Background: The role of different TLRs in response to pathogen infection in the gut remains elusive.
Results: TLR11 works as a blocker of Salmonella penetration. Remarkably, in mice lacking TLR11, highly invasive Salmonella induced apparent hemorrhage at Peyer patches.
Conclusion: TLR11 plays an important role in preventing murine intestinal infection.
Significance: TLR11 knock-out mice can serve as a good animal model to study Salmonella infection.
Keywords: Innate Immunity, Intestine, Pathogen-associated Molecular Pattern (PAMP), Signaling, Toll-like Receptors (TLRs), Animal Model, Peyer Patches, Salmonella Infection, TLR11
Abstract
Toll-like receptors (TLRs) are key molecular sensors used by the mammalian innate immune system to detect microorganisms. Although TLR functions in colonic immune homeostasis and tolerance to commensal bacteria have been intensively researched, the precise roles of different TLRs in response to pathogen infection in the gut remain elusive. Peyer patches are the major entrance of Salmonella infection and antigen transportation in intestine. Here, we report that, in contrast to TLR5 as a “carrier of Salmonella,” TLR11 works as a “blocker of Salmonella” to prevent highly invasive Salmonella from penetrating into the murine Peyer patches and spreading systemically. TLR11 plays an important role in mediating TNF-α induction and systemic inflammation in response to Salmonella infection. Remarkably, in mice lacking TLR11, apparent hemorrhages at Peyer patches are induced by highly invasive Salmonella, a phenotype resembling human Salmonella infection. Therefore, our results indicate a potentially important role for TLR11 in preventing murine intestinal infection and modulating antigen transportation in the gut and imply an important role for various TLRs in cooperation with tight control of pathogens penetrating into Peyer patches. The TLR11 knock-out mouse can serve as a good animal model to study Salmonella infection.
Introduction
To elicit prompt and proper immune response, the host relies on evolutionarily ancient, germ line-encoded pattern-recognition receptors (PRRs)2 to recognize the molecular patterns of invading microorganisms (1–3). PRRs generally identify highly conserved invariable molecular patterns which are often essential for the survival of microorganisms (4). The best characterized family of PRRs is the Toll-like receptors (TLRs), which recognize and bind their cognate ligands to the leucine-rich repeat domain, recruit adaptor molecules to their intracellular signaling domain, and subsequently induce a series of signaling cascades that ultimately result in the activation of NF-κB and other immune responsive genes (5). Currently 13 TLRs have been identified in mammals (6). The contribution of different TLRs to infection depends on the site of the infection and the pathogen (1, 7).
The intestinal tract is constantly exposed to an abundance of commensal bacteria and regularly encounters pathogenic microbes in contaminated food and water. Under physiological conditions the intestinal tract displays immune tolerance to commensal microorganisms but retains its ability to mount an effective immune response to invading pathogens (8). Recently, TLR functions in colonic homeostasis and tolerance to commensal bacteria have been studied in great detail (9–11), in particular, how the apical and basolateral epithelia of the colon respond differently to TLR ligands (11). In contrast to the role of TLRs in colonic homeostasis and tolerance to commensal bacteria, the precise role of different TLRs in response to pathogen infection in different organs such as the intestine is still unclear (8, 12).
Salmonella, the most common enteric pathogen, are facultative intracellular Gram-negative bacteria. Approximately 1.4 million cases (sporadic form or outbreaks) per year of salmonellosis occur in the United States alone (13). Salmonella enterica serotype Typhimurium infection in humans causes gastroenteritis and remains localized in the intestine, whereas mice infected with this species get a systemic infection with pathogenesis resembling that of typhoid fever in humans (13–15). Thus, a murine infection model is a useful tool to study systemic Salmonella infection. The understanding of the complex host response to Salmonella infection in humans and other animal species has advanced considerably through the use of mouse models of infection. Among the TLR family members, TLR2, 4, 5, and 9 are most likely to be involved in intestinal infections as they recognize bacterial lipoprotein, LPS, flagellin, and CpG DNA, respectively (8, 12). TLR11 plays a critical role not only in parasitic infection through the recognition of profilin from parasites (16) but also in preventing bacterial infection in the urinary tract (17). With the consideration that the bacteria in the urinary tract infection are mainly from gastrointestinal tract, we investigate the role of TLR11 in murine Salmonella infection.
EXPERIMENTAL PROCEDURES
Bacterial Strains
S. enteric serotype Typhimurium LT2 (laboratory strain) and SR11 (highly invasive pathogen) (18) were inoculated in Luria-Bertani (LB) broth medium and cultured at 37 °C overnight without shaking.
Animals
TLR11 and TLR5 knock-out mice were generated by Dr. Ghosh and Dr. Flavell, respectively, on C57/BL6 × 129 background at Yale University and completed 12 and 15 backcross generations with C57/BL6 mice, respectively. The MyD88, TLR2, and TLR4 knock-out mice on a C57/BL6 background were originally generated by Dr. S. Akira (Osaka University, Japan), and MyD88 was kindly provided by Dr. R. Medzhitov (Yale University). All of the mice were bred under specific pathogen-free conditions at the Institute of Biosciences and Technology, Texas A&M University System Health Science Center (Houston, TX).
NF-κB Luciferase Assays
Chinese hamster ovary (CHO)-K1 cells (from ATCC) were transiently transfected with a 0.05 μg of NF-κB luciferase reporter construct pBIIX and 0.2 μg of a construct directing expression of Renilla luciferase under the control of the constitutively active thymidine kinase promoter (pRL-TK; Promega), together with 1 μg of different TLR expression constructs and plated into 24-well tissue culture plates. Twenty-four hours after transfection, cells were stimulated, where indicated, with 40 μl/ml heat-killed bacterial lysates for 6 h, and luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer's instructions. Data are presented as the mean ± S.D. of triplicate samples and are representative of three independent experiments.
ELISA
Peritoneal macrophages were prepared as described by Takeuchi et al. (19). The macrophages were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum. Production of TNF-α from peritoneal macrophages was measured by ELISA according to the manufacturer's instructions (BD Biosciences)
Salmonella Infection and Analysis
Age- and sex-matched mice were used for all experiments. Mice were starved for 4 h and then given 100 μl of 10% NaHCO3 using an oral gavage needle to neutralize stomach acid. Mice were then infected orally with 200 μl of 109 Salmonella in sterile PBS using a gavage needle, and the food and water were returned. After 3 days, the Peyer patches, spleen, and liver were harvested from each mouse that had been euthanized by cervical dislocation. Half of the organs were fixed in neutral buffered formalin for histological analysis, and the remaining half was homogenized in sterile 0.025% Triton X-100/PBS and used for surviving bacterial titer. For histological analysis, the formalin-fixed organs were embedded in paraffin, and sections were stained with hematoxylin & eosin. For intraperitoneal infections, mice were injected with the same amount of Salmonella in 500 μl of sterile PBS.
Statistics
The Mann-Whitney U test was used for calculating the statistically significant differences in the mean bacterial burden in different organs of wild-type compared with knock-out animals.
RESULTS
Bleeding of Peyer Patches in TLR11−/− Mice after Salmonella Infection
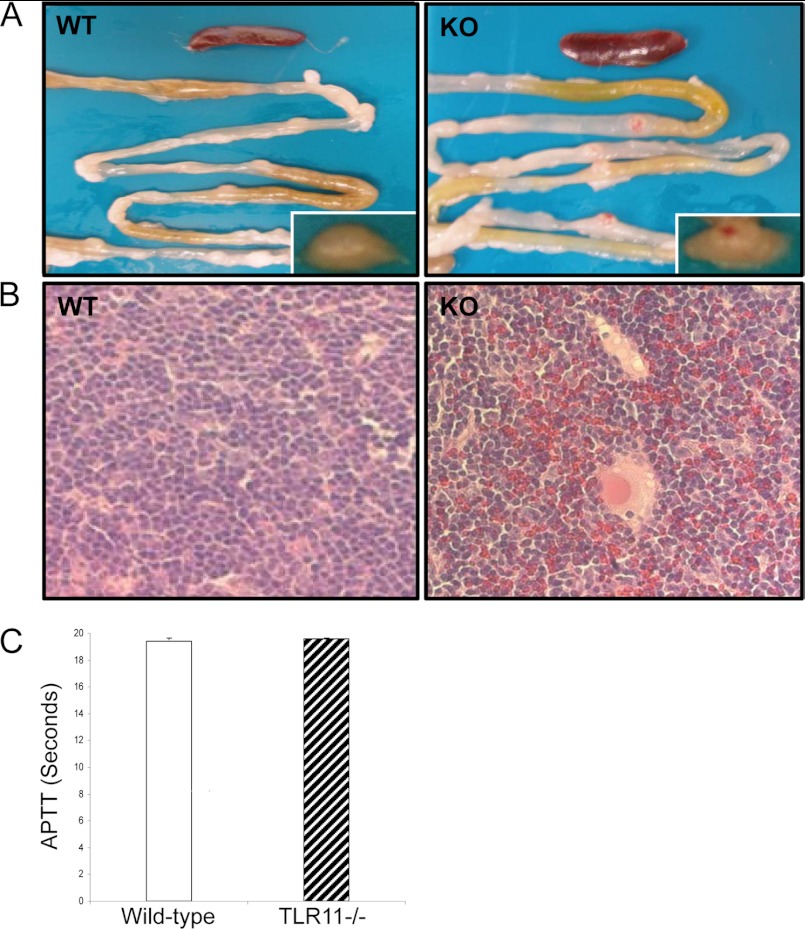
We realized that TLR11 is expressed in the epithelial cell not only in the urinary tract but also in the gastrointestinal tract (17); therefore, we investigated the role of TLR11 in Salmonella infection by using its knock-out mouse. The highly invasive Salmonella strain was used in the murine infection model. TLR11-deficient mice and wild-type littermates were challenged orally with 109 highly invasive Salmonella typhimurium strain SR11 (18). Because the Peyer patches are the main portal of entry for invasive enteric Salmonella infections (19, 20), we wanted first to determine whether there is a bacterial load difference between the wild-type and TLR11-deficient mice in this organ. Before we did that, however, we observed that the Peyer patches exhibited noticeable bleeding in TLR11-deficient mice, but not in any wild-type littermates after 72 h of infection (Fig. 1A). Histological examination revealed red blood cell leaking in Peyer patches of TLR11−/− mice (Fig. 1B). We also tested the parameters related to coagulation before and after infection. There was no significant difference in the partial thromboplastin time between wild-type and TLR11−/− mice before and after infection (Fig. 1C). In line with previous reports, the ability of Salmonella to invade the enteric tissues determines whether they will induce damage and bleeding in the Peyer patches (21). Thus, the Peyer patch bleeding in TLR11−/− mice is most likely due to their increased susceptibility to Salmonella invasion.
FIGURE 1.
Bleeding of Peyer patches in TLR11−/− mice after Salmonella infection. A, photograph of representative Peyer patches and small intestine from 10 TLR11−/− mice (KO), 10 wild-type (WT) littermates after 3 days of oral infection by Salmonella. Enlargements of the photo in the typical Peyer patches are at right bottom corner. B, tissue sections with H&E staining showing the histology of infected Peyer patches from the 50% TLR11 knock-out mice with red blood cell leaking. C, partial thromboplastin time (APTT) in wild-type and TLR11−/− mice before and after infection.
TLR11 Prevents Systemic Salmonella Infection
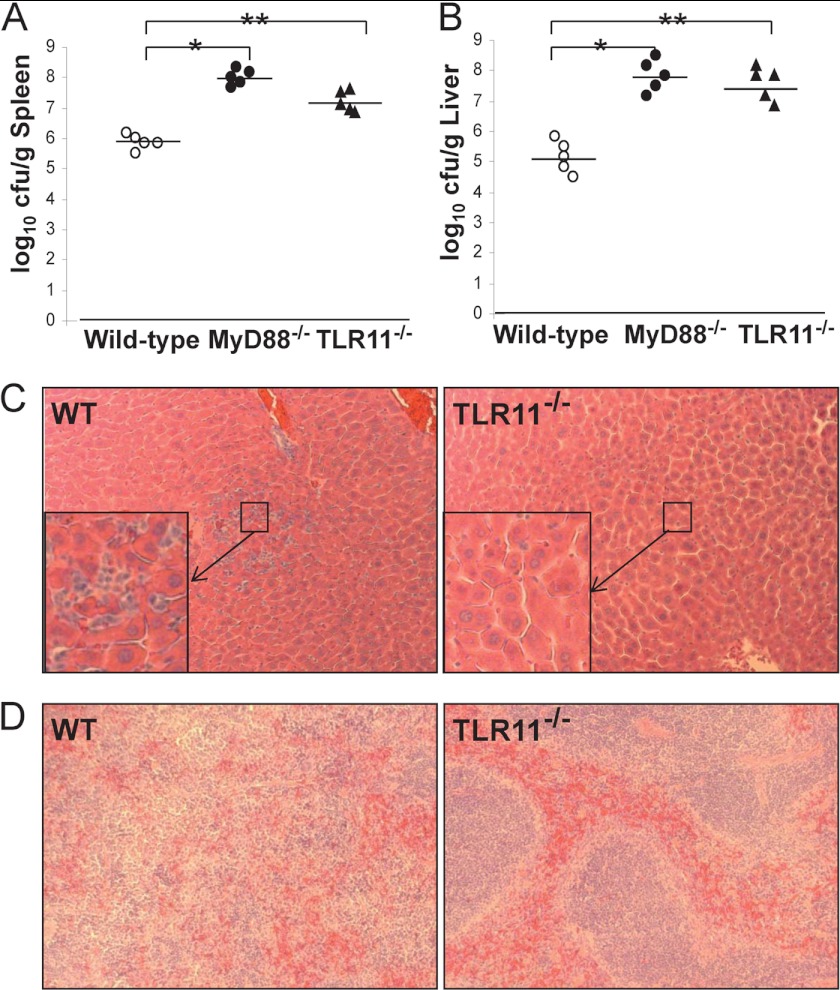
Susceptible mouse strains challenged orally with S. typhimurium usually develop a systemic, typhoid-like illness where the bacteria can be recovered from organs of the reticuloendothelial system. Generally, the hosts' immune system is able to control the systemic spread of the bacteria, indicating that even in susceptible mouse strains, an effective immune response is elicited to control the systemic spread of the bacteria. TLR11−/−, MyD88−/−, and the same background wild-type mice were inoculated with 109 S. typhimurium. Three days after challenge, liver and spleens, the major systemic sites of Salmonella replication (14), from all three groups were compared for bacterial titers and cellular pathology (Fig. 2). Similar to the results seen in the Peyer patches, the number of bacteria found in the spleens and livers of infected animals was significantly higher in TLR11 knock-out mice compared with wild-type control animals (Fig. 2, A and B). Quantitation revealed that there were at least 10-fold and 30-fold more bacteria found in the spleens and livers of infected TLR11 knock-out animals, respectively (Fig. 2, A and B). These results demonstrate the enhanced ability of Salmonella to disseminate to systemic sites in TLR11 knock-out mice. Compared with the WT mice, there was less inflammation in the livers and spleens of TLR11 knock-out mice as determined by the reduced infiltration of leukocytes and necrotic damage (Fig. 2, C and D), even though there was a more significant bacterial burden (Fig. 2, A and B). This may indicate that TLR11 is able to recognize a molecular component of Salmonella as its ligand to activate an effective immune response to reduce the initial systemic spread of S. typhimurium. Consistent with previous reports (19), organs taken from MyD88−/− mice harbored 30–300-fold more bacteria (Fig. 2, A and B), but also showed reduced inflammation compared with the spleens and livers of wild-type mice (data not shown).
FIGURE 2.
TLR11 prevent Salmonella for systemic infection. Bacterial burden was examined in the spleens (A) and livers (B) of MyD88−/− mice (n = 5), TLR11−/− mice (n = 5), and wild-type littermates (n = 5) 72 h after oral infection with 1 × 109 S. typhimurium. The tissue sections with H&E staining showed the histology of infected livers and spleens from the MyD88 knock-out mice and wild-type animals (data not shown) and from the TLR11 knock-out mice and wild-type littermates (C and D). The enlargement of the photo in the center square box is shown at left bottom corner in C. The sections of liver and spleen from wild-type mice showed significantly greater inflammatory response, estimated by the amount of liver necrosis, numbers of infiltrating leukocytes (C), as well as spleen tissue architecture damage (D) compared with TLR11 knock-out mice. *, p < 0.005 as MyD88−/− (filled circles) compared with wild-type (open circles). **, p < 0.01 as TLR11−/− (filled triangles) compared with wild-type littermates. Data are representative of three independent experiments (two-tailed Mann-Whitney test).
TLR11 Is Capable of Recognizing Salmonella to Activate TLR Signaling Pathway
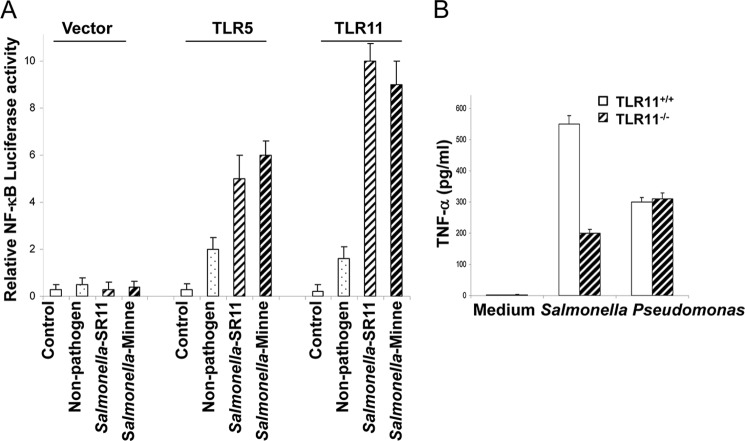
Signaling by the TLRs following ligand engagement generally induces NF-κB activation via a series of signaling cascades, which then activates the innate immunity and further modulates adaptive immunity (22–25). Whereas the TLR11 ligand in bacteria is still unidentified, the reduced infiltration of inflammatory cells seen in liver and spleen of TLR11−/− mice indicates that this molecule is critical to the activation of the murine innate immune response to Salmonella. To determine whether TLR11 activated by Salmonella also induces NF-κB activation, we used heat-killed lysates from two different pathogenic strains of Salmonella, Salmonella minnesota and highly invasive S. typhimurium SR11, along with a nonpathogenic strain of S. typhimurium LT2 to stimulate TLR11-expressing CHO cells with NF-κB luciferase reporter pBIIX. Remarkably, lysates of the two pathogenic strains showed a striking activation of NF-κB at levels significantly greater than that seen with lysates from the nonpathogenic strain in CHO cells transiently transfected with TLR11 (Fig. 3A). We also observed similar phenomena for TLR5, which is consistent with previous reports (26). We found that the peritoneal macrophages from wild-type mice expressed high levels of TLR11 detected by RT-PCR. We also confirmed that there is no TLR11 expression in the macrophages from TLR11 knock-out mice (data not shown). Murine peritoneal macrophages were also stimulated with heat-killed pathogenic S. typhimurium SR11, nonpathogenic S. typhimurium LT2, and Pseudomonas (as control). Macrophages from the TLR11−/− mice showed a dramatically reduced response to S. typhimurium compared with wild-type cells (Fig. 3B), whereas the response to nonpathogenic S. typhimurium LT2 and Pseudomonas was almost unaffected (Fig. 3B and data not shown) (27). These results strongly suggest that TLR11 recongnizes a component in pathogenic S. typhimurium.
FIGURE 3.
Salmonella activates TLR11 signaling pathway to induce NF-κB activation. A, CHO cells were transiently transfected with TLR5, TLR11, or empty expression vectors together with a NF-κB luciferase reporter pBIIX for 24 h. Luciferase activity was measured in the cells treated with 40 μl ml−1 bacterial lysates from the indicated saturated bacterial cultures or LB alone (Control). Data are representative of three independent experiments. B, thioglycollate-elicited peritoneal macrophages were isolated from TLR11 knock-out mice and wild-type littermates. The macrophages were tested for response to S. typhimurium and Pseudomonas by measuring secretion TNF-α with ELISA.
TLR11−/− Mice Are Resistant to Lethal Shock
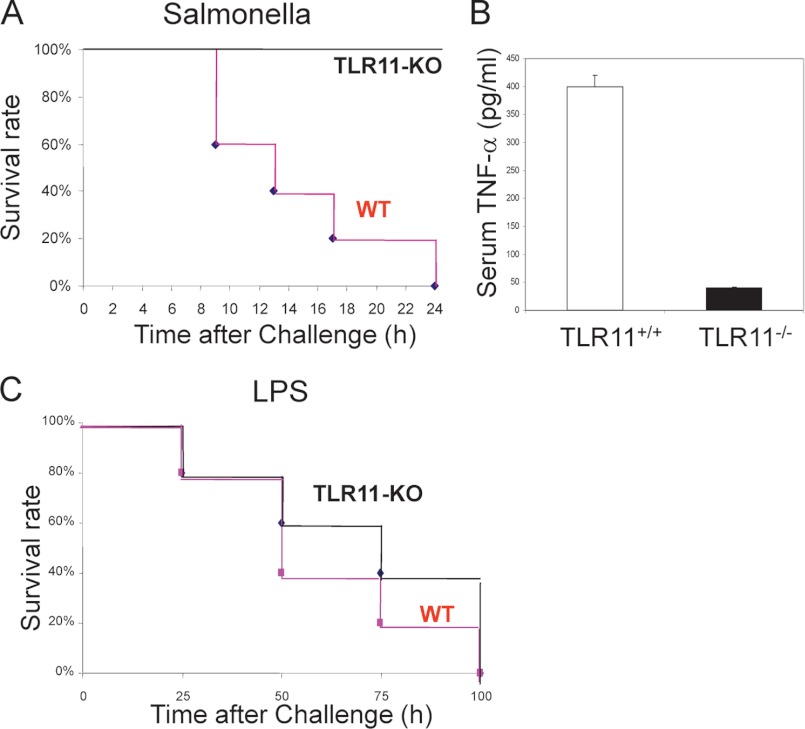
We then investigated whether the knockout of TLR11 would affect Salmonella-inducing lethal shock. The highly invasive Salmonella strain SR11 was injected into TLR11 knock-out mice and wild-type control mice intraperitoneally. TLR11−/− mice were resistant to rapid death following challenge with Salmonella at doses (109 cfu/mouse) that are sufficient to cause rapid death in wild-type mice (Fig. 4A). We realized that at 2 h after the intraperitoneal injection, wild-type mice had the highest production of serum TNF-α, the proinflammatory cytokine associated with early innate immune responses. Therefore, we challenged all the mice with Salmonella by intraperitoneal injection for 2 h to check TNF-α production in serum. Remarkably, TLR11−/− mice infected with Salmonella displayed dramatically deficient TNF-α production (Fig. 4B). In contrast, there is almost no difference between TLR11 wild-type and knock-out mice when LPS was injected intraperitoneally to induce septic shock (Fig. 4C). These results strongly imply that TLR11 plays a critical role in murine Salmonella infection.
FIGURE 4.
TLR11−/− mice are resistant to lethal shock. A, TLR11-deficient mice are resistant to rapidly lethal shock by Salmonella. Survival of sex-matched TLR11-deficient mice (n = 5) and wild-type littermates (n = 5) after intraperitoneal injection with 109 S. typhimurium is shown. The survival rate was monitored hourly. p = 0.001, TLR11−/− mice versus wild-type littermates (log-rank test). B, serum TNF-α level of TLR11 knock-out mice and wild-type littermates (five animals/group) was measured by ELISA at 2 h after intraperitoneal infection with Salmonella (109 cfu/mouse). TLR11−/− mice infected with Salmonella display deficient TNF-α production. The data shown are representative of three independent experiments (error bars, S.D. derived from an average of three mice). C, survival of sex-matched TLR11-deficient mice (n = 5) and wild-type littermates (n = 5) after intraperitoneal injection with 1.0 mg of LPS.
TLR11 and TLR4 Block the Penetration of Invasive Salmonella into the Murine Peyer Patches
To investigate the role of various TLRs in Salmonella infection, we utilized different TLR knockouts, including TLR2, TLR4, TLR5, and TLR11 as well as MyD88. Different knock-out mice and wild-type controls were challenged orally with 109 highly invasive S. typhimurium strain SR11 (18). Because the Peyer patches are the main portal of entry for invasive enteric Salmonella infections (20), we only focused on this organ to determine whether the bacterial load differed between the wild-type and TLR-deficient mice. We found that TLR11 and TLR4 knock-out mice had the bacterial titers of the highly invasive strain SR11 in Peyer patches more than 10 times higher than the wild-type animals (Fig. 5). Not surprisingly, we also found that there were more bacteria in the Peyer patches of MyD88−/− mice than in TLR11−/− animals (Fig. 5). Consistent with previous report (10), there was less bacteria burden in TLR5−/− Peyer patches and no difference in TLR2−/− Peyer patches by compared with their wild-type controls. Therefore, different TLRs cooperate to control Salmonella penetration into murine Peyer patches (Fig. 6).
FIGURE 5.
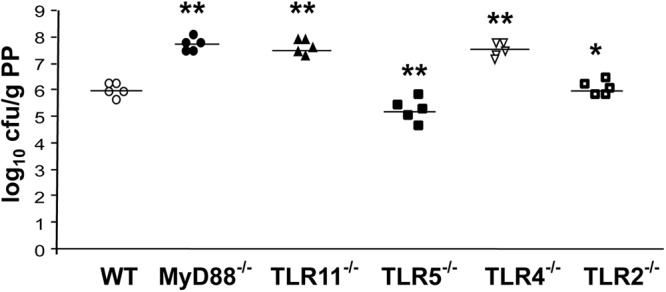
Blocker-TLR and carrier-TLR cooperate to control Salmonella penetration into murine Peyer patches (PP). Bacterial burden in the Peyer patches of MyD88−/− mice (n = 5), wild-type littermates (n = 5), and different TLR-deficient mice (n = 5, respectively) is shown. Data are representative of three independent experiments. *, p > 0.05; **, p < 0.05 (compared with WT) using the Mann-Whitney U test.
FIGURE 6.
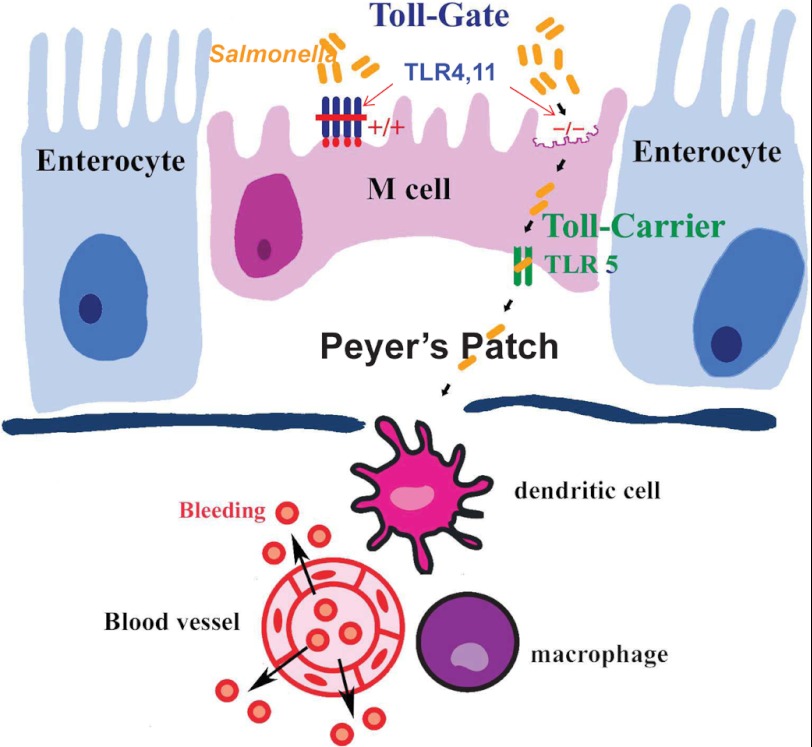
A proposed model for TLRs to control Salmonella penetration into murine Peyer patches. In the mouse models of S. enterica serovar Typhimurium infection, pathogens perfer to penetrate the M cell of Peyer patches. TLRs play a critical role to control Salmonella infection. In contrast to TLR5 as a carrier-TLR of Salmonella (10), TLR4 and TLR11 may work as blocker-TLRs to prevent highly invasive Salmonella from penetrating into Peyer patches and spreading systemically. Remarkably and surprisingly, in mice lacking MyD88 obvious hemorrhages are induced only at Peyer patches by highly invasive Salmonella.
DISCUSSION
As the key sensors for pathogen associate molecular patterns, TLRs play a critical role in preventing host infection (1–3). How different TLRs function depends on the site of the infection and the pathogen (7). Recently, TLR functions in colonic homeostasis and tolerance to commensal bacteria have been intensively studied (9–11), particularly, how the apical and basolateral epithelia of the colon respond to CpG DNA, the ligand of TLR9 (11). TLRs are expressed not only in immune cells like macrophage and dendritic cells but also in epithelial or endothelial cells which are in direct contact with pathogens and commensal bacteria. In contrast to the role of TLRs in colonic homeostasis and tolerance to commensal bacteria, the precise role of different TLRs in response to pathogen infection in different organs such as the intestine is still unclear (8, 12). It appears that some TLRs function as primary sensors, whereas others may act as secondary (or later) sensors in intestinal infection (23). It has been reported that the timely response to oral infections with Salmonella requires TLR2, TLR4 (23), and TLR5 (10, 28, 29), when mice are challenged with a lethal dose. In the mesenteric lymph modes, TLR4-deficient mice harbor between 10- and 100-fold more bacteria than wild-type mice (23, 30). Surprisingly, TLR2 knockout has no phenotype, but the bacterial burden of the TLR2/TLR4 double knockouts is synergistically increased compared with the bacterial burden of the TLR4 single knockout (23), indicating that TLR2 responses are relevant only if TLR4 is active (7). In line with this report, another report showed that TLR2 did not activate after being challenged for 6 h, but activated after being challenged for 24 h (23). A similar phenomenon was reported in the TLR5 knock-out animal (28). Taken together, these results suggest that TLR4 is the dominant TLR involved in the host response to Salmonella infection (7). However, the role of TLR11 in Salmonella infection is independent of TLR4 because there was no difference between TLR11−/− mice and wild-type littermates to LPS challenge (Fig. 4C). TLR11−/− mice are independent of but similar to TLR4 in increased bacterial burden in Peyer patches, as well as in the spleen and liver. Thus, in addition to TLR4, TLR11 is another primary dominant TLR involved in the murine response to Salmonella infection.
Surprisingly but not completely unexpected, there is more bacterial burden but less inflammatory reaction in both the livers and spleens of TLR11-deficient mice. Most likely, TLR11 not only functions to control and clear bacteria but also is able to recognize its ligand in Salmonella to induce inflammatory signaling. This hypothesis was further confirmed by reducing serum TNF-α secretion in TLR11−/− mice dramatically by intraperitoneal injection of a lethal dose of Salmonella. To find out the TLR11 natural ligand in bacteria will significantly advance our understanding of TLR11 biological function. Although we have already figured out that the profilin-like protein in parasites is the ligand of TLR11 (16), there is no homology to profilin in the bacteria database. We believe that TLR11 recognizes a different structural molecular pattern in bacteria and parasites. Our results indicate that perhaps the ligand of TLR11, other than the known ligands of TLRs, also contributes to the effective murine immune response to Salmonella.
TLR11 prevents not only intestinal but also systemic Salmonella infection because there is more bacterial burden in Peyer patches as well as in spleens and livers of TLR11−/− mice. More importantly, because TLR11 has a close similarity to TLR5 by phylogenic tree analysis (17) and TLR11 seems to recognize a protein ligand such as profilin in parasites (16), TLR11 may play a role as a blocker in Salmonella infection (Fig. 5), working against the TLR5 function as Salmonella carrier (10). As we know, the phenotype of increasing bacteria burden to induce Peyer patch bleeding has not been observed in other TLRs or other PRRs knock-out mice infected by Salmonella. Currently, we are still not sure of the exact mechanism of inducing only visible Peyer patch bleeding. It will be interesting to see the phenotype of Peyer patches to Salmonella infection in TLR5/TLR11 and TLR4/TLR11 double-knock-out mice and to see the possible balance of both TLR5 and TLR11 single knock-out phenotype or the matching of TLR4/TLR11 double knock-out mice to the phenotype from MyD88-deficient animals.
In summary, we found that highly invasive Salmonella induced Peyer patch bleeding in the TLR11 knock-out mouse, a phenotype resembling human Salmonella infection. Thus, this can be a useful murine infection model in the study of Salmonella infection. Our results also define that different TLRs play different role in murine Peyer patches to control Salmonella infection, in which TLR5 appears to work as a “carrier-TLR” of Salmonella (10), whereas TLR4 and TLR11 may work as “blocker-TLRs” of Salmonella to prevent highly invasive Salmonella from penetration into the murine Peyer patches and spreading systemically (Fig. 6). These results collectively indicate an important role for various TLRs in cooperation to tightly control of pathogen penetration into Peyer patches and therefore might provide a new perspective on the host-pathogen interactions.
Acknowledgment
We thank Lida M. Keene (Texas A&M University Health Science Center-IBT) for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R56AI085164. This work was also supported by the Texas A&M University Health Science Center.
- PRR
- pattern-recognition receptor
- CHO
- Chinese hamster ovary
- TLR
- Toll-like receptor.
REFERENCES
- 1. Akira S., Uematsu S., Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell 124, 783–801 [DOI] [PubMed] [Google Scholar]
- 2. West A. P., Koblansky A. A., Ghosh S. (2006) Recognition and signaling by Toll-like receptors. Annu. Rev. Cell Dev. Biol. 22, 409–437 [DOI] [PubMed] [Google Scholar]
- 3. Beutler B., Jiang Z., Georgel P., Crozat K., Croker B., Rutschmann S., Du X., Hoebe K. (2006) Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 24, 353–389 [DOI] [PubMed] [Google Scholar]
- 4. Janeway C. A., Jr., Medzhitov R. (2002) Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 [DOI] [PubMed] [Google Scholar]
- 5. Takeda K., Kaisho T., Akira S. (2003) Toll-like receptors. Annu. Rev. Immunol. 21, 335–376 [DOI] [PubMed] [Google Scholar]
- 6. Tabeta K., Georgel P., Janssen E., Du X., Hoebe K., Crozat K., Mudd S., Shamel L., Sovath S., Goode J., Alexopoulou L., Flavell R. A., Beutler B. (2004) Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. U.S.A. 101, 3516–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerold G., Zychlinsky A., de Diego J. L. (2007) What is the role of Toll-like receptors in bacterial infections? Semin. Immunol. 19, 41–47 [DOI] [PubMed] [Google Scholar]
- 8. Eckmann L. (2006) Sensor molecules in intestinal innate immunity against bacterial infections. Curr. Opin. Gastroenterol. 22, 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. (2004) Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 [DOI] [PubMed] [Google Scholar]
- 10. Uematsu S., Jang M. H., Chevrier N., Guo Z., Kumagai Y., Yamamoto M., Kato H., Sougawa N., Matsui H., Kuwata H., Hemmi H., Coban C., Kawai T., Ishii K. J., Takeuchi O., Miyasaka M., Takeda K., Akira S. (2006) Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat. Immunol. 7, 868–874 [DOI] [PubMed] [Google Scholar]
- 11. Lee J., Mo J. H., Katakura K., Alkalay I., Rucker A. N., Liu Y. T., Lee H. K., Shen C., Cojocaru G., Shenouda S., Kagnoff M., Eckmann L., Ben-Neriah Y., Raz E. (2006) Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat. Cell. Biol. 8, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 12. Abreu M. T., Fukata M., Arditi M. (2005) TLR signaling in the gut in health and disease. J. Immunol. 174, 4453–4460 [DOI] [PubMed] [Google Scholar]
- 13. Dougan G., John V., Palmer S., Mastroeni P. (2011) Immunity to salmonellosis. Immunol. Rev. 240, 196–210 [DOI] [PubMed] [Google Scholar]
- 14. Ohl M. E., Miller S. I. (2001) Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52, 259–274 [DOI] [PubMed] [Google Scholar]
- 15. Hapfelmeier S., Hardt W. D. (2005) A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol. 13, 497–503 [DOI] [PubMed] [Google Scholar]
- 16. Yarovinsky F., Zhang D., Andersen J. F., Bannenberg G. L., Serhan C. N., Hayden M. S., Hieny S., Sutterwala F. S., Flavell R. A., Ghosh S., Sher A. (2005) TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science 308, 1626–1629 [DOI] [PubMed] [Google Scholar]
- 17. Zhang D., Zhang G., Hayden M. S., Greenblatt M. B., Bussey C., Flavell R. A., Ghosh S. (2004) A Toll-like receptor that prevents infection by uropathogenic bacteria. Science 303, 1522–1526 [DOI] [PubMed] [Google Scholar]
- 18. Tükel C., Raffatellu M., Humphries A. D., Wilson R. P., Andrews-Polymenis H. L., Gull T., Figueiredo J. F., Wong M. H., Michelsen K. S., Akçelik M., Adams L. G., Bäumler A. J. (2005) CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol. Microbiol. 58, 289–304 [DOI] [PubMed] [Google Scholar]
- 19. Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., Takeda K., Akira S. (1999) Differential roles of TLR2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity 11, 443–451 [DOI] [PubMed] [Google Scholar]
- 20. Jones B. D., Ghori N., Falkow S. (1994) Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180, 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao L., Ezak T., Li Z. Y., Kawamura Y., Hirose K., Watanabe H. (2001) Vi-suppressed wild strain Salmonella typhi cultured in high osmolarity is hyperinvasive toward epithelial cells and destructive of Peyer's patches. Microbiol. Immunol. 45, 149–158 [DOI] [PubMed] [Google Scholar]
- 22. Hapfelmeier S., Stecher B., Barthel M., Kremer M., Müller A. J., Heikenwalder M., Stallmach T., Hensel M., Pfeffer K., Akira S., Hardt W. D. (2005) The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J. Immunol. 174, 1675–1685 [DOI] [PubMed] [Google Scholar]
- 23. Weiss D. S., Raupach B., Takeda K., Akira S., Zychlinsky A. (2004) Toll-like receptors are temporally involved in host defense. J. Immunol. 172, 4463–4469 [DOI] [PubMed] [Google Scholar]
- 24. Akira S. (2006) TLR signaling. Curr. Top. Microbiol. Immunol. 311, 1–16 [DOI] [PubMed] [Google Scholar]
- 25. Iwasaki A., Medzhitov R. (2004) Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5, 987–995 [DOI] [PubMed] [Google Scholar]
- 26. Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., Aderem A. (2001) The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 27. Mathur R., Oh H., Zhang D., Park S. G., Seo J., Koblansky A., Hayden M. S., Ghosh S. (2012) A mouse model of Salmonella typhi infection. Cell 151, 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feuillet V., Medjane S., Mondor I., Demaria O., Pagni P. P., Galán J. E., Flavell R. A., Alexopoulou L. (2006) Involvement of Toll-like receptor 5 in the recognition of flagellated bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 12487–12492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galán J. E. (2001) Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17, 53–86 [DOI] [PubMed] [Google Scholar]
- 30. Chabot S., Wagner J. S., Farrant S., Neutra M. R. (2006) TLRs regulate the gatekeeping functions of the intestinal follicle-associated epithelium. J. Immunol. 176, 4275–4283 [DOI] [PubMed] [Google Scholar]
- 31.Deleted in proof