Background: Proteasomal degradation of PP2A regulatory subunits has been described, but responsible E3 ubiquitin ligases have remained elusive.
Results: KLHL15 is an E3 ubiquitin ligase adaptor targeting the B′/B56β regulatory subunit for proteasomal degradation, promoting formation of alternative PP2A holoenzymes.
Conclusion: KLHL15 contributes to brain-specific expression of B′β and modifies PP2A holoenzyme composition.
Significance: E3 ligase-mediated B subunit degradation is a novel mechanism to remodel the PP2A heterotrimer.
Keywords: Phosphoprotein Phosphatase, PP2A, Protein Turnover, Serine Threonine Protein Phosphatase, Signal Transduction, Ubiquitin Ligase, Ubiquitylation, Proteasomal Degradation, Protein Phosphatase 2A
Abstract
Protein phosphatase 2A (PP2A), a ubiquitous and pleiotropic regulator of intracellular signaling, is composed of a core dimer (AC) bound to a variable (B) regulatory subunit. PP2A is an enzyme family of dozens of heterotrimers with different subcellular locations and cellular substrates dictated by the B subunit. B′β is a brain-specific PP2A regulatory subunit that mediates dephosphorylation of Ca2+/calmodulin-dependent protein kinase II and tyrosine hydroxylase. Unbiased proteomic screens for B′β interactors identified Cullin3 (Cul3), a scaffolding component of E3 ubiquitin ligase complexes, and the previously uncharacterized Kelch-like 15 (KLHL15). KLHL15 is one of ∼40 Kelch-like proteins, many of which have been identified as adaptors for the recruitment of substrates to Cul3-based E3 ubiquitin ligases. Here, we report that KLHL15-Cul3 specifically targets B′β to promote turnover of the PP2A subunit by ubiquitylation and proteasomal degradation. Comparison of KLHL15 and B′β tissue expression profiles suggests that the E3 ligase adaptor contributes to selective expression of the PP2A/B′β holoenzyme in the brain. We mapped KLHL15 residues critical for homodimerization as well as interaction with Cul3 and B′β. Explaining PP2A subunit selectivity, the divergent N terminus of B′β was found necessary and sufficient for KLHL15-mediated degradation, with Tyr-52 having an obligatory role. Although KLHL15 can interact with the PP2A/B′β heterotrimer, it only degrades B′β, thus promoting exchange with other regulatory subunits. E3 ligase adaptor-mediated control of PP2A holoenzyme composition thereby adds another layer of regulation to cellular dephosphorylation events.
Introduction
PP2A3 is one of four major serine/threonine phosphatases (in addition to PP1, PP2B, and PP2C) (1) and comprises up to 1% of total protein in mammalian cells (2). Predominantly found as a heterotrimer containing a core dimer of catalytic (C) and scaffolding (A) subunits, PP2A is a widely distributed and critical mediator of many signaling pathways. A third, variable B subunit defines the substrates and localization of the holoenzyme in the cell (3–5). PP2A B regulatory subunits are subdivided into three gene families, B (PR55/PPP2R2), B′ (B56/PR61/PPP2R5), and B″ (PR72/PPP2R3), with little sequence or structural similarity between them. In mammals each gene family is comprised of four to five highly similar genes. Additional complexity arises through alternative splicing. The five mammalian B′ gene products (α/β/γ/δ/ϵ) feature a conserved, tandem α-helical central domain that binds to the AC dimer (6, 7). The variable N and C termini contain subcellular localization sequences and phosphorylation sites for isoform-specific regulation (8). Upon down-regulation of the A or C subunit, PP2A B and B′ subunits are rapidly degraded by the proteasome (9–11). However, whether proteasomal degradation constitutes a physiological PP2A regulatory mechanism involving dedicated E3 ligases has remained unclear.
We previously identified PP2A/B′β as a brain-specific PP2A holoenzyme that catalyzes the dephosphorylation and inactivation of tyrosine hydroxylase, the rate-limiting enzyme in dopamine synthesis (12), and uncovered B′β residues that mediate substrate specificity (7). Because PP2A/B′β could be a target for intervention into Parkinson disease, a hallmark of which is loss of dopamine in the striatum, we set out to discover novel interactors of this regulatory subunit by proteomic approaches.
These screens identified the E3 ubiquitin ligase scaffold cullin3 (Cul3) and an uncharacterized protein, KLHL15. KLHL15 belongs to a large family of proteins with a characteristic domain architecture consisting of an N-terminal BTB (broad-complex, tram track, and bric-a-brac) domain, a C-terminal Kelch β-propeller, and a middle BACK (between BTB and C-terminal Kelch) domain (13, 14). Several KLHL proteins have been shown to bridge Cul3-containing E3 ligases to specific substrates (15–21). Here we report on the characterization of the KLHL15-Cul3 complex as a B subunit-specific regulator of PP2A. We propose E3 ligase-mediated exchange of regulatory subunits as a novel mechanism to control PP2A holoenzyme composition and downstream dephosphorylation events.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293 cells were cultured (37 °C and 5% CO2) in Dulbecco's modified Eagle's medium (Invitrogen) with 10% (v/v) fetal bovine serum. Cells were grown to 60% confluency on collagen-coated plates and transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol for transient transfection of adhered cells. 24–48 h post-transfection, cells were washed in phosphate-buffered saline and harvested. For experiments including MG132 (Sigma), cells were incubated with 50 μm concentrations of the drug (from a 50 mm stock in dimethyl sulfoxide, DMSO) for the final 12 h before lysis.
Plasmids
The green fluorescent protein (GFP)-tagged KLHL1 and KLHL24 plasmids were kindly provided by Dr. Michael Koob (University of Minnesota, Minneapolis, MN) and Dr. Fernanda Laezza (Washington University, St. Louis, MO), respectively. The HA-Cul2, HA-Cul3, and HA-Cul3Δroc constructs were kind gifts of Dr. Mark Hannink (University of Missouri, Colombia, MO). The coding sequence of KLHL15 was amplified from HEK293 RNA isolated with TRIzol reagent (Invitrogen) using the one-step RT-PCR kit from Qiagen. Both partial and full-length KLHL15 cDNAs were subcloned into mammalian (pEGFP-C1) and bacterial (pGEX-2T, pRSET) expression plasmids using compatible restriction sites. Plasmids expressing EE-tagged Aα subunit (22), GFP-Drp1 (23), wild-type and mutant FLAG-B′β (PPP2R5B) (7, 12), and triple HA-tagged B′/B56 isoforms (24) were described previously. N- and C-terminal fragments of B′β and B′ϵ were amplified by PCR from the full-length cDNAs and ligated into pEGFP-C1 and pGEX-4T1 using compatible restriction sites.
To silence endogenous KLHL15, four shRNAs were selected (25), subcloned downstream of the H1 promoter (26), and tested for efficient knockdown by coexpression with GFP-KLHL15. To replace endogenous with mutant GFP-KLHL15, the most effective H1-shRNA cassette (target, GGACAGTGCTCAATAACAA) was subcloned into the Pci1 site of pEGFP-C1 KLHL15 after rendering the KLHL15 cDNA RNAi-resistant by introducing four silent nucleotide changes into the target site (23).
Site-directed mutagenesis of KLHL15 and B′β was carried out using the QuikChange protocol (Invitrogen). KLHL15 mutations were introduced into the gene replacement plasmid expressing shRNA and RNAi-resistant GFP-KLHL15. All constructs were verified by sequencing.
Antibodies
Antibodies from commercial sources include: HA epitope (mouse monoclonal, Santa Cruz Biotechnology), GFP (rabbit polyclonal, Abcam), FLAG (rabbit polyclonal, Cell Signaling; mouse monoclonal M2, Sigma), EE epitope (mouse monoclonal, Covance), PP2A/A subunit (rabbit polyclonal, Cell Signaling), PP2A/C subunit (mouse monoclonal, BD Biosciences). The rabbit polyclonal antibody against B′β was described previously (12). To generate antisera specific to KLHL15, a GST fusion of the Kelch domain (amino acids 255–604) was expressed in Escherichia coli and purified on glutathione-agarose (Pierce/Thermo Scientific). Rabbit polyclonal antisera were raised at the Iowa State Hybridoma Facility (Ames, IA) and affinity-purified over an antigen column. To this end, GST-KLHL15255–604 was coupled to diaminodipropylamine (DADPA) resin using 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) cross-linking agent (Pierce/Thermo Scientific) according to the manufacturer's instructions. Antisera diluted 1:1 in PBS were passed over the antigen column, and antibody was eluted with 20 mm glycine, pH 2.5. The eluate was neutralized with Tris base and concentrated using centrifugal filter columns (Amicon/Millipore).
Affinity Purification Coupled to Mass Spectrometry
pcDNA5-FLAG-B′β (PPP2R5B, BC045619) was stably transfected in T-REx Flp-In 293 cells (Invitrogen) and submitted to affinity purification with FLAG antibody coupled to magnetic beads as described previously (27). pcDNA3-FLAG-B′β was stably transfected into HEK293 cells and submitted to affinity purification with FLAG antibodies coupled to agarose beads (M2, Sigma) as described previously (28). Two biological replicates of the samples from the T-REx Flp-In 293 cells were analyzed on a LTQ mass spectrometer, and one HEK293 sample was analyzed on an LTQ-Orbitrap mass spectrometer. In parallel several negative control runs (cells expressing the tag alone and/or the tag fused to GFP) were analyzed on the respective mass spectrometers. All acquisition conditions have been described previously (29). A database search was performed using the Mascot search engine against the human and adenovirus complements of the RefSeq library (v45), allowing for deamidation (NQ) and oxidation (M) with trypsin fixed as an enzyme, and one missed cleavage allowed. For LTQ data, the MS tolerance was fixed at 3 Da, and the MS/MS tolerance was fixed at 0.6 Da. For LTQ-Orbitrap data, the MS tolerance was fixed at 12 ppm, whereas the MS/MS tolerance was fixed at 0.6 Da. Hits identified in any of the negative control runs were removed from the list of putative interactors, and only those hits detected in all three of the biological replicates with at least 2 unique peptides were kept. Furthermore, proteins identified in ≥4% of 275 AP-MS analyses performed under the same conditions were deemed potential frequent fliers and removed from further analysis. With the exception of the core PP2A enzymes, only two proteins were identified, CUL3 and KLHL15.
Immunoprecipitation and Immunoblot Analyses
For immunoprecipitation, cells were lysed in buffer containing 20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 mm benzamidine, and 1% (v/v) Triton X-100. Lysates were allowed to rotate at 4 °C for 30 min to complete solubilization. Samples were then centrifuged at 13,000 × g for 15 min to pellet debris. A portion of the supernatant was added directly to SDS-PAGE sample buffer to provide an input sample. HA-tagged and FLAG-tagged proteins were immunoprecipitated as indicated for each experiment from HEK293 cells overexpressing the indicated proteins via HA or FLAG antibody EZ-view Red Agarose gel (Sigma) for 4 h rotating at 4 °C. EE-tagged proteins were immunoprecipitated with protein A/G-Sepharose (Santa Cruz Biotechnology) preincubated with 4 μg of EE antibody. V5-tagged proteins were immunoprecipitated with goat anti-V5-agarose (Abcam). Beads were washed 4 times by centrifugation in lysis buffer and eluted by boiling in SDS-PAGE sample buffer. Samples were separated on 10% (w/v) SDS-PAGE gels, electrotransferred to nitrocellulose, and immunoblotted as indicated. Proteins were visualized using species-specific secondary antibodies conjugated to IRDye 680/800 (LI-COR) and a LI-COR Odyssey infrared scanner. Signals were quantified by densitometry using the gel analysis plugin of ImageJ (National Institutes of Health).
B′β Turnover Assays
Cycloheximide (Sigma) was dissolved in DMSO to a concentration of 100 mm. Cycloheximide was added directly to cultured HEK293 cells to a final concentration of 100 μg/ml 1–10 h before lysis. Cells were washed in phosphate-buffered saline and lysed simultaneously in SDS-PAGE sample buffer containing 5% (v/v) β-mercaptoethanol. Lysates were sonicated with a probe tip to shear DNA and boiled for 5–10 min. Samples were then subjected to immunoblotting and analysis as described above.
GST Pulldown Assays
GST-B′β1–82, GST-B′ϵ1–68, and His6-KLHL15255–604 were expressed in E. coli and purified according to standard protocols. GST-B′β1–82 and GST-B′ϵ1–68 remained immobilized on glutathione-agarose, whereas His6-KLHL15255–604 was eluted from a nickel-nitrilotriacetic acid-agarose column (Qiagen) using 150 mm imidazole in 50 mm Tris, pH 7.5, 150 mm KCl. Immobilized GST-B′β/ϵ and soluble His6-KLHL15 were incubated at ∼5 μm each (4 h, 4 °C), rotating end over end. After four washes in immunoprecipitation lysis buffer (see above), samples were subjected to Western blotting and analysis as described above.
Quantitative RT-PCR
Total RNA was isolated from freshly dissected or flash-frozen rat tissues with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA concentrations were determined using a Nanodrop 1000 Spectrophotometer (Thermo Scientific). Quantitative RT-PCR was performed with using the SYBR Green One-Step RT-PCR kit from Qiagen on a CFX96 Touch Real-time PCR Detection System from Bio-Rad. The following primers were used to amplify 136–180-bp amplicons from mRNA (listed 5′ to 3′): B′β forward: GGCAAGTTCCTGGGTCTCCG; B′β reverse: AGCGCAAAGCCATTGATGATGC; KLHL15 forward: CAGGGGACGTGGAAGGATTC; KLHL15 reverse: GCTTTATGGGCCTGGAACTG; Pgk1 forward: CTGGAGAACCTCCGCTTTCATG; Pgk1 reverse: CAAAAGCATCAT TGACATAGACATC; Hprt1 forward: CAGTCAACGGGGGACATAAAAG; Hprt1 reverse: GGTCCTTTTCACCAGCAAGC; β-actin forward: TGACCCAGATCATGTTTGAGACC; β-actin reverse: TAGATGGGCACAGTGTGGGTGA.
All but the β-actin primers span introns, thus disfavoring amplification of any contaminating genomic DNA. Relative B′β and KLHL15 mRNA levels were determined using the ΔΔCt method using Pgk1, hypoxanthine phosphoribosyltransferase 1, and β-actin as internal controls with similar results.
RESULTS
KLHL15 Specifically Associates with B′β
Mass spectrometry was carried out to identify proteins that associate with B′β. To this end, FLAG epitope-tagged B′β was stably expressed in HEK293 cells followed by FLAG immunoprecipitation and elution of precipitated proteins with ammonium carbonate. Tryptic peptides were then subjected to liquid chromatography and tandem mass spectroscopy. B′β-interacting proteins reliably identified in three independent experiments were the PP2A A and C subunits and Cul3 as well as KLHL15 (Table 1). KLHL15 is a member of the Kelch-like or BTB-Kelch protein family that is encoded by 39 genes in humans. As first shown for Keap1/KLHL19 (16, 17, 30), these proteins function as substrate adaptors in Cul3-based E3 ubiquitin ligases (13, 31, 32).
TABLE 1.
Mass spectrometry results
Mascot scores, spectral counts, number of unique peptides, and sequence coverage for the indicated proteins are listed (PPP2R5B, B′β). In each cell, “|” delimitates data from different biological replicates.
| Gene name | Mascot score | Spectral counts | Unique peptides | Coverage |
|---|---|---|---|---|
| % | ||||
| PPP2R5B | 1218|919|933 | 86|82|45 | 18|13|13 | 54|36|35 |
| KLHL15 | 258|197|269 | 12|7|5 | 5|7|5 | 12|9|10 |
| CUL3 | 375|381|395 | 8|9|8 | 5|5|7 | 11|9|12 |
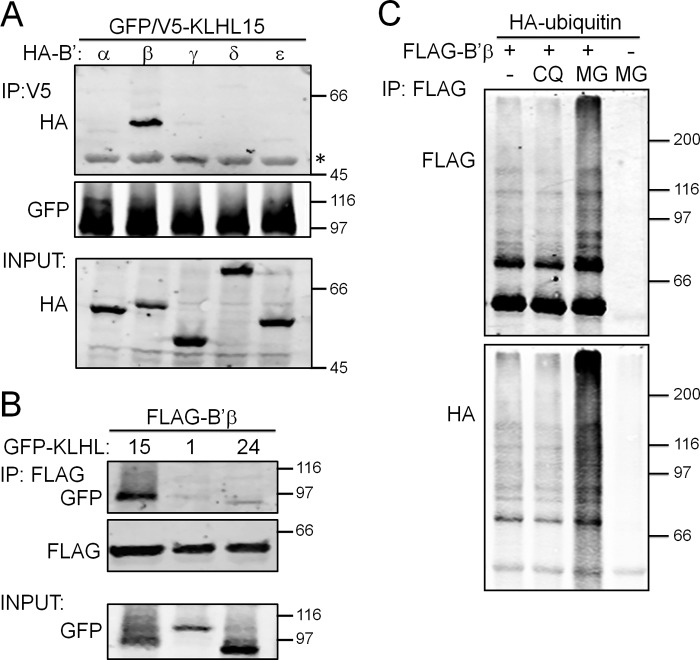
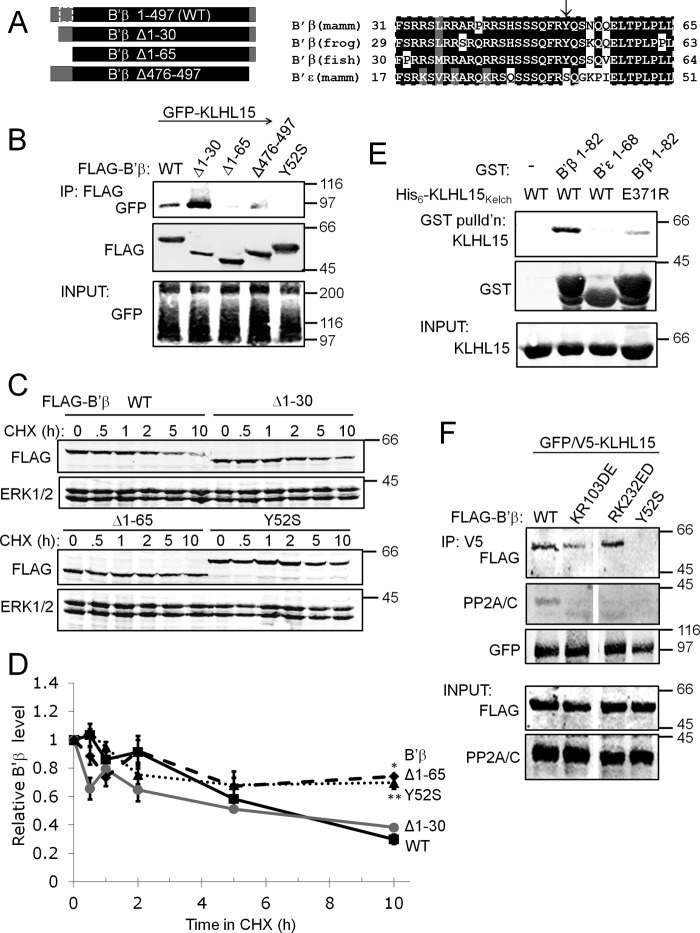
To assess the specificity of the interaction between B′β and KLHL15, the human KLHL15 cDNA was isolated by RT-PCR, tagged at the N terminus with GFP and V5 epitopes, and transiently coexpressed with the five HA-tagged B′ family subunits (α-ϵ). Remarkable considering the high sequence similarity between B′ isoforms (60–80%), KLHL15 selectively coimmunoprecipitated B′β (Fig. 1A). Further analysis confirmed that members of the distantly related B and B″ subunits of PP2A do not interact with KLHL15 (data not shown). To test whether B′β discriminates among KLHL-family members, FLAG-B′β was coexpressed with GFP-tagged KLHL1, KLHL15, or KLHL24. Only KLHL15 coimmunoprecipitated robustly with B′β (Fig. 1B), suggesting that KLHL15 functions as a highly selective E3 ubiquitin ligase adaptor for B′β. Interrogating the fate of ubiquitylated B′β, we immunoisolated the FLAG-tagged protein from HEK293 cells also expressing HA-tagged ubiquitin after incubation with chloroquine or MG132 (50 μm,12 h) to inhibit lysosomal or proteasomal degradation, respectively. Proteasome, but not lysosome inhibition, resulted in the accumulation of heterogeneous, high molecular weight species positive for both FLAG and HA epitopes (Fig. 1C).
FIGURE 1.
Selective association between KLHL15 and B′β. A, HEK293 cells were cotransfected with GFP/V5-tagged KLHL15 and the indicated, HA-tagged B′ regulatory subunits followed by KLHL15 immunoprecipitation (IP) via the V5 tag. Only B′β associates with KLHL15. B, HEK293 cells coexpressing FLAG-tagged B′β as well as GFP-tagged KLHL1, KLHL15, or KLHL24 were subjected to B′β immunoprecipitation via the FLAG tag, resulting in selective KLHL15 recovery. C, FLAG-B′β was immunoprecipitated from HEK293 cells also transfected with HA-ubiquitin after incubation with chloroquine (CQ) or MG132 (MG, 50 μm, 12 h). Proteasome inhibition by MG132 results in accumulation of polyubiquitylated B′β. The position of molecular mass markers (kDa) is shown to the right of the blots. The asterisk indicates IgG.
KLHL15 Promotes B′β Ubiquitylation and Degradation
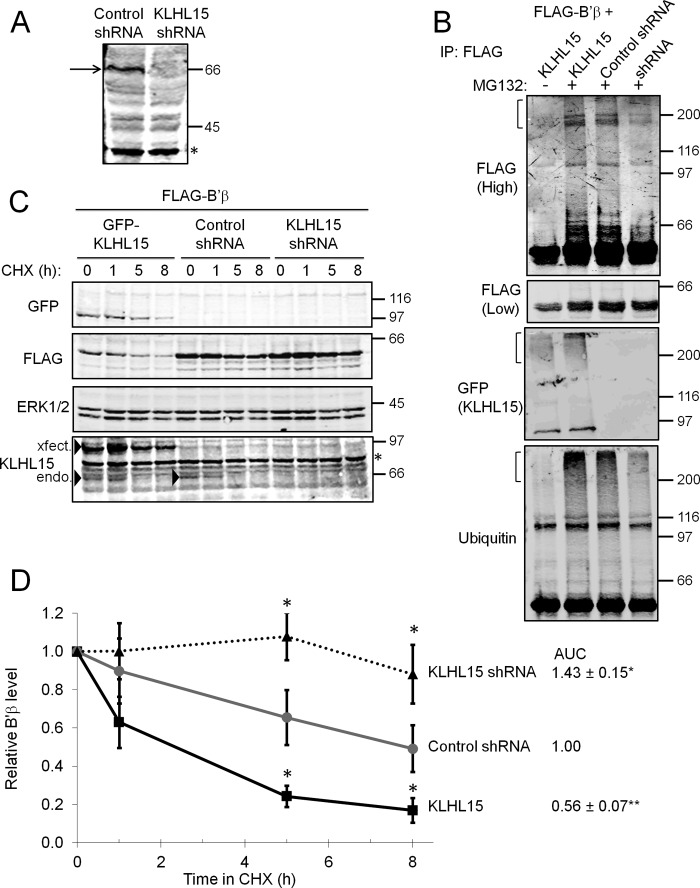
Because other KLHL proteins act as substrate recognition modules for an E3 ligase core complex composed of Cul3 and Roc1 (13), we sought to determine whether KLHL15 regulates B′β levels via ubiquitylation and proteasomal degradation. First, we generated KLHL15-directed polyclonal antibodies and effective shRNAs to confirm expression of endogenous KLHL15 in HEK293 cells (Fig. 2A). To assess whether B′β is ubiquitylated in a KLHL15-dependent manner, FLAG-B′β was immunoprecipitated from HEK293 cells also transfected with GFP-KLHL15, KLHL15-directed shRNAs, or control, scrambled shRNAs. Proteasome inhibition by MG132 before cell lysis resulted in the appearance of multiple ubiquitin- and FLAG-immunoreactive species larger than unmodified B′β. Compared with control shRNA-transfected cells, KLHL15 knockdown decreased the abundance of higher molecular weight forms of B′β, whereas GFP-KLHL15 overexpression enhanced B′β ubiquitylation (Fig. 2B). GFP-KLHL15 recovered in FLAG-B′β immunoprecipitates migrated close to its predicted molecular weight, as well as a smear near the stacker/running gel interface. This suggests that KLHL15 undergoes autoubiquitylation, as was reported for other KLHL proteins (15, 33–36).
FIGURE 2.
KLHL15 promotes B′β ubiquitylation and degradation. A, silencing of endogenous KLHL15 (arrow) in HEK293 cells transiently transfected with plasmids expressing shRNA from a H1 promoter is shown. B, HEK293 cells co-expressing FLAG-B′β and the indicated plasmids were treated with vehicle (−) or MG132 (+, 50 μm) for 12 h before lysis and FLAG immunoprecipitation (IP). Compared with control shRNA-expressing cells and in the presence of MG132, KLHL15 overexpression promotes the appearance of large, ubiquitylated B′β species (FLAG and ubiquitin-positive, indicated by brackets), whereas KLHL15 shRNA decreases B′β ubiquitylation. C and D, FLAG-B′β was coexpressed with GFP-KLHL15, non-targeting, control shRNA, or KLHL15-directed shRNA-expressing plasmids (1:1 mass ratio) followed by inhibition of protein synthesis by CHX (100 μg/ml) for 0–8 h. Total cell lysates were probed for the indicated proteins (representative blots in C), and B′β degradation was quantified by dividing FLAG signals by ERK1/2 signals in the same lane and normalizing to the zero time point (D). Both individual time points and area under the curve (normalized to control) are shown as the means ± S.E. of four independent experiments. *, p < 0.05; **, p < 0.01 by Student's t test. Molecular mass marker positions are shown in kDa. Asterisks on blots indicate nonspecific bands. Arrowheads in C point to transfected (xfect.), GFP-tagged, and endogenous (endo.) KLHL15.
To test whether KLHL15-mediated ubiquitylation promotes B′β protein turnover, we assessed B′β levels after inhibition of protein synthesis by cycloheximide (CHX) for up to 8 h (Fig. 2, C and D). As a loading control, B′β signals were normalized to extracellular signal-regulated kinase (ERK1/2) signals in the same lane, which were stable under these conditions (as compared with Ponceau S-stained total protein). Compared with cotransfection with a non-targeting shRNA, overexpression of KLHL15 lowered steady-state levels of B′β (0 h CHX) as well as decreased the half-life of B′β from ∼8 to ∼2.5 h. Conversely, silencing of endogenous KLHL15 increased B′β levels at steady state and stabilized the protein such that ∼90% of the protein remained after 8 h in CHX (Fig. 2, C and D). Comparison of the area under the curve (AUC) of the B′β degradation time courses as well as the 5- and 8-h time points revealed significant differences between KLHL15 overexpression, silencing, and control (Fig. 2D). Together these data indicate that KLHL15 recruits a Cul3-based E3 ligase to promote polyubiquitylation and proteasomal degradation of specifically the B′β subunit of PP2A. Near complete stabilization of B′β in the absence of KLHL15 further demonstrates that B′β degradation is principally mediated by ubiquitylation via KLHL15, at least in HEK293 cells.
KLHL15 May Contribute to Brain-specific Expression of B′β
We previously showed that B′β protein is abundantly expressed in the brain but undetectable in other tissues even after microcystin affinity purification of PP2A holoenzymes (12). We, therefore, wondered whether posttranslational regulation by KLHL15 may contribute to selective expression of B′β in the nervous system. Quantitative RT-PCR analysis showed that B′β mRNA is fairly uniformly expressed in a panel of nine rat tissues (Fig. 3A). KLHL15 mRNA expression is also ubiquitous, with the highest levels in lung, muscle, and spleen (Fig. 3A). Transcript profiles were similar regardless of whether β-actin, phosphoglycerate kinase 1 (Pgk1) or hypoxanthine phosphoribosyltransferase 1 mRNA was used for normalization (Fig. 3A and not shown). Plotting the ratio of B′β to KLHL15 mRNA levels revealed that forebrain, brainstem, and cerebellum contain more B′β than KLHL15 mRNA, whereas the opposite holds for all non-neuronal tissues examined (Fig. 3B). Because B′β transcripts are ubiquitous, brain-specific expression of the PP2A/B′β protein (12) may, therefore, at least in part result from relatively low levels of KLHL15 in the brain.
FIGURE 3.
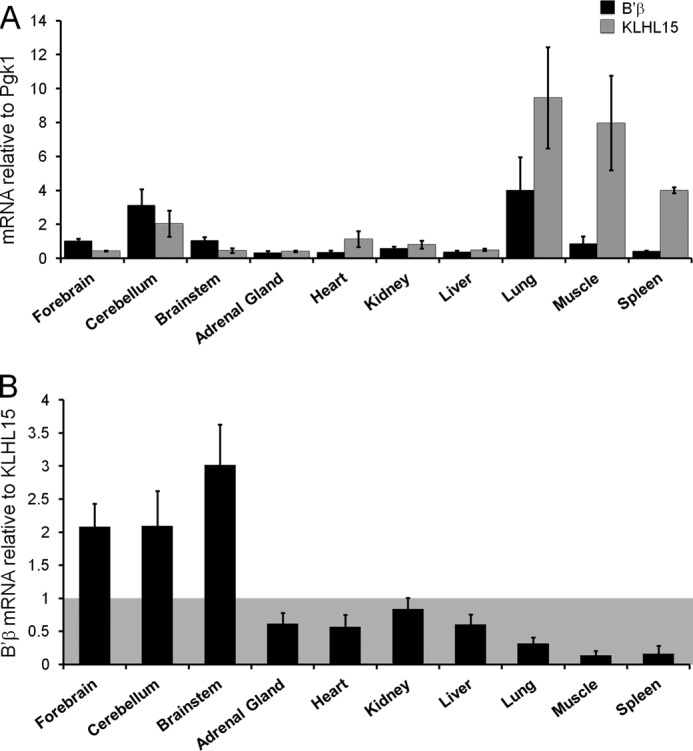
Brain-specific expression of PP2A/B′β may arise from low KLHL15 mRNA levels in the brain. A and B, shown is quantitative RT-PCR analysis of KLHL15 (gray) and B′β (black) mRNA levels in the indicated rat tissues. In A, KLHL15 and B′β mRNA levels were normalized to phosphoglycerate kinase 1 (Pgk1) mRNA levels. In B, B′β is plotted relative to KLHL15 mRNA (means ± S.E. of 4–8 independent experiments). Essentially identical results were obtained with a second set of KLHL15 primers (not shown).
The BTB Domain Mediates KLHL15 Dimerization and Cul3-dependent B′β Degradation
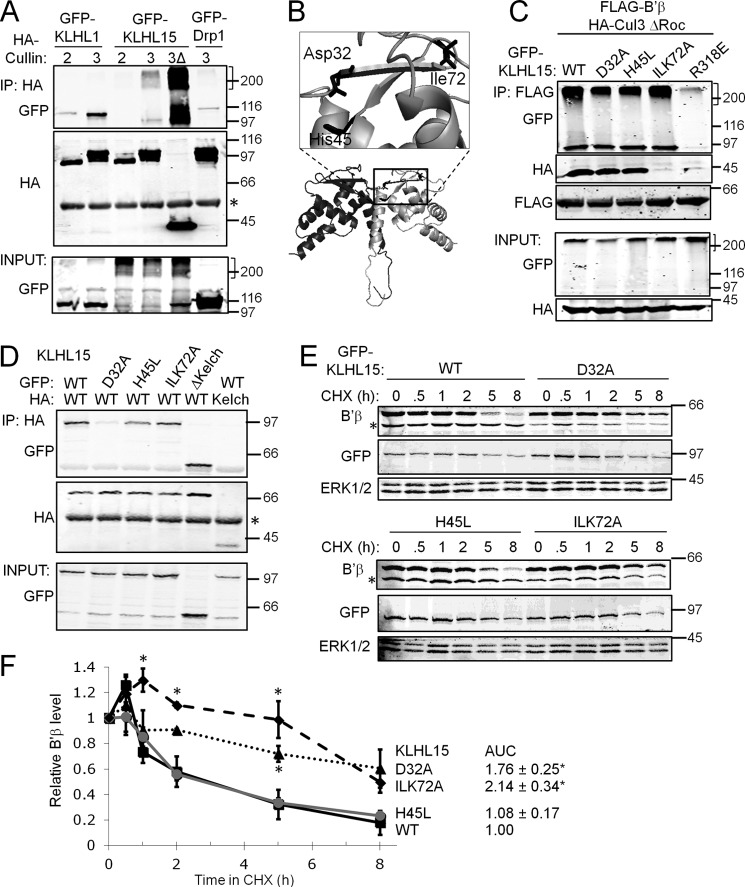
The mass spectrometry-based identification of both KLHL15 and Cul3 in B′β immunoprecipitates is in line with previous evidence that BTB-Kelch proteins tether Cul3-containing E3 ubiquitin ligases to their targets (13, 31, 32). To confirm an interaction between Cul3 and KLHL15, immunoprecipitation was carried out from transfected HEK293 cells. HA-tagged Cul3, but not Cul2, immunoprecipitated GFP-KLHL15 (Fig. 4A). Even larger amounts of GFP-KLHL15 were recovered in immunoprecipitates of Cul3 ΔRoc, a dominant-negative C-terminal deletion mutant unable to bind the E3 core component Roc1. GFP-KLHL1 also preferentially associated with Cul3 over Cul2, underscoring Cul3 preference as a feature shared among KLHL family proteins.
FIGURE 4.
Cul3 recruitment and dimerization of KLHL15 via the BTB domain is necessary for B′β down-regulation. A, HA-tagged Cul2, Cul3, or Cul3 ΔRoc (3Δ) were coexpressed with GFP-tagged KLHL1, KLHL15, or Drp1 (control) in HEK293 cells and analyzed by HA immunoprecipitation (IP). Cul3 and Cul3 ΔRoc, but not Cul2, associate with KLHL15. B, shown is a structure model of the KLHL15 BTB domain dimer (threading prediction by PHYRE2 (45)). Critical residues identified here are shown in stick representation. C, HEK293 cells triple-transfected with FLAG-B′β, HA-Cul3 ΔRoc, and the indicated GFP-KLHL15 plasmids were subjected to FLAG immunoprecipitation. KLHL15 ILK72A does not associate with Cul3. D, GFP- and HA-tagged KLHL15 were coexpressed in HEK293 cells and analyzed for homodimerization by HA immunoprecipitation. The Kelch domain is dispensable and Asp-32 in the BTB domain is required for KLHL15 dimerization. E and F, FLAG-B′β coexpressed with the indicated GFP-KLHL15 constructs was assessed for degradation in the presence of CHX for up to 8 h. E shows representative blots, whereas F shows quantification of B′β normalized to ERK1/2 levels in the same lane (means ± S.E., n = 3 experiments, see Fig. 2 legend). *, p < 0.05 by Student's t test. Molecular mass marker positions are shown in kDa. The brackets indicate polyubiquitylated KLHL15 that migrates at the interface between stacking and running gel. AUC, area under the curve.
To explore the role of the KLHL15 BTB domain in regulation of PP2A/B′β, conserved amino acids were targeted by site-directed mutagenesis. Guided by a threading model of the BTB domain dimer (Fig. 4B), we mutated two buried residues near the predicted dimerization interface (D32A, H45L) as well as three adjacent exposed residues (Ile-72, Leu-73, Lys-74; ILK72AAA). To avoid confounds due to dimerization of endogenous with transfected, mutant KLHL15, we used a single plasmid, RNAi-based gene replacement strategy (23). To this end, H1 promoter-driven shRNA targeting KLHL15 (Fig. 2A) was expressed from the same plasmid as the mutant GFP-KLHL15 cDNA, which was rendered RNAi-resistant by silent mutagenesis of the RNAi target site. HEK293 cells coexpressing FLAG-B′β, HA-Cul3, and GFP-KLHL15 were subjected to FLAG immunoprecipitation to elucidate sequence determinants for recruitment of Cul3 to B′β. Both KLHL15 and Cul3 were detected in B′β immune complexes when wild-type, D32A, or H45L mutant KLHL15 was expressed (Fig. 4C). In contrast, KLHL15 ILK72AAA interacted with B′β but did not recruit Cul3 into the complex. As a specificity control, neither KLHL15 nor Cul3 associated with B′β when the KLHL15 Kelch domain was mutated (R318E, see below).
Similar to other KLHL family proteins including Drosophila Kelch (37), KLHL15 dimerizes in a Kelch domain-independent manner according to coimmunoprecipitation analyses of HEK293 cells transfected with differentially (HA- and GFP-) tagged KLHL15 (Fig. 4D). Neither the mutation that disrupted Cul3 association (ILK72AAA) nor His-45 substitution compromised the ability of KLHL15 to associate with itself. However, Asp-32 proved necessary for KLHL15 homodimerization (Fig. 4D).
Next, KLHL15 BTB domain mutants were tested for the effects on B′β half-life in the presence of CHX. H45L, the substitution that did not affect Cul3 binding and dimerization, destabilized B′β similarly to wild-type KLHL15 (t½ ∼ 2 h, Fig. 4, E and F). In contrast, blocking either Cul3 recruitment (ILK72AAA) or dimerization (D32A) of KLHL15 significantly slowed turnover of B′β (t½ ≥ 8 h, Fig. 4, E and F). These results indicate that KLHL15 mediates proteasomal degradation of B′β via Ile-72, Leu-73, Lys-74-dependent scaffolding of the Cul3-Roc1 E3 core complex, with Asp-32-dependent homodimerization also contributing to KLHL15 activity.
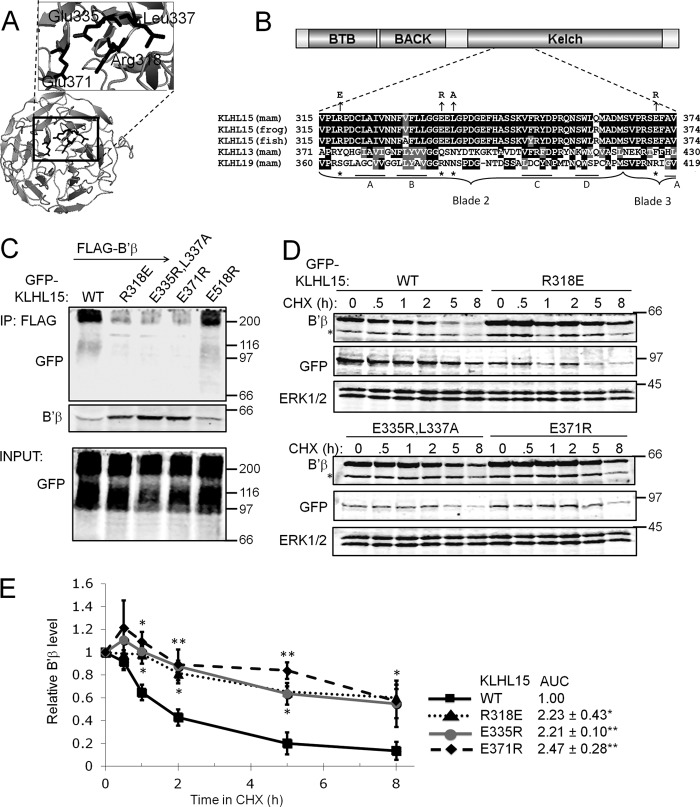
The Top of the KLHL15 β-Propeller Mediates B′β Association
According to high confidence structure modeling templated on the Kelch repeats of Keap1/KLHL19 (38, 39), the C terminus of KLHL15 folds into a cone-shaped β-propeller with six propeller blades composed of four antiparallel β-strands each (Fig. 5A). Keap1/KLHL19 was previously shown to interact with a peptide derived from its substrate Nrf2 via multiple, blade-spanning residues that form the top cavity of the Kelch β-propeller (39). The corresponding residues are perfectly conserved among KLHL15 orthologs in vertebrates but are distinct in other KLHL family members (Fig. 5B) and thus may function as substrate specificity determinants. A subset of these putative substrate binding residues mapping to Kelch blades 1 and 2 (asterisks in Fig. 5B) was assessed for effects on B′β interaction and down-regulation. As with BTB domain mutants, Kelch domain mutant GFP-KLHL15 replaced endogenous KLHL15 via shRNA expressed from the same plasmid. R318E, E335R/L337A, and E371R substitution strongly impaired association of GFP-KLHL15 with FLAG-B′β by coimmunoprecipitation (Fig. 5C) and KLHL15-mediated degradation of B′β (t½ > 8 h, Fig. 5, D and E). On the other hand, mutation of a residue of similar evolutionary conservation but predicted localization at the bottom of the KLHL15 β-propeller (E518R) had no interaction phenotype (Fig. 5C). Thus, conserved residues unique to KLHL15 form a substrate binding pocket predicted to reside on top of the Kelch β-propeller.
FIGURE 5.
Identification of a B′β docking site on the KLHL15 C-terminal β-propeller. A, a structure model of the KLHL15 Kelch repeats generated by PHYRE2 (45) is shown viewed from the top of the β-propeller. Residues necessary for B′β interaction are depicted in stick representation. B, shown is a KLHL15 domain diagram and sequence alignment of β-propeller blades 2/3, with component β-strands (A–D) indicated. Residues in KLHL19/Keap1 that interact with a substrate peptide derived from Nrf2 (39) are indicated by asterisks. The aligning residues in KLHL15 are perfectly conserved in orthologs from mammals (mam), Xenopus laevis (frog), and Danio rerio (fish) and were substituted as indicated by arrows. C, FLAG-B′β and Kelch domain-mutant GFP-KLHL15 were coexpressed in HEK293 cells and assessed for association by FLAG immunoprecipitation (IP). Mutation of residues predicted to line the top cavity of the β-propeller compromise the interaction with B′β, whereas Glu-518 is dispensable. D and E, degradation of B′β was measured in the presence of the indicated GFP-KLHL15 constructs. Mutations that interfere with B′β binding also attenuate B′β turnover. D shows representative blots, whereas E shows quantification of B′β normalized to ERK1/2 levels (means ± S.E., n = 3 experiments, see Fig. 2 legend). *, p < 0.05; **, p < 0.01 by Student's t test. Molecular mass marker positions are shown in kDa. Asterisks on blots indicate nonspecific bands. AUC, area under the curve.
The B′β N Terminus Interacts Directly with KLHL15
B′ subunits share a highly conserved central portion composed of tandem α-helical repeats that interacts with the PP2A A and C subunit (6). Because KLHL15 associates with B′β, but not other B′ isoforms (Fig. 1A), we surmised that instead, the divergent N- or C-terminal extensions of B′β contain the KLHL15 binding domain. In support, initial coimmunoprecipitation experiments with truncated B′β cDNAs mapped critical residues to the N terminus (amino acids 31–65, Fig. 6A). Interestingly, truncation of the first 30 residues consistently enhanced the interaction of B′β with KLHL15, suggesting perhaps that the extreme N terminus folds back to occlude an adjacent KLHL15 binding domain.
FIGURE 6.
Residues within the divergent N terminus of B′β mediate direct association with the KLHL15 β-propeller. A, shown is a diagram of B′β and N- and C-terminal truncation mutants, with the region sufficient for PP2A holoenzyme association shown in black (7). The KLHL15 binding domain in B′β (stippled box, amino acids 31–65) is aligned with the corresponding region of B′ϵ. Tyr-52 (arrow) is conserved in B′β orthologs from mammals (mam), Xenopus laevis (frog), and Danio rerio (fish) but is replaced with a Ser in B′ϵ. B, the indicated FLAG-B′β truncation and point mutants were coexpressed with GFP-KLHL15 in HEK293 cells and tested for association by FLAG immunoprecipitation (IP). Deletion of the N terminus (Δ1–65) and Tyr-52 substitution abrogates the interaction. C and D, the indicated B′β mutants were expressed in HEK293 cells and assessed for turnover by endogenous KLHL15 in the presence of CHX (0–10 h). C shows representative blots, and D shows quantification (means ± S.E., n = 3 experiments, see the Fig. 2 legend). E, GST tagged B′β and B′ϵ N termini as well as hexahistidine (His6)-tagged KLHL255–604 (Kelch repeats) were purified from E. coli and used for GST pulldown (pulld'n) analysis. Shown are GST and KLHL15 immunoblots. GST-B′β1–82, but not B′ϵ1–68, interacts directly with wild type but to a lesser extent with E371R mutant KLHL15Kelch. F, GFP/V5-KLHL15 was coexpressed with the indicated point mutants of FLAG-B′β and subjected to immunoprecipitation via the V5 tag (the left two and right two lanes are from the same blots but not adjacent). KLHL15 interacts with PP2A holoenzyme-incorporated B′β as indicated by coimmunoprecipitation of endogenous PP2A/C when wild-type B′β is expressed as well as with monomeric (KR103DE, RK232ED) B′β. Molecular mass marker positions are shown in kDa. *, p < 0.05; **, p < 0.01 by Student's t test.
Sequence alignments pointed to B′β Tyr-52 as a probable binding determinant, as this residue is perfectly conserved among B′β orthologs from fish to man but is divergent within the B′ family (Fig. 6A). Indeed, replacement of Tyr-52 with the corresponding residue in B′ϵ (Y52S), the isoform most similar to B′β, eliminated KLHL15 association (Fig. 6A). B′β degradation assays corroborated these findings, demonstrating enhanced stability of B′β Δ1–65 and Y52S compared with wild-type B′β or the Δ1–30 truncation (∼80% versus ∼40% remaining after 10 h in CHX; Fig. 6, B and C).
To confirm that KLHL15 and B′β interact directly via the Kelch domain and the divergent N terminus, respectively, we expressed and purified from E. coli His6-KLHL15255–604 and GST-B′β1–82 (GST-B′ϵ1–68 as a negative control). In glutathione-agarose pulldown experiments, GST-B′β1–82 precipitated wild-type KLHL15255–604 effectively, whereas GST-B′ϵ1–68 displayed no detectable interaction. Paralleling association studies in intact cells (Fig. 5C), the E371R substitution weakened the association between the KLHL15 β-propeller and the B′β N terminus (Fig. 6D).
Because the first 65 residues are dispensable for incorporation of B′β into the PP2A heterotrimer (7), we next asked whether KLHL15 discriminates between free and holoenzyme-associated B′β. To this end, charge reversal substitutions were incorporated into B′β at tandem α-helical repeat 1 (KR103DE) and 4 (RK232ED) to block A and C subunit association (Fig. 6F and data not shown). Highly conserved across species and B′ subunit isoforms, but not contacting A or C subunits directly (40, 41), these residues instead appear to be necessary for proper folding of the two tandem α-helical repeats. Both monomeric B′ mutants efficiently associated with KLHL15; furthermore, the endogenous PP2A C subunit was detected in KLHL15 immunoprecipitations when wild-type B′β was coexpressed (Fig. 6F). Therefore, KLHL15 targets B′β regardless of whether or not the regulatory subunit is incorporated into the PP2A heterotrimer.
The B′β N terminus Is Sufficient for KLHL15-mediated Degradation
We next inquired whether the B′β N terminus contains a degron, i.e. sequence determinants sufficient for degradation. To this end we assessed turnover of the B′β N terminus (amino acids 1–82) fused to GFP after overexpression or knockdown of KLHL15 (Fig. 7A). About 40% of GFP-B′β1–82 degraded when protein synthesis was inhibited by CHX for 8 h. In contrast, GFP fusions of the B′ϵ N terminus (amino acids 1–68) or the B′β C terminus (amino acids 476–497) were stable over this time course (Fig. 7, A–D). KLHL15 silencing prevented degradation of GFP-Bβ1–82, whereas KLHL15 overexpression accelerated degradation. Neither silencing nor forced expression of KLHL15 had an impact on the stability of GFP-B′β476–497 or GFP-B′ϵ1–68. Therefore, the N terminus contains both E3 ligase docking and ubiquitylation sites sufficient for degradation of B′β.
FIGURE 7.
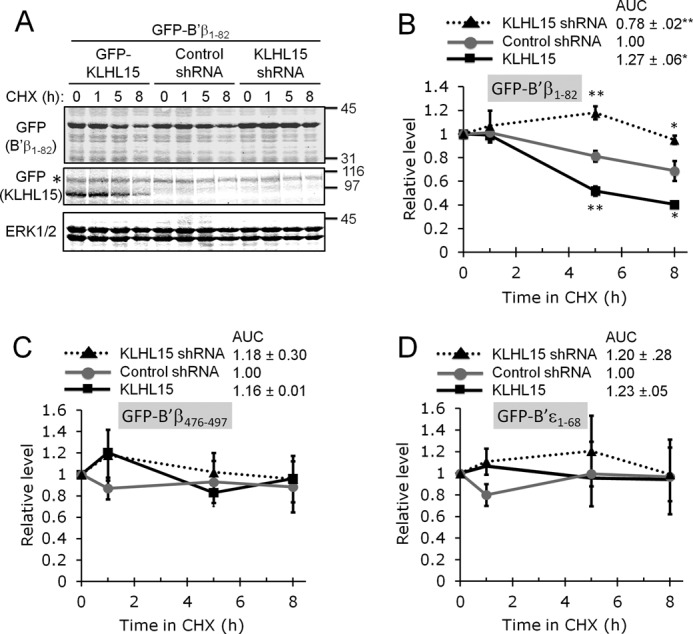
The B′β N terminus is sufficient for KLHL15-mediated turnover. A–D, the B′β N (residues 1–82) and C terminus (476–497) and the B′ϵ N terminus (1–68) were fused to GFP and cotransfected with plasmids expressing GFP-KLHL15, control shRNA, or shRNA targeting KLHL15. Stability of GFP-B′ fusion proteins was then assessed in the presence of CHX for up to 8 h. A shows representative blots from a B′β1–82 turnover experiment, whereas panels B–D show quantification of B′β1–82 (B), B′β476–497 (C), and B′ϵ1–68 (D) turnover (means ± S.E., n = 3 experiments, see Fig. 2 legend). GFP-B′β1–82 is less stable than the other GFP fusion proteins and was selectively regulated by KLHL15 overexpression and silencing. Molecular mass marker positions are shown in kDa. *, p < 0.05; **, p < 0.01 by Student's t test. AUC, area under the curve.
KLHL15 Does Not Degrade the PP2A Holoenzyme but Influences Its Composition
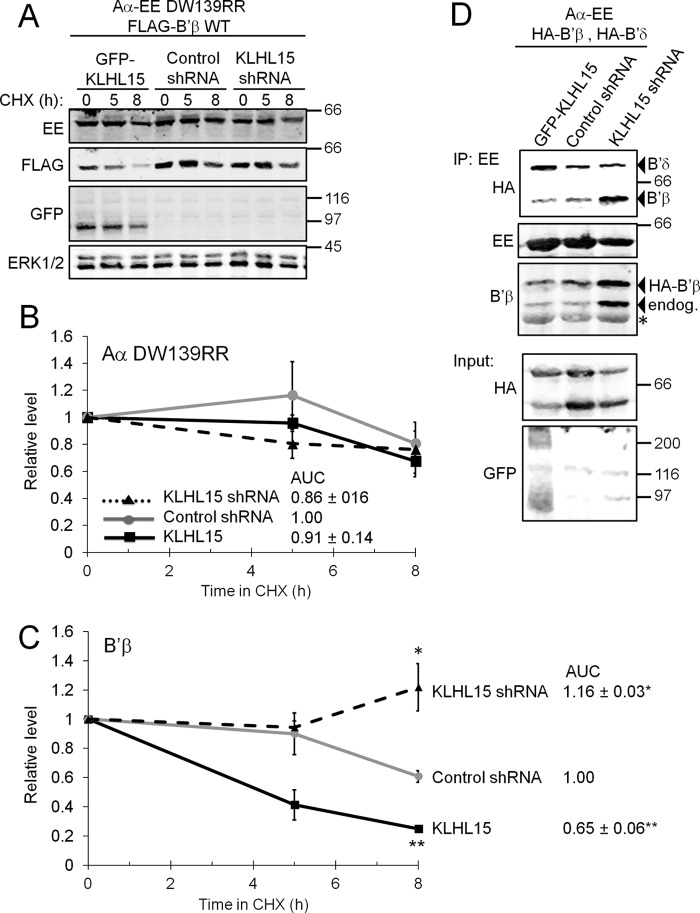
Because KLHL15 can associate with the B′β-containing PP2A holoenzyme (Fig. 6F), we examined whether KLHL15 mediates wholesale degradation of the PP2A/B′β heterotrimer. In cells transfected with B′β, neither endogenous nor epitope-tagged A and C subunit levels were modulated by KLHL15 overexpression or knockdown (data not shown). However, this negative result could be explained by B′β competing with endogenous regulatory subunits for association with A and C subunits such that only a small pool of A and C subunits becomes subject to proteasomal degradation via KLHL15. To provide B′β with privileged access to the A subunit, we employed an A subunit point mutant (DW139RR) that interacts with B′ but not B or B″ subunits (22). EE-tagged Aα DW139RR, FLAG-B′β, and either GFP-KLHL15, control shRNA, or KLHL15-directed shRNA were coexpressed in HEK293 cells. Immunoblotting for the EE epitope revealed little to no effect of KLHL15 levels on A subunit stability in the presence of CHX (Fig. 8, A and B), whereas an inverse relationship between KLHL15 and B′β levels was confirmed by blotting for FLAG (Fig. 8, A and C). Thus, even though KLHL15 can bind to the PP2A/B′β holoenzyme, the E3 adaptor only ubiquitylates and degrades the regulatory subunit.
FIGURE 8.
KLHL15 modulates PP2A holoenzyme composition. A and B, FLAG-B′β and an EE-tagged PP2A/A subunit that only associates with B′ regulatory subunits (Aα DW139RR (22)) were coexpressed with GFP-KLHL15, control shRNA, or KLHL15-directed shRNA and assessed for stability in CHX (0–8 h). Representative blots are shown in A, whereas quantification of Aα and B′β levels are shown in B and C, respectively (means ± S.E., n = 3 experiments, see Fig. 2 legend). Although B′β stability is modulated by KLHL15, the stability of the scaffolding A subunit is not. AUC, area under the curve. D, HEK293 cells coexpressing HA-tagged B′β and B′δ, EE-tagged Aα, and either GFP-KLHL15, control shRNA, or KLHL15-directed shRNA were subjected to immunoprecipitation (IP) with EE-tag antibodies to isolate PP2A holoenzymes containing the transfected A subunit. KLHL15 silencing promotes B′β, whereas KLHL15 overexpression promotes B′δ incorporation into the PP2A heterotrimer. Molecular mass marker positions are shown in kDa. The asterisks indicates the IgG heavy chain. *, p < 0.05; **, p < 0.01 by Student's t test.
To provide direct evidence that KLHL15 can impact PP2A holoenzyme composition, we assessed competition of B′β and B′δ for incorporation into the PP2A heterotrimer as a function of KLHL15 levels. HA-tagged B′ isoforms were coexpressed with wild-type EE-tagged A subunit, and cell lysates were subjected to EE-tag immunoprecipitation and immunoblotting (Fig. 8D). Association of both endogenous and HA-tagged B′β with the A subunit increased when KLHL15 was silenced (1.4–2.2-fold, n = 2), whereas PP2A holoenzyme incorporation of B′δ increased when KLHL15 was overexpressed (1.6–2.0-fold, n = 2). Therefore, by virtue of degrading B′β, KLHL15 creates a window of opportunity for other regulatory subunits with distinct subcellular localization and substrate specificity to incorporate into the PP2A heterotrimer.
DISCUSSION
BTB/Kelch domain-containing (KLHL, Kelch-like, BBK) proteins comprise a large family of evolutionarily conserved proteins with a characteristic domain architecture. Although Kelch and BTB domains are common protein-protein interaction modules found in proteins ranging from metabolic enzymes to transcription factors and ion channels, their presence in the same polypeptide chain defines the largest group of substrate adaptors for E3 ubiquitin ligases, with 39 Kelch-like genes in the human genome (14). The present report assigns a function to a previously uncharacterized member of this gene family, identifying the KLHL15-Cul3-Roc1 complex as the first E3 ligase for PP2A regulatory subunits.
The characterization of KLHL15 as an E3 ligase adaptor adds to a growing list of reports establishing KLHL family proteins as specificity determinants for the ubiquitin-proteasome system. Best characterized at both structural and functional levels is proteasomal degradation of Nrf2, which mediates transcriptional responses to oxidative stress via Keap1/KLHL19 (16). Similarly, KLHL9 and KLHL13 degrade Aurora B, the kinase component of the chromosomal passenger complex, to enable normal mitotic progression (21). Other examples of critical KLHL proteins include Gigaxonin/KLHL16, a ubiquitously expressed protein mutated in giant axonal neuropathy that binds and promotes ubiquitylation of MAP1B (42). KLHL12 targets Dishevelled, an essential mediator of Wnt/β-catenin signaling, antagonizing the pathway in cultured cells (15).
Site-directed mutagenesis guided by structure prediction supports the consensus view of KLHL proteins as modular adaptors, with the N-terminal BTB domain mediating homodimerization of KLHL15 and Cul3 recruitment via distinct interaction surfaces and the top cavity of the C-terminal Kelch β-propeller docking to the substrate, B′β. Predictably, interfering with either B′β or Cul3 interaction abrogated KLHL15-mediated down-regulation of B′β. Unexpectedly, a point mutation in the BTB domain that specifically blocked KLHL15 self-association (D32A) had the same effect (Fig. 4, E and F). Consistent with this finding, missense mutations in KLHL10 linked to male sterility were recently shown to interfere with homodimerization of the protein (43). Perhaps substrate ubiquitylation by KLHL proteins is an obligatory trans reaction, with one subunit of the dimer directing Cul3-Roc1 for ubiquitin transfer to a substrate bound by the other subunit.
This report also defines a novel regulatory mechanism for PP2A, one of the major Ser/Thr phosphatases. With at least 12 genes encoding regulatory subunit in mammals, PP2A comprises a family of signaling enzymes with unique substrate specificity, localization, and regulation by second messengers (3–5, 8). We found that KLHL15 displays exquisite specificity for binding and down-regulation of B′β, one of five members of the B′/B56 family of regulatory subunits. PP2A/B′β is enriched in the nervous system, where it was shown to dephosphorylate and inactivate two important enzymes, 1) calcium/calmodulin-dependent protein kinase II, a key mediator of long term potentiation and spatial memory (44), and 2) tyrosine hydroxylase, the rate-limiting enzyme in dopamine synthesis (12). According to qRT-PCR, B′β mRNA is relatively uniformly expressed among across tissues (Fig. 3A), whereas B′β protein is only detectable in the brain (12). Because brain is the only tissue with higher B′β than KLHL15 mRNA levels, we propose that the brain-specific expression of the B′β protein is at least in part a result of efficient KLHL15-mediated proteasomal degradation in other tissues.
KLHL15 likely not only determines cell and tissue specificity of B′β expression but may also regulate abundance of this PP2A regulatory subunit dynamically and in response to signaling events. This could for instance occur through B′β phosphorylation at Tyr-52, an essential KLHL15 binding determinant (Fig. 6, B–D), or at nearby Ser-44, Ser-46, and Ser-47, Ser residues that have been identified in unbiased phosphoproteomic screens (PhosphoSitePlus).
The β-propeller of KLHL15 directly engages the divergent N terminus of B′β, a region of the regulatory subunit necessary and sufficient for proteasomal degradation (Figs. 6, B and E, and 7, A and B). Accordingly, KLHL15 targets both monomeric and holoenzyme-incorporated B′β (Fig. 6F). However, we found no evidence that KLHL15 degrades the A and C subunit within the PP2A/B′β heterotrimer. Instead, the E3 ligase adaptor appears to abstract B′β from the AC dimer, allowing other regulatory subunits to take the place of B′β (Fig. 8D). Selective ubiquitylation and proteasomal degradation of B′β by KLHL15 may, therefore, be a mechanism for dynamic regulation of PP2A holoenzyme composition and specific substrate dephosphorylation.
On a final note, our results raise the intriguing possibility that each of the dozen or more PP2A regulatory subunit is paired with a dedicated E3 ubiquitin ligase. Indeed, substrates for the majority of the 39 human KLHL family proteins remain to be identified. KLHL protein-catalyzed exchange of PP2A regulatory subunits could thus shape dephosphorylation profiles in every cell.
Acknowledgments
We thank Kevin Barnum and Larisa Greve for generating several KLHL15 mutants, Mark Hannink, Fernanda Laezza, and Michael Koob for generous gifts of cDNAs, Ted Wilson for providing RNA from rat tissues, Beatriz Badillo for subcloning B′β and establishing stable cell lines, Zhen-Yuan Lin and Michael Mullin for help with affinity purification and mass spectrometry, and all members of the Strack laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants NS043254, NS056244, and NS057714 (to S. S.). This work was also supported by the Canadian Institutes of Health Research Grant MOP-84314 (to A.-C. G.) and American Heart Association Predoctoral Fellowship 0715634Z (to S. K. N.).
- PP2A
- protein phosphatase 2A
- Cul3
- Cullin3
- KLHL15
- Kelch-like 15
- BTB
- broad-complex, tram track and bric-a-brac
- CHX
- cycloheximide.
REFERENCES
- 1. Cohen P. T. (1997) Novel protein serine/threonine phosphatases. Variety is the spice of life. Trends Biochem. Sci. 22, 245–251 [DOI] [PubMed] [Google Scholar]
- 2. Kremmer E., Ohst K., Kiefer J., Brewis N., Walter G. (1997) Separation of PP2A core enzyme and holoenzyme with monoclonal antibodies against the regulatory A subunit. Abundant expression of both forms in cells. Mol. Cell. Biol. 17, 1692–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janssens V., Goris J. (2001) Protein phosphatase 2A. A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353, 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Janssens V., Longin S., Goris J. (2008) PP2A holoenzyme assembly. In cauda venenum (the sting is in the tail). Trends Biochem. Sci. 33, 113–121 [DOI] [PubMed] [Google Scholar]
- 5. Virshup D. M., Shenolikar S. (2009) From promiscuity to precision. Protein phosphatases get a makeover. Mol. Cell 33, 537–545 [DOI] [PubMed] [Google Scholar]
- 6. Shi Y. (2009) Serine/threonine phosphatases. Mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 7. Saraf A., Oberg E. A., Strack S. (2010) Molecular determinants for PP2A substrate specificity. Charged residues mediate dephosphorylation of tyrosine hydroxylase by the PP2A/B' regulatory subunit. Biochemistry 49, 986–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang J., Phiel C. (2010) Functions of B56-containing PP2As in major developmental and cancer signaling pathways. Life Sci. 87, 659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silverstein A. M., Barrow C. A., Davis A. J., Mumby M. C. (2002) Actions of PP2A on the MAP kinase pathway and apoptosis are mediated by distinct regulatory subunits. Proc. Natl. Acad. Sci. U.S.A. 99, 4221–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strack S., Cribbs J. T., Gomez L. (2004) Critical role for protein phosphatase 2A heterotrimers in mammalian cell survival. J. Biol. Chem. 279, 47732–47739 [DOI] [PubMed] [Google Scholar]
- 11. Strack S., Ruediger R., Walter G., Dagda R. K., Barwacz C. A., Cribbs J. T. (2002) Protein phosphatase 2A holoenzyme assembly. Identification of contacts between B-family regulatory and scaffolding A subunits. J. Biol. Chem. 277, 20750–20755 [DOI] [PubMed] [Google Scholar]
- 12. Saraf A., Virshup D. M., Strack S. (2007) Differential expression of the B'β regulatory subunit of protein phosphatase 2A modulates tyrosine hydroxylase phosphorylation and catecholamine synthesis. J. Biol. Chem. 282, 573–580 [DOI] [PubMed] [Google Scholar]
- 13. Stogios P. J., Downs G. S., Jauhal J. J., Nandra S. K., Privé G. G. (2005) Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prag S., Adams J. C. (2003) Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC bioinformatics 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Angers S., Thorpe C. J., Biechele T. L., Goldenberg S. J., Zheng N., MacCoss M. J., Moon R. T. (2006) The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-β-catenin pathway by targeting Dishevelled for degradation. Nat. Cell Biol. 8, 348–357 [DOI] [PubMed] [Google Scholar]
- 16. Cullinan S. B., Gordan J. D., Jin J., Harper J. W., Diehl J. A. (2004) The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase. Oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24, 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kobayashi A., Kang M. I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24, 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Laezza F., Wilding T. J., Sequeira S., Craig A. M., Huettner J. E. (2008) The BTB/kelch protein, KRIP6, modulates the interaction of PICK1 with GluR6 kainate receptors. Neuropharmacology 55, 1131–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rondou P., Haegeman G., Vanhoenacker P., Van Craenenbroeck K. (2008) BTB Protein KLHL12 targets the dopamine D4 receptor for ubiquitination by a Cul3-based E3 ligase. J. Biol. Chem. 283, 11083–11096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salinas G. D., Blair L. A., Needleman L. A., Gonzales J. D., Chen Y., Li M., Singer J. D., Marshall J. (2006) Actinfilin is a Cul3 substrate adaptor, linking GluR6 kainate receptor subunits to the ubiquitin-proteasome pathway. J. Biol. Chem. 281, 40164–40173 [DOI] [PubMed] [Google Scholar]
- 21. Sumara I., Peter M. (2007) A Cul3-based E3 ligase regulates mitosis and is required to maintain the spindle assembly checkpoint in human cells. Cell Cycle 6, 3004–3010 [DOI] [PubMed] [Google Scholar]
- 22. Ruediger R., Fields K., Walter G. (1999) Binding specificity of protein phosphatase 2A core enzyme for regulatory B subunits and T antigens. J. Virol. 73, 839–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cribbs J. T., Strack S. (2007) Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 8, 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCright B., Rivers A. M., Audlin S., Virshup D. M. (1996) The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J. Biol. Chem. 271, 22081–22089 [DOI] [PubMed] [Google Scholar]
- 25. Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W. S., Khvorova A. (2004) Rational siRNA design for RNA interference. Nat. Biotechnol. 22, 326–330 [DOI] [PubMed] [Google Scholar]
- 26. Brummelkamp T. R., Bernards R., Agami R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 [DOI] [PubMed] [Google Scholar]
- 27. Kean M. J., Ceccarelli D. F., Goudreault M., Sanches M., Tate S., Larsen B., Gibson L. C., Derry W. B., Scott I. C., Pelletier L., Baillie G. S., Sicheri F., Gingras A. C. (2011) Structure-function analysis of core STRIPAK proteins. A signaling complex implicated in Golgi polarization. J. Biol. Chem. 286, 25065–25075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen G. I., Gingras A. C. (2007) Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods 42, 298–305 [DOI] [PubMed] [Google Scholar]
- 29. Dunham W. H., Larsen B., Tate S., Badillo B. G., Goudreault M., Tehami Y., Kislinger T., Gingras A. C. (2011) A cost-benefit analysis of multidimensional fractionation of affinity purification-mass spectrometry samples. Proteomics 11, 2603–2612 [DOI] [PubMed] [Google Scholar]
- 30. Zhang D. D., Lo S. C., Cross J. V., Templeton D. J., Hannink M. (2004) Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24, 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pintard L., Willems A., Peter M. (2004) Cullin-based ubiquitin ligases. Cul3-BTB complexes join the family. EMBO J. 23, 1681–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van den Heuvel S. (2004) Protein degradation. CUL-3 and BTB, partners in proteolysis. Curr. Biol. 14, R59–R61 [PubMed] [Google Scholar]
- 33. Zhang D. D., Lo S. C., Sun Z., Habib G. M., Lieberman M. W., Hannink M. (2005) Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem. 280, 30091–30099 [DOI] [PubMed] [Google Scholar]
- 34. Sumara I., Quadroni M., Frei C., Olma M. H., Sumara G., Ricci R., Peter M. (2007) A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev. Cell 12, 887–900 [DOI] [PubMed] [Google Scholar]
- 35. Kigoshi Y., Tsuruta F., Chiba T. (2011) Ubiquitin ligase activity of Cul3-KLHL7 protein is attenuated by autosomal dominant retinitis pigmentosa causative mutation. J. Biol. Chem. 286, 33613–33621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nam S., Min K., Hwang H., Lee H. O., Lee J. H., Yoon J., Lee H., Park S., Lee J. (2009) Control of rapsyn stability by the CUL-3-containing E3 ligase complex. J. Biol. Chem. 284, 8195–8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson D. N., Cooley L. (1997) Drosophila kelch is an oligomeric ring canal actin organizer. J. Cell Biol. 138, 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li X., Zhang D., Hannink M., Beamer L. J. (2004) Crystal structure of the Kelch domain of human Keap1. J. Biol. Chem. 279, 54750–54758 [DOI] [PubMed] [Google Scholar]
- 39. Lo S. C., Li X., Henzl M. T., Beamer L. J., Hannink M. (2006) Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 25, 3605–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu Y., Xing Y., Chen Y., Chao Y., Lin Z., Fan E., Yu J. W., Strack S., Jeffrey P. D., Shi Y. (2006) Structure of the protein phosphatase 2A holoenzyme. Cell 127, 1239–1251 [DOI] [PubMed] [Google Scholar]
- 41. Cho U. S., Xu W. (2007) Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445, 53–57 [DOI] [PubMed] [Google Scholar]
- 42. Allen E., Ding J., Wang W., Pramanik S., Chou J., Yau V., Yang Y. (2005) Gigaxonin-controlled degradation of MAP1B light chain is critical to neuronal survival. Nature 438, 224–228 [DOI] [PubMed] [Google Scholar]
- 43. Yatsenko A. N., Roy A., Chen R., Ma L., Murthy L. J., Yan W., Lamb D. J., Matzuk M. M. (2006) Non-invasive genetic diagnosis of male infertility using spermatozoal RNA: KLHL10 mutations in oligozoospermic patients impair homodimerization. Hum. Mol. Genet. 15, 3411–3419 [DOI] [PubMed] [Google Scholar]
- 44. Fukunaga K., Muller D., Ohmitsu M., Bakó E., DePaoli-Roach A. A., Miyamoto E. (2000) Decreased protein phosphatase 2A activity in hippocampal long-term potentiation. J. Neurochem. 74, 807–817 [DOI] [PubMed] [Google Scholar]
- 45. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]