorari F., Minard P., Desmadril M., Yon J. M. (1996) Occurrence of transient multimeric species during the refolding of a monomeric protein. J. Biol. Chem. 271, 5270–5276 [DOI] [PubMed] [Google Scholar]
- 18. Das U., Hariprasad G., Ethayathulla A. S., Manral P., Das T. K., Pasha S., Mann A., Ganguli M., Verma A. K., Bhat R., Chandrayan S. K., Ahmed S., Sharma S., Kaur P., Singh T. P., Srinivasan A. (2007) Inhibition of protein aggregation. Supramolecular assemblies of arginine hold the key. PLoS One 2, e1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Golovanov A. P., Hautbergue G. M., Wilson S. A., Lian L. Y. (2004) A simple method for improving protein solubility and long-term stability. J. Am. Chem. Soc. 126, 8933–8939 [DOI] [PubMed] [Google Scholar]
- 20. Terradot L., Durnell N., Li M., Li M., Ory J., Labigne A., Legrain P., Colland F., Waksman G. (2004) Biochemical characterization of protein complexes from the Helicobacter pylori protein interaction map. Strategies for complex formation and evidence for novel interactions within type IV secretion systems. Mol. Cell Proteomics 3, 809–819 [DOI] [PubMed] [Google Scholar]
- 21. Minamino T., Kinoshita M., Hara N., Takeuchi S., Hida A., Koya S., Glenwright H., Imada K., Aldridge P. D., Namba K. (2012) Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Mol. Microbiol. 83, 775–788 [DOI] [PubMed] [Google Scholar]
- 22. Henche A. L., Koerdt A., Ghosh A., Albers S. V. (2012) Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environ. Microbiol. 14, 779–793 [DOI] [PubMed] [Google Scholar]
- 23. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN. Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 24. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) EMAN2. An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 25. van Heel M., Harauz G., Orlova E. V., Schmidt R., Schatz M. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 [DOI] [PubMed] [Google Scholar]
- 26. Lupas A. (1996) Coiled coils. New structures and new functions. Trends Biochem. Sci. 21, 375–382 [PubMed] [Google Scholar]
- 27. Wu J., Filutowicz M. (1999) Hexahistidine (His6)-tag dependent protein dimerization. A cautionary tale. Acta Biochim. Pol. 46, 591–599 [PubMed] [Google Scholar]
- 28. Henche A. L., Ghosh A., Yu X., Jeske T., Egelman E., Albers S. V. (2012) Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius. Environ. Microbiol., doi: 10.1111/j.1462-2920.2012.02898.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berg H. C., Anderson R. A. (1973) Bacteria swim by rotating their flagellar filaments. Nature 245, 380–382 [DOI] [PubMed] [Google Scholar]
- 30. Berg H. C. (2003) The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54 [DOI] [PubMed] [Google Scholar]
- 31. Suzuki H., Yonekura K., Namba K. (2004) Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J. Mol. Biol. 337, 105–113 [DOI] [PubMed] [Google Scholar]
- 32. Paul K., Gonzalez-Bonet G., Bilwes A. M., Crane B. R., Blair D. (2011) Architecture of the flagellar rotor. EMBO J. 30, 2962–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Erhardt M., Namba K., Hughes K. T. (2010) Bacterial nanomachines. The flagellum and type III injectisome. Cold Spring Harbor Perspect. Biol. 2, a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thomas D. R., Morgan D. G., DeRosier D. J. (1999) Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad Sci. U.S.A. 96, 10134–10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Young H. S., Dang H. Y., Lai Y. M., DeRosier D. J., Khan S. (2003) Variable symmetry in Salmonella typhimurium flagellar motors. Biophys. J. 84, 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. González-Pedrajo B., Minamino T., Kihara M., Namba K. (2006) Interactions between C ring proteins and export apparatus components. A possible mechanism for facilitating type III protein export. Mol. Microbiol. 60, 984–998 [DOI] [PubMed] [Google Scholar]