Background: Type I toxin-antitoxins are widespread modules with unclear functions.
Results: The three-dimensional structure of PepA1, a toxin expressed upon acidic and oxidative bursts of Staphylococcus aureus, has been solved.
Conclusion: S. aureus has a functional type I TA system that induces lytic cell death.
Significance: Inducing PepA1 expression in S. aureus could be an effective antibiotic.
Keywords: Cell Death, NMR, Peptide Conformation, RNA, Staphylococcus aureus, Cell Death, NMR, Peptide Structure, Toxin-Antitoxin RNA Pair
Abstract
We report a functional type I toxin-antitoxin (TA) module expressed by a human pathogen, Staphylococcus aureus. TA systems consist of stable toxins and labile antitoxins encoded within small genetic modules widespread in eubacteria and archaea. TA genes provide stress adaptation and protection against DNA loss or invasion. The genes encoding the SprA1 toxic peptide (PepA1) and the SprA1AS RNA antitoxin are within a pathogenicity island on opposite strands and possess a 3′ overlap. To prevent peptide toxicity during S. aureus growth, PepA1 expression from stable (half-life > 3 h) SprA1 is repressed by elevated amounts of unstable (half-life = ∼10 mn) SprA1AS. In vivo, PepA1 localizes at the bacterial membrane and triggers S. aureus death. Based on NMR and CD data, its solution structure was solved and is a long bent, interrupted helix. Molecular dynamics simulations indicate that PepA1 compaction and helical content fluctuate in accordance with its cytoplasm or membrane location. When inserted into the S. aureus membrane, the PepA1 conformation switches to a ∼7-nm-long continuous helix, presumably forming pores to alter membrane integrity. PepA1 expression is induced upon acidic and oxidative stresses by reducing SprA1AS levels. As an altruistic behavior during infection, some cells may induce the expression of that toxin that would facilitate departure from the host immune cells for spreading.
Introduction
Toxin-antitoxin (TA)4 systems are widespread in bacteria and archae. They are composed of a stable toxin and a labile antitoxin (1). When the short-lived antitoxin gets degraded, the stable toxin is no longer neutralized and causes cell death. Their abundance and presence in multiple copies indicate that they provide benefits for bacterial growth and survival, involved not only in ordinary physiology but also in bacterial pathogenicity (2). Three types of TA modules have been described. Type I TA pairs consist of a toxic peptide and a convergent antisense RNA that inhibits toxin translation (3). The protein toxin and RNA antitoxin are usually encoded on opposite strands, with the overlap occurring at either the 5′ or the 3′ end of the mRNA. Type II TA systems consist of an antitoxin protein that binds to and inactivates a toxin protein. Type III TA pairs encode protein toxins inhibited by an RNA antitoxin encoded by tandem repeats upstream of the toxin gene (4). Their functions, however, remain hypothetical and controversial (5). Therefore, the clarification of their cellular functions and regulations is essential to better understand bacterial physiology under diverse stress conditions. In this report, the focus is on Type I TA modules in which the expression of the toxin is regulated by an antisense RNA transcribed in reverse orientation. To date, type I TA loci have been experimentally shown only in a few lineages of Enteroproteobacteria and Firmicutes (Bacillus and Enterococcus), but computer predictions suggest that they are widely scattered among bacteria, including in Staphylococcus aureus (6).
S. aureus is an opportunistic Gram-positive bacterium responsible for nosocomial and community-acquired infections in humans, ranging from localized to life-threatening diseases. The adaptability of this bacterium to drastic environmental changes depends on complicated interplays between two component systems, transcription regulatory proteins and nearly a hundred regulatory RNAs. These small RNAs includes cis- and trans-acting RNAs, antisense RNAs, noncoding RNAs and small mRNAs-encoding peptides (1, 7, 8). In this bacterium, however, the demarcation between cis- and trans-tRNAs can sometimes be fuzzy, as recently shown for the SprA1/SprA1AS pair, in which SprA1 expresses a peptide that is repressed, in trans, by cis-SprA1AS (9). SprA1 translation is inhibited through pairing interactions with SprA1AS hiding the SprA1 translation initiation signals. The genetic locus containing sprA1/sprA1AS lies on a pathogenicity island and produces two tRNAs, SprA1 and SprA1AS, transcribed in opposite directions from a bidirectional terminator. Additional copies of the RNA pair are detected within the core genomes of numerous strains from the Staphylococcus genus, in some plasmids, and in several strains of the Enterococcus genus. Sequence comparisons have suggested that this tRNA pair might form a type I toxin-antitoxin module (6).
In this report, experimental data are provided to support the existence of a functional Type I TA module expressed in S. aureus, in which the toxin is a 30-amino acid-long peptide named PepA1. Cell fractionation assays indicate that PepA1 localizes at the S. aureus membrane. Based on mutational analysis and cell mortality experiments, PepA1 but not SprA1 causes cell death by affecting S. aureus membrane organization. Oxidative and acidic stresses trigger PepA1 expression by lowering SprA1AS levels, in turn relieving the constitutive translational repression of SprA1. The PepA1 structure was solved by NMR spectroscopy and adopts a slightly bent, interrupted, helix fold. It represents the first atomic resolution structure of a type I TA module in S. aureus. CD spectra were collected in both aqueous and nonpolar environments and indicate that when the lipid content increases, the helical content of PepA1 rises. These structural data, together with molecular dynamics simulations computed in an artificial lipid bilayer, suggest that PepA1 compaction and helical content both vary according to its cytoplasm or membrane location. Our structural investigations indicate that when PepA1 is inserted into a membrane bilayer, it adopts an extended, continuous helix as for the “pore-forming” peptides, consistent with its membrane positioning in vivo.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Genetic Manipulations
The primers used for all the constructions are listed in supplemental Table S1. Strains and plasmids are listed in supplemental Table S2. S. aureus strains were grown at 37 °C in brain-heart infusion broth (Oxoid) or in Roswell Park Memorial Institute 1640 medium (GlutaMAXTM; Invitrogen). When necessary, chloramphenicol was added at 10 μg ml−1. To generate the anhydrotetracycline (aTc)-inducible construct for SprA1 (pAT12ΩsprA1), the SprA1 sequence (from nucleotides +1 to +208) was amplified from Newman genomic DNA and inserted into PstI/EcoRV-digested pAT12 vector. To generate an inducible construct containing the SprA1-STOP mutant (pAT12ΩsprA1 STOP), nucleotides T64 and C65 were mutated into two adenines to create a premature termination codon. In pCN35ΩsprA1FLAG/sprA1AS, both RNAs are expressed from their endogenous promoters. Three PCRs allowed the amplification of sprA1/sprA1AS locus from Newman genomic DNA (with 171 nucleotides upstream and 105 nucleotides downstream) with 3× FLAG incorporated in frame at the C-terminal end of SprA1 ORF. 3× FLAG (66-bp, 22 amino acids) is detected using monoclonal anti-FLAG® M2-Peroxidase (HRP). The resulting amino acid sequence of the SprA1 fusion peptide is MLIFVHIIAPVISGCAIAFFSYWLSRRNTKDYKDHDGDYKDHDIDYKDDDDK.
RNA Extractions, Northern Blots, and RNA Half-life Determinations
RNA extractions and Northern blots were performed as described (9). For the RNA half-life measurements, S. aureus was cultured overnight, and the cultures were diluted 1:100 and grown for an additional 3 h at 37 °C. Rifampicin was added to the culture at a final concentration of 200 μg ml−1. Samples (2 ml each) were taken after rifampicin addition at 0, 15, 30, 60, 120, and 200 min. The samples were centrifuged, and the pellets were shock frozen in liquid nitrogen and stored at −80 °C. RNAs were then extracted, and Northern blot assays were performed.
SprA1 WT and Mutant Inductions, Growth, and Cell Death Experiments
Strains containing the relevant plasmids were grown overnight in brain-heart infusion broth. Overnight cultures were diluted 1/100 in brain-heart infusion broth containing 1 μm of aTc and grown in 96-well plate. A600 nm were measured at 5-min intervals for 20 h in Synergy HT (Bioteck). For the viability assays, the S. aureus strains were cultured and induced with aTc for 1 h. The cultures were then washed with PBS and stained with LIVE/DEAD bacterial viability kits (Invitrogen), following the manufacturer's instructions. That strain contains SYTO 9 (green fluorescent) and propidium iodide (red fluorescent), both binding DNAs. When used alone, the SYTO 9 stains all the bacteria with intact or damaged membranes. In contrast, propidium iodide only penetrates bacteria with damaged membranes, causing a reduction in the SYTO 9 stain green fluorescence. Pictures of fluorescence-labeled cells were captured with a DMRXA2 Leica microscope and a COOLSNAP HQ charge-coupled device camera, using Metavue software (Molecular Devices, Sunnyvale, CA). To determine the effect of SprA1 WT or mutant induction on growth, staphylococcal cultures were prepared as follows: 2-fold serial dilutions of exponential cultures were spotted on brain-heart infusion broth plates containing 1 μm aTc and 10 μg ml−1 chloramphenicol and incubated at 37 °C for 24 h.
Protein Extractions and Western Blots
Protein extracts are performed as described previously (9). For the secreted protein preparation, culture supernatants are collected and precipitated with 10% trichloroacetic acid. The precipitates are washed with ice-cold acetone and dissolved into a Tris-Tricine loading buffer. For fractionated cell wall, cytoplasm, or membrane proteins, bacterial pellets were resuspended into 50 mm Tris-HCl, pH 7.5, 20 mm MgCl2, supplemented with 30% sucrose and lysostaphin (0.1 mg/ml). The suspension was incubated 10 min at room temperature and then centrifuged at 4000 rpm at 4 °C for 8 min. The supernatant contains the cell wall protein fraction. The pellet (protoplasts) was dissolved into 50 mm Tris-HCl, pH 7.5, 20 mm MgCl2 and sonicated for a 10-s pulse and centrifuged at 19,000 rpm for 60 min. The supernatant contained the cytoplasmic protein fraction, whereas the pellet included the membrane protein fraction. The membrane fraction was dissolved into 50 mm Tris-HCl, pH 7.5, 20 mm MgCl2. The samples were separated on Tricine-SDS-PAGE 16% gel, transferred onto Westran S PVDF membrane (Whatman), revealed using the Amersham Biosciences ECLTM Plus detection kit, and scanned with the LAS4000 imager (GE Healthcare).
Stress Conditions Assays
Strains were grown overnight in RPMI. The cultures were diluted 1/100 in RPMI and grown until an A600 nm of 1. For the acid stress, hydrochloric acid was added to the culture to reach a pH of 4.5. For the saline stress, sodium chloride was added to the culture at a final concentration of 2 m. For the oxidative stress, hydrogen peroxide was added to the culture at a final concentration of 10 mm. After the addition of the stress agents, samples (2 ml each) were taken at 15 and 30 min. The samples were centrifuged, and the pellets were stocked at −80 °C until use.
Peptide Synthesis
PepA1 peptide synthesis was performed by Proteogenix (France). All of the dilutions of PepA1 in trifluoroethanol (TFE) and SDS and in the ternary NMR mixtures were freshly prepared from dissolving the weighed dry peptide into the adequate buffer.
CD Experiments
Far ultraviolet CD spectra were recorded in a 0.1-cm-path length quartz cell at 21 °C on a Jobin Yvon CD6 spectrometer over a 185–260-nm range, with a 2-nm bandwidth, a step size of 1 nm, and an integration time of 2 s/point. The spectra were averaged over five records. Buffer contributions were removed by subtracting the CD spectra of the control samples. PepA1 aggregates in aqueous solutions below 40% (v/v) TFE and concentrations of SDS below the critical micellar concentration (CMC of ∼8 mm). PepA1 CD spectra were recorded in SDS solutions with concentrations well above the CMC.
NMR Experiments
NMR spectra were recorded on a 600MHz Avance Bruker spectrometer equipped with a triple resonance cryo-probe and Z-axis gradient capability. Weighed peptide was dissolved in CD3OH/CDCl3/H2O buffer at pH 3.8 for a final concentration of 5 mm. The signal was field locked on CD3OH. All of the spectra were recorded in Shigemi 5-mm tubes, plugged, and sealed to avoid buffer evaporation, at temperatures ranging from 283 to 293 K. TOCSY (tm = 80 ms), NOESY (tm = 60, 90, 200, and 350 ms), and COSY experiments were collected using the Watergate pulse sequence for water signal suppression. The data were processed using Topsin 3.1 (Bruker). The chemical shift of the water signal was used as a reference (4.91 ppm at 288 K). Cross-peaks assignments were carried out in an iterative way using the network anchorage procedure and distance conversions provided by the CCPN software (10).
PepA1 Three-dimensional Model Constructions
1000 models were computed starting from a randomized extended peptide, using the integrated CCPN/ARIA2.3.1 packages (10–12) constrained by 313 NOE-derived distances sorted into four binned classes ([1.7–2.7], [1.7–3.3], [2.5–4.5], and [3.0–6.0] Å). Dihedral and J-coupling were not restrained in the computations. Me2SO is an aprotic and nonbiomimetic solvent, and our mixed buffer contains water and methanol. Purified PepA1, however, is neither soluble in pure water, MeOH or Me2SO. So neither computational refinement in boxes of each of these three solvents is acceptable because we measured “interproton” distances in a mixed buffer with different biophysical properties. We thus decided to refine the top 20 structures of lowest energy in explicit Me2SO, because its dielectric constant is the closest to that of the NMR buffer. An energy-minimized average structure was computed. Violations were checked both with CCPN and MOLMOL (13). The 10 best refined structures of lowest energy and with restraint violation of <0.2 Å were checked using MOLPROBITY 3.19 (14). The structures were displayed in PYMOL (DeLano Scientific LLC). The atomic coordinates have been deposited in the Protein Data Bank (code 4B19) and the proton chemical shifts to the Biological Magnetic Resonance Bank (accession code r4b19nmr).
Molecular Dynamics Simulations
The average energy-minimized structure was submitted to molecular dynamics using the GROMACS package (15). Molecular dynamics (MD) was carried in a simulated lipid bilayer of dipalmitoyl phosphatidylcholine (DPPC) according to the protocol described by Kandasamy and Larson (16). The peptide was inserted perpendicularly to the DPPC bilayer and submitted to several cycles of energy minimization and positional restraints cycles using a modified GROMOS96 53A6 force field extended to include Berger lipid parameters. The simulation was carried out with periodic boundary conditions. The DPPC bilayer was solvated with SPC16 water molecules, and chloride ions were added to have a value of 0 as a net electrostatic charge of the system. The bond lengths were constrained by the all atoms LINCS algorithm. A particle mesh Ewald algorithm was used for the electrostatic interactions with a cut-off of 0.9 nm. The simulations were performed at neutral pH with runs of 50 ns at 323 K coupling the system to an external bath. GROMACS routines were utilized to check the trajectories and the quality of the simulations. The final structures were again checked using MOLPROBITY 3.19.
RESULTS
SprA1 Inhibits S. aureus Growth by Its Encoded PepA1 Peptide, Triggering Death by Membrane Damage
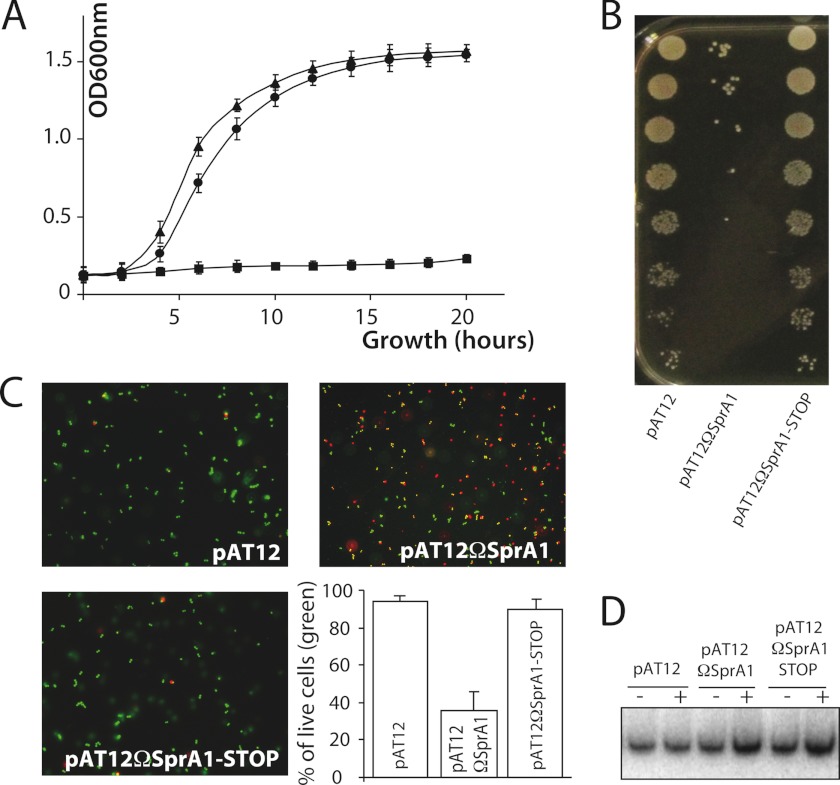
Based on computer searches, sprA1 was proposed to be the toxic component of a type I TA pair (6, 17). Recent data showed that a PepA1 fusion peptide, encoded by SprA1, is expressed in vivo and that a synthetic PepA1 lyses mammalian erythrocytes (9). Therefore, we sought to determine whether PepA1 is a toxin that can influence S. aureus growth. To test that hypothesis, the SprA1 sequence, from G1889597 to U1889804 in strain Newman, was cloned into an aTc-inducible pAT12 vector (18). A SprA1 mutant with a termination codon replacing the fourth codon of the internal PepA1 coding sequence (pAT12ΩSprA1-STOP) containing two point mutations (T64A and C65A) to fabricate a premature UAA termination codon (supplemental Fig. S1) was also constructed. Without aTc, S. aureus cells containing the constructs expressing either SprA1 WT or SprA1-STOP grow perfectly well, as for the cells containing the empty vector (not shown). In the presence of aTc, the induced transcription of SprA1 inhibits S. aureus growth in liquid cultures (pAT12ΩSprA1; Fig. 1A). Such effect is also evident with the addition of aTc on a solid rich medium and by serial dilution assays (Fig. 1B). Remarkably, the aTc induction of the SprA1-STOP mutant that is unable to express PepA1 restores S. aureus growth near to WT levels in both liquid and solid cultures (Fig. 1). It provides experimental evidence that SprA1 inhibits S. aureus growth because it encodes and expresses PepA1.
FIGURE 1.
The peptide encoded by SprA1, PepA1, inhibits S. aureus growth and induces death by membrane permeabilization. A, comparative growth curves, after aTc induction, of strain Newman containing either an empty pAT12 plasmid (triangles), pAT12ΩSprA1 (squares), or pAT12ΩsprA1-STOP (circles). B, serial dilutions of strain Newman containing empty pAT12, pAT12ΩsprA1, or pAT12ΩsprA1-STOP, deposited on medium containing 1 μm aTc, and incubated overnight. C, pictures of fluorescent-labeled S. aureus cells after LIVE/DEAD experiments demonstrating that PepA1 encoded by SprA1 induces S. aureus death. Newman cells containing empty pAT12 plasmid are alive (green fluorescence), whereas those containing pAT12ΩsprA1 have damaged membranes (red fluorescence). Most cells containing pAT12ΩsprA1-STOP are alive. S. aureus living cells were counted for each of the three constructs, and the results are expressed as percentages of living cells. The error bars correspond to standard deviations inferred from three independent experiments. D, Northern blot detection of SprA1 RNA levels in the three strains before (−) and after (+) a 1-h induction with 1 μm aTc.
Is SprA1, via PepA1, able to trigger S. aureus death? To address that question, bacteria containing empty pAT12, pAT12ΩSprA1, or a pAT12ΩSprA1-STOP peptide mutant were stained with fluorescent dyes discriminating alive (intact membranes) from dead cells (disrupted membranes) and visualized by fluorescent microscopy. Cells transformed with an empty pAT12 vector are intact (stained green) 1 h after the aTc induction, whereas about half the cells transformed with pAT12ΩSprA1 (stained red) die (Fig. 1C). Cells expressing SprA1-STOP display similar viability to wild type cells (Fig. 1C). Therefore, PepA1 causes S. aureus death by disrupting membrane integrity. Northern blots were performed to verify the expression of SprA1 (WT or mutated) in the three constructs, before and after aTc induction (Fig. 1D). The aTc induction increases ∼3-fold the SprA1 WT or mutated RNA levels. During S. aureus growth, PepA1 toxicity is constitutively repressed by a tight translation control of SprA1, performed by SprA1AS.
Stable SprA1 Expresses PepA1 Early during Growth Counteracted by Unstable SprA1AS
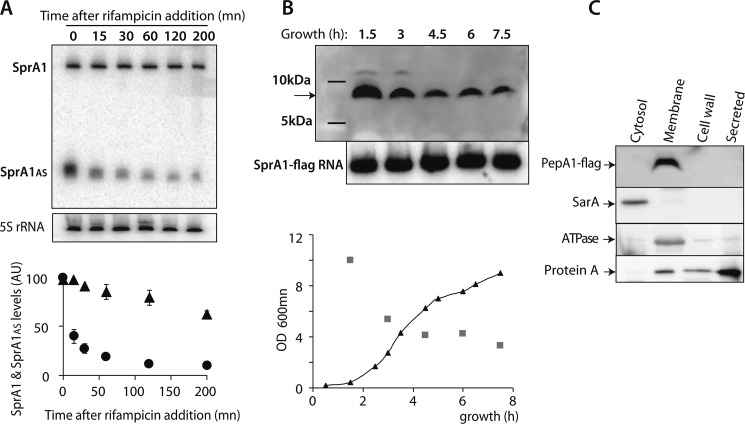
If the SprA1AS RNAs were an antidote to PepA1 toxicity, acted as antitoxins, and were part of a type I TA system, their in vivo stability should be lower than SprA1. SprA1AS degradation should allow PepA1 expression, in turn inducing S. aureus cell death. To determine SprA1 and SprA1AS stabilities in vivo, total RNAs were purified from S. aureus strain Newman at various times after rifampicin exposure. SprA1AS levels continually decline after rifampicin addition, whereas SprA1 levels remain essentially constant (Fig. 2A). Half-life determinations of the two RNAs showed a ∼20-fold shorter half-life for SprA1AS (∼10 min) compared with that of SprA1 (∼3 h). During S. aureus growth, SprA1AS is produced at a ∼30–90-fold excess relative to SprA1 (9), probably to balance its instability. Therefore, it is anticipated that PepA1 translational derepression, in response to specific environmental signals, occurs by lowering SprA1AS levels.
FIGURE 2.
Stable SprA1 expresses PepA1 at the membrane early during S. aureus growth counteracted by unstable SprA1AS. A, Northern blot analysis of SprA1 and SprA1AS in vivo stabilities in S. aureus strain Newman and their half-life determinations. Total RNAs were extracted at the indicated times after rifampicin addition. Quantifications of SprA1 (triangles) and SprA1AS (circles) levels relative to the amount of 5 S rRNA from identical RNA extractions. The error bars correspond to standard deviations inferred from two independent experiments. The 5 S rRNA is the loading control. B, in vivo detection of a ∼7-kDa flagged PepA1 at various times during S. aureus growth in Newman strain ΔsprA1-ΔsprA1ASpCN35ΩΔsprA1FLAG-sprA1AS by immunoblots using anti-FLAG antibodies. The arrow points to the flagged PepA1. The faint upper bands at the 1.5- and 3-h incubation times are also detected with the empty plasmid. Total proteins were extracted, purified, and quantified from the bacterial cells. Northern blot analysis of the SprA1-flag RNA levels during growth are internal controls indicating plasmid maintenance over time, despite PepA1 toxicity. Lower panel, growth curve of Newman strain ΔsprA1-ΔsprA1ASpCN35ΩΔsprA1FLAG-sprA1AS (black triangles) with the quantification of the flagged PepA1 levels (gray squares). In the immunoblots, equal amounts of protein extracts were deposited and verified on a Coomassie-stained gel. C, in vivo subcellular detection of the flagged PepA1 peptide by immunoblots using anti-FLAG antibodies in strain Newman. Total proteins were extracted and purified from the different S. aureus cellular compartments. Immunoblots against SarA, ATPase, and protein A are internal controls.
After determining SprA1 and SprA1AS expression levels (9) and stabilities in vivo (this report), we aimed at following PepA1 expression levels during S. aureus growth. Despite several attempts, we were unable to produce polyclonal antibodies directed against chemically synthesized and purified PepA1, to monitor the in vivo expression of native, untagged, PepA1 peptide. Therefore, a reporter peptide construct was designed by adding, in frame, a 3× FLAG sequence ahead of the termination codon of the PepA1 ORF in strain Newman (supplemental Fig. S1). Flagged PepA1 expression in vivo might be less toxic than WT PepA1. S. aureus cells expressing pCN35-SprA1wt could not be obtained (9). Indeed, a S. aureus Newman strain deleted for the native sprA1/sprA1AS locus and containing pCN35-SprA1FLAG could be successfully obtained (ΔsprA1-ΔsprA1AS pCN35ΩΔsprA1FLAG-sprA1AS). Immunoblots using anti-FLAG antibodies reveal that the SprA1 fusion peptide is expressed in vivo at all times during S. aureus growth (Fig. 2B), with an apparent molecular mass of ∼7 kDa, in agreement with the predicted 52-amino acid-long fusion peptides inferred from the SprA1 nucleotide sequence (supplemental Fig. S1). Flagged PepA1 is highly expressed very early during S. aureus growth in a rich medium (lag phase). Its production then decreases later on throughout growth. A similar PepA1 expression pattern was observed when growing S. aureus in a restricted, minimal medium (not shown). To rule out that the decrease of PepA1 expression over time might be due to a progressive reduction of plasmid number per cell caused by peptide toxicity, Northern blot analysis of SprA1-FLAG RNA levels over growth were performed and indicate that the SprA1 RNA levels do not decline over time (Fig. 2B).
The PepA1 Peptide Is Detected at the S. aureus Membrane
The sequence alignments and amino acid composition of PepA1 both suggest it is hydrophobic and contains a cationic domain. Because cationic peptides can interact with bacterial cytoplasmic membranes (19) and because PepA1 induces S. aureus membrane damage (this report), subcellular localization of flagged PepA1, within the S. aureus cells, was performed. Membrane, cell wall, cytoplasm and extracellular fractions were prepared from S. aureus Newman cells. The purity of our cell fractionations was then analyzed by western immunoblot analysis, using anti-SarA, anti-ATPase, and anti-protein A antibodies. SarA is a cytoplasmic transcription factor and is only detected in the cytoplasmic fraction, as expected. The ATPase is a membrane protein and is indeed only identified in the membrane fraction. Protein A is a secreted protein and is mainly identified in the exoproteins fraction (Fig. 2C). Interestingly, immunoblots with anti-FLAG antibodies indicate that PepA1 is exclusively detected in the membrane fraction (Fig. 2C). It indicates that once PepA1 is translated from SprA1 within the cytoplasm, it goes into the S. aureus membranes. We cannot rule out, however, that the presence of a FLAG within PepA1 amino acid sequence could influence its cellular location.
PepA1 High Resolution Structure Inferred from the NMR Data
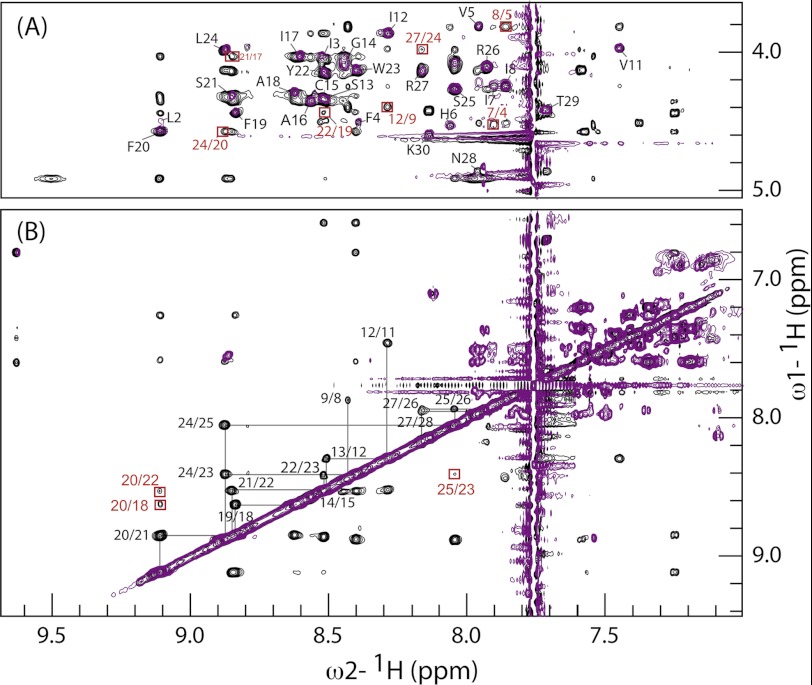
PepA1 three-dimensional structure was probed by NMR in a ternary mixture of CD3OH/CDCl3/H2O that mimics the membrane hydrophobicity (20–22). The peptide solubility in this buffer exceeds 5 mm allowing the NMR spectra at such PepA1 concentration to be recorded. Several CD3OH/CDCl3 ratios ranging from 0.5 to 6, at pHs of 3.8 or 4.5, were tested. At these acidic pHs, the backbone NH exchange rates are slow, preventing signal loss from the corresponding protons. The 6–11-ppm region of the one-dimensional spectrum displays a good dispersion of the NH proton resonance peaks over a wide range of CD3OH/CDCl3 ratios, consistent with a folded peptide structure. Peaks widths, however, disclose for some ratios conformation equilibrium and/or aggregation. For a CD3OH/CDCl3 ratio close to 1 and at pH 3.8, the one-dimensional spectrum discloses a unique and homogeneous conformation (supplemental Fig. S2), which was analyzed further by NMR. The spectra are independent of the PepA1 concentration, pointing to the existence of monomers. Spectra recorded at other ratios and various temperatures were also assigned to solve resonance overlaps. An iterative assignment procedure from the CCPN/ARIA2.3.1 package was used to solve the PepA1 tertiary structure. The assignment of 29 out 30 spin systems was achieved by analyzing the TOCSY, COSY, and NOESY spectra (see the assignment graph in supplemental Fig. S3). The N-terminal Met amino proton was not detected because of its fast exchange with water molecules. The fingerprint (NH-Hα and NH-NH) regions display many strong inter-residue (i, i+1), NH/NH and medium range (i, i+1), and (i, i+3) NH-Hα cross-peaks (Fig. 3), indicative of a high α-helical content within PepA1 structure. In particular, the NHi/NHi+1 connectivity pathway can be traced without interruption from Ala-18 to Asn-28, evidencing a highly structured and extended C-terminal helix. Such pathway between Val-5 and Ile-8 is elusive because of poor signal dispersion of their NH resonances. Prolines are usually helices breakers. However, NHi/Hαi+3 cross-peaks characteristics of α-helices are detected on either sides of Pro-10. Therefore, part of the N-terminal Leu-2 to Ala-9 sequence folds also as an α-helix. Spectra recorded at other CD3OH/CDCl3/H2O ratios all provide support to the presence of an α-helix. However, they display variations of chemical shifts and differences in the pattern of cross-peaks, indicating that the structure of the PepA1 N-terminal portion and its overall helical content vary with the buffer dielectric constant (supplemental Fig. S4).
FIGURE 3.
Assignments of PepA1 NMR resonances. Superposition of extended regions of TOCSY (purple) and NOESY (black) spectra recorded in a 1:1.2 CDCl3/CD3OH ratio at 288 K, used to solve PepA1 three-dimensional structure. A, the NH-Hα cross-peaks are labeled in black when intraresidual and boxed in red for i/i+3 connectivities. B, the NH-NH assignment walk is displayed, showing that most of the C-terminal residues can be connected. Some i/i+2 NH-NH cross-peaks are boxed in red.
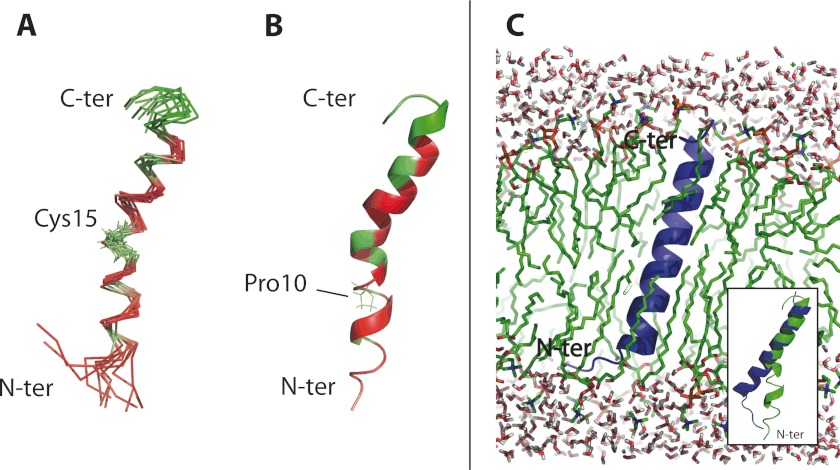
Based on a set of 311 NMR-derived constrained distances (supplemental Table S3), 1000 PepA1 structures were computed using the integrated CCPN/ARIA2.3.1 packages (11). A superposition of the top 10 PepA1 structures of lower energy, which display no distance violations above 0.2 Å, and the best structure are respectively displayed in Fig. 4 (A and B). The refinement procedure was carried out in Me2SO, a solvent less polar than water but with a dielectric constant similar to that of the NMR sample buffer. According to the NMR data, about two-thirds of PepA1 sequence folds into an extended helix that is interrupted and slightly bent in the vicinity of Cys-15. At the N terminus of PepA1, the helical fold disappears.
FIGURE 4.
PepA1 three-dimensional structure and molecular dynamics simulation of insertion into the S. aureus membrane. A, superimposition of 10 converged lowest energy NMR structures of PepA1 based on 313 NMR-derived distance restraints, refined in Me2SO, using ARIA 2.3.1. The hydrophobic residues are in red, and the polar and charged amino acids are in green. B, ribbon representation of the structure of lowest energy. PepA1 solution structure is mainly helical with an interruption at Pro-10. The helix is kinked at Cys-15. C, PepA1 structure after a 50-ns MD simulation in a hydrated DPPC bilayer. White and red, water molecules; green, DPPC lipid chains; blue, PepA1 structure with the MS simulation. PepA1 structure, within the membrane, is predicted to fold as an extended, uninterrupted, helix that runs across the membrane. Inset, superimposition of the pepA1 structure before (green) and after (blue) a 50-ns MD simulation into an artificial lipid bilayer. C-ter, C-terminal; N-ter, N-terminal.
NMR Spectra Recorded in SDS Solutions
The one-dimensional spectra obtained after dissolution of PepA1 in either 100 or 400 mm SDS are complex, with a superposition of peaks of different line widths, preventing the record of interpretable NOESY spectra (not shown). Nevertheless, the dispersion of the amide proton resonance shows that PepA1 is, at least, partly folded. The complexity of the spectra suggests that PepA1 adopts several conformations, including multimers. The broad resonances could also arise from peptide adsorbed or inserted into the SDS bilayer.
To overcome these problems, we carried out a second set of experiments in which SDS-d25 is gradually added to the CD3OH/CDCl3/H2O mixed buffer to reach final concentrations of 1.5, 5, 15, 50, and 100 mm. The pH was also gradually varied all along the titration to reach pH 6.8. The titration starts with a homogeneous population of monomers. It then shows that the PepA1 structure is maintained up to the highest SDS concentration (supplemental Fig. S5, left panel). A two-dimensional NOESY spectrum was also recorded, evidencing structural changes concerning, for the most part, the pepA1 N-terminal domain (supplemental Fig. S5, right panel). The spectra recorded in the 100 mm SDS mixed buffer and for pH levels ranging from 3.0 to 6.8 display very similar patterns, apart from the broadening of some NH resonances because of faster exchange with water at a higher pH and chemical shift variation of the terminal Lys-30 NH resonance. They do not disclose strong structural rearrangements (not shown).
Molecular Dynamics Simulations of PepA1 Structure in a Lipid Bilayer and CD Analyses
The affinity of α-helices for membrane lipids depends on the distributions of the hydrophobic and charged residues. A cluster of hydrophilic residues is located at the PepA1 C terminus (Fig. 4A, green), whereas the long interrupted helix and the N terminus both display patches of hydrophobic side chains (Fig. 4A, red). PepA1 solubility in a membrane-mimicking solvent as well as the helical segment of ∼7 nm in length containing polar and nonpolar domains both suggest that PepA1 structure will be modified when inserted into the S. aureus membranes. Bacterial membranes contain anionic lipids, as phosphatidic glycerols. To get insights into the structure of PepA1 when surrounded by lipids, the former average PepA1 NMR structure was submitted to a 50-ns molecular dynamic simulation in the presence of an artificial bilayer composed of DPPC solvated by water molecules. PepA1 structure was manually placed into the membrane perpendicularly to the surface, with its charged C terminus toward the solvent. The analysis of the 50-ns MD trajectory is displayed in supplemental Fig. S6. During the simulation, PepA1 rapidly wrapped itself and straightened into an uninterrupted, extended helix crossing the lipid bilayer (Fig. 4C, blue). In such an artificial system, PepA1 N terminus was trapped into the bilayer and was not able to displace some phospholipids to achieve a complete refolding. Strikingly, PepA1 estimated length is consistent with the 7–9 Å width of cell membranes. The simulations suggest that after its adhesion to the membrane surface, PepA1 undergoes a conformational rearrangement, allowing its insertion into the membrane, probably leading to membrane disruption and, ultimately, cell death.
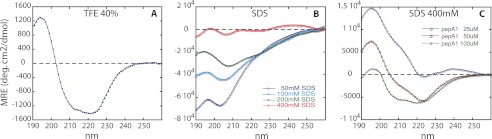
CD spectra were recorded in membrane mimicking environments, in either 40% TFE or in SDS aqueous solutions of various concentrations. PepA1 concentrations were respectively set to 175 μm in TFE and varied from 10 to 50 μm in SDS.
The data obtained in TFE (Fig. 5A) confirm the elevated ability of PepA1 to form a helix, as independently shown by the NMR spectra. Interestingly, when the SDS concentration increases, i.e., when the SDS to peptide ratio increases, progressively mimicking a membrane environment, the helical content of PepA1 rises (Fig. 5B), in agreement with the results inferred from the dynamic simulations. Fig. 5C shows that the CD signature stills varies with the SDS/peptide ratio even for concentration of SDS well above the CMC. The NMR experiments recorded in SDS indicate that PepA1 structure and oligomerization state both depend on the surrounding. Therefore the differences observed between spectra from panels B and C of Fig. 5, recorded at 400 mm SDS, are probably due to various amounts of oligomers. The reverse experiment monitoring the influence of decreasing the SDS concentrations suggests that once inserted into the membranes, PepA1 retains its helical structure and that the “inserted state” is stable (supplemental Fig. S7).
FIGURE 5.
Evolution of PepA1 as a function of SDS and peptide concentrations followed by circular dichroism. A, CD spectrum of PepA1 175 μm in 40% TFE, with the hallmark of the presence of a helix in its structure (positive peak at 195 nm and two negative peaks at 208 and 222 nm). B, PepA1 (10 μm) CD spectra in increasing concentrations of SDS above the CMC (the negative charges of SDS mimic those of the bacterial membranes). The helical content of PepA1 increases with the SDS/peptide ratio. C, PepA1 CD spectra recorded at a fixed SDS concentration (400 mm) with increasing PepA1 concentrations.
PepA1 Expression Increases upon Oxidative and Acidic Stresses by Lowering SprA1AS Levels
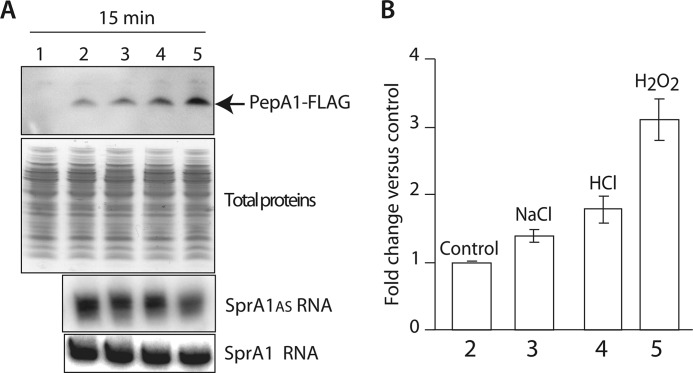
S. aureus cell fractionations indicated that PepA1 is a membrane peptide, further supported by structural data. We hypothesized that PepA1 might be an environmental sensor. Preliminary experiments were performed to identify experimental conditions that could influence PepA1 expression levels. S. aureus Newman strain ΔsprA1-ΔsprA1AS pCN35ΩSprA1FLAG/sprA1AS expressing flagged PepA1 was subjected to either an oxidative stress by adding 10 mm hydrogen peroxide, a saline stress by supplementing 2 m sodium chloride, or an acidic stress by lowering the pH to 4.5, for 15 min. Immunoblots against PepA1 reveal that flagged PepA1 production reproducibly rises ∼2-fold when the pH decreases, whereas PepA1 induction by the saline stress is less convincing (Fig. 6). Interestingly, compared with control, nonstressed cells, a ∼3-fold PepA1 induction was shown when applying oxidative stress (Fig. 6). Before and after each stress, the bacteria were plated, and colony-forming units were counted. The colony-forming units numbers were similar between the stressed and nonstressed cells (not shown), indicating that the S. aureus cells survive to 15 min of treatment with each of the three conditions tested.
FIGURE 6.
PepA1 expression increases upon oxidative and acidic stresses by lowering SprA1AS levels. A, top panel, in vivo monitoring of flagged PepA1 expression levels by immunoblots using anti-FLAG antibodies when the S. aureus cells are subjected to selected environmental changes. Newman strain ΔsprA1-ΔsprA1ASpCN35ΩΔsprA1FLAG-sprA1AS compared with an isogenic strain harboring an empty pCN35 vector as a negative control (lane 1), Newman strain ΔsprA1-ΔsprA1ASpCN35ΩΔsprA1FLAG-sprA1AS without stresses (lane 2), in the presence of 2 m NaCl (lane 3), after a pH drop to 4.5 (lane 4), or in the presence of 10 mm H2O2 (lane 5). Each stress was performed for 15 min. Middle panel, Coomassie-stained gel of the total proteins as loading controls. Bottom two panels, Northern blot analysis of SprA1 and SprA1AS expression levels without stresses, in the presence of 2 m NaCl, after a pH drop to 4.5 or in the presence of 10 mm H2O2. B, quantification of flagged PepA1 expression levels when applying each of the three stresses on strain ΔsprA1-sprA1ASpCN35ΩsprA1FLAG-sprA1AS, relative to the amount nonstressed cells. The error bars correspond to standard deviations inferred from three independent experiments.
What is the mechanism underlying stress-induced PepA1 increase in vivo? Because SprA1AS represses PepA1 translation initiation from SprA1 (9), a temporary decrease in the SprA1AS levels in vivo, triggered by specific environmental signals could, in turn, increase PepA1 expression levels. To test that hypothesis, similar stress conditions were performed, total RNAs were extracted immediately after, and Northern blots were conducted to monitor SprA1 and SprA1AS levels before and after the exposure of the S. aureus cells to the various stresses. Whereas the SprA1 levels remains stable in all the stresses, the levels of SprA1AS are reproducibly decreased by ∼50% after the oxidative stress and also by ∼25% after the acidic stress (Fig. 6). It demonstrates that the stress-induced increase of PepA1 levels in vivo occurs by lowering the SprA1AS levels, in turn reducing its translational repression onto SprA1-mediated PepA1 translation.
DISCUSSION
This report provides experimental support that S. aureus expresses a functional type I toxin-antitoxin pair. Because these toxins kill the cells expressing them, their expression is constitutively repressed by an unstable antisense RNA until triggered by specific environmental signals, presumably for an altruistic behavior for the overall bacterial community. Until recently (6), their abundance within the bacterial genomes was largely underestimated, probably because they are small and lack sequence signatures. Little is known about the functions and advantages of expressing type I toxins during bacterial growth in their natural habitats. Indeed, the deletion of genes encoding type I TA modules usually lacks associated phenotypes, but they may contribute to lower bacterial fitness under specific environments. Type I toxins are small hydrophobic proteins or peptides thought to insert and oligomerize in cell membranes to form pores or to act as detergents, in turn leading to cell death. These molecules exert toxicity when produced above a certain threshold. At lower levels, however, they may possess subtle physiological functions such as targeted protein degradations or interactions with dedicated membrane proteins or with two-component systems (23).
In this report, evidence is provided to support that the SprA1-SprA1AS RNA pair functions as a type I TA module in S. aureus. By a multidisciplinary approach, it was demonstrated that PepA1 is a toxic peptide. Type 1 TA systems encode a stable toxin and a labile RNA antitoxin counteracting toxicity. Within the SprA1-SprA1AS pair, stable (half-life exceeding 3 h) SprA1 expresses a toxic peptide, PepA1, and unstable (∼10 min half-life) SprA1AS is an antisense RNA antitoxin preventing, in trans, PepA1 translation initiation (9). Hydrophobic PepA1, a small peptide encoded by SprA1, inserts within the S. aureus membranes and induces apoptosis-like death in bacteria by perturbing membrane integrity. Preliminary data addressing the role of PepA1 once inserted within the S. aureus membranes indicate that, at low (1 μg/ml) concentration, the peptide strongly increases the electric conductance of artificial membranes, leading to their collapses probably by forming conductive peptide channels. This may provide a rationale for PepA1 lytic activity against mammalian erythrocytes (9). Therefore, PepA1 damages erythrocytes and S. aureus cell membranes either by forming pores or by interfering with membrane-associated functions.
During S. aureus growth, PepA1 expression is avoided by the SprA1AS antitoxin to prevent membrane damages but is induced upon oxidative and, to a lesser extent, acidic stresses. The NMR spectra recorded in SDS support PepA1 conformational changes, which could influence its ability to bind S. aureus membranes. S. aureus internalization into the phagolysosome of the host immune cells triggers an oxidative burst to kill the bacteria. One hypothesis to propose why PepA1 is induced upon oxidative and acidic bursts might be to kill most of the rapidly dividing internalized bacteria, to allow a minor fraction of slowly diving bacteria to persist and escape the phagolysosomes whose membranes will also be damaged by PepA1, to ultimately escape the immune cells for spreading into the host. Alternatively, the induction of PepA1 expression upon oxidative and acidic signals could be explained by its ability to modulate the activity of membrane proteins involved in iron transport. Indeed, iron homeostasis deregulation leads to an oxidative stress, and the efficient regulation of iron metabolism is essential for survival to lower oxygen toxicity (24). Consistently with our previous observation that it is hemolytic, PepA1 could participate in this regulation and drive lysis of host erythrocytes during the infection when iron supplies are scarce. When synthetic and purified PepA1 is externally provided to liquid cultures of S. aureus (other bacterial species were also tested), it has no influence on bacterial growth (not shown), suggesting that PepA1 might be unable to cross the cell wall from the exterior to reach the bacterial membranes. Alternatively, it may require intracellular post-translational modifications to become cytolytic.
We bring the first high resolution three-dimensional structure of a type I toxin expressed by S. aureus. The NMR-constrained PepA1 structure computations, as well as the molecular dynamics studies, indicate that PepA1 adopts a discontinuous helical structure capable of condensing into an extended, straight helix when inserted into a membrane, as shown in the S. aureus cells. The PepA1 helical structure forms a scaffold characteristic of an amphipathic peptide. Its structure is flexible enough to adapt to its environment within the bacteria. The NMR spectra collected in various experimental conditions all suggest that whereas the pepA1 C-terminal domain is folded, its N-terminal zone is less structured and very sensitive to its environment. The SDS CMC increases with the MeOH concentration, and above 30% MeOH (25), the micelles disaggregate. In the mixed CD3OH/CDCl3/H2O 100 mm SDS buffer (supplemental Fig. S5), the SDS micelles are probably not formed. This is consistent with the thin line widths of the resonances. This set of experiments aimed at mimicking an environment adjoining the membrane. The experiments revealed that PepA1 structure is slightly influenced by the presence of SDS. MD simulations in artificial membranes provide a snapshot and some insights of what happens when inserted into the membrane. They are consistent with the experimental flexibility observed by NMR and suggest that once inserted into a membrane, the conformation of PepA1 stretches and is able to span the lipid bilayer. The CD data also support these observations as the PepA1 helical content is raised in the presence of increasing concentrations of SDS, considered as a negatively charged bacterial membrane mimic. PepA1 arginine-rich C terminus is proposed to interact with the anionic groups of membrane glycerol residues. Such interactions were previously shown by NMR performed on a peptide in complex with membrane lipids (26). In summary, PepA1 membrane localization, its hemolytic activity (9), and its ability to induce S. aureus death when expressed in response to specific environmental signals are biological properties consistent with its solution tertiary structure.
The only solved atomic structure of a Type I toxin expressed by a Gram-positive organism is that of Fst expressed from a plasmid from Enterococcus faecalis. Fst forms a membrane-binding α-helix with an unstructured C terminus, possessing structural similarities with PepA1 but lacking antimicrobial or hemolytic activities (27), contrary to PepA1 (9). Fst has no cysteine residue, whereas PepA1 contains one located at a flexible hinge in the structure. PepA1 might form dimers by intermolecular disulfide linkages, whereas Fst cannot. Peptide dimer formation induces oligomerization at and within cellular membranes. Among the identified type-I toxins (i.e., Hok, Ldr, Fst, EHEC, YhzE, TxpA, YonT, Ibs, and ShoB), PepA1 and Hok are the only two possessing a conserved cysteine that could trigger dimerization. In conclusion, an RNA pair that acts as a functional type I TA module, expressed by numerous S. aureus clinical isolates, is reported, and the structure of the toxin was solved. The stable RNA from the pair produces a peptide that fits into the bacterial and cell host membranes and induces death. The upcoming challenge is to unravel its functions during staphylococcal growth, colonization, and infection.
Acknowledgments
We are thankful to Drs S. Chabelskaya, M. Hallier, and M. L. Pinel-Marie for critical reading of the manuscript and comments; Dr. E. Camberlein for help in the Northern blot analyses and to Sarah Elaid; and Dr. Serge Bouaziz from LCRB for sharing their expertise on CD and structural investigations on membrane peptides.
This work was supported by Grant ANR-09-MIEN-030-01 from the Agence Nationale pour la Recherche (to B. F. and S. N.-L.), funds from the INSERM and the CNRS, a doctoral grant from Brittany (to N. S.), a grant from the Fondation pour la Recherche Médicale (to N. S.), and funds from the French Department of Research and Education.

This article contains supplemental Tables S1–S3 and Figs. S1–S7.
The atomic coordinates and structure factors (code 4B19) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TA
- toxin-antitoxin
- TOCSY
- total correlation spectroscopy
- aTc
- anhydrotetracycline
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- TFE
- trifluoroethanol
- CMC
- critical micellar concentration
- MD
- molecular dynamics
- DPPC
- dipalmitoyl phosphatidylcholine.
REFERENCES
- 1. Van Melderen L., Saavedra De Bast M. (2009) Bacterial toxin-antitoxin systems. More than selfish entities? PLoS Genet. 5, e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yamaguchi Y., Park J. H., Inouye M. (2011) Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79 [DOI] [PubMed] [Google Scholar]
- 3. Fozo E. M., Hemm M. R., Storz G. (2008) Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 72, 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fineran P. C., Blower T. R., Foulds I. J., Humphreys D. P., Lilley K. S., Salmond G. P. (2009) The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U.S.A. 106, 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Magnuson R. D. (2007) Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189, 6089–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fozo E. M., Makarova K. S., Shabalina S. A., Yutin N., Koonin E. V., Storz G. (2010) Abundance of type I toxin-antitoxin systems in bacteria. Searches for new candidates and discovery of novel families. Nucleic Acids Res. 38, 3743–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Felden B., Vandenesch F., Bouloc P., Romby P. (2011) The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog. 7, e1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Romilly C., Caldelari I., Parmentier D., Lioliou E., Romby P., Fechter P. (2012) Current knowledge on regulatory RNAs and their machineries in Staphylococcus aureus. RNA Biol. 9, 402–413 [DOI] [PubMed] [Google Scholar]
- 9. Sayed N., Jousselin A., Felden B. (2012) A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat. Struct. Mol. Biol. 19, 105–112 [DOI] [PubMed] [Google Scholar]
- 10. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) The CCPN data model for NMR spectroscopy. Development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 11. Rieping W., Habeck M., Bardiaux B., Bernard A., Malliavin T. E., Nilges M. (2007) ARIA2. Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics 23, 381–382 [DOI] [PubMed] [Google Scholar]
- 12. Nilges M., Bernard A., Bardiaux B., Malliavin T., Habeck M., Rieping W. (2008) Accurate NMR structures through minimization of an extended hybrid energy. Structure 16, 1305–1312 [DOI] [PubMed] [Google Scholar]
- 13. Koradi R., Billeter M., Wuthrich K. (1996) MOLMOL. A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 14. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berendsen H. J., van der Spoel D., van Drunen R. (1995) GROMACS: A message-passing parallel molecular dynamics implementation. Comp. Phys. Comm. 91, 43–56 [Google Scholar]
- 16. Kandasamy S. K., Larson R. G. (2006) Molecular dynamics simulations of model trans-membrane peptides in lipid bilayers. A systematic investigation of hydrophobic mismatch. Biophys. J. 90, 2326–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beaume M., Hernandez D., Farinelli L., Deluen C., Linder P., Gaspin C., Romby P., Schrenzel J., Francois P. (2010) Cartography of methicillin-resistant S. aureus transcripts. Detection, orientation and temporal expression during growth phase and stress conditions. PLoS One 5, e10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bohn C., Rigoulay C., Chabelskaya S., Sharma C. M., Marchais A., Skorski P., Borezée-Durant E., Barbet R., Jacquet E., Jacq A., Gautheret D., Felden B., Vogel J., Bouloc P. (2010) Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 38, 6620–6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hancock R. E., Rozek A. (2002) Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 206, 143–149 [DOI] [PubMed] [Google Scholar]
- 20. Pashkov V. S., Maslennikov I. V., Tchikin L. D., Efremov R. G., Ivanov V. T., Arseniev A. S. (1999) Spatial structure of the M2 transmembrane segment of the nicotinic acetylcholine receptor α-subunit. FEBS Lett. 457, 117–121 [DOI] [PubMed] [Google Scholar]
- 21. Burkhart B. M., Gassman R. M., Langs D. A., Pangborn W. A., Duax W. L., Pletnev V. (1999) Gramicidin D conformation, dynamics and membrane ion transport. Biopolymers 51, 129–144 [DOI] [PubMed] [Google Scholar]
- 22. Standberg E., Ulrich A. S. (2004) NMR methods for antimicrobial peptides. Concepts Magn. Reson. 23A, 89–120 [Google Scholar]
- 23. Alix E., Blanc-Potard A. B. (2009) Hydrophobic peptides. Novel regulators within bacterial membrane. Mol. Microbiol. 72, 5–11 [DOI] [PubMed] [Google Scholar]
- 24. Touati D. (2000) Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373, 1–6 [DOI] [PubMed] [Google Scholar]
- 25. Fischer J., Jandera P. (1996) Chromatographic behaviour in reversed-phase high-performance liquid chromatography with micellar and submicellar mobile phases. Effects of the organic modifier. J. Chromatogr. B Biomed Appl. 681, 3–19 [DOI] [PubMed] [Google Scholar]
- 26. Wang G. (2008) NMR studies of a model antimicrobial peptide in the micelles of SDS, dodecylphosphocholine, or dioctanoylphosphatidylglycerol. Open Magn. Reson. J. 1, 9–15 [Google Scholar]
- 27. Göbl C., Kosol S., Stockner T., Rückert H. M., Zangger K. (2010) Solution structure and membrane binding of the toxin fst of the par addiction module. Biochemistry 49, 6567–6575 [DOI] [PMC free article] [PubMed] [Google Scholar]